Nanopore Sequencing Resolves Elusive Long Tandem-Repeat Regions in Mitochondrial Genomes
Abstract
1. Introduction
2. Results
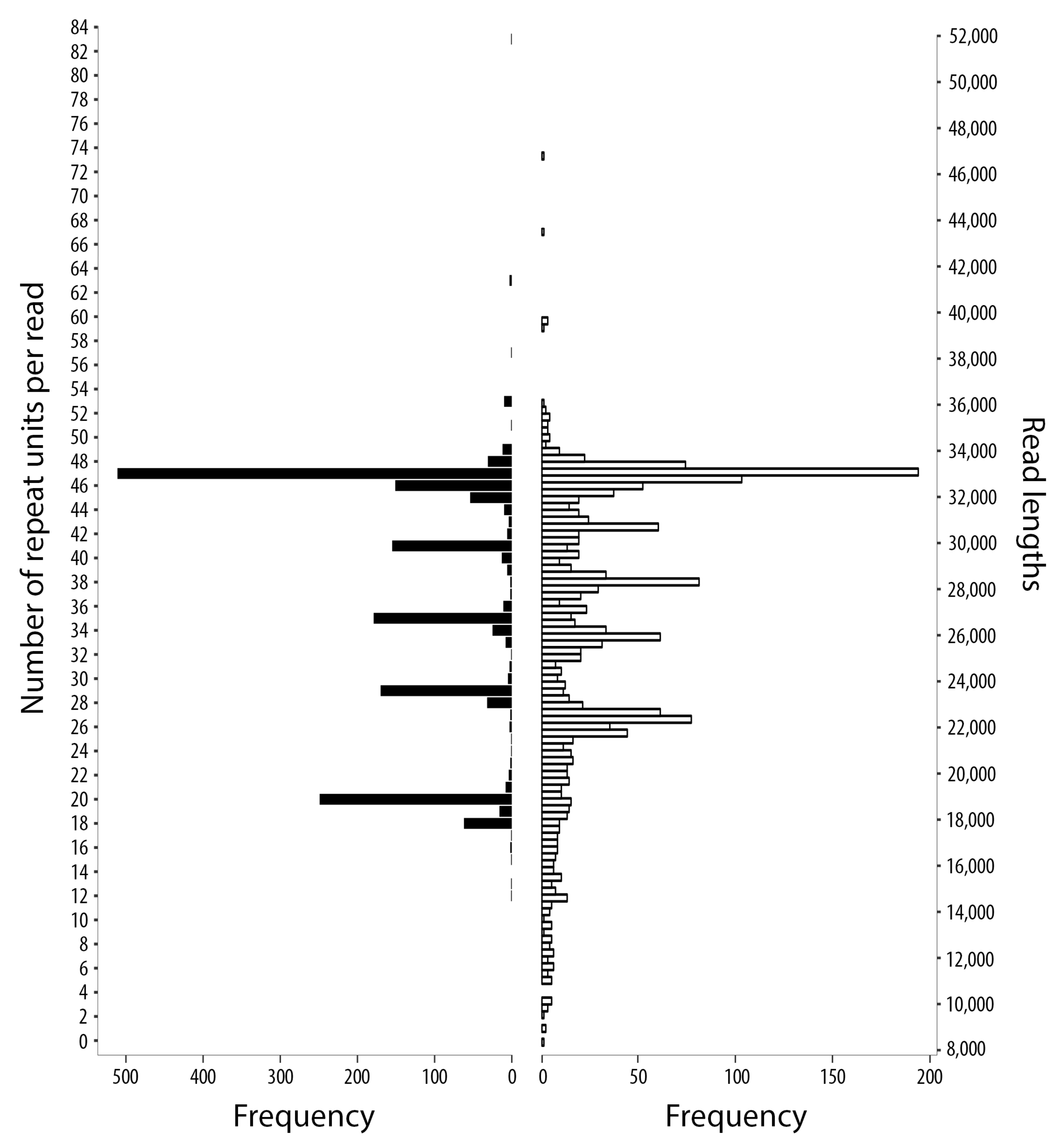
2.1. Sequence Data Sets and Mapping Results
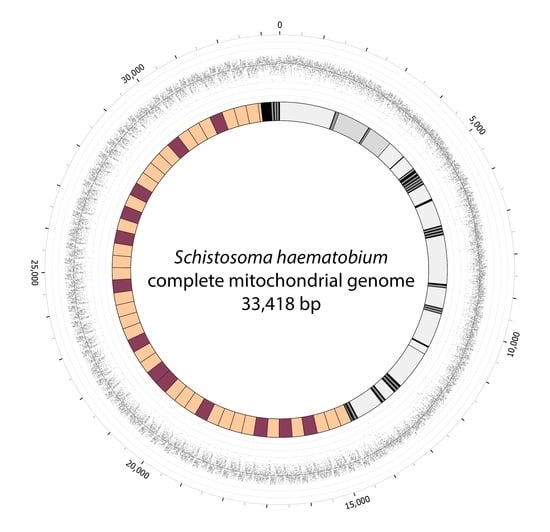
2.2. Complete mt Genomes for S. haematobium, with a Marked Variation in the Number of Repeat Units within the Non-Coding Region
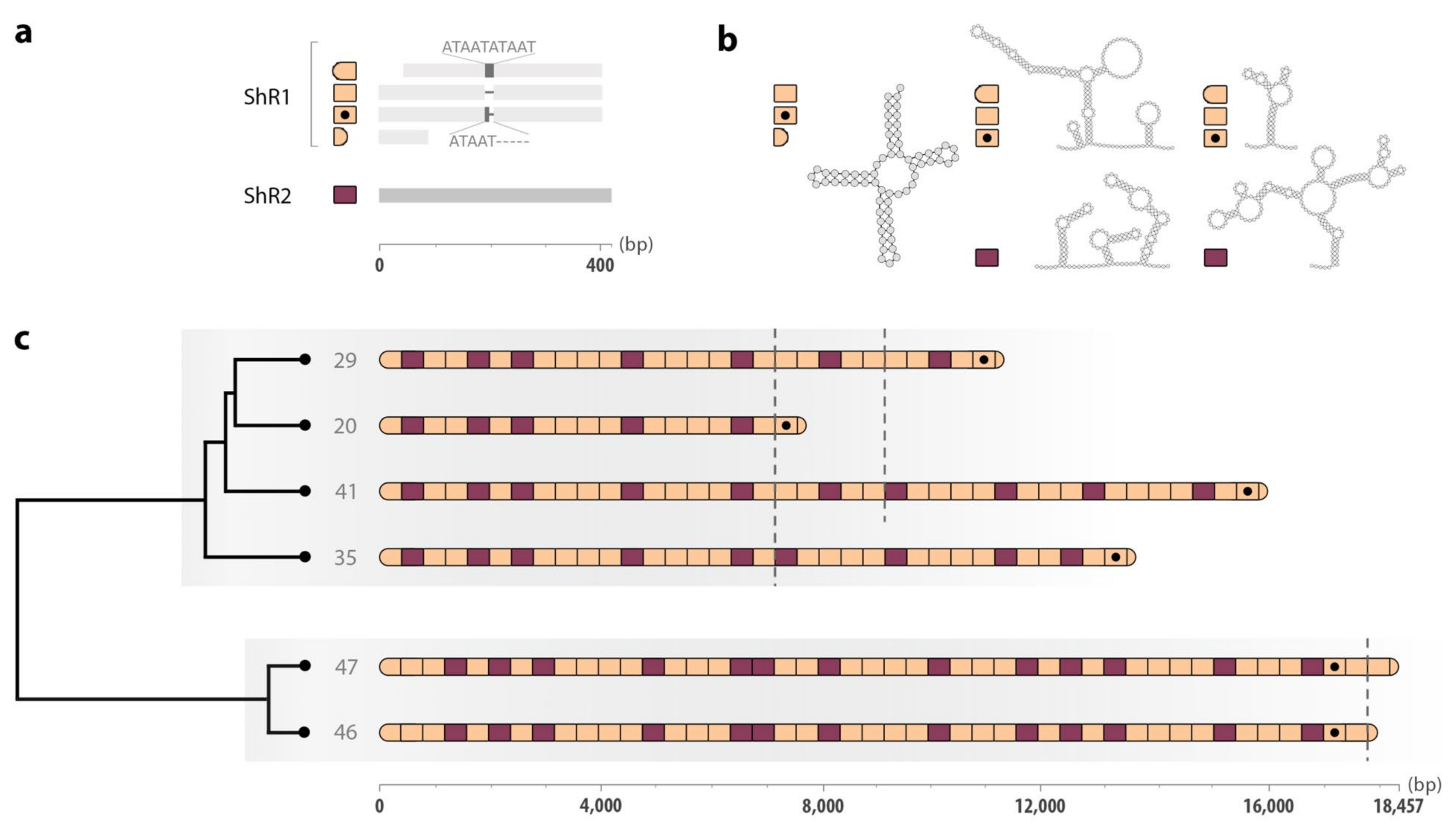
2.3. Structural Features of the Non-Coding Regions
2.4. Tandem-Repeat Patterns in the Non-Coding Region
3. Discussion
4. Materials and Methods
4.1. Parasite Material
4.2. Isolation of High Molecular Weight Genomic DNA, Library Construction and Sequencing
4.3. Defining the mt Genomes
4.4. Annotation of the mt Genomes and Characterisation of the Tandem-Repeat Regions
4.5. Hierarchical Cluster Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, I.; Rawat, D.S.; Pasha, S.T.; Biswas, S.; Sharma, Y.D. Complete nucleotide sequence of the 6 kb element and conserved cytochrome b gene sequences among Indian isolates of Plasmodium falciparum. Int. J. Parasitol. 2001, 31, 1107–1113. [Google Scholar] [CrossRef]
- Sloan, D.B.; Alverson, A.J.; Chuckalovcak, J.P.; Wu, M.; McCauley, D.E.; Palmer, J.D.; Taylor, D.R. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012, 10, e1001241. [Google Scholar] [CrossRef] [PubMed]
- Gissi, C.; Iannelli, F.; Pesole, G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity 2008, 101, 301–320. [Google Scholar] [CrossRef] [PubMed]
- Lavrov, D.V.; Pett, W. Animal mitochondrial DNA as we do not know it: Mt-genome organization and evolution in nonbilaterian lineages. Genome Biol. Evol. 2016, 8, 2896–2913. [Google Scholar] [CrossRef] [PubMed]
- Zíková, A.; Hampl, V.; Paris, Z.; Týč, J.; Lukeš, J. Aerobic mitochondria of parasitic protists: Diverse genomes and complex functions. Mol. Biochem. Parasitol. 2016, 209, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Sandor, S.; Zhang, Y.; Xu, J. Fungal mitochondrial genomes and genetic polymorphisms. Appl. Microbiol. Biotechnol. 2018, 102, 9433–9448. [Google Scholar] [CrossRef]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019, 15, e1008373. [Google Scholar] [CrossRef]
