The Distribution of Several Genomic Virulence Determinants Does Not Corroborate the Established Serotyping Classification of Bacillus thuringiensis
Abstract
1. Introduction
2. Results
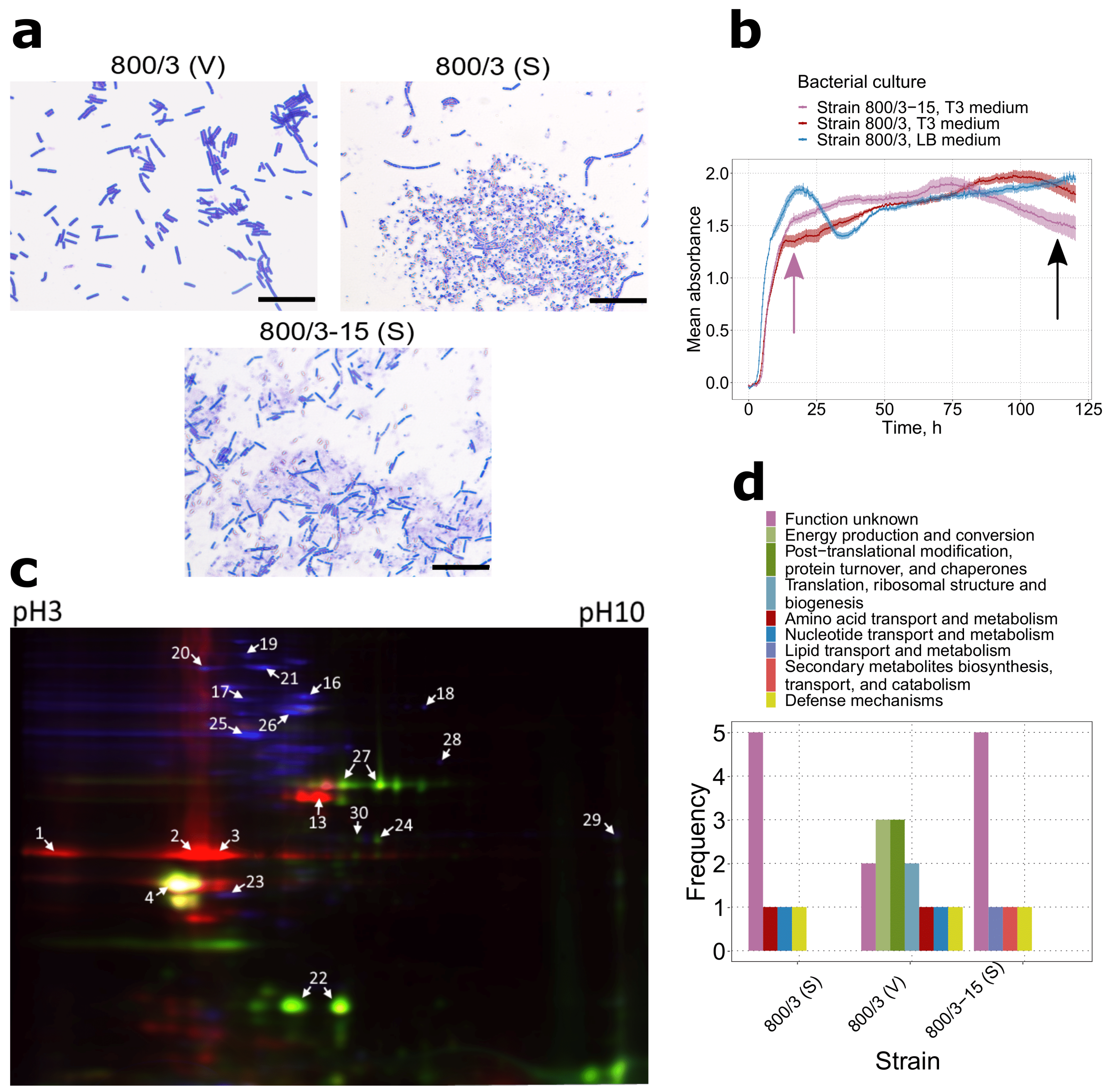
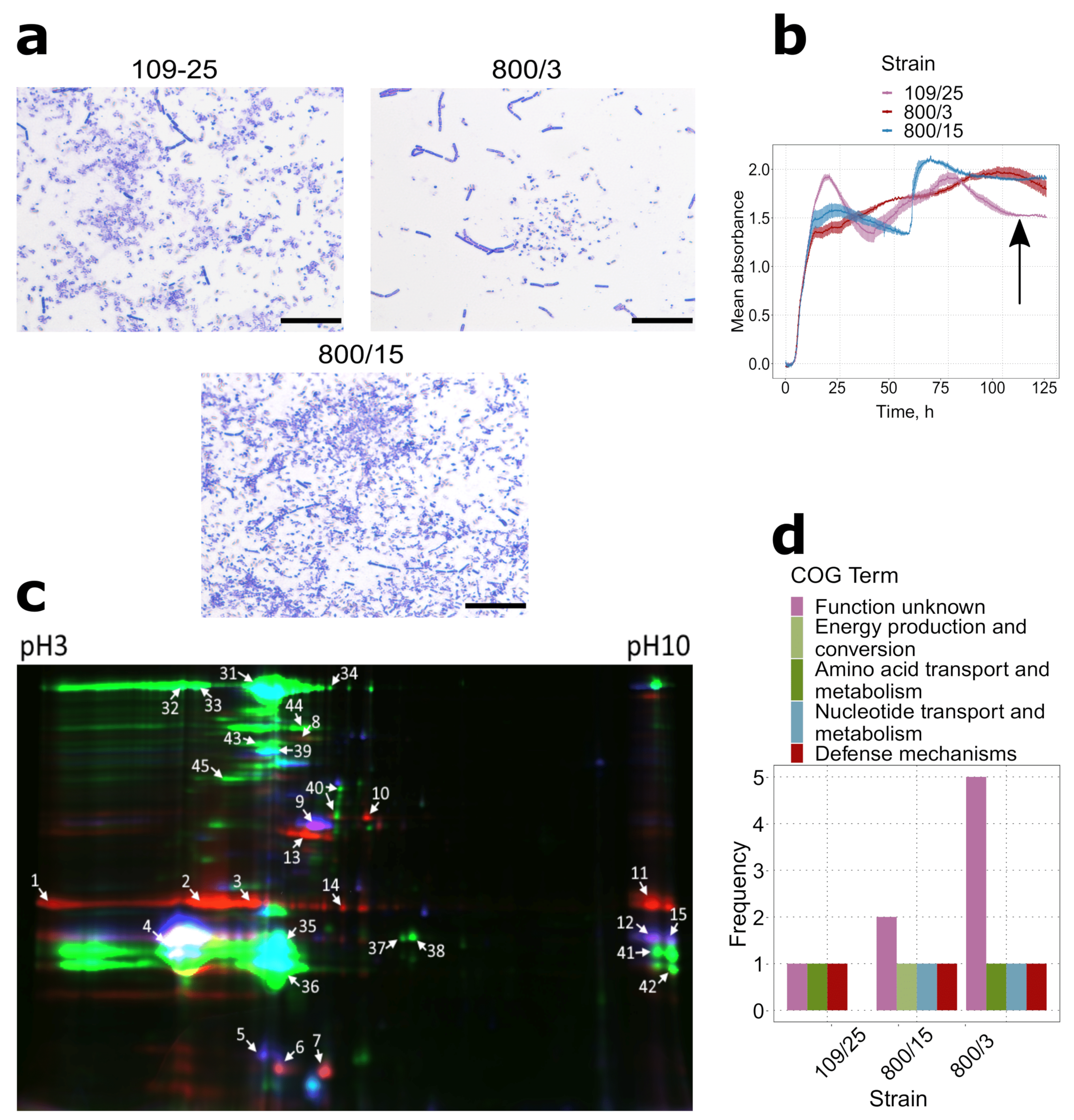
2.1. Virulence Factors Are Enriched in the Proteomes of Bt var. israelensis Virulent Sporulating Cultures Compared to the Avirulent and Vegetative Ones
2.2. Spores of Serovars Israelenses, Darmstatdiensis, and Thuringiensis Demonstrate Distinct Patterns of Protein Presence
2.3. The Distribution of Genes Corresponding to Products Identified with Proteomics Assay among Bt Genomes Does Not Support Strains’ Serovar Attribution
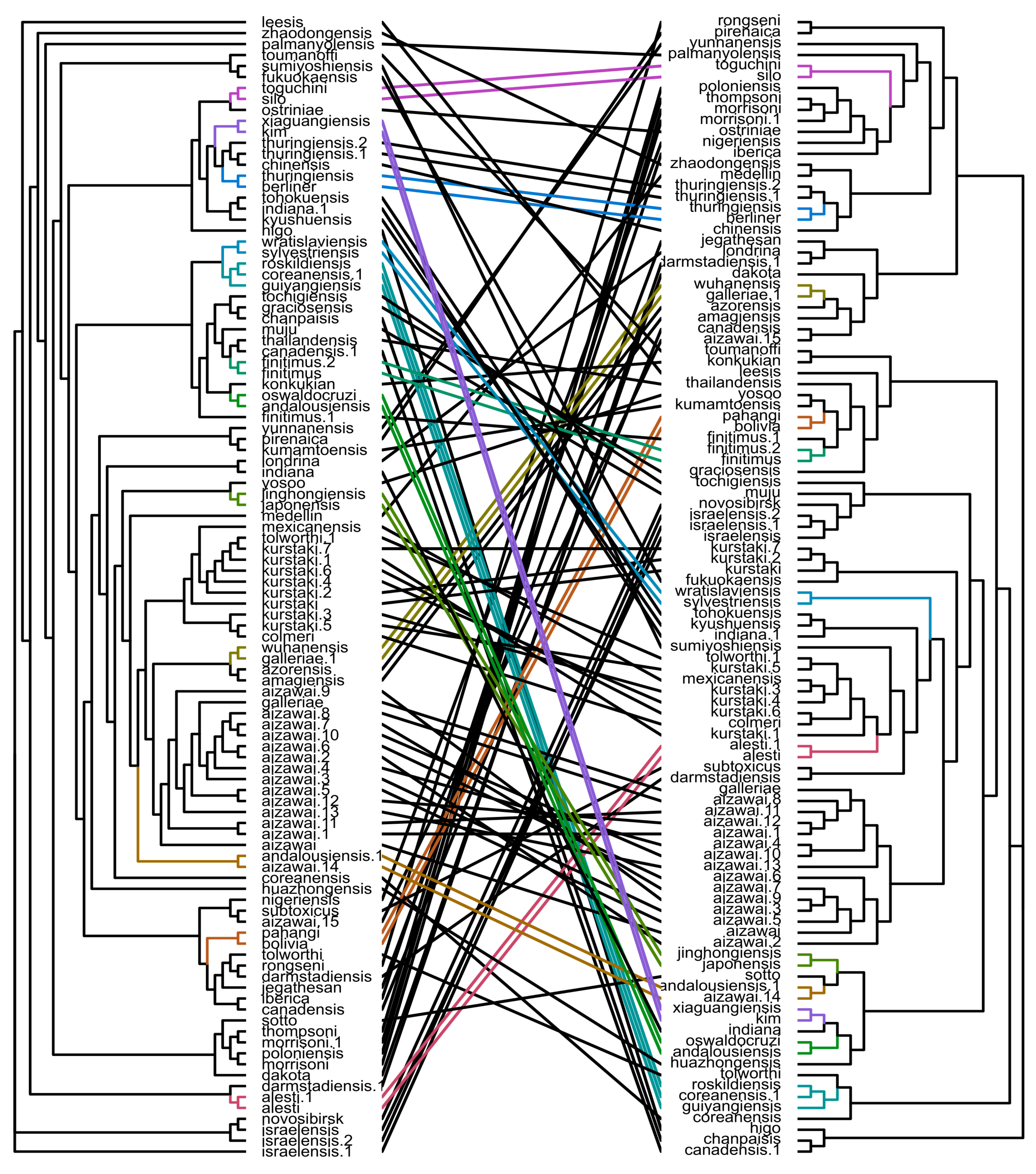
2.4. Pangenome-Wise Phylogeny Does Not Also Correlate with Serotyping Classification
2.4.1. Pangenome Reconstruction
2.4.2. Pangenome-Derived Phylogeny
2.5. Flagellin-Based Phylogeny Is Remarkably Distinct from the Pangenome-Based Inference
2.6. Single-Loci and Genome-Wise Phylogenetic Trees Are Consistent with Each Other and Serotyping Classification at Different Degree
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growing Conditions
4.2. Protein Extraction, Two-Dimensional Fluorescent Difference Gel Electrophoresis, and Protein Mass-Spectrometry
4.3. Protein Functional Annotation
4.4. Flagellin Sequence Search in the Genomic Data
4.5. Bt Genomes Acquisition and Pan-Genome Reconstruction
4.5.1. Data Acquisition
4.5.2. Gene Presence Analysis
4.5.3. Pangenome Reconstruction
4.6. Phylogeny Reconstruction
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Glazer, A.; Nikaido, H. Microbial Biotechnology: Fundamentals of Applied Microbiology, 2nd ed.; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Norris, J.R. The Classification of Bacillus thuringiensis. J. Appl. Bacteriol. 1964, 27, 439–447. [Google Scholar] [CrossRef]
- Heimpel, A.M.; Angus, T.A. The Taxonomy of Insect Pathogens Related to Bacillus cereus Frankland and Frankland. Can. J. Microbiol. 1958, 4, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Barjac, H.; Frachon, E. Classification of Bacillus thuringiensis strains. BioControl 1990, 35, 233–240. [Google Scholar] [CrossRef]
- Lecadet, M.-M.; Frachon, E.; Dumanoir, V.C.; Ripouteau, H.; Hamon, S.; Laurent, P.; Thiéry, I. Updating the H-antigen classification of Bacillus thuringiensis. J. Appl. Microbiol. 1999, 86, 660–672. [Google Scholar] [CrossRef] [PubMed]
- Sanchis, V.; Chaufaux, J.; Lereclus, D. Biotechnological improvement of Bacillus Thuringiensis: Dangers and risks. Ann. Inst. Pasteur Actual. 1996, 7, 271–284. [Google Scholar] [CrossRef]
- Jakhar, A.; Kashyap, L.; Nath Goswami, T.; Kumar Patel, V.; Kumar Sharma, R. Bacillus thuringiensis and insect pest management. In Biopesticides and Bioagents; Apple Academic Press: Palm Bay, FL, USA, 2017; pp. 331–369. [Google Scholar]
- Xu, D.; Côté, J.C. Sequence diversity of the Bacillus thuringiensis and B. cereus sensu lato flagellin (H antigen) protein: Comparison with H serotype diversity. Appl. Environ. Microbiol. 2006, 72, 4653–4662. [Google Scholar] [CrossRef] [PubMed]
- Malovichko, Y.V.; Nizhnikov, A.A.; Antonets, K.S. Repertoire of the Bacillus thuringiensis Virulence Factors Unrelated to Major Classes of Protein Toxins and Its Role in Specificity of Host-Pathogen Interactions. Toxins 2019, 11, 347. [Google Scholar] [CrossRef]
