Differential Response of Lung Cancer Cells, with Various Driver Mutations, to Plant Polyphenol Resveratrol and Vitamin D Active Metabolite PRI-2191
Abstract
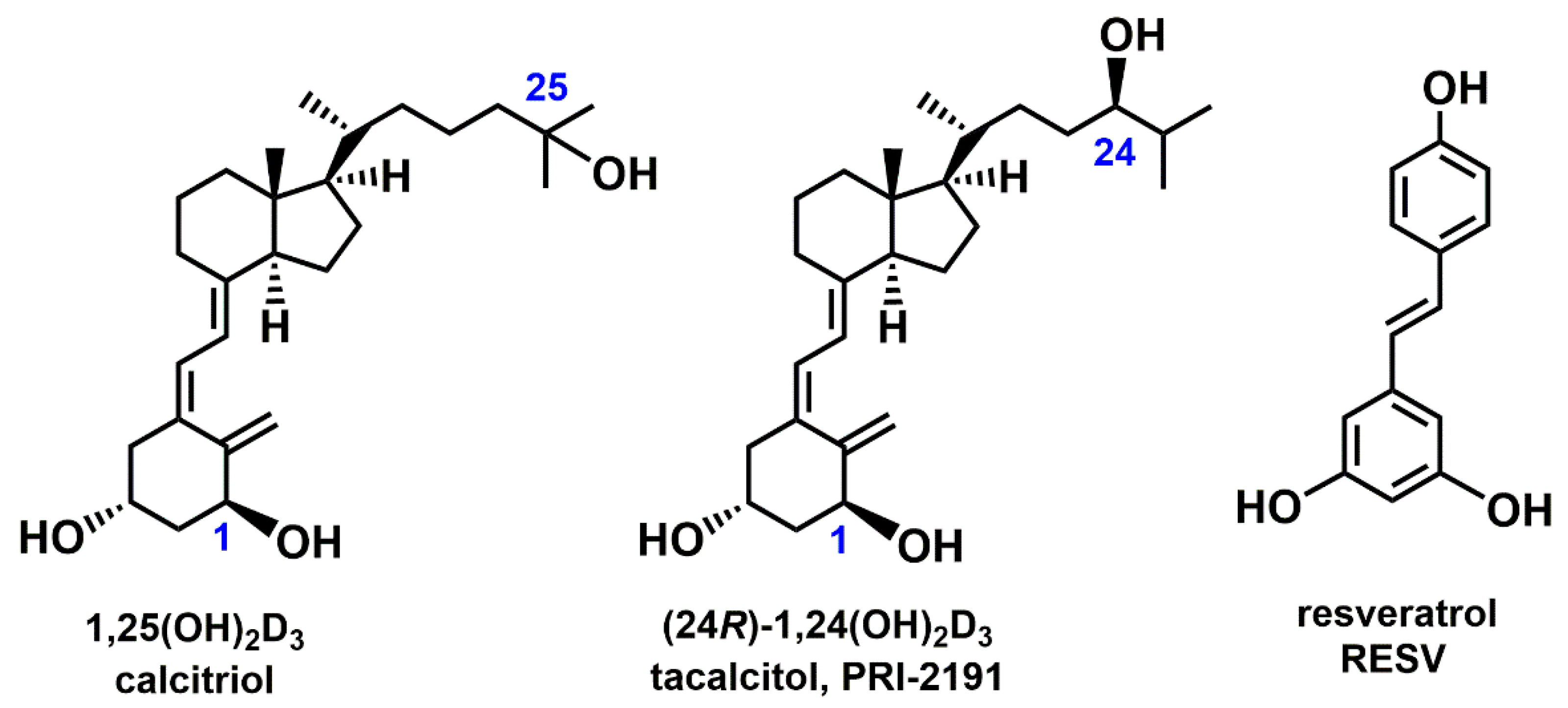
1. Introduction
2. Results
2.1. Antiproliferative Activity of RESV and PRI-2191 in Lung Cancer Cell Lines
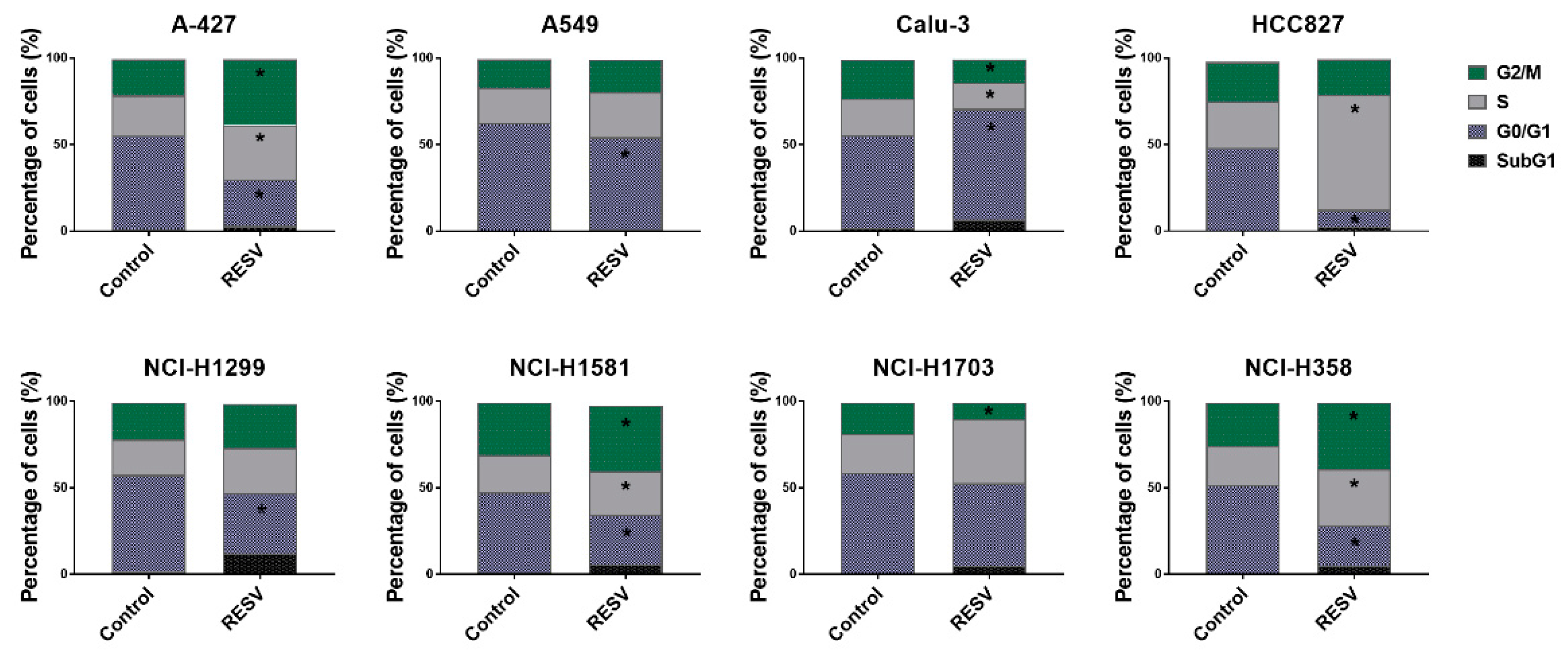
2.2. Cell Cycle Analysis of Lung Cancer Cells Treated with RESV and PRI-2191
2.3. Induction of Caspase-3 Activity by RESV and PRI-2191
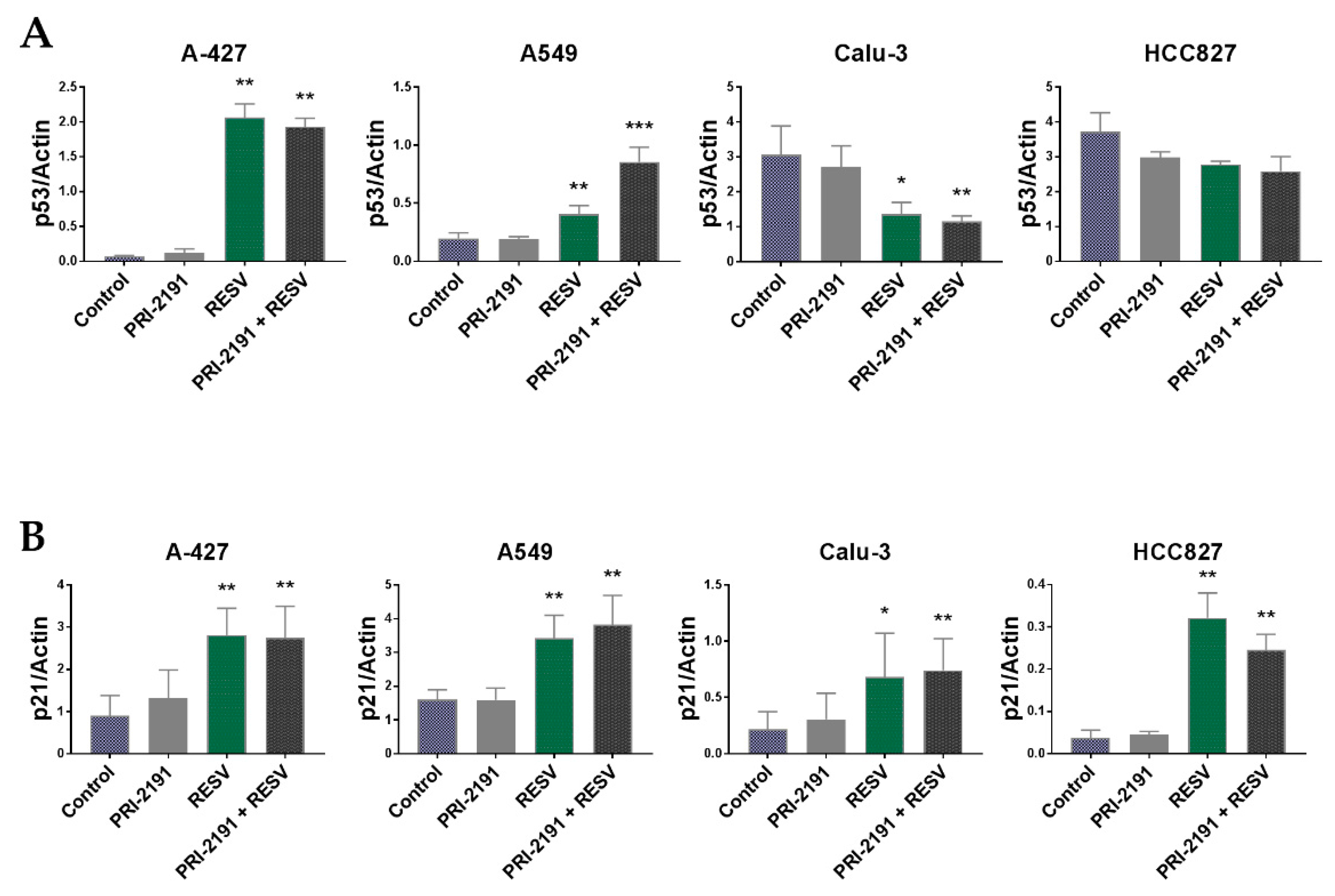
2.4. Changes in p53 and p21 Expression in Lung Cancer Cells after RESV and PRI-2191 Treatment
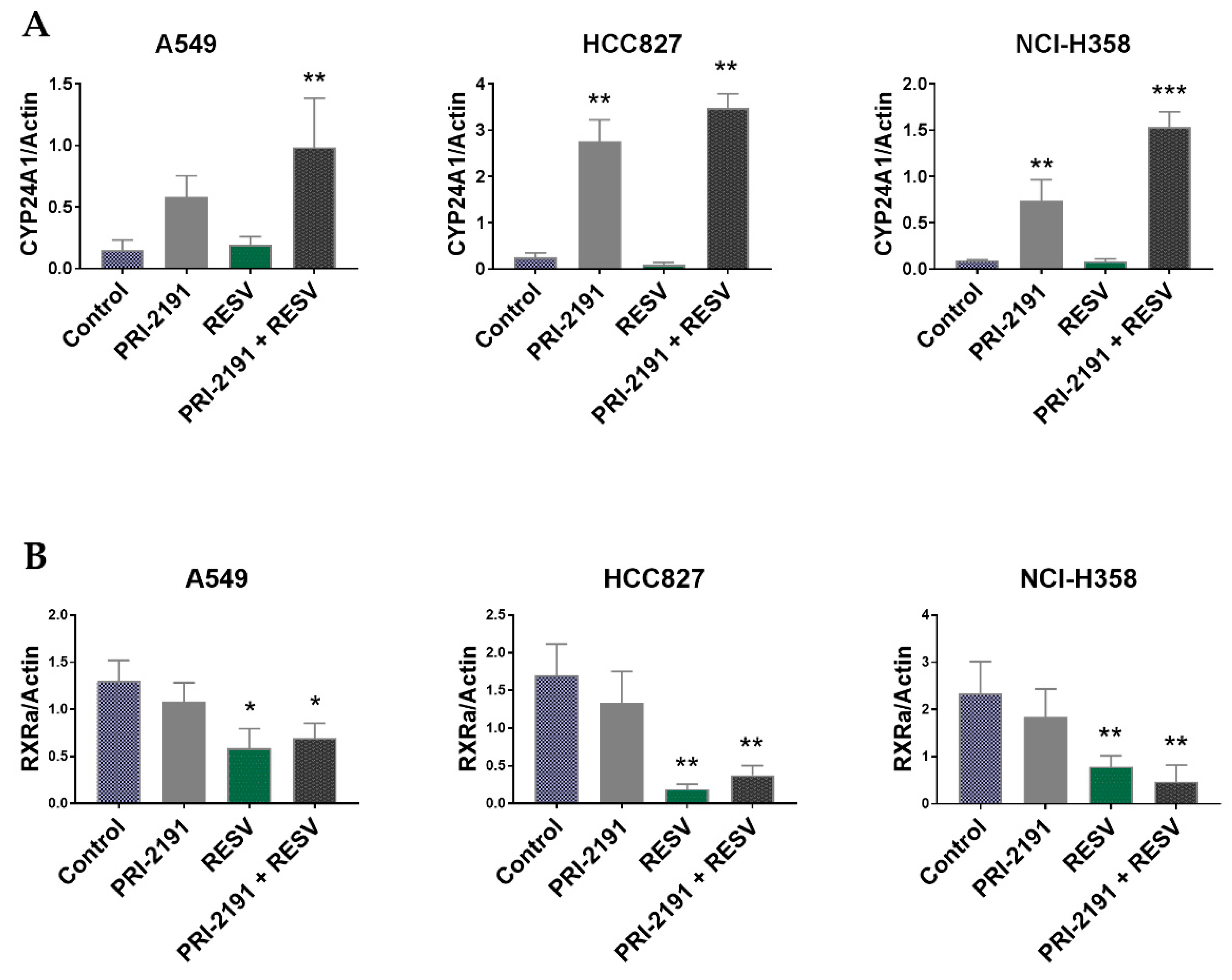
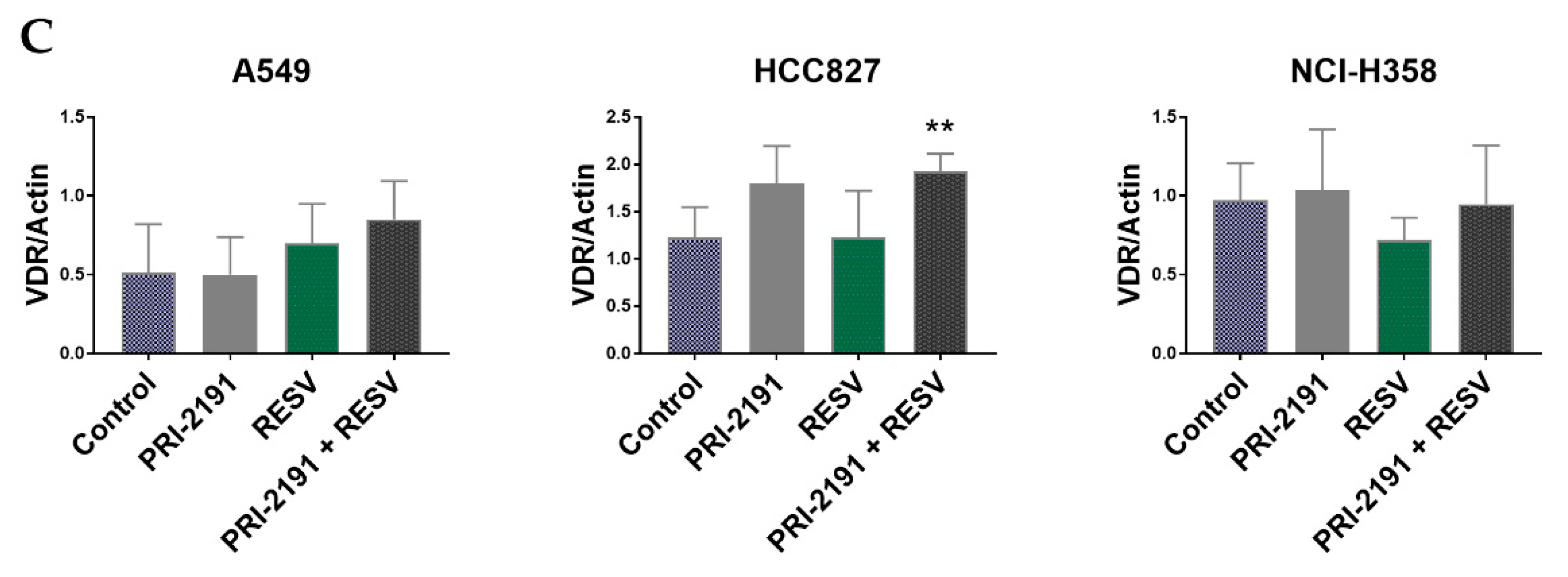
2.5. Differential Expression of CYP24A1, RXRα, and VDR in Lung Cancer Cells after PRI-2191 and RESV Treatment
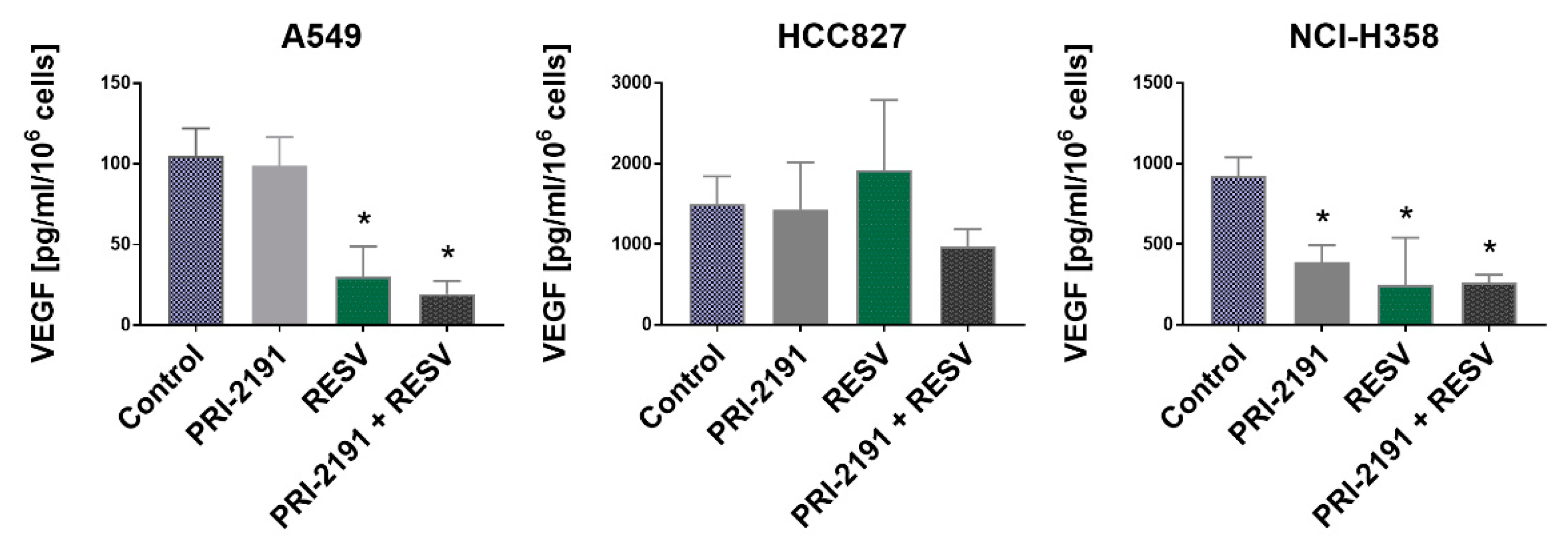
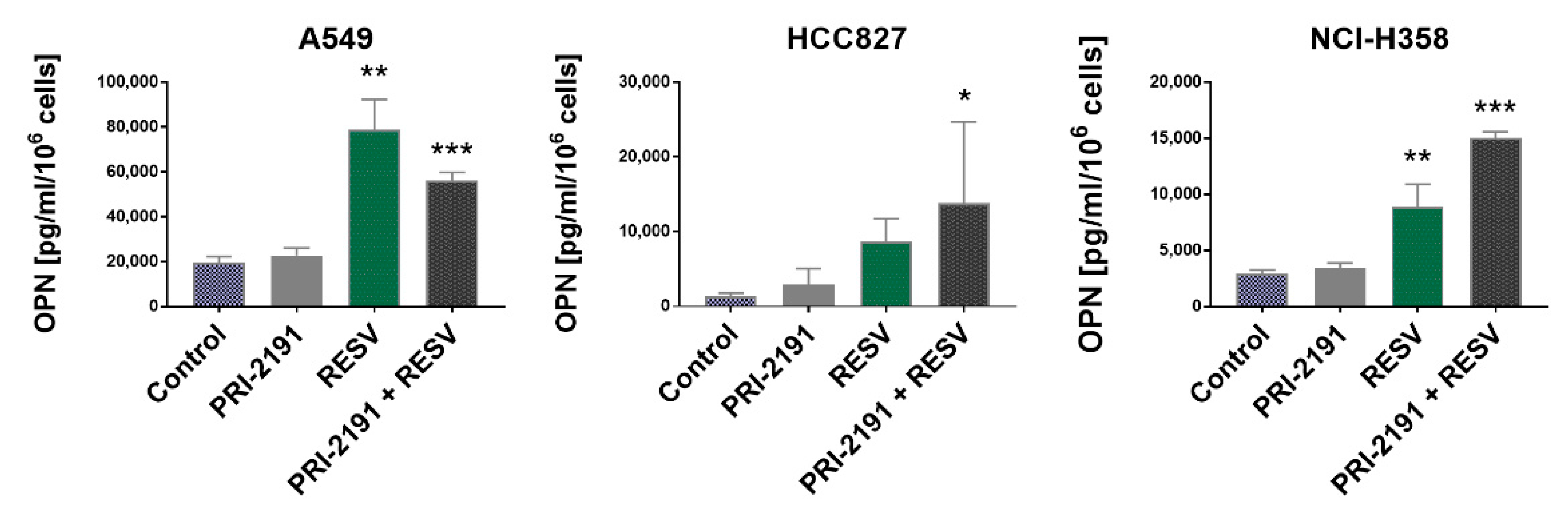
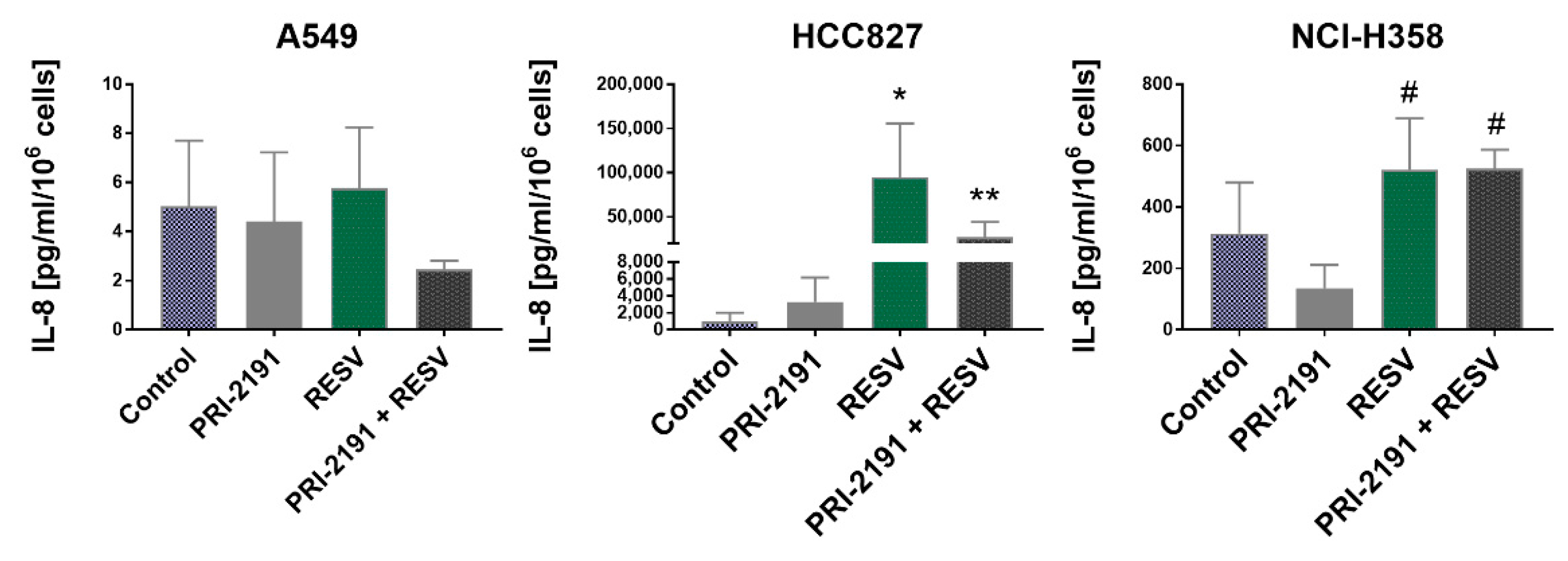
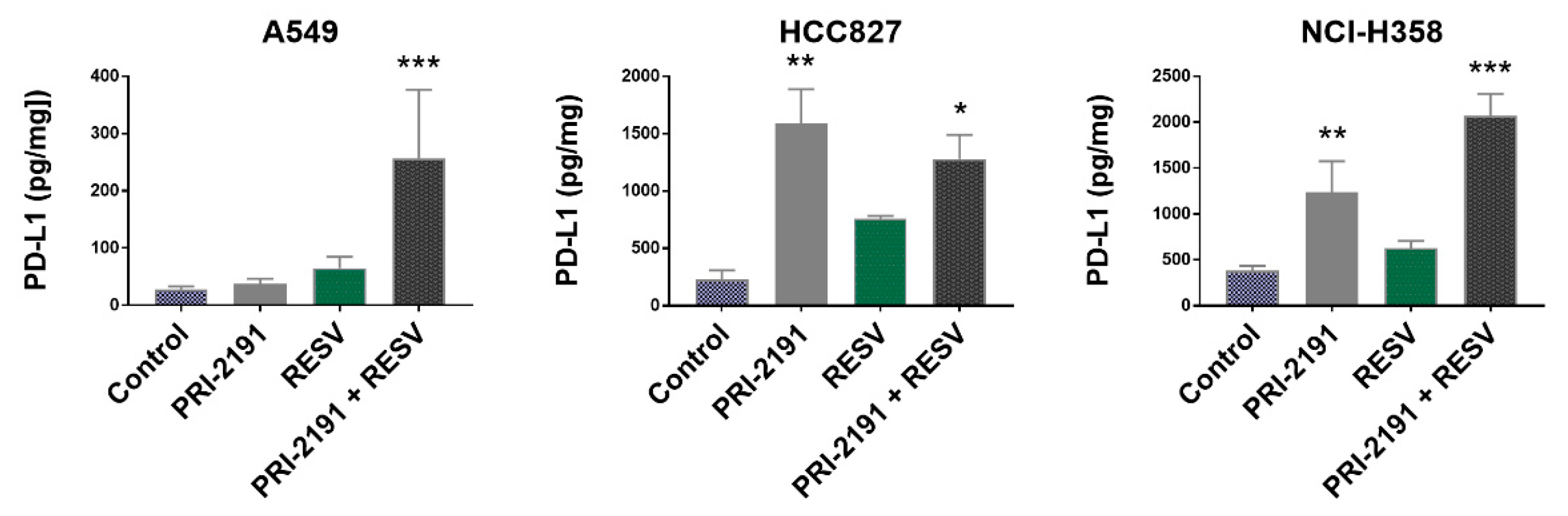
2.6. Impact of RESV and -2191 on VEGF, PD-L1, IL-8, and OPN Expression
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culturing Conditions
4.2. Compounds
4.3. Antiproliferative Activity
4.4. Cell Cycle Analysis
4.5. Caspase-3 Activity
4.6. Western Blot
4.7. qPCR Analysis
4.8. ELISA Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fresco, P.; Borges, F.I.G.M.; Diniz, C.G.; Marques, M. New insights on the anticancer properties of dietary polyphenols. Med. Res. Rev. 2006, 26, 747–766. [Google Scholar] [CrossRef]
- Li, M.; Zhang, Z.; Hill, D.L.; Wang, H.; Zhang, R. Curcumin, a Dietary Component, Has Anticancer, Chemosensitization, and Radiosensitization Effects by Down-regulating the MDM2 Oncogene through the PI3K/mTOR/ETS2 Pathway. Cancer Res. 2007, 67, 1988–1996. [Google Scholar] [CrossRef]
- Pesakhov, S.; Khanin, M.; Studzinski, G.P.; Danilenko, M. Distinct Combinatorial Effects of the Plant Polyphenols Curcumin, Carnosic Acid, and Silibinin on Proliferation and Apoptosis in Acute Myeloid Leukemia Cells. Nutr. Cancer 2010, 62, 811–824. [Google Scholar] [CrossRef]
- Wang, Z.; Dabrosin, C.; Yin, X.; Fuster, M.M.; Arreola, A.; Rathmell, W.K.; Generali, D.; Nagaraju, G.P.; El-Rayes, B.; Ribatti, D.; et al. Broad targeting of angiogenesis for cancer prevention and therapy. Semin. Cancer Biol. 2015, 35, S224–S243. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, S.; Badoux, J.K.; Scott-Boyer, M.-P.; Parolo, S.; Matone, A.; Priami, C.; Morine, M.J.; Kaput, J.; Moco, S. A computationally driven analysis of the polyphenol-protein interactome. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Fulda, S.; Debatin, K.-M. Sensitization for anticancer drug-induced apoptosis by the chemopreventive agent resveratrol. Oncogene 2004, 23, 6702–6711. [Google Scholar] [CrossRef]
- Kubota, T.; Uemura, Y.; Kobayashi, M.; Taguchi, H. Combined effects of resveratrol and paclitaxel on lung cancer cells. Anticancer. Res. 2003, 23, 4039–4046. [Google Scholar]