- Lunt, D.H.; Whipple, L.E.; Hyman, B.C. Mitochondrial DNA variable number tandem repeats (VNTRs): Utility and problems in molecular ecology. Mol. Ecol. 1998, 7, 1441–1455. [Google Scholar] [CrossRef]
- Boyce, T.M.; Zwick, M.E.; Aquadro, C.F. Mitochondrial DNA in the bark weevils: Size, structure and heteroplasmy. Genetics 1989, 123, 825–836. [Google Scholar] [CrossRef]
- Okimoto, R.; Chamberlin, H.M.; Macfarlane, J.L.; Wolstenholme, D.R. Repeated sequence sets in mitochondrial DNA molecules of root knot nematodes (Meloidogyne): Nucleotide sequences, genome location and potential for host-race identification. Nucleic Acids Res. 1991, 19, 1619–1626. [Google Scholar] [CrossRef]
- Després, L.; Imbert-Establet, D.; Monnerot, M. Molecular characterization of mitochondrial DNA provides evidence for the recent introduction of Schistosoma mansoni into America. Mol. Biochem. Parasitol. 1993, 60, 221–229. [Google Scholar] [CrossRef]
- Tørresen, O.K.; Star, B.; Mier, P.; Andrade-Navarro, M.A.; Bateman, A.; Jarnot, P.; Gruca, A.; Grynberg, M.; Kajava, A.V.; Promponas, V.J.; et al. Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases. Nucleic Acids Res. 2019, 47, 10994–11006. [Google Scholar] [CrossRef]
- Monnens, M.; Thijs, S.; Briscoe, A.G.; Clark, M.; Frost, E.J.; Littlewood, D.T.J.; Sewell, M.; Smeets, K.; Artois, T.; Vanhove, M.P.M. The first mitochondrial genomes of endosymbiotic rhabdocoels illustrate evolutionary relaxation of atp8 and genome plasticity in flatworms. Int. J. Biol. Macromol. 2020, 162, 454–469. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Jex, A.R.; Campbell, B.E.; Gasser, R.B. Long PCR amplification of the entire mitochondrial genome from individual helminths for direct sequencing. Nat. Protoc. 2007, 2, 2339–2344. [Google Scholar] [CrossRef] [PubMed]