- Wang, J.; Mei, H.; Qian, H.; Tang, Q.; Liu, X.; Yu, Z.; He, J. Expression Profile and Regulation of Spore and Parasporal Crystal Formation-Associated Genes in Bacillus thuringiensis. J. Proteome Res. 2013, 12, 5487–5501. [Google Scholar] [CrossRef]
- Huang, S.; Ding, X.; Sun, Y.; Yang, Q.; Xiao, X.; Cao, Z.; Xia, L. Proteomic Analysis of Bacillus thuringiensis at Different Growth Phases by Using an Automated Online Two-Dimensional Liquid Chromatography-Tandem Mass Spectrometry Strategy. Appl. Environ. Microbiol. 2012, 78, 5270–5279. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Gao, Q.; Liu, L.; Liu, H.; Wang, Y.; Peng, D.; Ruan, L.; Raymond, B.; Sun, M. Comparative Genomics of Bacillus thuringiensis Reveals a Path to Specialized Exploitation of Multiple Invertebrate Hosts. mBio 2017, 8. [Google Scholar] [CrossRef]
- Baek, I.; Lee, K.; Goodfellow, M.; Chun, J. Comparative Genomic and Phylogenomic Analyses Clarify Relationships Within and Between Bacillus cereus and Bacillus thuringiensis: Proposal for the Recognition of Two Bacillus thuringiensis Genomovars. Front. Microbiol. 2019, 10, 1978. [Google Scholar] [CrossRef]
- Alves, G.B.; Melo, F.L.; Oliveira, E.E.; Haddi, K.; Costa, L.T.M.; Dias, M.L.; Campos, F.S.; Pereira, E.J.G.; Corrêa, R.F.T.; Ascêncio, S.D.; et al. Comparative genomic analysis and mosquito larvicidal activity of four Bacillus thuringiensis serovar israelensis strains. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hollensteiner, J.; Poehlein, A.; Spröer, C.; Bunk, B.; Sheppard, A.E.; Rosentstiel, P.; Schulenburg, H.; Liesegang, H. Complete Genome sequence of the nematicidal Bacillus thuringiensis MYBT18246. Stand. Genomic Sci. 2017, 12, 48. [Google Scholar] [CrossRef] [PubMed]
- Rang, J.; He, H.; Wang, T.; Ding, X.; Zuo, M.; Quan, M.; Sun, Y.; Yu, Z.; Hu, S.; Xia, L. Comparative Analysis of Genomics and Proteomics in Bacillus thuringiensis 4.0718. PLoS ONE 2015, 10, e0119065. [Google Scholar] [CrossRef] [PubMed]
- Quan, M.; Xie, J.; Liu, X.; Li, Y.; Rang, J.; Zhang, T.; Zhou, F.; Xia, L.; Hu, S.; Sun, Y.; et al. Comparative Analysis of Genomics and Proteomics in the New Isolated Bacillus thuringiensis X022 Revealed the Metabolic Regulation Mechanism of Carbon Flux Following Cu2+ Treatment. Front. Microbiol. 2016, 7, 792. [Google Scholar] [CrossRef][Green Version]
- Sajid, M.; Geng, C.; Li, M.; Wang, Y.; Liu, H.; Zheng, J.; Peng, D.; Sun, M. Whole-Genome Analysis of Bacillus thuringiensis Revealing Partial Genes as a Source of Novel Cry Toxins. Appl. Environ. Microbiol. 2018, 84, e00277-18. [Google Scholar] [CrossRef]
- Thomas, D.J.I.; Morgan, J.A.W.; Whipps, J.M.; Saunders, J.R. Plasmid transfer between the Bacillus thuringiensis subspecies kurstaki and tenebrionis in laboratory culture and soil and in lepidopteran and coleopteran larvae. Appl. Environ. Microbiol. 2000, 66, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Méric, G.; Mageiros, L.; Pascoe, B.; Woodcock, D.J.; Mourkas, E.; Lamble, S.; Bowden, R.; Jolley, K.A.; Raymond, B.; Sheppard, S.K. Lineage-specific plasmid acquisition and the evolution of specialized pathogens in Bacillus thuringiensis and the Bacillus cereus group. Mol. Ecol. 2018, 27, 1524–1540. [Google Scholar] [CrossRef] [PubMed]