- Baatout, S.; Derradji, H.; Jacquet, P.; Ooms, D.; Michaux, A.; Mergeay, M. Enhanced radiation-induced apoptosis of cancer cell lines after treatment with resveratrol. Int. J. Mol. Med. 2004, 13, 895–902. [Google Scholar] [CrossRef]
- Niedzwiecki, A.; Roomi, M.W.; Kalinovsky, T.; Rath, M. Anticancer Efficacy of Polyphenols and Their Combinations. Nutrients 2016, 8, 552. [Google Scholar] [CrossRef] [PubMed]
- Amararathna, M.; Johnston, M.R.; Rupasinghe, H.P.V. Plant Polyphenols as Chemopreventive Agents for Lung Cancer. Int. J. Mol. Sci. 2016, 17, 1352. [Google Scholar] [CrossRef] [PubMed]
- Feldman, D.; Krishnan, A.V.; Swami, S.; Giovannucci, E.; Feldman, B.J. The role of vitamin D in reducing cancer risk and progression. Nat. Rev. Cancer 2014, 14, 342–357. [Google Scholar] [CrossRef]
- Okamoto, R.; Delansorne, R.; Wakimoto, N.; Doan, N.B.; Akagi, T.; Shen, M.; Ho, Q.H.; Said, J.W.; Koeffler, H.P. Inecalcitol, an analog of 1α,25(OH)2D3, induces growth arrest of androgen-dependent prostate cancer cells. Int. J. Cancer 2012, 130, 2464–2473. [Google Scholar] [CrossRef]
- Trynda, J.; Turlej, E.; Milczarek, M.; Pietraszek, A.; Chodyński, M.; Kutner, A.; Wietrzyk, J. Antiproliferative Activity and in Vivo Toxicity of Double-Point Modified Analogs of 1,25-Dihydroxyergocalciferol. Int. J. Mol. Sci. 2015, 16, 24873–24894. [Google Scholar] [CrossRef] [PubMed]
- Maj, E.; Trynda, J.; Maj, B.; Gębura, K.; Bogunia-Kubik, K.; Chodyński, M.; Kutner, A.; Wietrzyk, J. Differential response of lung cancer cell lines to vitamin D derivatives depending on EGFR, KRAS, p53 mutation status and VDR polymorphism. J. Steroid Biochem. Mol. Biol. 2019, 193, 105431. [Google Scholar] [CrossRef] [PubMed]
- Milczarek, M.; Rossowska, J.; Klopotowska, D.; Stachowicz, M.; Kutner, A.; Wietrzyk, J. Tacalcitol increases the sensitivity of colorectal cancer cells to 5-fluorouracil by downregulating the thymidylate synthase. J. Steroid Biochem. Mol. Biol. 2019, 190, 139–151. [Google Scholar] [CrossRef]
- Milczarek, M.; Psurski, M.; Kutner, A.; Wietrzyk, J. Vitamin D analogs enhance the anticancer activity of 5-fluorouracil in an in vivomouse colon cancer model. BMC Cancer 2013, 13, 294. [Google Scholar] [CrossRef] [PubMed]
- Leyssens, C.; Verlinden, L.; Verstuyf, A. The future of vitamin D analogs. Front. Physiol. 2014, 5, 18. [Google Scholar] [CrossRef]
- Maj, E.; Filip-Psurska, B.; Świtalska, M.; Kutner, A.; Wietrzyk, J. Vitamin D Analogs Potentiate the Antitumor Effect of Imatinib Mesylate in a Human A549 Lung Tumor Model. Int. J. Mol. Sci. 2015, 16, 27191–27207. [Google Scholar] [CrossRef]
