Pax5 Negatively Regulates Osteoclastogenesis through Downregulation of Blimp1
Abstract
1. Introduction
2. Results
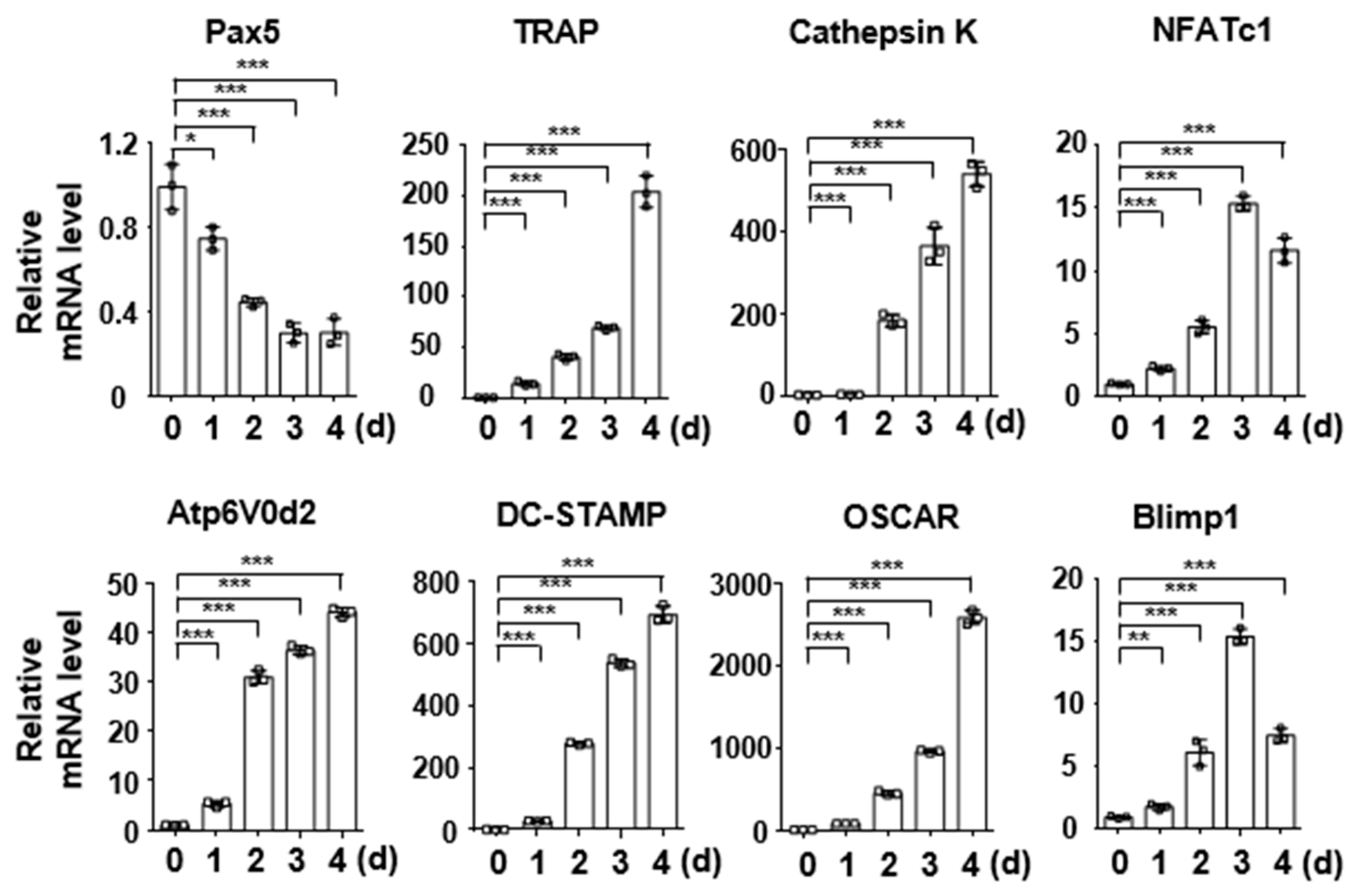
2.1. Pax5 Expression Is Downregulated during RANKL-Induced Osteoclastogenesis
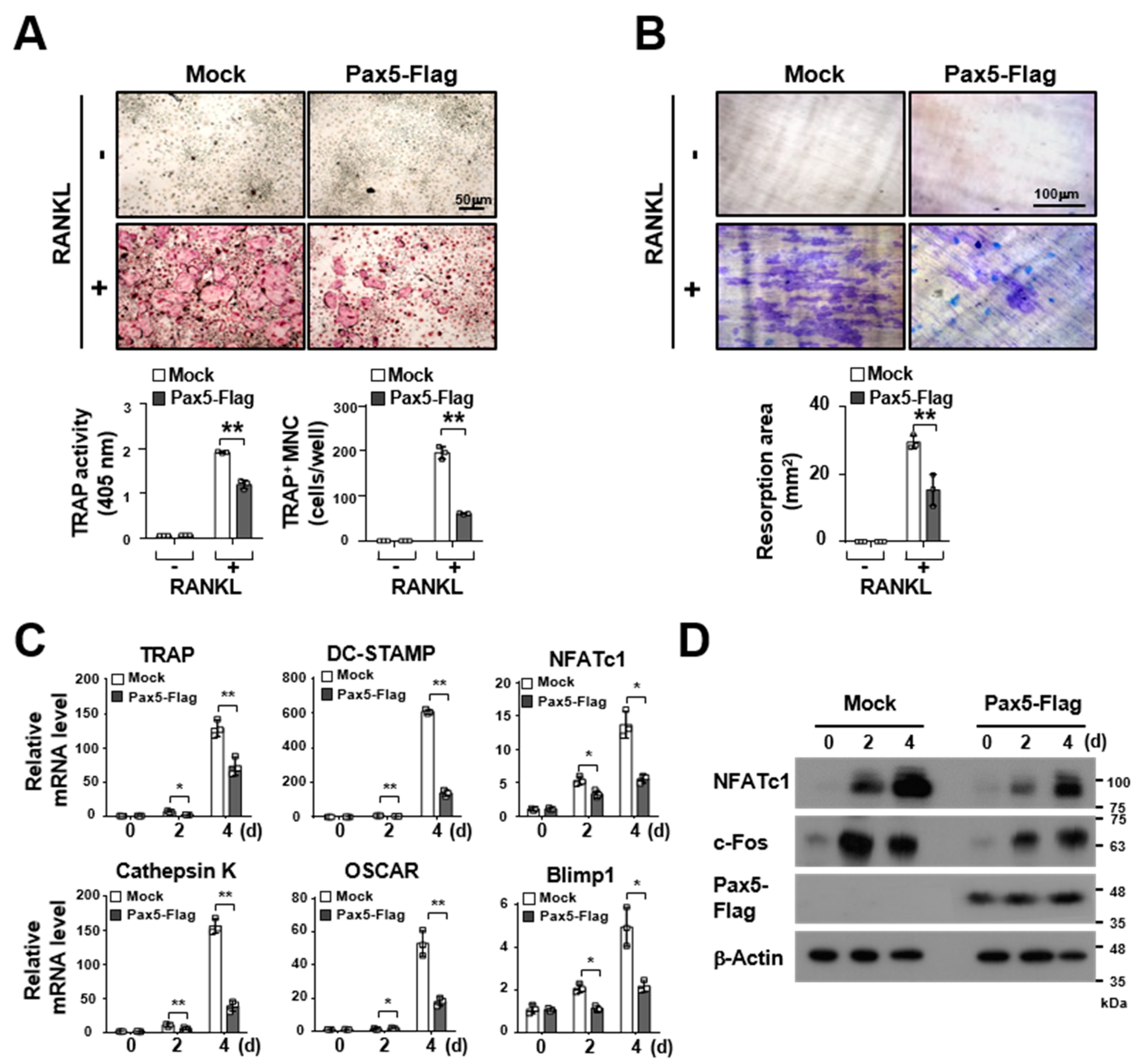
2.2. Pax5 Negatively Regulates RANKL-Induced Osteoclastogenesis
2.3. Pax5 Enhances the Expression of Antiosteoclastogenic Factors through Downregulation of Blimp1
2.4. Osteoclastogenesis Is Reduced by Pax5 Transgene Expression in OC Lineage Cells
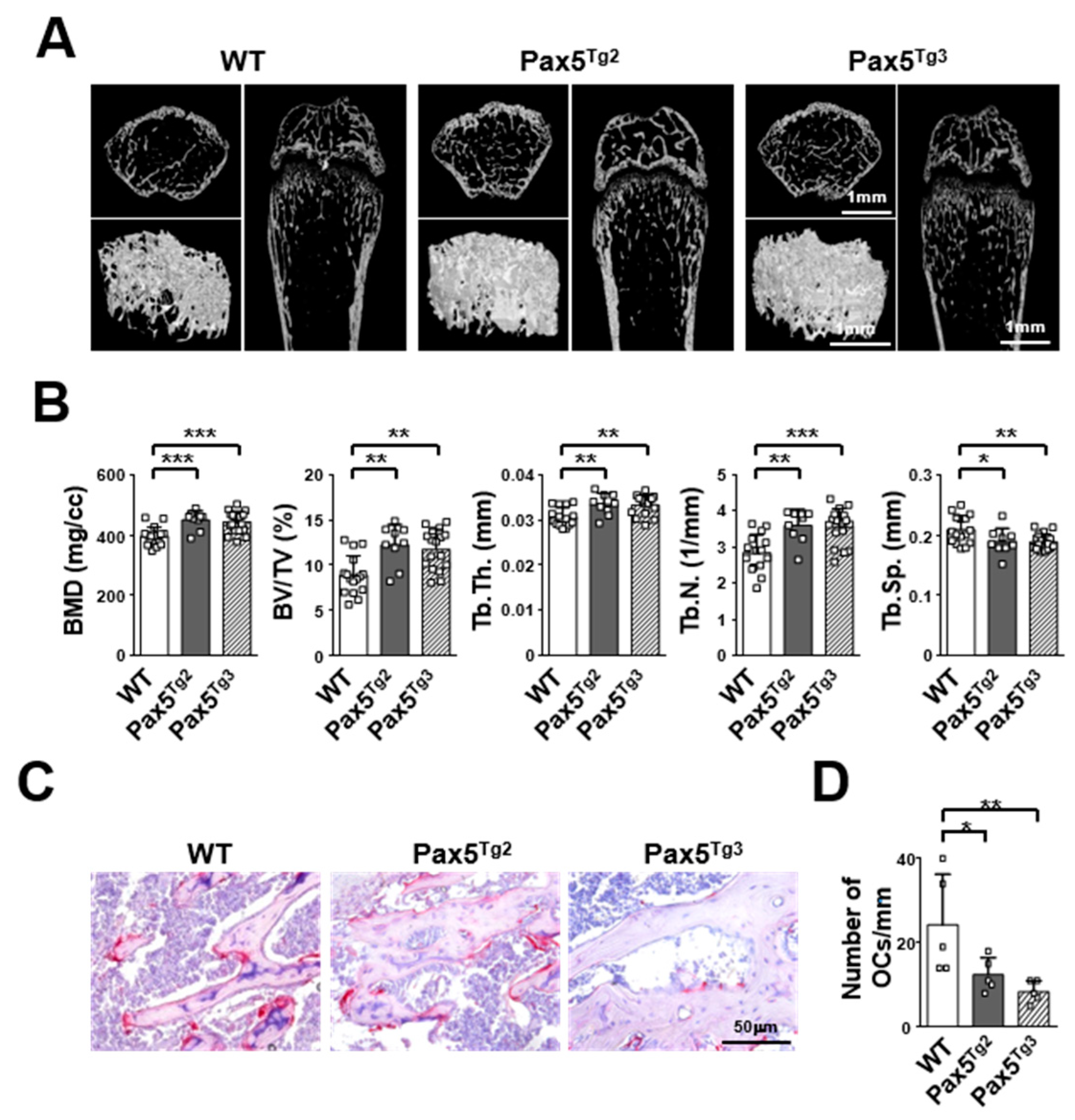
2.5. Mice Expressing the Pax5 Transgene Show an Osteopetrotic-Like Bone Phenotype Caused by Reduced OC Formation
3. Discussion
4. Materials and Methods
4.1. Antibodies, Reagents, Cell Lines, and Mice
4.2. Plasmids
4.3. Osteoclastogenesis and OC Analysis
4.4. Luciferase Reporter Assay
4.5. Micro-CT and Histological Analysis
4.6. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Pax5 | Paired box protein 5 |
| Blimp1 | B lymphocyte-induced maturation protein 1 |
| RANKL | Receptor activator of nuclear factor kappa B ligand |
| M-CSF | Macrophage-colony stimulating factor |
| BMM | Bone marrow-derived macrophage |
| NFATc1 | Nuclear factor of activated T cells c1 |
| TRAP | Tartrate-resistant acid phosphatase |
| DC-STAMP | Dendritic cell-specific transmembrane protein |
| OSCAR | Osteoclast-associated receptor |
| Bcl6 | B-cell lymphoma 6 |
| IRF8 | Interferon regulatory factor 8 |
| MafB | v-Maf musculoaponeurotic fibrosarcoma oncogene family homolog B |
References
- Amarasekara, D.S.; Yun, H.; Kim, S.; Lee, N.; Kim, H.; Rho, J. Regulation of Osteoclast Differentiation by Cytokine Networks. Immune Netw. 2018, 18, e8. [Google Scholar] [CrossRef]