- Jensen, G.B.; Hansen, B.M.; Eilenberg, J.; Mahillon, J. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 2003, 5, 631–640. [Google Scholar] [CrossRef]
- Asano, S.I.; Nukumizu, Y.; Bando, H.; Iizuka, T.; Yamamoto, T. Cloning of novel enterotoxin genes from Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 1997, 63, 1054–1057. [Google Scholar] [CrossRef] [PubMed]
- Unlü, M.; Morgan, M.E.; Minden, J.S. Difference gel electrophoresis. A single gel method for detecting changes in protein extracts. Electrophoresis 1997, 18, 2071–2077. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fonslow, B.R.; Shan, B.; Baek, M.-C.; Yates, J.R. Protein Analysis by Shotgun/Bottom-up Proteomics. Chem. Rev. 2013, 113, 2343–2394. [Google Scholar] [CrossRef] [PubMed]
- Maltseva, A.L.; Varfolomeeva, M.A.; Lobov, A.A.; Tikanova, P.; Panova, M.; Mikhailova, N.A.; Granovitch, A.I. Proteomic similarity of the Littorinid snails in the evolutionary context. PeerJ 2020, 8, e8546. [Google Scholar] [CrossRef] [PubMed]
- Ermolova, V.P.; Grishechkina, S.D.; Belousova, M.E.; Antonets, K.S.; Nizhnikov, A.A. Insecticidal properties of Bacillus thuringiensis var. israelensis. II. Comparative morphological and molecular genetic analysis of the crystallogenic and acrystallogenic strains. Sel’skokhozyaistvennaya Biol. 2019, 54, 1281–1289. [Google Scholar] [CrossRef]
- Ermolova, V.P.; Grishechkina, S.D.; Rakhman, A.M.; Antonets, K.S.; Belousova, M.E.; Yakhno, V.V.; Nizhnikov, A.A. Insecticidal properties of Bacillus thuringiensis var. israelensis. I. The activity spectrum of a larvicidal preparation based on industrial strain 7-1/23A. Sel’skokhozyaistvennaya Biol. 2019, 54, 1268–1280. [Google Scholar] [CrossRef]
- Tikhonovich, I.A.; Grishechkina, S.D.; Ermolova, V.P.; Romanova, T.A. Strain Bacillus thuringiensis var darmstadiensis n25 as means of integrated effect on harmful coleopteran insects and phytopathogenic fungi 2014. Patent RU2514023, 27 April 2014. [Google Scholar]
- Tikhonovich, I.A.; Romanova, T.A.; Ermolova, V.P.; Grishechkina, S.D. Bacterial strain Bacillus thuringiensis var thuringiensis n800/15 as agent for preparing entomocidal biopreparation 2012. Patent RU2012143320, 10 October 2012. [Google Scholar]
- Soufiane, B.; Côté, J.-C. Discrimination among Bacillus thuringiensis H serotypes, serovars and strains based on 16S rRNA, gyrB and aroE gene sequence analyses. Antonie Van Leeuwenhoek 2008, 95, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Nisnevitch, M.; Cohen, S.; Ben-Dov, E.; Zaritsky, A.; Sofer, Y.; Cahan, R. Cyt2Ba of Bacillus thuringiensis israelensis: Activation by putative endogenous protease. Biochem. Biophys. Res. Commun. 2006, 344, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Nisnevitch, M.; Sigawi, S.; Cahan, R.; Nitzan, Y. Isolation, characterization and biological role of camelysin from Bacillus thuringiensis subsp. israelensis. Curr. Microbiol. 2010, 61, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tang, M.; Liao, Y.; Xu, W. Interactions between molecular chaperone P20 and Cyt2Ba7 toxin in Bacillus thuringiensis. bioRxiv 2017, 129833. [Google Scholar] [CrossRef]