- Maj, E.; Filip-Psurska, B.; Milczarek, M.; Psurski, M.; Kutner, A.; Wietrzyk, J. Vitamin D derivatives potentiate the anticancer and anti-angiogenic activity of tyrosine kinase inhibitors in combination with cytostatic drugs in an A549 non-small cell lung cancer model. Int. J. Oncol. 2017, 52, 337–366. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, A.; Anisiewicz, A.; Filip-Psurska, B.; Klopotowska, D.; Maciejewska, M.; Mazur, A.; Wietrzyk, J. Divergent Effect of Tacalcitol (PRI-2191) on Th17 Cells in 4T1 Tumor Bearing Young and Old Ovariectomized Mice. Aging Dis. 2020, 11, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.J.; Trump, D.L. Vitamin D Receptor Signaling and Cancer. Endocrinol. Metab. Clin. North Am. 2017, 46, 1009–1038. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Getz, G.; Wheeler, D.A.; Mardis, E.R.; McLellan, M.D.; Cibulskis, K.; Sougnez, C.; Greulich, H.; Muzny, D.M.; Morgan, M.B.; et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nat. Cell Biol. 2008, 455, 1069–1075. [Google Scholar] [CrossRef]
- Bacchi, C.E.; Ciol, H.; Queiroga, E.M.; Benine, L.C.; Silva, L.H.; Ojopi, E.B. Epidermal growth factor receptor and KRAS mutations in Brazilian lung cancer patients. Clinics 2012, 67, 419–424. [Google Scholar] [CrossRef]
- Robles, A.I.; Linke, S.P.; Harris, C.C. The p53 network in lung carcinogenesis. Oncogene 2002, 21, 6898–6907. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Sur, S.; Prives, C. p53: The Most Frequently Altered Gene in Human Cancers. Nat. Educ. 2010, 3, 6. [Google Scholar]
- Nachliely, M.; Sharony, E.; Bolla, N.R.; Kutner, A.; Danilenko, M. Prodifferentiation Activity of Novel Vitamin D2 Analogs PRI-1916 and PRI-1917 and Their Combinations with a Plant Polyphenol in Acute Myeloid Leukemia Cells. Int. J. Mol. Sci. 2016, 17, 1068. [Google Scholar] [CrossRef] [PubMed]
- Howitz, K.T.; Bitterman, K.J.; Cohen, H.Y.; Lamming, D.W.; Lavu, S.; Wood, J.G.; Zipkin, R.E.; Chung, P.; Kisielewski, A.; Zhang, L.-L.; et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 2003, 425, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Carafa, V.; Rotili, D.; Forgione, M.; Cuomo, F.; Serretiello, E.; Hailu, G.S.; Jarho, E.; Lahtela-Kakkonen, M.; Maija, L.-K.; Altucci, L. Sirtuin functions and modulation: from chemistry to the clinic. Clin. Epigenet. 2016, 8, 1–21. [Google Scholar] [CrossRef]
- Langley, E.; Pearson, M.; Faretta, M.; Bauer, U.; Frye, R.A.; Minucci, S.; Pelicci, P.G.; Kouzarides, T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002, 21, 2383–2396. [Google Scholar] [CrossRef]
- Sabir, M.S.; Khan, Z.; Hu, C.; Galligan, M.A.; Dussik, C.M.; Mallick, S.; Stone, A.D.; Batie, S.F.; Jacobs, E.T.; Whitfield, G.K.; et al. SIRT1 enzymatically potentiates 1,25-dihydroxyvitamin D3 signaling via vitamin D receptor deacetylation. J. Steroid Biochem. Mol. Biol. 2017, 172, 117–129. [Google Scholar] [CrossRef]
- Ohyama, Y.; Ozono, K.; Uchida, M.; Shinki, T.; Kato, S.; Suda, T.; Yamamoto, O.; Noshiro, M.; Kato, Y. Identification of a vitamin D-responsive element in the 5’-flanking region of the rat 25-hydroxyvitamin D3 24-hydroxylase gene. J. Biol. Chem. 1994, 269, 10545–10550. [Google Scholar] [CrossRef]
- Sharabani, H.; Izumchenko, E.; Wang, Q.; Kreinin, R.; Steiner, M.; Barvish, Z.; Kafka, M.; Sharoni, Y.; Levy, J.; Uskokovic, M.; et al. Cooperative antitumor effects of vitamin D3 derivatives and rosemary preparations in a mouse model of myeloid leukemia. Int. J. Cancer 2006, 118, 3012–3021. [Google Scholar] [CrossRef]
