Current Understanding of Role of Vesicular Transport in Salt Secretion by Salt Glands in Recretohalophytes
Abstract
1. Introduction
2. Structural Features of Salt Glands
3. Physiological Studies of Salt Glands
4. Cytological Studies of Salt Glands
5. Insights from Omics Applications
6. Mechanism of Vesicular Transport in Salt Secretion
7. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Morton, M.J.L.; Awlia, M.; Al-Tamimi, N.; Saade, S.; Pailles, Y.; Negrão, S.; Tester, M. Salt stress under the scalpel—Dissecting the genetics of salt tolerance. Plant J. 2019, 97, 148–163. [Google Scholar] [CrossRef]
- Shabala, S.; Wu, H.H.; Bose, J. Salt stress sensing and early signalling events in plant roots: Current knowledge and hypothesis. Plant Sci. 2015, 241, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Leng, B.; Wang, B. Progress in Studying Salt Secretion from the Salt Glands in Recretohalophytes: How Do Plants Secrete Salt? Front. Plant Sci. 2016, 7, 977. [Google Scholar] [CrossRef]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef]
- Zelm, E.V.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef]
- Flowers, T.J.; Glenn, E.P.; Volkov, V. Could vesicular transport of Na+ and Cl− be a feature of salt tolerance in halophytes? Ann. Bot. 2019, 123, 1–18. [Google Scholar] [CrossRef]
- Flowers, T.J.; Munns, R.; Colmer, T.D. Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann. Bot. 2015, 115, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Ashihara, H.; Adachi, K.; Otawa, M.; Yasumoto, E.; Fukushima, Y.; Kato, M.; Sano, H.; Sasamoto, H.; Baba, S. Compatible Solutes and Inorganic Ions in the Mangrove Plant Avicennia marina and Their Effects on the Activities of Enzymes. Z. Natrforsch. C 1997, 52, 433–440. [Google Scholar] [CrossRef]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.-Y.; Li, J.; Wang, P.-Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Zhu, J.J.C. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) Pathway: Established and Emerging Roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wang, B. Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Ann. Bot. 2015, 115, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Yang, Z.; Liu, J.; Zhu, T.; Wei, X.; Fan, H.; Wang, B. Adaptation Mechanism of Salt Excluders under Saline Conditions and Its Applications. Int. J. Mol. Sci. 2018, 19, 3668. [Google Scholar] [CrossRef] [PubMed]
- Breckle, S.W. Salinity tolerance of different halophyte types. In Genetic Aspects of Plant Mineral Nutrition; El Bassam, N., Dambroth, M., Loughman, B.C., Eds.; Springer: Dordrecht, The Netherland, 1990; Volume 42, pp. 167–175. [Google Scholar] [CrossRef]
- Liphschitz, N.; Waisel, Y. Existence of Salt Glands in Various Genera of the Gramineae. New Phytol. 1974, 73, 507–513. [Google Scholar] [CrossRef]
- Meng, X.; Zhou, J.; Sui, N. Mechanisms of salt tolerance in halophytes: Current understanding and recent advances. Cen. Eur. J. Biol. 2018, 13, 149–154. [Google Scholar] [CrossRef]
- Dassanayake, M.; Larkin, J.C. Making Plants Break a Sweat: The Structure, Function, and Evolution of Plant Salt Glands. Front. Plant Sci. 2017, 8, 406. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Arisz, W.; Camphuis, I.; Heikens, H.; Van Tooren, A.V. The secretion of the salt glands of Limonium latifolium Ktze. Acta Bot. Neerl. 1955, 4, 322–338. [Google Scholar] [CrossRef]