- Diaz-Mendoza, M.; Bideshi, D.K.; Ortego, F.; Farinós, G.P.; Federici, B.A. The 20-kDa chaperone-like protein of Bacillus thuringiensis ssp. israelensis enhances yield, crystal size and solubility of Cry3A. Lett. Appl. Microbiol. 2012, 54, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Chitlaru, T.; Gat, O.; Gozlan, Y.; Ariel, N.; Shafferman, A. Differential Proteomic Analysis of the Bacillus anthracis Secretome: Distinct Plasmid and Chromosome CO2-Dependent Cross Talk Mechanisms Modulate Extracellular Proteolytic Activities. J. Bacteriol. 2006, 188, 3551–3571. [Google Scholar] [CrossRef] [PubMed]
- Gohar, M.; Økstad, O.A.; Gilois, N.; Sanchis, V.; Kolstø, A.B.; Lereclus, D. Two-dimensional electrophoresis analysis of the extracellular proteome of Bacillus cereus reveals the importance of the PlcR regulon. Proteomics 2002, 2, 784–791. [Google Scholar] [CrossRef]
- Li, X.; Ding, X.; Xia, L.; Sun, Y.; Yuan, C.; Yin, J. Proteomic Analysis of Bacillus thuringiensis Strain 4.0718 at Different Growth Phases. Sci. World J. 2012, 2012, 1–10. [Google Scholar] [CrossRef]
- Song, S.; Xia, L.Q.; Huang, J.L.; Sun, Y.J.; Ding, X.Z. Analysis of insecticidal crystal proteins from Bacillus thuringiensis strain 4.0718 through two-dimensional gel electrophoresis and MALDI-TOF-mass spectrometry. Wei Sheng Wu Xue Bao 2005, 45, 467–471. [Google Scholar]
- Ding, X.; Huang, J.; Xia, L.; Li, X.; Yuan, C.; Dan, S. A proteomic analysis approach to study insecticidal crystal proteins from different strains of Bacillus thuringiensis. Biocontrol Sci. Technol. 2009, 19, 289–299. [Google Scholar] [CrossRef]
- Martínez-Gomariz, M.; Hernáez, M.L.; Gutiérrez, D.; Ximénez-Embún, P.; Preéstamo, G. Proteomic Analysis by Two-Dimensional Differential Gel Electrophoresis (2D DIGE) of a High-Pressure Effect in Bacillus cereus. J. Agric. Food Chem. 2009, 57, 3543–3549. [Google Scholar] [CrossRef]
- Bardot, C.; Besse-Hoggan, P.; Carles, L.; Le Gall, M.; Clary, G.; Chafey, P.; Federici, C.; Broussard, C.; Batisson, I. How the edaphic Bacillus megaterium strain Mes11 adapts its metabolism to the herbicide mesotrione pressure. Environ Pollut. 2015, 199, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Cancino-Rodezno, A.; Lozano, L.; Oppert, C.; Castro, J.I.; Lanz-Mendoza, H.; Encarnación, S.; Evans, A.E.; Gill, S.S.; Soberón, M.; Jurat-Fuentes, J.L.; et al. Comparative Proteomic Analysis of Aedes aegypti Larval Midgut after Intoxication with Cry11Aa Toxin from Bacillus thuringiensis. PLoS ONE 2012, 7, e37034. [Google Scholar] [CrossRef] [PubMed]
- Hoch, P.G.; Burenina, O.Y.; Weber, M.H.; Elkina, D.A.; Nesterchuk, M.V.; Sergiev, P.V.; Hartmann, R.K.; Kubareva, E.A. Phenotypic characterization and complementation analysis of Bacillus subtilis 6S RNA single and double deletion mutants. Biochimie 2015, 117, 87–99. [Google Scholar] [CrossRef]
- Dalhammar, G.; Steiner, H. Characterization of inhibitor A, a protease from Bacillus thuringiensis which degrades attacins and cecropins, two classes of antibacterial proteins in insects. JBIC J. Biol. Inorg. Chem. 1984, 139, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Dammak, I.; Dammak, M.; Tounsi, S. Histopathological and combinatorial effects of the metalloprotease InhA1 and Cry proteins of Bacillus thuringiensis against Spodoptera littoralis. Int. J. Biol. Macromol. 2015, 81, 759–762. [Google Scholar] [CrossRef] [PubMed]