- Bobilev, I.; Novik, V.; Levi, I.; Shpilberg, O.; Levy, J.; Sharoni, Y.; Studzinski, G.P.; Danilenko, M. The Nrf2 transcription factor is a positive regulator of myeloid differentiation of acute myeloid leukemia cells. Cancer Biol. Ther. 2011, 11, 317–329. [Google Scholar] [CrossRef]
- Bhatia, V.; Falzon, M. Restoration of the anti-proliferative and anti-migratory effects of 1,25-dihydroxyvitamin D by silibinin in vitamin D-resistant colon cancer cells. Cancer Lett. 2015, 362, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Wassermann, R.; Novik, V.; Danilenko, M. Cell-Type-Specific Effects of Silibinin on Vitamin D-Induced Differentiation of Acute Myeloid Leukemia Cells Are Associated with Differential Modulation of RXRalpha Levels. Leuk. Res. Treat. 2012, 2012, 401784. [Google Scholar] [CrossRef]
- Inoue, J.; Choi, J.-M.; Yoshidomi, T.; Yashiro, T.; Sato, R. Quercetin enhances VDR activity, leading to stimulation of its target gene expression in Caco-2 cells. J. Nutr. Sci. Vitaminol. 2010, 56, 326–330. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Haussler, M.R.; Saini, R.K.; Sabir, M.S.; Dussik, C.M.; Khan, Z.; Whitfield, G.K.; Griffin, K.P.; Kaneko, I.; Jurutka, P.W. Vitamin D Nutrient-Gene Interactions and Healthful Aging. In Molecular Basis of Nutrition and Aging; Elsevier: Amsterdam, The Netherlands, 2016; pp. 449–471. [Google Scholar]
- Benitez, D.A.; Pozo-Guisado, E.; Alvarez-Barrientos, A.; Fernandez-Salguero, P.M.; Castellón, E.A. Mechanisms Involved in Resveratrol-Induced Apoptosis and Cell Cycle Arrest in Prostate Cancer-Derived Cell Lines. J. Androl. 2006, 28, 282–293. [Google Scholar] [CrossRef]
- Pozo-Guisado, E.; Alvarez-Barrientos, A.; Mulero-Navarro, S.; Santiago-Josefat, B.; Fernandez-Salguero, P.M. The antiproliferative activity of resveratrol results in apoptosis in MCF-7 but not in MDA-MB-231 human breast cancer cells: cell-specific alteration of the cell cycle. Biochem. Pharmacol. 2002, 64, 1375–1386. [Google Scholar] [CrossRef]
- Da Costa, D.C.F.; Campos, N.P.C.; Santos, R.A.; Guedes-Da-Silva, F.H.; Martins-Dinis, M.M.D.C.; Zanphorlin, L.; Ramos, C.; Rangel, L.P.; Silva, J.L. Resveratrol prevents p53 aggregation in vitro and in breast cancer cells. Oncotarget 2018, 9, 29112–29122. [Google Scholar] [CrossRef]
- Oren, M.; Rotter, V. Mutant p53 Gain-of-Function in Cancer. Cold Spring Harb. Perspect. Biol. 2009, 2, a001107. [Google Scholar] [CrossRef]
- Pedrote, M.M.; Motta, M.F.; Ferretti, G.D.; Norberto, D.R.; Spohr, T.C.; Lima, F.R.; Gratton, E.; Silva, J.L.; de Oliveira, G.A. Oncogenic Gain of Function in Glioblastoma Is Linked to Mutant p53 Amyloid Oligomers. iScience 2020, 23, 100820. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.W.; Kang, H.J.; Kim, H.J.; Kong, Y.; Brown, M.L.; Bae, I. Targeting Mutant p53 by a SIRT1 Activator YK-3-237 Inhibits the Proliferation of Triple-Negative Breast Cancer Cells. Oncotarget 2013, 4, 984–994. [Google Scholar] [CrossRef]