- Hommelsheim, C.M.; Frantzeskakis, L.; Huang, M.; Ülker, B. PCR amplification of repetitive DNA: A limitation to genome editing technologies and many other applications. Sci. Rep. 2014, 4, 5052. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The third revolution in sequencing technology. Trends Genet. 2018, 34, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Kono, N.; Arakawa, K. Nanopore sequencing: Review of potential applications in functional genomics. Dev. Growth Differ. 2019, 61, 316–326. [Google Scholar] [CrossRef]
- Oey, H.; Zakrzewski, M.; Narain, K.; Devi, K.R.; Agatsuma, T.; Nawaratna, S.; Gobert, G.N.; Jones, M.K.; Ragan, M.A.; McManus, D.P.; et al. Whole-genome sequence of the oriental lung fluke Paragonimus westermani. GigaScience 2019, 8, giy146. [Google Scholar] [CrossRef] [PubMed]
- Oey, H.; Zakrzewski, M.; Gravermann, K.; Young, N.D.; Korhonen, P.K.; Gobert, G.N.; Nawaratna, S.; Hasan, S.; Martínez, D.M.; You, H.; et al. Whole-genome sequence of the bovine blood fluke Schistosoma bovis supports interspecific hybridization with S. haematobium. PLoS Pathog. 2019, 15, e1007513. [Google Scholar] [CrossRef] [PubMed]
- Kinkar, L.; Korhonen, P.K.; Cai, H.; Gauci, C.G.; Lightowlers, M.W.; Saarma, U.; Jenkins, D.J.; Li, J.; Li, J.; Young, N.D.; et al. Long-read sequencing reveals a 4.4 kb tandem repeat region in the mitogenome of Echinococcus granulosus (sensu stricto) genotype G1. Parasit. Vectors 2019, 12, 238. [Google Scholar] [CrossRef]
- Kinkar, L.; Young, N.D.; Sohn, W.-M.; Stroehlein, A.J.; Korhonen, P.K.; Gasser, R.B. First record of a tandem-repeat region within the mitochondrial genome of Clonorchis sinensis using a long-read sequencing approach. PLoS Negl. Trop. Dis. 2020, 14, e0008552. [Google Scholar] [CrossRef]
- Littlewood, D.T.J.; Lockyer, A.E.; Webster, B.L.; Johnston, D.A.; Le, T.H. The complete mitochondrial genomes of Schistosoma haematobium and Schistosoma spindale and the evolutionary history of mitochondrial genome changes among parasitic flatworms. Mol. Phylogenet. Evol. 2006, 39, 452–467. [Google Scholar] [CrossRef]