- Grass, G.; Schierhorn, A.; Sorkau, E.; Müller, H.; Rücknagel, P.; Nies, D.H.; Fricke, B. Camelysin Is a Novel Surface Metalloproteinase from Bacillus cereus. Infect. Immun. 2004, 72, 219–228. [Google Scholar] [CrossRef][Green Version]
- Slamti, L.; Perchat, S.; Huillet, E.; Lereclus, D. Quorum sensing in Bacillus thuringiensis is required for completion of a full infectious cycle in the insect. Toxins 2014, 6, 2239–2255. [Google Scholar] [CrossRef] [PubMed]
- Pomerantsev, A.P.; Pomerantseva, O.M.; Camp, A.S.; Mukkamala, R.; Goldman, S.; Leppla, S.H. PapR peptide maturation: Role of the NprB protease in Bacillus cereus 569 PlcR/PapR global gene regulation. FEMS Immunol. Med Microbiol. 2009, 55, 361–377. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.-C.; Popova, T.G.; Millis, B.A.; Mukherjee, D.V.; Zhou, W.; Liotta, L.A.; Petricoin, E.F.; Chandhoke, V.; Bailey, C.; Popov, S.G. Secreted Neutral Metalloproteases of Bacillus anthracis as Candidate Pathogenic Factors. J. Biol. Chem. 2006, 281, 31408–31418. [Google Scholar] [CrossRef] [PubMed]
- Ceuppens, S.; Rajkovic, A.; Heyndrickx, M.; Tsilia, V.; Van De Wiele, T.; Boon, N.; Uyttendaele, M. Regulation of toxin production by Bacillus cereus and its food safety implications. Crit. Rev. Microbiol. 2011, 37, 188–213. [Google Scholar] [CrossRef]
- Cadot, C.; Tran, S.-L.; Vignaud, M.-L.; De Buyser, M.-L.; Kolstø, A.-B.; Brisabois, A.; Nguyen-Thé, C.; Lereclus, D.; Guinebretière, M.-H.; RamaRao, N. InhA1, NprA, and HlyII as Candidates for Markers To Differentiate Pathogenic from Nonpathogenic Bacillus cereus Strains. J. Clin. Microbiol. 2010, 48, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Rasigade, J.-P.; Hollandt, F.; Wirth, T. Genes under positive selection in the core genome of pathogenic Bacillus cereus group members. Infect. Genet. Evol. 2018, 65, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Ramirez, A.; Ibarra, J.E. Fingerprinting of Bacillus thuringiensis Type Strains and Isolates by Using Bacillus cereus Group-Specific Repetitive Extragenic Palindromic Sequence-Based PCR Analysis. Appl. Environ. Microbiol. 2005, 71, 1346–1355. [Google Scholar] [CrossRef] [PubMed]
- Ohba, M.; Aizawa, K. Frequency of acrystalliferous spore-forming bacteria possessing flagellar antigens of Bacillus thuringiensis. J. Basic Microbiol. 1986, 26, 185–188. [Google Scholar] [CrossRef]
- Miteva, V.; Abadjieva, A.; Grigorova, R. Differentiation among strains and serotypes of Bacillus thuringiensis by M13 DNA fingerprinting. J. Gen. Microbiol. 1991, 137, 593–600. [Google Scholar] [CrossRef][Green Version]
- Chaves, J.Q.; Cavados CF, G.; Rabinovitch, L. Phenotypic and genotypic features of new autoagglutinating Bacillus thuringiensis strains. J. Invertebr. Pathol. 2008, 98, 85–92. [Google Scholar] [CrossRef]
- Sacchi, C.T.; Whitney, A.M.; Mayer, L.W.; Morey, R.; Steigerwalt, A.; Boras, A.; Weyant, R.S.; Popovic, T. Sequencing of 16S rRNA Gene: A Rapid Tool for Identification of Bacillus anthracis. Emerg. Infect. Dis. 2002, 8, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Tsen, H. Discrimination of Bacillus cereus and Bacillus thuringiensis with 16S rRNA and gyrB gene based PCR primers and sequencing of their annealing sites. J. Appl. Microbiol. 2002, 92, 912–919. [Google Scholar] [CrossRef] [PubMed]