- Willis, A.; Jung, E.J.; Wakefield, T.; Chen, X. Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes. Oncogene 2004, 23, 2330–2338. [Google Scholar] [CrossRef]
- Zhao, Y.; Lu, S.; Wu, L.; Chai, G.; Wang, H.; Chen, Y.; Sun, J.; Yu, Y.; Zhou, W.; Zheng, Q.; et al. Acetylation of p53 at Lysine 373/382 by the Histone Deacetylase Inhibitor Depsipeptide Induces Expression of p21Waf1/Cip1. Mol. Cell. Biol. 2006, 26, 2782–2790. [Google Scholar] [CrossRef] [PubMed]
- Rokudai, S.; Laptenko, O.; Arnal, S.M.; Taya, Y.; Kitabayashi, I.; Prives, C. MOZ increases p53 acetylation and premature senescence through its complex formation with PML. PNAS 2013, 110, 3895–3900. [Google Scholar] [CrossRef]
- Brochier, C.; Dennis, G.; Rivieccio, M.A.; McLaughlin, K.; Coppola, G.; Ratan, R.R.; Langley, B. Specific Acetylation of p53 by HDAC Inhibition Prevents DNA Damage-Induced Apoptosis in Neurons. J. Neurosci. 2013, 33, 8621–8632. [Google Scholar] [CrossRef]
- Ekongsbak, M.; Levring, T.B.; Egeisler, C.; Von Essen, M.R. The Vitamin D Receptor and T Cell Function. Front. Immunol. 2013, 4, 148. [Google Scholar] [CrossRef]
- Haussler, M.R.; Whitfield, G.K.; Kaneko, I.; Haussler, C.A.; Hsieh, D.; Hsieh, J.-C.; Jurutka, P.W. Molecular Mechanisms of Vitamin D Action. Calcif. Tissue Int. 2013, 92, 77–98. [Google Scholar] [CrossRef] [PubMed]
- Dampf-Stone, A.; Batie, S.F.; Sabir, M.S.; Jacobs, E.T.; Lee, J.H.; Whitfield, G.K.; Haussler, M.R.; Jurutka, P.W. Resveratrol Potentiates Vitamin D and Nuclear Receptor Signaling. J. Cell. Biochem. 2015, 116, 1130–1143. [Google Scholar] [CrossRef]
- Singh, C.K.; Kumar, A.; Lavoie, H.A.; DiPette, D.J.; Singh, U.S. Resveratrol Prevents Impairment in Activation of Retinoic Acid Receptors and MAP Kinases in the Embryos of a Rodent Model of Diabetic Embryopathy. Reprod. Sci. 2012, 19, 949–961. [Google Scholar] [CrossRef]
- García-Quiroz, J.; García-Becerra, R.; Santos-Cuevas, C.; Ramírez-Nava, G.J.; Morales-Guadarrama, G.; Cárdenas-Ochoa, N.; Segovia-Mendoza, M.; Prado-Garcia, H.; Ordaz-Rosado, D.; Avila, E.; et al. Synergistic Antitumorigenic Activity of Calcitriol with Curcumin or Resveratrol is Mediated by Angiogenesis Inhibition in Triple Negative Breast Cancer Xenografts. Cancers 2019, 11, 1739. [Google Scholar] [CrossRef]
- Tang, Z.; Liu, X.-Y.; Zou, P. Resveratrol inhibits the secretion of vascular endothelial growth factor and subsequent proliferation in human leukemia U937 cells. Acta Acad. Med. Wuhan 2007, 27, 508–512. [Google Scholar] [CrossRef]
- Núñez, M.J.; Novío, S.; Balboa, J.; Seoane, J.; Suárez, J.A.; Freire-Garabal, M. Effects of resveratrol on expression of vascular endothelial growth factor in human gingival fibroblasts stimulated by periodontal pathogens. Acta Odontol. Scand. 2010, 68, 239–247. [Google Scholar] [CrossRef]