- Hemmi, K.; Kakehashi, R.; Kambayashi, C.; Du Preez, L.; Minter, L.; Furuno, N.; Kurabayashi, A. Exceptional enlargement of the mitochondrial genome results from distinct causes in different rain frogs (Anura: Brevicipitidae: Breviceps). Int. J. Genom. 2020, 2020, e6540343. [Google Scholar] [CrossRef]
- Diaz, F.; Bayona-Bafaluy, M.P.; Rana, M.; Mora, M.; Hao, H.; Moraes, C.T. Human mitochondrial DNA with large deletions repopulates organelles faster than full-length genomes under relaxed copy number control. Nucleic Acids Res. 2002, 30, 4626–4633. [Google Scholar] [CrossRef] [PubMed]
- Shao, R.; Barker, S.C.; Mitani, H.; Aoki, Y.; Fukunaga, M. Evolution of duplicate control regions in the mitochondrial genomes of metazoa: A case study with Australasian Ixodes ticks. Mol. Biol. Evol. 2005, 22, 620–629. [Google Scholar] [CrossRef]
- Havird, J.C.; Forsythe, E.S.; Williams, A.M.; Werren, J.H.; Dowling, D.K.; Sloan, D.B. Selfish mitonuclear conflict. Curr. Biol. 2019, 29, R496–R511. [Google Scholar] [CrossRef] [PubMed]
- Van den Ameele, J.; Li, A.Y.Z.; Ma, H.; Chinnery, P.F. Mitochondrial heteroplasmy beyond the oocyte bottleneck. Semin. Cell Dev. Biol. 2020, 97, 156–166. [Google Scholar] [CrossRef]
- Lee, W.-J.; Conroy, J.; Howell, W.H.; Kocher, T.D. Structure and evolution of teleost mitochondrial control regions. J. Mol. Evol. 1995, 41, 54–66. [Google Scholar] [CrossRef]
- Zhang, D.-X.; Hewitt, G.M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 1997, 25, 99–120. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Ruokonen, M.; Kvist, L. Structure and evolution of the avian mitochondrial control region. Mol. Phylogenet. Evol. 2002, 23, 422–432. [Google Scholar] [CrossRef]
- Pereira, F.; Soares, P.; Carneiro, J.; Pereira, L.; Richards, M.B.; Samuels, D.C.; Amorim, A. Evidence for variable selective pressures at a large secondary structure of the human mitochondrial DNA control region. Mol. Biol. Evol. 2008, 25, 2759–2770. [Google Scholar] [CrossRef]