- Walsh, M.C.; Kim, N.; Kadono, Y.; Rho, J.; Lee, S.Y.; Lorenzo, J.; Choi, Y. Osteoimmunology: Interplay between the immune system and bone metabolism. Annu. Rev. Immunol. 2006, 24, 33–63. [Google Scholar] [CrossRef]
- Amarasekara, D.S.; Yu, J.; Rho, J. Bone Loss Triggered by the Cytokine Network in Inflammatory Autoimmune Diseases. J. Immunol. Res. 2015, 2015, 832127. [Google Scholar] [CrossRef] [PubMed]
- Rho, J.; Takami, M.; Choi, Y. Osteoimmunology: Interactions of the immune and skeletal systems. Mol. Cells 2004, 17, 1–9. [Google Scholar]
- Miyauchi, Y.; Ninomiya, K.; Miyamoto, H.; Sakamoto, A.; Iwasaki, R.; Hoshi, H.; Miyamoto, K.; Hao, W.; Yoshida, S.; Morioka, H.; et al. The Blimp1-Bcl6 axis is critical to regulate osteoclast differentiation and bone homeostasis. J. Exp. Med. 2010, 207, 751–762. [Google Scholar] [CrossRef]
- Takayanagi, H. Osteoimmunology: Shared mechanisms and crosstalk between the immune and bone systems. Nat. Rev. Immunol. 2007, 7, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Cobaleda, C.; Schebesta, A.; Delogu, A.; Busslinger, M. Pax5: The guardian of B cell identity and function. Nat. Immunol. 2007, 8, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.; Dorfler, P.; Aguzzi, A.; Kozmik, Z.; Urbanek, P.; Maurer-Fogy, I.; Busslinger, M. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 1992, 6, 1589–1607. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.S.; Wasserman, R.; Hayakawa, K.; Hardy, R.R. Identification of the earliest B lineage stage in mouse bone marrow. Immunity 1996, 5, 527–535. [Google Scholar] [CrossRef]
- Lin, K.I.; Angelin-Duclos, C.; Kuo, T.C.; Calame, K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol. Cell Biol. 2002, 22, 4771–4780. [Google Scholar] [CrossRef] [PubMed]
- Mora-Lopez, F.; Reales, E.; Brieva, J.A.; Campos-Caro, A. Human BSAP and BLIMP1 conform an autoregulatory feedback loop. Blood 2007, 110, 3150–3157. [Google Scholar] [CrossRef] [PubMed]