- Sahin, E.; Bayçu, C.; Koparal, A.T.; Donmez, D.B.; Bektur, E. Resveratrol reduces IL-6 and VEGF secretion from co-cultured A549 lung cancer cells and adipose-derived mesenchymal stem cells. Tumor Biol. 2015, 37, 7573–7582. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.; Iyer, A.K.V.; Yakisich, J.S.; Azad, N. Anti-Tumorigenic Effects of Resveratrol in Lung Cancer Cells Through Modulation of c-FLIP. Curr. Cancer Drug Targets 2017, 17, 669–680. [Google Scholar] [CrossRef]
- Reinmuth, N.; Jauch, A.; Xu, E.C.; Muley, T.; Granzow, M.; Hoffmann, H.; Dienemann, H.; Herpel, E.; Schnabel, P.A.; Herth, F.J.; et al. Correlation of EGFR mutations with chromosomal alterations and expression of EGFR, ErbB3 and VEGF in tumor samples of lung adenocarcinoma patients. Lung Cancer 2008, 62, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Wong, J.P.C.; Kwok, H.F. Osteopontin—A promising biomarker for cancer therapy. J. Cancer 2017, 8, 2173–2183. [Google Scholar] [CrossRef]
- Jin, Y.; Tong, D.-Y.; Chen, J.-N.; Feng, Z.-Y.; Yang, J.-Y.; Shao, C.-K.; Li, J.-P. Overexpression of Osteopontin, αvβ3 and Pim-1 Associated with Prognostically Important Clinicopathologic Variables in Non-Small Cell Lung Cancer. PLoS ONE 2012, 7, e48575. [Google Scholar] [CrossRef]
- Noda, M.; Vogel, R.L.; Craig, A.M.; Prahl, J.; DeLuca, H.F.; Denhardt, D.T. Identification of a DNA sequence responsible for binding of the 1,25-dihydroxyvitamin D3 receptor and 1,25-dihydroxyvitamin D3 enhancement of mouse secreted phosphoprotein 1 (SPP-1 or osteopontin) gene expression. PNAS 1990, 87, 9995–9999. [Google Scholar] [CrossRef]
- Shen, Q.; Christakos, S. The Vitamin D Receptor, Runx2, and the Notch Signaling Pathway Cooperate in the Transcriptional Regulation of Osteopontin. J. Biol. Chem. 2005, 280, 40589–40598. [Google Scholar] [CrossRef] [PubMed]
- Boissy, P.; Andersen, T.L.; Abdallah, B.M.; Kassem, M.; Plesner, T.; Delaissé, J.-M. Resveratrol Inhibits Myeloma Cell Growth, Prevents Osteoclast Formation, and Promotes Osteoblast Differentiation. Cancer Res. 2005, 65, 9943–9952. [Google Scholar] [CrossRef]
- Lee, Y.-M.; Shin, S.-I.; Shin, K.-S.; Park, B.-H.; Kim, E.-C. The role of sirtuin 1 in osteoblastic differentiation in human periodontal ligament cells. J. Periodontal Res. 2011, 46, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jiang, Z.; Li, X.; Zhang, X. SIRT1 overexpression protects non-small cell lung cancer cells against osteopontin-induced epithelial-mesenchymal transition by suppressing NF-κB signaling. OncoTargets Ther. 2018, 11, 1157–1171. [Google Scholar] [CrossRef] [PubMed]
- Rangaswami, H.; Bulbule, A.; Kundu, G.C. Osteopontin: role in cell signaling and cancer progression. Trends Cell Biol. 2006, 16, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, L.E.; Newton, R.; Kennedy, G.E.; Fenwick, P.S.; Leung, R.H.F.; Ito, K.; Russell, R.E.K.; Barnes, P.J. Anti-inflammatory effects of resveratrol in lung epithelial cells: molecular mechanisms. Am. J. Physiol. Cell. Mol. Physiol. 2004, 287, L774–L783. [Google Scholar] [CrossRef]