- Bavykin, S.G.; Lysov, Y.P.; Zakhariev, V.; Kelly, J.J.; Jackman, J.; Stahl, D.A.; Cherni, A. Use of 16S rRNA, 23S rRNA, and gyrB gene sequence analysis to determine phylogenetic relationships of Bacillus cereus group microorganisms. J. Clin. Microbiol. 2004, 42, 3711–3730. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lai, Q.; Göker, M.; Meier-Kolthoff, J.P.; Wang, M.; Sun, Y.; Wang, L.; Shao, Z. Genomic insights into the taxonomic status of the Bacillus cereus group. Sci. Rep. 2015, 5, srep14082. [Google Scholar] [CrossRef]
- Xu, D.; Côté, J.C. Sequence Diversity of Bacillus thuringiensis Flagellin (H Antigen) Protein at the Intra-H Serotype Level. Appl. Environ. Microbiol. 2008, 74, 5524–5532. [Google Scholar] [CrossRef]
- La Duc, M.T.; Satomi, M.; Agata, N.; Venkateswaran, K. gyrB as a phylogenetic discriminator for members of the Bacillus anthracis-cereus-thuringiensis group. J. Microbiol. Methods 2004, 56, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Lopez de la Cruz, D.; Valencia-Castro, C.M.; Hernandez-Teran, F.; Barboza Corona, J.E.; de la Fuente Salcido, N.M. Antibacterial Activity of Native Bacillus thuringiensis Strains from Fernandez Canyon State Park, Mexico. J. Antimicrob. Agents 2018, 4, 1–5. [Google Scholar] [CrossRef]
- Reinoso-Pozo, Y.; Del Rincón-Castro, M.C.; Ibarra, J.E. Characterization of a highly toxic strain of Bacillus thuringiensis serovar kurstaki very similar to the HD-73 strain. FEMS Microbiol. Lett. 2016, 363, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ruiu, L.; Falchi, G.; Floris, I.; Marche, M.G.; Mura, M.E.; Satta, A. Pathogenicity and characterization of a novel Bacillus cereus sensu lato isolate toxic to the Mediterranean fruit fly Ceratitis capitata Wied. J. Invertebr. Pathol. 2015, 126, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zuo, M.; Wang, T.; Sun, Y.; Liu, S.; Hu, S.; He, H.; Yang, Q.; Rang, J.; Quan, M.; et al. Proteomic analysis of the influence of Cu2+ on the crystal protein production of Bacillus thuringiensis X022. Microb. Cell Factories 2015, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Koh, I.; Lim, M.Y.; Chung, W.-H.; Rho, M. Pan-genome analysis of Bacillus for microbiome profiling. Sci. Rep. 2017, 7, 10984. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wang, D.; Gao, F. Toward a high-quality pan-genome landscape of Bacillus subtilis by removal of confounding strains. Briefings Bioinform. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bazinet, A.L. Pan-genome and phylogeny of Bacillus cereus sensu lato. BMC Evol. Biol. 2017, 17, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Li, Z.; Liu, J.; Shu, C.; Wang, X.; Zhang, X.; Yu, X.; Zhao, D.; Liu, G.; Hu, S.; et al. A pangenomic study of Bacillus thuringiensis. J. Genet. Genom. 2011, 38, 567–576. [Google Scholar] [CrossRef]
- Wang, A.; Ash, G.J. Whole Genome Phylogeny of Bacillus by Feature Frequency Profiles (FFP). Sci. Rep. 2015, 5, 5. [Google Scholar] [CrossRef]