- Levering, C.A.; Thomson, W.W. The ultrastructure of the salt gland of Spartina foliosa. Planta 1971, 97, 183–196. [Google Scholar] [CrossRef]
- Ziegler, H.; Lüttge, U. Die Salzdrüsen von Limonium vulgare. Planta 1967, 74, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Shimony, C.; Fahn, A. Light- and electron-microscopical studies on the structure of salt glands of Tamarix aphylla L. Bot. J. Linn. Soc. 1968, 60, 283–288. [Google Scholar] [CrossRef]
- Ding, F.; Yang, J.; Yuan, F.; Wang, B.S. Progress in mechanism of salt excretion in recretohalopytes. Front. Biol. 2010, 5, 164–170. [Google Scholar] [CrossRef]
- Semenova, G.A.; Fomina, I.R.; Biel, K.Y. Structural features of the salt glands of the leaf of Distichlis spicata ‘Yensen 4a’ (Poaceae). Protoplasma 2010, 240, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Tian, C.; Feng, G.; Yuan, J. Ability of multicellular salt glands in Tamarix species to secrete Na+ and K+ selectively. Sci. China Life Sci. 2011, 54, 282–289. [Google Scholar] [CrossRef]
- Laursen, M.; Gregersen, J.L.; Yatime, L.; Nissen, P.; Fedosova, N.U. Structures and characterization of digoxin- and bufalin-bound Na+,K+-ATPase compared with the ouabain-bound complex. Proc. Natl. Acad. Sci. USA 2015, 112, 1755–1760. [Google Scholar] [CrossRef]
- Feng, Z.; Sun, Q.; Deng, Y.; Sun, S.; Zhang, J.; Wang, B. Study on pathway and characteristics of ion secretion of salt glands of Limonium bicolor. Acta Physiol. Plant 2014, 36, 2729–2741. [Google Scholar] [CrossRef]
- Wilson, H.; Mycock, D.; Weiersbye, I.M. The salt glands of Tamarix usneoides E. Mey. ex Bunge (South African Salt Cedar). Int. J. Phytoremediation 2017, 19, 587–595. [Google Scholar] [CrossRef]
- Yun, H.S.; Kwon, C. Vesicle trafficking in plant immunity. Curr. Opin. Plant Biol. 2017, 40, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, M.; Gao, C.; Zeng, Y.; Cui, Y.; Shen, W.; Jiang, L. The roles of endomembrane trafficking in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 55–69. [Google Scholar] [CrossRef]
- Qu, X.; Zhang, R.; Zhang, M.; Diao, M.; Xue, Y.; Huang, S. Organizational Innovation of Apical Actin Filaments Drives Rapid Pollen Tube Growth and Turning. Mol. Plant 2017, 10, 930–947. [Google Scholar] [CrossRef]
- Echeverria, E. Vesicle-mediated solute transport between the vacuole and the plasma membrane. Plant Physiol. 2000, 123, 1217–1226. [Google Scholar] [CrossRef]
- Van Damme, D.; Inzé, D.; Russinova, E. Vesicle Trafficking during Somatic Cytokinesis. Plant Physiol. 2008, 147, 1544–1552. [Google Scholar] [CrossRef]
- Yuan, F.; Lyu, M.J.A.; Leng, B.Y.; Zheng, G.Y.; Feng, Z.T.; Li, P.H.; Zhu, X.G.; Wang, B.S. Comparative transcriptome analysis of developmental stages of the Limonium bicolor leaf generates insights into salt gland differentiation. Plant Cell Environ. 2015, 38, 1637–1657. [Google Scholar] [CrossRef]
- Deng, Y.; Feng, Z.; Yuan, F.; Guo, J.; Suo, S.; Wang, B. Identification and functional analysis of the autofluorescent substance in Limonium bicolor salt glands. Plant Physiol. Biochem. 2015, 97, 20–27. [Google Scholar] [CrossRef]
- Tan, W.K.; Lin, Q.; Lim, T.M.; Kumar, P.; Loh, C.S. Dynamic secretion changes in the salt glands of the mangrove tree species Avicennia officinalis in response to a changing saline environment. Plant Cell Environ. 2013, 36, 1410–1422. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Bose, J.; Hedrich, R. Salt bladders: Do they matter? Trends Plant Sci. 2014, 19, 687–691. [Google Scholar] [CrossRef]