- Tino, A.B.; Chitcholtan, K.; Sykes, P.H.; Garrill, A. Resveratrol and acetyl-resveratrol modulate activity of VEGF and IL-8 in ovarian cancer cell aggregates via attenuation of the NF-κB protein. J. Ovarian Res. 2016, 9, 1–12. [Google Scholar] [CrossRef]
- Pastore, S.; Lulli, D.; Maurelli, R.; Dellambra, E.; De Luca, C.; Korkina, L.G. Resveratrol Induces Long-Lasting IL-8 Expression and Peculiar EGFR Activation/Distribution in Human Keratinocytes: Mechanisms and Implications for Skin Administration. PLoS ONE 2013, 8, e59632. [Google Scholar] [CrossRef]
- Fan, X.-X.; Yao, X.-J.; Xu, S.W.; Wong, V.K.-W.; He, J.-X.; Ding, J.; Xue, W.-W.; Mujtaba, T.; Michelangeli, F.; Huang, M.; et al. (Z) 3,4,5,4′-trans-tetramethoxystilbene, a new analogue of resveratrol, inhibits gefitinb-resistant non-small cell lung cancer via selectively elevating intracellular calcium level. Sci. Rep. 2015, 5, 16348. [Google Scholar] [CrossRef]
- Cordero, J.B.; Cozzolino, M.; Lu, Y.; Vidal, M.; Slatopolsky, E.; Stahl, P.D.; Barbieri, M.A.; Dusso, A. 1,25-Dihydroxyvitamin D Down-regulates Cell Membrane Growth- and Nuclear Growth-promoting Signals by the Epidermal Growth Factor Receptor. J. Biol. Chem. 2002, 277, 38965–38971. [Google Scholar] [CrossRef]
- McGaffin, K.R.; A Chrysogelos, S. Identification and characterization of a response element in the EGFR promoter that mediates transcriptional repression by 1,25-dihydroxyvitamin D3 in breast cancer cells. J. Mol. Endocrinol. 2005, 35, 117–133. [Google Scholar] [CrossRef]
- Shen, Z.; Zhang, X.; Tang, J.; Kasiappan, R.; Jinwal, U.; Li, P.; Hann, S.; Nicosia, S.V.; Wu, J.; Zhang, X.; et al. The coupling of epidermal growth factor receptor down regulation by 1alpha,25-dihydroxyvitamin D3 to the hormone-induced cell cycle arrest at the G1-S checkpoint in ovarian cancer cells. Mol. Cell. Endocrinol. 2011, 338, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Ostrand-Rosenberg, S.; Horn, L.A.; Haile, S.T. The Programmed Death-1 Immune-Suppressive Pathway: Barrier to Antitumor Immunity. J. Immunol. 2014, 193, 3835–3841. [Google Scholar] [CrossRef]
- Dong, H.; Strome, S.E.; Salomao, D.R.; Tamura, H.; Hirano, F.; Flies, D.B.; Roche, P.C.; Lu, J.; Zhu, G.; Tamada, K.; et al. Tumor-associated B7-H1 promotes T-cell apoptosis: A potential mechanism of immune evasion. Nat. Med. 2002, 8, 793–800. [Google Scholar] [CrossRef]
- Dimitrov, V.; Bouttier, M.; Boukhaled, G.; Salehi-Tabar, R.; Avramescu, R.G.; Memari, B.; Hasaj, B.; Lukacs, G.L.; Krawczyk, C.M.; White, J.H. Hormonal vitamin D up-regulates tissue-specific PD-L1 and PD-L2 surface glycoprotein expression in humans but not mice. J. Biol. Chem. 2017, 292, 20657–20668. [Google Scholar] [CrossRef] [PubMed]
- Bendix, M.; Greisen, S.; Dige, A.; Hvas, C.L.; Bak, N.; Jørgensen, S.P.; Dahlerup, J.F.; Deleuran, B.; Agnholt, J. Vitamin D increases programmed death receptor-1 expression in Crohn’s disease. Oncotarget 2017, 8, 24177–24186. [Google Scholar] [CrossRef]