- Shakya, M.; Ahmed, S.A.; Davenport, K.W.; Flynn, M.C.; Lo, C.-C.; Chain, P.S.G. Standardized phylogenetic and molecular evolutionary analysis applied to species across the microbial tree of life. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sahl, J.W.; Vazquez, A.J.; Hall, C.M.; Busch, J.D.; Tuanyok, A.; Mayo, M.; Schupp, J.M.; Lummis, M.; Pearson, T.; Shippy, K.; et al. The Effects of Signal Erosion and Core Genome Reduction on the Identification of Diagnostic Markers. mBio 2016, 7, e00846-16. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.J.A.; Tasara, T.; Klumpp, J.; Stephan, R.; Ehling-Schulz, M.; Johler, S. Whole-genome-based phylogeny of Bacillus cytotoxicus reveals different clades within the species and provides clues on ecology and evolution. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Keshri, J.; Ramirez, R.; Berrang, M.E.; Oakley, B.B. Draft Genome Sequences of Two Potentially Novel Bacillus Isolates from Backyard and Commercial Chicken Gastrointestinal Tracts. Microbiol. Resour. Announc. 2020, 9, 9. [Google Scholar] [CrossRef]
- Carroll, L.M.; Wiedmann, M.; Kovac, J. Proposal of a Taxonomic Nomenclature for the Bacillus cereus Group Which Reconciles Genomic Definitions of Bacterial Species with Clinical and Industrial Phenotypes. mBio 2020, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, M.B.; Martin, P.A.W.; Kuhar, D.; Farrar, R.R.; Gundersen-Rindal, D.E. Phylogenetic Distribution of Phenotypic Traits in Bacillus thuringiensis Determined by Multilocus Sequence Analysis. PLoS ONE 2013, 8, e66061. [Google Scholar] [CrossRef] [PubMed]
- Schnepf, E.; Crickmore, N.; Van Rie, J.; Lereclus, D.; Baum, J.; Feitelson, J.; Zeigler, D.R.; Dean, D.H. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 1998, 62, 775–806. [Google Scholar] [CrossRef] [PubMed]
- González, J.M., Jr.; Carlton, B.C. A large transmissible plasmid is required for crystal toxin production in Bacillus thuringiensis variety israelensis. Plasmid 1984, 11, 28–38. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, C.; Zhu, Y.; Deng, Y.; Guo, S.; Peng, D.; Ruan, L.; Sun, M. The resolution and regeneration of a cointegrate plasmid reveals a model for plasmid evolution mediated by conjugation and oriT site-specific recombination. Environ. Microbiol. 2013, 15, 3305–3318. [Google Scholar] [CrossRef]
- Wang, Y.; Peng, D.; Dong, Z.; Zhu, L.; Guo, S.; Sun, M. Cloning and analysis of a large plasmid pBMB165 from Bacillus thuringiensis revealed a novel plasmid organization. PLoS ONE 2013, 8, 1–7. [Google Scholar] [CrossRef][Green Version]
- Lechuga, A.; Lood, C.; Salas, M.; Van Noort, V.; Lavigne, R.; Redrejo-Rodríguez, M. Completed Genomic Sequence of Bacillus thuringiensis HER1410 Reveals a Cry-Containing Chromosome, Two Megaplasmids, and an Integrative Plasmidial Prophage. G3 Genes Genomes Genet. 2020, 10, 2927–2939. [Google Scholar] [CrossRef] [PubMed]
- Travers, R.S.; Martin, P.A.W.; Reichelderfer, C.F. Selective Process for Efficient Isolation of Soil Bacillus spp. Appl. Environ. Microbiol. 1987, 53, 1263–1266. [Google Scholar] [CrossRef]
- Ginestet, C. ggplot2: Elegant Graphics for Data Analysis. J. R. Stat. Soc. Ser. A 2011, 174, 245–246. [Google Scholar] [CrossRef]
- Rampersad, J.; Khan, A.; Ammons, D. Usefulness of staining parasporal bodies when screening for Bacillus thuringiensis. J. Invertebr. Pathol. 2002, 79, 203–204. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nat. Cell Biol. 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bourexis, D.; Brister, J.R.; Bryant, S.H.; Canese, K.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar]
- Bateman, A. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef] [PubMed]