- Shapiro-Shelef, M.; Lin, K.I.; McHeyzer-Williams, L.J.; Liao, J.; McHeyzer-Williams, M.G.; Calame, K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity 2003, 19, 607–620. [Google Scholar] [CrossRef]
- Nera, K.P.; Kohonen, P.; Narvi, E.; Peippo, A.; Mustonen, L.; Terho, P.; Koskela, K.; Buerstedde, J.M.; Lassila, O. Loss of Pax5 promotes plasma cell differentiation. Immunity 2006, 24, 283–293. [Google Scholar] [CrossRef]
- Nutt, S.L.; Heavey, B.; Rolink, A.G.; Busslinger, M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 1999, 401, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, M.C.; Xi, Y.; Pflugh, D.L.; Hesslein, D.G.; Schatz, D.G.; Lorenzo, J.A.; Bothwell, A.L. Pax5-deficient mice exhibit early onset osteopenia with increased osteoclast progenitors. J. Immunol. 2004, 173, 6583–6591. [Google Scholar] [CrossRef]
- Chiu, W.S.; McManus, J.F.; Notini, A.J.; Cassady, A.I.; Zajac, J.D.; Davey, R.A. Transgenic mice that express Cre recombinase in osteoclasts. Genesis 2004, 39, 178–185. [Google Scholar] [CrossRef]
- Nutt, S.L.; Kee, B.L. The transcriptional regulation of B cell lineage commitment. Immunity 2007, 26, 715–725. [Google Scholar] [CrossRef]
- Cotta, C.V.; Zhang, Z.; Kim, H.G.; Klug, C.A. Pax5 determines B- versus T-cell fate and does not block early myeloid-lineage development. Blood 2003, 101, 4342–4346. [Google Scholar] [CrossRef]
- Enver, T. B-cell commitment: Pax5 is the deciding factor. Curr. Biol. 1999, 9, R933–R935. [Google Scholar] [CrossRef][Green Version]
- Xu, W.; Rould, M.A.; Jun, S.; Desplan, C.; Pabo, C.O. Crystal structure of a paired domain-DNA complex at 2.5 A resolution reveals structural basis for Pax developmental mutations. Cell 1995, 80, 639–650. [Google Scholar] [CrossRef]