- Falkenberg, M. Mitochondrial DNA replication in mammalian cells: Overview of the pathway. Essays Biochem. 2018, 62, 287–296. [Google Scholar] [CrossRef]
- Casane, D.; Dennebouy, N.; de Rochambeau, H.; Mounolou, J.C.; Monnerot, M. Nonneutral evolution of tandem repeats in the mitochondrial DNA control region of lagomorphs. Mol. Biol. Evol. 1997, 14, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Jannotti-Passos, L.K.; Souza, C.P.; Parra, J.C.; Simpson, A.J.G. Biparental mitochondrial DNA inheritance in the parasitic trematode Schistosoma mansoni. J. Parasitol. 2001, 87, 79–82. [Google Scholar] [CrossRef]
- Guerra, D.; Plazzi, F.; Stewart, D.T.; Bogan, A.E.; Hoeh, W.R.; Breton, S. Evolution of sex-dependent mtDNA transmission in freshwater mussels (Bivalvia: Unionida). Sci. Rep. 2017, 7, 1551. [Google Scholar] [CrossRef] [PubMed]
- Levinson, G.; Gutman, G.A. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol. Biol. Evol. 1987, 4, 203–221. [Google Scholar] [CrossRef]
- Lunt, D.H.; Hyman, B.C. Animal mitochondrial DNA recombination. Nature 1997, 387, 247. [Google Scholar] [CrossRef]
- Boore, J.L.; Brown, W.M. Big trees from little genomes: Mitochondrial gene order as a phylogenetic tool. Curr. Opin. Genet. Dev. 1998, 8, 668–674. [Google Scholar] [CrossRef]
- Pâques, F.; Leung, W.-Y.; Haber, J.E. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol. 1998, 18, 2045–2054. [Google Scholar] [CrossRef]
- Rokas, A.; Ladoukakis, E.; Zouros, E. Animal mitochondrial DNA recombination revisited. Trends Ecol. Evol. 2003, 18, 411–417. [Google Scholar] [CrossRef]
- Lewis, S.C.; Joers, P.; Willcox, S.; Griffith, J.D.; Jacobs, H.T.; Hyman, B.C. A rolling circle replication mechanism produces multimeric lariats of mitochondrial DNA in Caenorhabditis elegans. PLoS Genet. 2015, 11, e1004985. [Google Scholar] [CrossRef]
- Després, L.; Imbert-Establet, D.; Combes, C.; Bonhomme, F.; Monnerot, M. Isolation and polymorphism in mitochondrial DNA from Schistosoma mansoni. Mol. Biochem. Parasitol. 1991, 47, 139–141. [Google Scholar] [CrossRef]