- Lucas, J.; Hsieh, T.-C.; Halicka, H.D.; Darzynkiewicz, Z.; Wu, J.M. Upregulation of PD-L1 expression by resveratrol and piceatannol in breast and colorectal cancer cells occurs via HDAC3/p300-mediated NF-κB signaling. Int. J. Oncol. 2018, 53, 1469–1480. [Google Scholar] [CrossRef]
- Verdura, S.; Cuyàs, E.; Cortada, E.; Brunet, J.; Lopez-Bonet, E.; Martin-Castillo, B.; Bosch-Barrera, J.; Encinar, J.A.; Menendez, J.A. Resveratrol targets PD-L1 glycosylation and dimerization to enhance antitumor T-cell immunity. Aging 2020, 12, 8–34. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, S.; Yang, Y.; Liu, T. Resveratrol induces immunogenic cell death of human and murine ovarian carcinoma cells. Infect. Agents Cancer 2019, 14, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Nevozhay, D. Cheburator Software for Automatically Calculating Drug Inhibitory Concentrations from In Vitro Screening Assays. PLoS ONE 2014, 9, e106186. [Google Scholar] [CrossRef] [PubMed]

| RESV | RESV + PRI-2191 1 | |
|---|---|---|
| NCI-H1703 | 21.6 ± 5.9 | 17.8 ± 3.3 |
| NCI-H1581 | 31.9 ± 10.1 | 34.3 ± 11.8 |
| NCI-H358 | 39.8 ± 1.6 | 47.4 ± 2.9 |
| A549 | 43.9 ± 3.2 | 36.5 ± 7.4 |
| NCI-H1299 | 52.2 ± 7.0 | 48.8 ± 8.5 |
| HCC827 | 60.6 ± 12.0 | 51.9 ± 13.2 |
| A-427 | 60.8 ± 25.0 | 71.6 ± 35.0 |
| Calu-3 | 231.8 ± 110.3 | 184.1 ± 64.6 |
| PRI-2191 | ||
|---|---|---|
| 1000 nM | 100 nM | |
| A-427 | 3.37 ± 4.91 | 2.57 ± 3.36 |
| A549 | 2.09 ± 1.38 | 0.55 ± 1.82 |
| Calu-3 | 0.70 ± 0.99 | 1.28 ± 0.10 |
| HCC827 | 20.47 ± 6.88 | 7.52 ± 4.63 |
| NCI-H1299 | 0.61 ± 0.44 | 0.45 ± 0.54 |
| NCI-H1581 | 3.11 ± 1.50 | 0.60 ± 0.24 |
| NCI-H1703 | 11.50 ± 5.74 | 8.82 ± 1.72 |
| NCI-H358 | * 10.63 ± 0.91 | * 12.83 ± 2.69 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maj, E.; Maj, B.; Bobak, K.; Gos, M.; Chodyński, M.; Kutner, A.; Wietrzyk, J. Differential Response of Lung Cancer Cells, with Various Driver Mutations, to Plant Polyphenol Resveratrol and Vitamin D Active Metabolite PRI-2191. Int. J. Mol. Sci. 2021, 22, 2354. https://doi.org/10.3390/ijms22052354
Maj E, Maj B, Bobak K, Gos M, Chodyński M, Kutner A, Wietrzyk J. Differential Response of Lung Cancer Cells, with Various Driver Mutations, to Plant Polyphenol Resveratrol and Vitamin D Active Metabolite PRI-2191. International Journal of Molecular Sciences. 2021; 22(5):2354. https://doi.org/10.3390/ijms22052354
Chicago/Turabian StyleMaj, Ewa, Beata Maj, Klaudia Bobak, Michalina Gos, Michał Chodyński, Andrzej Kutner, and Joanna Wietrzyk. 2021. "Differential Response of Lung Cancer Cells, with Various Driver Mutations, to Plant Polyphenol Resveratrol and Vitamin D Active Metabolite PRI-2191" International Journal of Molecular Sciences 22, no. 5: 2354. https://doi.org/10.3390/ijms22052354
APA StyleMaj, E., Maj, B., Bobak, K., Gos, M., Chodyński, M., Kutner, A., & Wietrzyk, J. (2021). Differential Response of Lung Cancer Cells, with Various Driver Mutations, to Plant Polyphenol Resveratrol and Vitamin D Active Metabolite PRI-2191. International Journal of Molecular Sciences, 22(5), 2354. https://doi.org/10.3390/ijms22052354