- Garvie, C.W.; Hagman, J.; Wolberger, C. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol. Cell 2001, 8, 1267–1276. [Google Scholar] [CrossRef]
- Czerny, T.; Schaffner, G.; Busslinger, M. DNA sequence recognition by Pax proteins: Bipartite structure of the paired domain and its binding site. Genes Dev. 1993, 7, 2048–2061. [Google Scholar] [CrossRef]
- Kozmik, Z.; Wang, S.; Dorfler, P.; Adams, B.; Busslinger, M. The promoter of the CD19 gene is a target for the B-cell-specific transcription factor BSAP. Mol. Cell Biol. 1992, 12, 2662–2672. [Google Scholar] [CrossRef]
- Dorfler, P.; Busslinger, M. C-terminal activating and inhibitory domains determine the transactivation potential of BSAP (Pax-5), Pax-2 and Pax-8. EMBO J. 1996, 15, 1971–1982. [Google Scholar] [CrossRef]
- Eberhard, D.; Jimenez, G.; Heavey, B.; Busslinger, M. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 2000, 19, 2292–2303. [Google Scholar] [CrossRef] [PubMed]
- Libermann, T.A.; Pan, Z.; Akbarali, Y.; Hetherington, C.J.; Boltax, J.; Yergeau, D.A.; Zhang, D.E. AML1 (CBFalpha2) cooperates with B cell-specific activating protein (BSAP/PAX5) in activation of the B cell-specific BLK gene promoter. J. Biol. Chem. 1999, 274, 24671–24676. [Google Scholar] [CrossRef] [PubMed]
- Maitra, S.; Atchison, M. BSAP can repress enhancer activity by targeting PU.1 function. Mol. Cell Biol. 2000, 20, 1911–1922. [Google Scholar] [CrossRef]
- Soung, D.Y.; Kalinowski, J.; Baniwal, S.K.; Jacome-Galarza, C.E.; Frenkel, B.; Lorenzo, J.; Drissi, H. Runx1-mediated regulation of osteoclast differentiation and function. Mol. Endocrinol. 2014, 28, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Tondravi, M.M.; McKercher, S.R.; Anderson, K.; Erdmann, J.M.; Quiroz, M.; Maki, R.; Teitelbaum, S.L. Osteopetrosis in mice lacking haematopoietic transcription factor PU.1. Nature 1997, 386, 81–84. [Google Scholar] [CrossRef]