- Le, T.H.; Humair, P.F.; Blair, D.; Agatsuma, T.; Littlewood, D.T.; McManus, D.P. Mitochondrial gene content, arrangement and composition compared in African and Asian schistosomes. Mol. Biochem. Parasitol. 2001, 117, 61–71. [Google Scholar] [CrossRef]
- Protasio, A.V.; Tsai, I.J.; Babbage, A.; Nichol, S.; Hunt, M.; Aslett, M.A.; Silva, N.D.; Velarde, G.S.; Anderson, T.J.C.; Clark, R.C.; et al. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl. Trop. Dis. 2012, 6, e1455. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.J.; Wang, X.; Wang, J.; Tan, Y.; Tang, X.; Werren, J.H.; Zhang, D.; Wang, X. Comparative analysis reveals the expansion of mitochondrial DNA control region containing unusually high G-C tandem repeat arrays in Nasonia vitripennis. Int. J. Biol. Macromol. 2021, 166, 1246–1257. [Google Scholar] [CrossRef]
- Omote, K.; Nishida, C.; Dick, M.H.; Masuda, R. Limited phylogenetic distribution of a long tandem-repeat cluster in the mitochondrial control region in Bubo (Aves, Strigidae) and cluster variation in Blakiston’s fish owl (Bubo blakistoni). Mol. Phylogenet. Evol. 2013, 66, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liang, D.; Mao, R.-L.; Hillis, D.M.; Wake, D.B.; Cannatella, D.C. Efficient sequencing of Anuran mtDNAs and a mitogenomic exploration of the phylogeny and evolution of frogs. Mol. Biol. Evol. 2013, 30, 1899–1915. [Google Scholar] [CrossRef]
- Xia, Y.; Zheng, Y.; Miura, I.; Wong, P.B.; Murphy, R.W.; Zeng, X. The evolution of mitochondrial genomes in modern frogs (Neobatrachia): Nonadaptive evolution of mitochondrial genome reorganization. BMC Genom. 2014, 15, 691. [Google Scholar] [CrossRef][Green Version]
- Solà, E.; Álvarez-Presas, M.; Frías-López, C.; Littlewood, D.T.J.; Rozas, J.; Riutort, M. Evolutionary analysis of mitogenomes from parasitic and free-living flatworms. PLoS ONE 2015, 10, e0120081. [Google Scholar] [CrossRef]