- Agarie, S.; Shimoda, T.; Shimizu, Y.; Baumann, K.; Sunagawa, H.; Kondo, A.; Ueno, O.; Nakahara, T.; Nose, A.; Cushman, J.C. Salt tolerance, salt accumulation, and ionic homeostasis in an epidermal bladder-cell-less mutant of the common ice plant Mesembryanthemum crystallinum. J. Exp. Bot. 2007, 58, 1957–1967. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.K.; Lim, T.M.; Loh, C.S. A simple, rapid method to isolate salt glands for three-dimensional visualization, fluorescence imaging and cytological studies. Plant Methods 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- Thomson, W.W.; Berry, W.L.; Liu, L.L. Localization and secretion of salt by the salt glands of Tamarix aphylla. Proc. Natl. Acad Sci. USA 1969, 63, 310–317. [Google Scholar] [CrossRef]
- Shimony, C.; Fahn, A.; Reinhold, L. Ultrastructure and ion gradients in the salt glands of Avicennia marina (Forssk.) VIERH. New Phytol. 1973, 72, 27–36. [Google Scholar] [CrossRef]
- Naidoo, Y.; Naidoo, G.; Ajmal Khan, M.; Weber, D.J. Localization of potential ion transport pathways in the salt glands of the halophyte Sporobolus virginicus. Ecophysiol. High Salin. Toler. Plants 2008, 40, 173–185. [Google Scholar] [CrossRef]
- Smaoui, A.; Barhoumi, Z.; Rabhi, M.; Abdelly, C. Localization of potential ion transport pathways in vesicular trichome cells of Atriplex halimus L. Protoplasma 2011, 248, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Naidoo, Y.; Naidoo, G. Cytochemical localisation of adenosine triphosphatase activity in salt glands of Sporobolus virginicus (L.) Kunth. S. Afr. J. Bot. 1999, 65, 370–373. [Google Scholar] [CrossRef][Green Version]
- Thomson, W.W.; Liu, L.L. Ultrastructural features of the salt gland of Tamarix aphylla L. Planta 1967, 73, 201–220. [Google Scholar] [CrossRef]
- Zouhaier, B.; Abdallah, A.; Najla, T.; Wahbi, D.; Wided, C.; Aouatef, B.A.; Chedly, A.; Abderazzak, S. Scanning and transmission electron microscopy and X-ray analysisof leaf salt glands of Limoniastrum guyonianum Boiss. under NaCl salinity. Micron 2015, 78, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Vassilyev, A.E.; Stepanova, A.A. The Ultrastructure of Ion-Secreting and Non-Secreting Salt Glands of Limonium platyphyllum. J. Exp. Bot. 1990, 41, 41–46. [Google Scholar] [CrossRef]
- Feng, Z.; Deng, Y.; Zhang, S.; Liang, X.; Yuan, F.; Hao, J.L.; Zhang, J.C.; Sun, S.F.; Wang, B.S. K(+) accumulation in the cytoplasm and nucleus of the salt gland cells of Limonium bicolor accompanies increased rates of salt secretion under NaCl treatment using NanoSIMS. Plant Sci. 2015, 238, 286–296. [Google Scholar] [CrossRef]
- Barhoumi, Z.; Djebali, W.; Abdelly, C.; Chaïbi, W.; Smaoui, A. Ultrastructure of Aeluropus littoralis leaf salt glands under NaCl stress. Protoplasma 2008, 233, 195–202. [Google Scholar] [CrossRef]
- Balsamo, R.; Thomson, W. Ultrastructural Features Associated with Secretion in the Salt Glands of Frankenia grandifolia (Frankeniaceae) and Avicennia germinans (Avicenniaceae). Am. J. Bot. 1993, 80. [Google Scholar] [CrossRef]
- Campbell, N.; Thomson, W.W. The Ultrastructural Basis of Chloride Tolerance in the Leaf of Frankenia. Ann. Bot. 1976, 40, 687–693. [Google Scholar] [CrossRef]