- Nishikawa, K.; Nakashima, T.; Hayashi, M.; Fukunaga, T.; Kato, S.; Kodama, T.; Takahashi, S.; Calame, K.; Takayanagi, H. Blimp1-mediated repression of negative regulators is required for osteoclast differentiation. Proc. Natl. Acad. Sci. USA 2010, 107, 3117–3122. [Google Scholar] [CrossRef]
- Lee, S.H.; Rho, J.; Jeong, D.; Sul, J.Y.; Kim, T.; Kim, N.; Kang, J.S.; Miyamoto, T.; Suda, T.; Lee, S.K.; et al. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat. Med. 2006, 12, 1403–1409. [Google Scholar] [CrossRef] [PubMed]
- Shin, B.; Yu, J.; Park, E.S.; Choi, S.; Hwang, J.M.; Yun, H.; Chung, Y.H.; Hong, K.S.; Choi, J.S.; Takami, M.; et al. Secretion of a truncated osteopetrosis-associated transmembrane protein 1 (OSTM1) mutant inhibits osteoclastogenesis through down-regulation of the B lymphocyte-induced maturation protein 1 (BLIMP1)-nuclear factor of activated T cells c1 (NFATc1) axis. J. Biol. Chem. 2014, 289, 35868–35881. [Google Scholar] [CrossRef] [PubMed]
- Park, E.S.; Choi, S.; Shin, B.; Yu, J.; Yu, J.; Hwang, J.M.; Yun, H.; Chung, Y.H.; Choi, J.S.; Choi, Y.; et al. Tumor necrosis factor (TNF) receptor-associated factor (TRAF)-interacting protein (TRIP) negatively regulates the TRAF2 ubiquitin-dependent pathway by suppressing the TRAF2-sphingosine 1-phosphate (S1P) interaction. J. Biol. Chem. 2015, 290, 9660–9673. [Google Scholar] [CrossRef]
- Yu, J.; Yun, H.; Shin, B.; Kim, Y.; Park, E.S.; Choi, S.; Yu, J.; Amarasekara, D.S.; Kim, S.; Inoue, J.; et al. Interaction of Tumor Necrosis Factor Receptor-associated Factor 6 (TRAF6) and Vav3 in the Receptor Activator of Nuclear Factor kappaB (RANK) Signaling Complex Enhances Osteoclastogenesis. J. Biol. Chem. 2016, 291, 20643–20660. [Google Scholar] [CrossRef]