- Chen, Z.-T.; Yu, B.; Du, Y.-Z. The nearly complete mitochondrial genome of a snout weevil, Eucryptorrhynchus brandti (Coleoptera: Curculionidae). Mitochondrial DNA A DNA Map. Seq. Anal. 2016, 27, 2736–2737. [Google Scholar] [CrossRef] [PubMed]
- Prada, C.F.; Boore, J.L. Gene annotation errors are common in the mammalian mitochondrial genomes database. BMC Genom. 2019, 20, 73. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Li, X.; Yin, X.; Li, X.; Yin, S.; Yang, M. The mitochondrial genome of Apion squamigerum (Coleoptera, Curculionoidea, Brentidae) and the phylogenetic implications. PeerJ 2020, 8, e8386. [Google Scholar] [CrossRef]
- Hug, L.A.; Baker, B.J.; Anantharaman, K.; Brown, C.T.; Probst, A.J.; Castelle, C.J.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A new view of the tree of life. Nat. Microbiol. 2016, 1, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Young, N.D.; Jex, A.R.; Li, B.; Liu, S.; Yang, L.; Xiong, Z.; Li, Y.; Cantacessi, C.; Hall, R.S.; Xu, X.; et al. Whole-genome sequence of Schistosoma haematobium. Nat. Genet. 2012, 44, 221–225. [Google Scholar] [CrossRef]
- Stroehlein, A.J.; Korhonen, P.K.; Chong, T.M.; Lim, Y.L.; Chan, K.G.; Webster, B.; Rollinson, D.; Brindley, P.J.; Gasser, R.B.; Young, N.D. High-quality Schistosoma haematobium genome achieved by single-molecule and long-range sequencing. GigaScience 2019, 8, giz108. [Google Scholar] [CrossRef] [PubMed]
- Lewis, F.A.; Liang, Y.-S.; Raghavan, N.; Knight, M. The NIH-NIAID schistosomiasis resource center. PLoS Negl. Trop. Dis. 2008, 2, e267. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Chin, C.-S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org (accessed on 23 September 2020).
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Telford, M.J.; Herniou, E.A.; Russell, R.B.; Littlewood, D.T.J. Changes in mitochondrial genetic codes as phylogenetic characters: Two examples from the flatworms. Proc. Natl. Acad. Sci. USA 2000, 97, 11359–11364. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- Kerpedjiev, P.; Hammer, S.; Hofacker, I.L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 2015, 31, 3377–3379. [Google Scholar] [CrossRef]