- Ding, F.; Song, J.; Ruan, Y.; Wang, B.-s. Comparison of the effects of NaCl and KCl at the roots on seedling growth, cell death and the size, frequency and secretion rate of salt glands in leaves of Limonium sinense. Acta Physiol. Plant. 2009, 31, 343–350. [Google Scholar] [CrossRef]
- Leng, B.Y.; Yuan, F.; Dong, X.X.; Wang, J.; Wang, B.S. Distribution pattern and salt excretion rate of salt glands in two recretohalophyte species of Limonium (Plumbaginaceae). S. Afr. J. Bot. 2018, 115, 74–80. [Google Scholar] [CrossRef]
- Tyerman, S.D. The devil in the detail of secretions. Plant Cell Environ. 2013, 36, 1407–1409. [Google Scholar] [CrossRef]
- Sookbirsingh, R.; Castillo, K.; Gill, T.E.; Chianelli, R.R. Salt Separation Processes in the Saltcedar Tamarix ramosissima (Ledeb.). Commun. Soil Sci. Plant Anal. 2010, 41, 1271–1281. [Google Scholar] [CrossRef]
- Hill, A.E. Ion and water transport in limonium. II. Short-circuit analysis. Biochim. Biophys. Acta 1967, 135, 461–465. [Google Scholar] [CrossRef]
- Pollak, G.; Waisel, Y. Salt Secretion in Aeluropus litoralis (Willd.) Parl. Ann. Bot. 1970, 34, 879–888. [Google Scholar] [CrossRef]
- Kadukova, J.; Manousaki, E.; Kalogerakis, N. Pb and Cd accumulation and phyto-excretion by salt cedar (Tamarix smyrnensis Bunge). Int. J. Phytoremediation 2008, 10, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Manousaki, E.; Kadukova, J.; Papadantonakis, N.; Kalogerakis, N. Phytoextraction and phytoexcretion of Cd by the leaves of Tamarix smyrnensis growing on contaminated non-saline and saline soils. Environ. Res. 2008, 106, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Badri, M.A.; Moalla, S.N.; Pulford, I.D. Cycling of metals in desert soils: Effects of Tamarix nilotica and inundation by lake water. Environ. Geochem. Health 2001, 23, 369–378. [Google Scholar] [CrossRef]
- Soltan, M.E.; Moalla, S.; Rashed, M.N.; Mahmoud, E. Assessment of metals in soil extracts and their uptake and movement within Tamarix nilotica at Lake Nasser Banks, Egypt. Chem. Ecol. 2004, 20, 137–154. [Google Scholar] [CrossRef]
- Mahmoud, E.; Soltan, M.E.; Sirry, S. Mobilization of different metals between Tamarix parts and their crystal salts—Soil system at the banks of river Nile, Aswan, Egypt. Toxicol. Environ. Chem. 2006, 88, 603–618. [Google Scholar] [CrossRef]
- Storey, R.; Thomson, W.W. An X-ray Microanalysis Study of the Salt Glands and Intracellular Calcium Crystals of Tamarix. Ann. Bot. 1994, 73, 307–313. [Google Scholar] [CrossRef]
- Barhoumi, Z.; Djebali, W.; Smaoui, A.; Chaïbi, W.; Abdelly, C. Contribution of NaCl excretion to salt resistance of Aeluropus littoralis (Willd) Parl. J. Plant Physiol. 2007, 164, 842–850. [Google Scholar] [CrossRef] [PubMed]
- Campbell, N.; Thomson, W.W. Chloride localization in the leaf of Tamarix. Protoplasma 1975, 83, 1–14. [Google Scholar] [CrossRef]
- Lu, C.; Feng, Z.; Yuan, F.; Han, G.; Guo, J.; Chen, M.; Wang, B. The SNARE protein LbSYP61 participates in salt secretion in Limonium bicolor. Environ. Exp. Bot. 2020, 176, 104076. [Google Scholar] [CrossRef]
- Yuan, F.; Lyu, M.J.; Leng, B.Y.; Zhu, X.G.; Wang, B.S. The transcriptome of NaCl-treated Limonium bicolor leaves reveals the genes controlling salt secretion of salt gland. Plant Mol. Biol. 2016, 91, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.K.; Ang, Y.; Lim, T.K.; Lim, T.M.; Kumar, P.; Loh, C.S.; Lin, Q. Proteome profile of salt gland-rich epidermis extracted from a salt-tolerant tree species. Electrophoresis 2015, 36, 2473–2481. [Google Scholar] [CrossRef]