- Yun, H.; Park, E.S.; Choi, S.; Shin, B.; Yu, J.; Yu, J.; Amarasekara, D.S.; Kim, S.; Lee, N.; Choi, J.S.; et al. TDAG51 is a crucial regulator of maternal care and depressive-like behavior after parturition. PLoS Genet 2019, 15, e1008214. [Google Scholar] [CrossRef]
- Kim, S.; Lee, N.; Park, E.S.; Yun, H.; Ha, T.U.; Jeon, H.; Yu, J.; Choi, S.; Shin, B.; Yu, J.; et al. T-Cell Death Associated Gene 51 Is a Novel Negative Regulator of PPARgamma That Inhibits PPARgammaRXRalpha Heterodimer Formation in Adipogenesis. Mol. Cells 2020, 44, 1–12. [Google Scholar] [CrossRef]
- Kim, T.; Ha, H.; Kim, N.; Park, E.S.; Rho, J.; Kim, E.C.; Lorenzo, J.; Choi, Y.; Lee, S.H. ATP6v0d2 deficiency increases bone mass, but does not influence ovariectomy-induced bone loss. Biochem. Biophys. Res. Commun. 2010, 403, 73–78. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.; Kim, S.; Lee, N.; Jeon, H.; Lee, J.; Takami, M.; Rho, J. Pax5 Negatively Regulates Osteoclastogenesis through Downregulation of Blimp1. Int. J. Mol. Sci. 2021, 22, 2097. https://doi.org/10.3390/ijms22042097
Yu J, Kim S, Lee N, Jeon H, Lee J, Takami M, Rho J. Pax5 Negatively Regulates Osteoclastogenesis through Downregulation of Blimp1. International Journal of Molecular Sciences. 2021; 22(4):2097. https://doi.org/10.3390/ijms22042097
Chicago/Turabian StyleYu, Jiyeon, Sumi Kim, Nari Lee, Hyoeun Jeon, Jun Lee, Masamichi Takami, and Jaerang Rho. 2021. "Pax5 Negatively Regulates Osteoclastogenesis through Downregulation of Blimp1" International Journal of Molecular Sciences 22, no. 4: 2097. https://doi.org/10.3390/ijms22042097
APA StyleYu, J., Kim, S., Lee, N., Jeon, H., Lee, J., Takami, M., & Rho, J. (2021). Pax5 Negatively Regulates Osteoclastogenesis through Downregulation of Blimp1. International Journal of Molecular Sciences, 22(4), 2097. https://doi.org/10.3390/ijms22042097