- Shikov, A.E.; Malovichko, Y.V.; Skitchenko, R.K.; Nizhnikov, A.A.; Antonets, K.S. No More Tears: Mining Sequencing Data for Novel Bt Cry Toxins with CryProcessor. Toxins 2020, 12, 204. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Brückner, A.; Heethoff, M. A chemo-ecologists’ practical guide to compositional data analysis. Chemoecology 2016, 27, 33–46. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2018, 35, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Demidenko, E. The next-generationK-means algorithm. Stat. Anal. Data Mining: ASA Data Sci. J. 2018, 11, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Horikoshi, M.; Li, W. ggfortify: Unified Interface to Visualize Statistical Results of Popular R Packages. R J. 2016, 8, 474–485. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Škunca, N.; Dessimoz, C. Phylogenetic Profiling: How Much Input Data Is Enough? PLoS ONE 2015, 10, e0114701. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.i.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T. ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2016, 8, 28–36. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. SNP-sites: Rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2016, 2, e000056. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Harris, C.R.; Millman, K.J.; Van Der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nat. Cell Biol. 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Sand, A.; Holt, M.K.; Johansen, J.; Brodal, G.S.; Mailund, T.; Pedersen, C.N.S. tqDist: A library for computing the quartet and triplet distances between binary or general trees. Bioinformatics 2014, 30, 2079–2080. [Google Scholar] [CrossRef]
- Galili, T. dendextend: An R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.S. Using DECIPHER v2.0 to analyze big biological sequence data in R. R J. 2016, 8, 352–359. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shikov, A.E.; Malovichko, Y.V.; Lobov, A.A.; Belousova, M.E.; Nizhnikov, A.A.; Antonets, K.S. The Distribution of Several Genomic Virulence Determinants Does Not Corroborate the Established Serotyping Classification of Bacillus thuringiensis. Int. J. Mol. Sci. 2021, 22, 2244. https://doi.org/10.3390/ijms22052244
Shikov AE, Malovichko YV, Lobov AA, Belousova ME, Nizhnikov AA, Antonets KS. The Distribution of Several Genomic Virulence Determinants Does Not Corroborate the Established Serotyping Classification of Bacillus thuringiensis. International Journal of Molecular Sciences. 2021; 22(5):2244. https://doi.org/10.3390/ijms22052244
Chicago/Turabian StyleShikov, Anton E., Yury V. Malovichko, Arseniy A. Lobov, Maria E. Belousova, Anton A. Nizhnikov, and Kirill S. Antonets. 2021. "The Distribution of Several Genomic Virulence Determinants Does Not Corroborate the Established Serotyping Classification of Bacillus thuringiensis" International Journal of Molecular Sciences 22, no. 5: 2244. https://doi.org/10.3390/ijms22052244
APA StyleShikov, A. E., Malovichko, Y. V., Lobov, A. A., Belousova, M. E., Nizhnikov, A. A., & Antonets, K. S. (2021). The Distribution of Several Genomic Virulence Determinants Does Not Corroborate the Established Serotyping Classification of Bacillus thuringiensis. International Journal of Molecular Sciences, 22(5), 2244. https://doi.org/10.3390/ijms22052244