- Tan, W.K.; Lim, T.K.; Loh, C.S.; Kumar, P. Proteomic Characterisation of the Salt Gland-Enriched Tissues of the Mangrove Tree Species Avicennia officinalis. PLoS ONE 2015, 10, e0133386. [Google Scholar] [CrossRef]
- Oh, D.H.; Barkla, B.; Vera-Estrella, R.; Pantoja, O.; Lee, S.Y.; Bohnert, H.; Dassanayake, M. Cell type-specific responses to salinity—The epidermal bladder cell transcriptome of Mesembryanthemum crystallinum. New Phytol. 2015, 207, 627–644. [Google Scholar] [CrossRef]
- Zhang, X.; Yao, Y.; Li, X.; Zhang, L.; Fan, S. Transcriptomic analysis identifies novel genes and pathways for salt stress responses in Suaeda salsa leaves. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Gendre, D.; Oh, J.; Boutte, Y.; Best, J.G.; Samuels, L.; Nilsson, R.; Uemura, T.; Marchant, A.; Bennett, M.J.; Grebe, M.; et al. Conserved Arabidopsis ECHIDNA protein mediates trans-Golgi-network trafficking and cell elongation. Proc. Natl. Acad. Sci. USA 2011, 108, 8048–8053. [Google Scholar] [CrossRef] [PubMed]
- Gendre, D.; McFarlane, H.E.; Johnson, E.; Mouille, G.; Sjodin, A.; Oh, J.; Levesque-Tremblay, G.; Watanabe, Y.; Samuels, L.; Bhalerao, R.P. Trans-Golgi Network Localized ECHIDNA/Ypt Interacting Protein Complex Is Required for the Secretion of Cell Wall Polysaccharides in Arabidopsis. Plant Cell 2013, 25, 2633–2646. [Google Scholar] [CrossRef] [PubMed]
- Drakakaki, G.; van de Ven, W.; Pan, S.Q.; Miao, Y.S.; Wang, J.Q.; Keinath, N.F.; Weatherly, B.; Jiang, L.W.; Schumacher, K.; Hicks, G.; et al. Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res. 2012, 22, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Asaoka, R.; Uemura, T.; Ito, J.; Fujimoto, M.; Ito, E.; Ueda, T.; Nakano, A. Arabidopsis RABA1 GTPases are involved in transport between the trans-Golgi network and the plasma membrane, and are required for salinity stress tolerance. Plant J. 2013, 73, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Leshem, Y.; Golani, Y.; Kaye, Y.; Levine, A. Reduced expression of the v-SNAREs AtVAMP71/AtVAMP7C gene family in Arabidopsis reduces drought tolerance by suppression of abscisic acid-dependent stomatal closure. J. Exp. Bot. 2010, 61, 2615–2622. [Google Scholar] [CrossRef]
- Fecht-Bartenbach, J.; Bogner, M.; Krebs, M.; Stierhof, Y.-D.; Schumacher, K.; Ludewig, U. Function of the anion transporter AtCLC-d in the trans-Golgi network. Plant J. 2007, 50, 466–474. [Google Scholar] [CrossRef]
- Heard, W.; Sklenar, J.; Tome, D.F.A.; Robatzek, S.; Jones, A.M.E. Identification of Regulatory and Cargo Proteins of Endosomal and Secretory Pathways in Arabidopsis thaliana by Proteomic Dissection. Mol. Cell. Proteomics 2015, 14, 1796–1813. [Google Scholar] [CrossRef]
- Krebs, M.; Beyhl, D.; Gorlich, E.; Al-Rasheid, K.A.; Marten, I.; Stierhof, Y.D.; Hedrich, R.; Schumacher, K. Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc. Natl. Acad. Sci. USA 2010, 107, 3251–3256. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Bu, Y.; Takano, T.; Zhang, X.; Liu, S. Conserved V-ATPase c subunit plays a role in plant growth by influencing V-ATPase-dependent endosomal trafficking. Plant Biotechnol. J. 2016, 14, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhang, L.; Cui, Y.N.; Wang, S.M.; Bao, A.K. Identification of candidate genes related to salt tolerance of the secretohalophyte Atriplex canescens by transcriptomic analysis. BMC Plant Biol. 2019, 19, 213. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Takano, T.; Tanaka, K.; Ishige, T.; Terashima, S.; Endo, C.; Kurusu, T.; Yajima, S.; Yano, K.; Tada, Y. Comprehensive analysis of transcriptome response to salinity stress in the halophytic turf grass Sporobolus virginicus. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Dang, Z.h.; Zheng, L.l.; Wang, J.; Gao, Z.; Wu, S.b.; Qi, Z.; Wang, Y.C. Transcriptomic profiling of the salt-stress response in the wild recretohalophyte Reaumuria trigyna. BMC Genom. 2013, 14, 29. [Google Scholar] [CrossRef]
- Ruiz, K.B.; Maldonado, J.; Biondi, S.; Silva, H. RNA-seq Analysis of Salt-Stressed versus Non Salt-Stressed Transcriptomes of Chenopodium quinoa Landrace R49. Genes 2019, 10, 1042. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, S.; Li, J.; Deng, Y.; Zhang, Z.; Xu, S.; Guo, W.; Zhong, C.; Zhou, R.; Shi, S. Transcriptome analysis of the Holly mangrove Acanthus ilicifolius and its terrestrial relative, Acanthus leucostachyus, provides insights into adaptation to intertidal zones. BMC Genom. 2015, 16, 605. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, X.; Liu, K.; Zhang, X.J.; Zhang, L.Y.; Fan, S.J. Comparative Transcriptome Analysis of Halophyte Zoysia macrostachya in Response to Salinity Stress. Plants 2020, 9, 458. [Google Scholar] [CrossRef] [PubMed]
- MacRobbie, E.A.C. Vesicle trafficking: A role in trans-tonoplast ion movements? J. Exp. Bot. 1999, 50, 925–934. [Google Scholar] [CrossRef]
- Hamaji, K.; Nagira, M.; Yoshida, K.; Ohnishi, M.; Oda, Y.; Uemura, T.; Goh, T.; Sato, M.; Morita, M.; Tasaka, M.; et al. Dynamic Aspects of Ion Accumulation by Vesicle Traffic Under Salt Stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2023–2033. [Google Scholar] [CrossRef] [PubMed]
- Gendre, D.; Jonsson, K.; Boutté, Y.; Bhalerao, R.P. Journey to the cell surface—The central role of the trans-Golgi network in plants. Protoplasma 2015, 252, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Bassham, D.C. TNO1 Is Involved in Salt Tolerance and Vacuolar Trafficking in Arabidopsis. Plant Physiol. 2011, 156, 514–526. [Google Scholar] [CrossRef]
- Zhu, J.; Gong, Z.; Zhang, C.; Song, C.-P.; Damsz, B.; Inan, G.; Koiwa, H.; Zhu, J.-K.; Hasegawa, P.M.; Bressan, R.A. OSM1/SYP61: A syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell 2002, 14, 3009–3028. [Google Scholar] [CrossRef]
- Baral, A.; Irani, N.G.; Fujimoto, M.; Nakano, A.; Mayor, S.; Mathew, M.K. Salt-induced remodeling of spatially restricted clathrin-independent endocytic pathways in Arabidopsis root. Plant Cell 2015, 27, 1297–1315. [Google Scholar] [CrossRef]
- Xue, Y.; Zhao, S.S.; Yang, Z.J.; Guo, Y.; Yang, Y.Q. Regulation of plasma membrane H+-ATPase activity by the members of the V-SNARE VAMP7C family in Arabidopsis thaliana. Plant Signal. Behav. 2019, 14. [Google Scholar] [CrossRef]
- Leshem, Y.; Melamed-Book, N.; Cagnac, O.; Ronen, G.; Nishri, Y.; Solomon, M.; Cohen, G.; Levine, A. Suppression of Arabidopsis vesicle-SNARE expression inhibited fusion of H2O2-containing vesicles with tonoplast and increased salt tolerance. Proc. Natl. Acad. Sci. USA 2006, 103, 18008–18013. [Google Scholar] [CrossRef]
- Martín-Davison, A.S.; Pérez-Díaz, R.; Soto, F.; Madrid-Espinoza, J.; González-Villanueva, E.; Pizarro, L.; Norambuena, L.; Tapia, J.; Tajima, H.; Blumwald, E.; et al. Involvement of SchRabGDI1 from Solanum chilense in endocytic trafficking and tolerance to salt stress. Plant Sci. 2017, 263, 1–11. [Google Scholar] [CrossRef]
- Chen, C.; Heo, J. Overexpression of Constitutively Active OsRab11 in Plants Enhances Tolerance to High Salinity Levels. J. Plant Biol. 2018, 61, 169–176. [Google Scholar] [CrossRef]
- Khassanova, G.; Kurishbayev, A.; Jatayev, S.; Zhubatkanov, A.; Zhumalin, A.; Turbekova, A.; Amantaev, B.; Lopato, S.; Schramm, C.; Jenkins, C.; et al. Intracellular Vesicle Trafficking Genes, RabC-GTP, Are Highly Expressed under Salinity and Rapid Dehydration but Down-Regulated by Drought in Leaves of Chickpea (Cicer arietinum L.). Front. Genet. 2019, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Sweetman, C.; Khassanova, G.; Miller, T.; Booth, N.; Kurishbayev, A.; Jatayev, S.; Gupta, N.; Langridge, P.; Jenkins, C.; Soole, K.; et al. Salt-induced expression of intracellular vesicle trafficking genes, CaRab-GTP, and their association with Na+ accumulation in leaves of chickpea (Cicer arietinum L.). BMC Plant Biol. 2020, 20, 183. [Google Scholar] [CrossRef] [PubMed]
- Rosquete, M.R.; Drakakaki, G. Plant TGN in the stress response: A compartmentalized overview. Curr. Opin. Plant Biol. 2018, 46, 122–129. [Google Scholar] [CrossRef]
- Reguera, M.; Bassil, E.; Tajima, H.; Wimmer, M.; Chanoca, A.; Otegui, M.S.; Paris, N.; Blumwald, E. pH Regulation by NHX-Type Antiporters Is Required for Receptor-Mediated Protein Trafficking to the Vacuole in Arabidopsis. Plant Cell 2015, 27, 1200–1217. [Google Scholar] [CrossRef] [PubMed]
- Sze, H.; Chanroj, S. Plant Endomembrane Dynamics: Studies of K(+)/H(+) Antiporters Provide Insights on the Effects of pH and Ion Homeostasis. Plant Physiol. 2018, 177, 875–895. [Google Scholar] [CrossRef]
- Bassil, E.; Blumwald, E. The ins and outs of intracellular ion homeostasis: NHX-type cation/H(+) transporters. Curr. Opin. Plant Biol. 2014, 22, 1–6. [Google Scholar] [CrossRef]
- Dettmer, J.; Hong-Hermesdorf, A.; Stierhof, Y.-D.; Schumacher, K. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 2006, 18, 715–730. [Google Scholar] [CrossRef]
- Sanadhya, P.; Agarwal, P.; Agarwal, P.K. Ion homeostasis in a salt-secreting halophytic grass. AoB Plants 2015, 7. [Google Scholar] [CrossRef]
- Baral, A.; Shruthi, K.; Mathew, M. Vesicular trafficking and salinity responses in plants. IUBMB Life 2015, 67. [Google Scholar] [CrossRef]
- Bassil, E.; Ohto, M.A.; Esumi, T.; Tajima, H.; Zhu, Z.; Cagnac, O.; Belmonte, M.; Peleg, Z.; Yamaguchi, T.; Blumwald, E. The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 2011, 23, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Li, M.R.; Li, Y.; Li, H.Q.; Wu, G.J. Overexpression of AtNHX5 improves tolerance to both salt and drought stress in Broussonetia papyrifera (L.) Vent. Tree Physiol. 2011, 31, 349–357. [Google Scholar] [CrossRef]
- Shen, J.; Zeng, Y.; Zhuang, X.; Sun, L.; Yao, X.; Pimpl, P.; Jiang, L. Organelle pH in the Arabidopsis endomembrane system. Mol. Plant 2013, 6, 1419–1437. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yuan, F.; Liu, Y.; Zhang, M.; Liu, Y.; Zhao, Y.; Wang, B.; Chen, M. Exogenous melatonin enhances salt secretion from salt glands by upregulating the expression of ion transporter and vesicle transport genes in Limonium bicolor. BMC Plant Biol. 2020, 20, 493. [Google Scholar] [CrossRef]
- Kim, S.J.; Brandizzi, F. The plant secretory pathway for the trafficking of cell wall polysaccharides and glycoproteins. Glycobiology 2016, 26, 940–949. [Google Scholar] [CrossRef] [PubMed]
- Hachez, C.; Laloux, T.; Reinhardt, H.; Cavez, D.; Degand, H.; Grefen, C.; De Rycke, R.; Inze, D.; Blatt, M.R.; Russinova, E.; et al. Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. Plant Cell 2014, 26, 3132–3147. [Google Scholar] [CrossRef] [PubMed]
- Pou, A.; Jeanguenin, L.; Milhiet, T.; Batoko, H.; Chaumont, F.; Hachez, C. Salinity-mediated transcriptional and post-translational regulation of the Arabidopsis aquaporin PIP2;7. Plant Mol. Biol. 2016, 92, 731–744. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, R.; Lindeboom, J.J.; Paredez, A.R.; Emons, A.M.; Ehrhardt, D.W. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 2009, 11, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Somaru, R.; Naidoo, Y.; Naidoo, G. Morphology and ultrastructure of the leaf salt glands of Odyssea paucinervis (Stapf) (Poaceae). Flora 2002, 197, 67–75. [Google Scholar] [CrossRef]
- Yuan, F.; Chen, M.; Yang, J.; Leng, B.; Wang, B. A system for the transformation and regeneration of the recretohalophyte Limonium bicolor. In Vitro Cell. Dev. Biol. Plant 2014, 50, 610–617. [Google Scholar] [CrossRef]
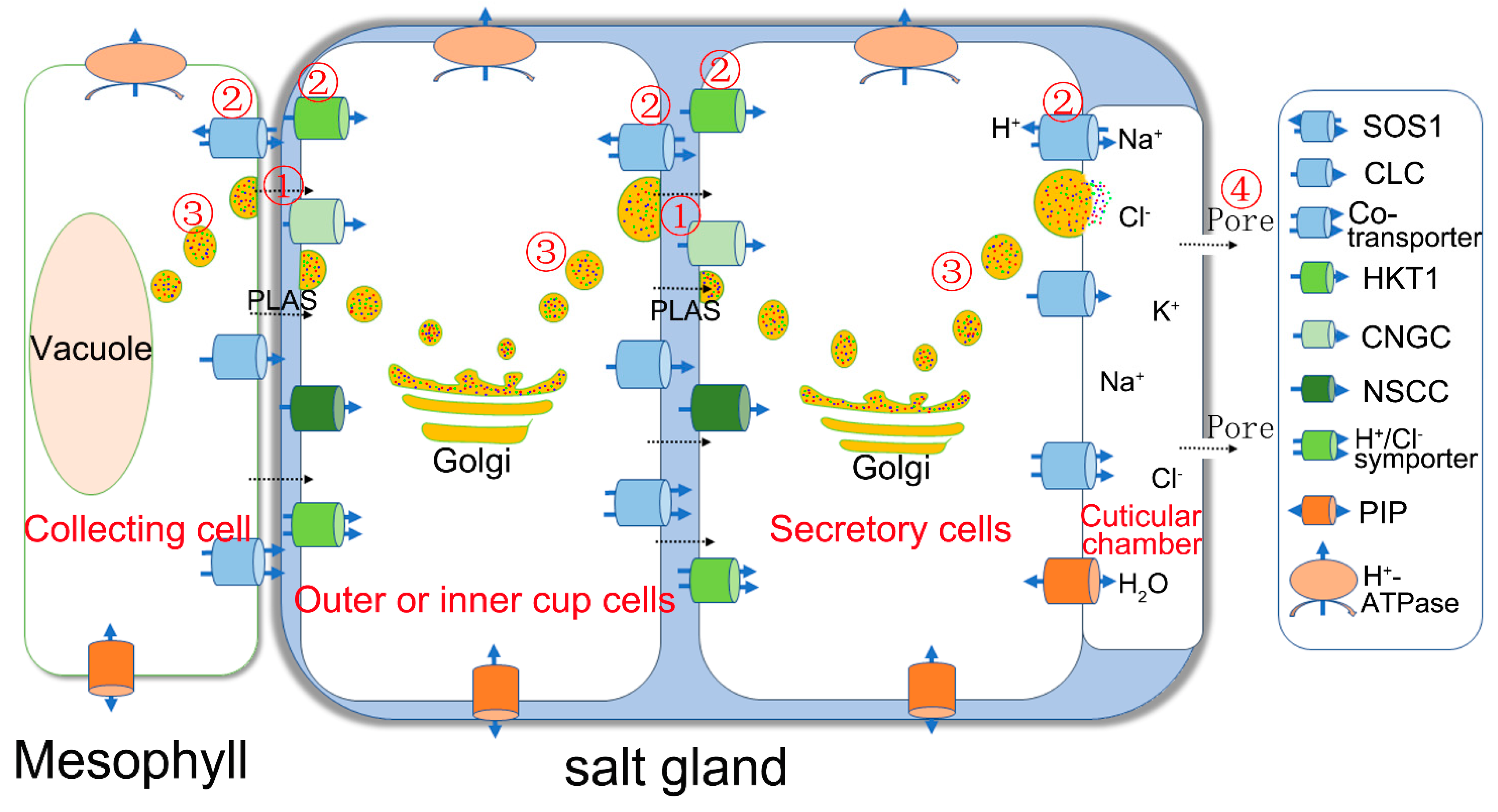
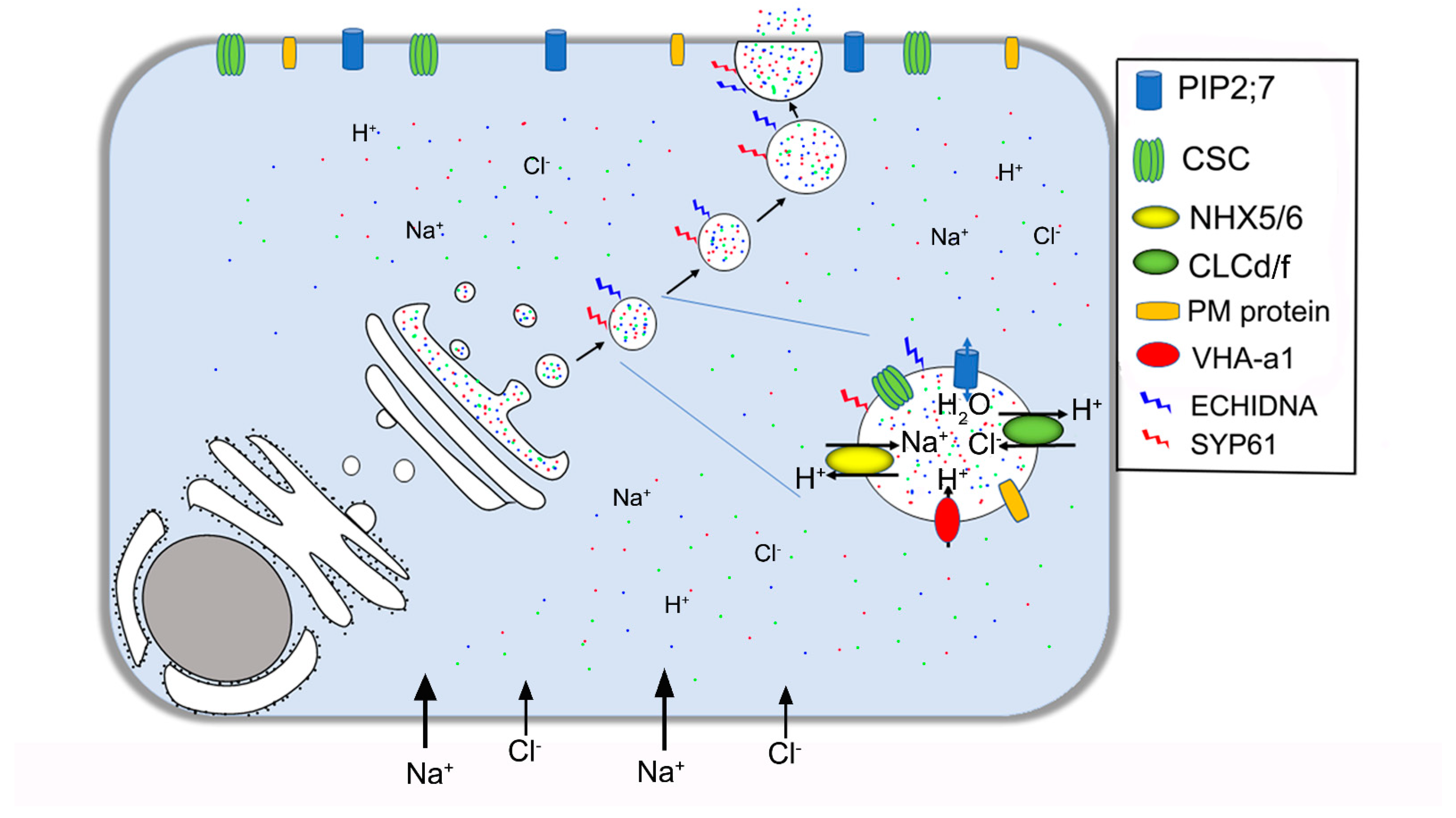
| Genes | Function | Location | Reference |
|---|---|---|---|
| ECHIDNA | Vesicle formation | TGN/EE | Gendre et al. 2011 [73]; Gendre et al. 2013 [74] |
| SYP61 | Membrane fusion | TGN/EE, PM | Drakakaki et al. 2012 [75]; Lu et al. 2020 [67] |
| RABA1A | Regulation of vesicle trafficking | TGN/EE | Asaoka et al. 2013 [76] |
| VAMP7C | Vesicle transport | TGN/EE | Leshem et al. 2010 [77] |
| NHX5/6 | Exchange H+/Na+ or K+ | TGN/EE, Golgi | Asaoka et al. 2013 [76] |
| CLCd | Exchange H+/Cl− or NO3− | TGN/EE, Golgi | Fecht–Bartenbach et al. 2007 [78]; Heard et al. 2015 [79] |
| VHA-a1 | H+-ATPase | TGN/EE | Krebs et al. 2010 [80]; Zhou et al. 2016 [81] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, C.; Yuan, F.; Guo, J.; Han, G.; Wang, C.; Chen, M.; Wang, B. Current Understanding of Role of Vesicular Transport in Salt Secretion by Salt Glands in Recretohalophytes. Int. J. Mol. Sci. 2021, 22, 2203. https://doi.org/10.3390/ijms22042203
Lu C, Yuan F, Guo J, Han G, Wang C, Chen M, Wang B. Current Understanding of Role of Vesicular Transport in Salt Secretion by Salt Glands in Recretohalophytes. International Journal of Molecular Sciences. 2021; 22(4):2203. https://doi.org/10.3390/ijms22042203
Chicago/Turabian StyleLu, Chaoxia, Fang Yuan, Jianrong Guo, Guoliang Han, Chengfeng Wang, Min Chen, and Baoshan Wang. 2021. "Current Understanding of Role of Vesicular Transport in Salt Secretion by Salt Glands in Recretohalophytes" International Journal of Molecular Sciences 22, no. 4: 2203. https://doi.org/10.3390/ijms22042203
APA StyleLu, C., Yuan, F., Guo, J., Han, G., Wang, C., Chen, M., & Wang, B. (2021). Current Understanding of Role of Vesicular Transport in Salt Secretion by Salt Glands in Recretohalophytes. International Journal of Molecular Sciences, 22(4), 2203. https://doi.org/10.3390/ijms22042203