- Suzuki, R.; Shimodaira, H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 2006, 22, 1540–1542. [Google Scholar] [CrossRef]
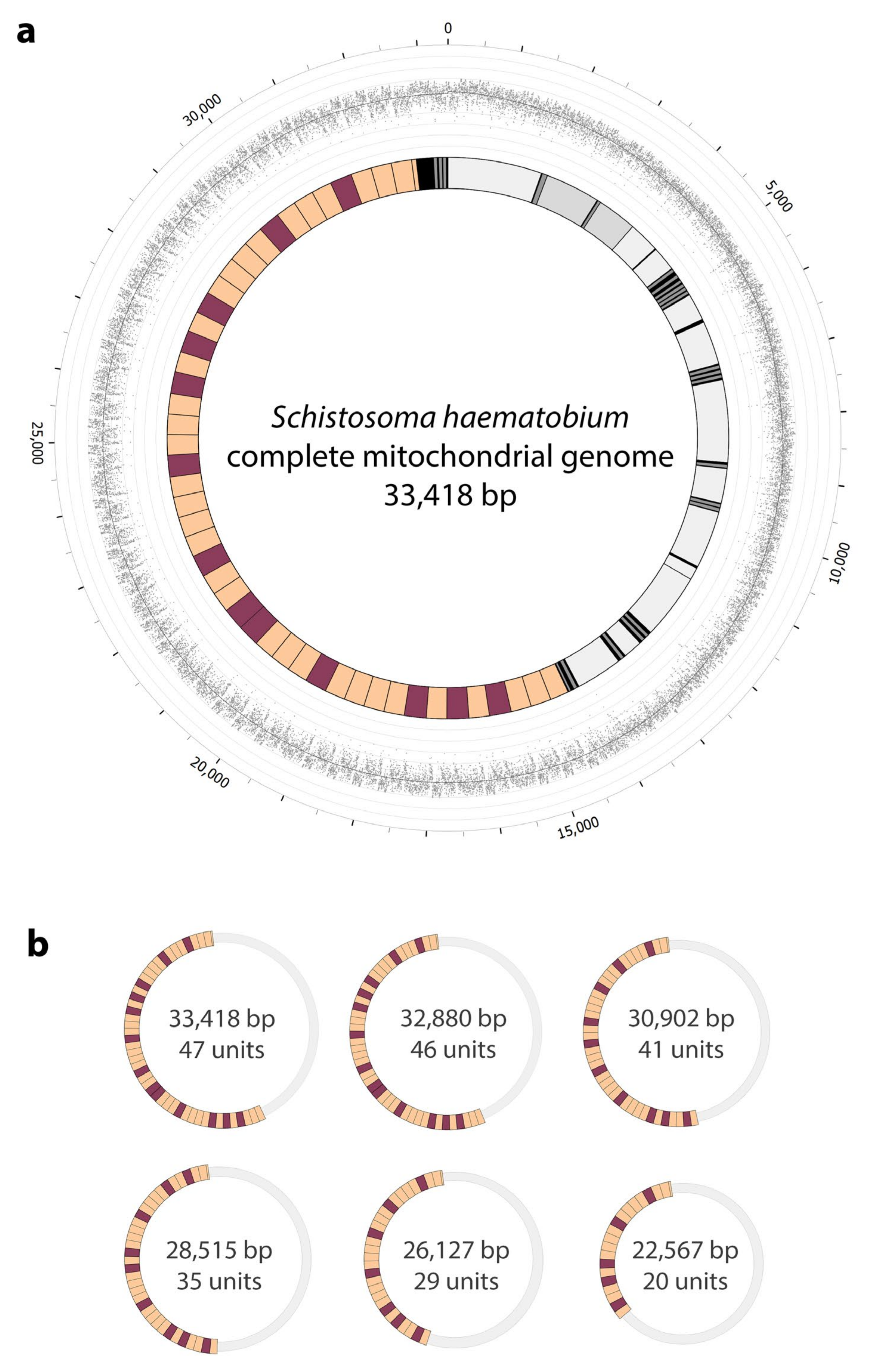
| Features (cf. Figure 2, Panel b) | ||||||
|---|---|---|---|---|---|---|
| Mt genome length/size (bp) | 33,418 | 32,880 | 30,902 | 28,515 | 26,127 | 22,567 |
| Length of tandem-repeat region (bp) | 18,458 | 17,920 | 15,942 | 13,555 | 11,167 | 7607 |
| Number of repeat units in this non-coding region Tandem-repeat region relative to mt genome length/size (%) No. of long-reads representing the tandem-repeat region | 47 55.2 511 | 46 54.5 151 | 41 51.6 155 | 35 47.5 179 | 29 42.7 170 | 20 33.7 249 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kinkar, L.; Gasser, R.B.; Webster, B.L.; Rollinson, D.; Littlewood, D.T.J.; Chang, B.C.H.; Stroehlein, A.J.; Korhonen, P.K.; Young, N.D. Nanopore Sequencing Resolves Elusive Long Tandem-Repeat Regions in Mitochondrial Genomes. Int. J. Mol. Sci. 2021, 22, 1811. https://doi.org/10.3390/ijms22041811
Kinkar L, Gasser RB, Webster BL, Rollinson D, Littlewood DTJ, Chang BCH, Stroehlein AJ, Korhonen PK, Young ND. Nanopore Sequencing Resolves Elusive Long Tandem-Repeat Regions in Mitochondrial Genomes. International Journal of Molecular Sciences. 2021; 22(4):1811. https://doi.org/10.3390/ijms22041811
Chicago/Turabian StyleKinkar, Liina, Robin B. Gasser, Bonnie L. Webster, David Rollinson, D. Timothy J. Littlewood, Bill C.H. Chang, Andreas J. Stroehlein, Pasi K. Korhonen, and Neil D. Young. 2021. "Nanopore Sequencing Resolves Elusive Long Tandem-Repeat Regions in Mitochondrial Genomes" International Journal of Molecular Sciences 22, no. 4: 1811. https://doi.org/10.3390/ijms22041811
APA StyleKinkar, L., Gasser, R. B., Webster, B. L., Rollinson, D., Littlewood, D. T. J., Chang, B. C. H., Stroehlein, A. J., Korhonen, P. K., & Young, N. D. (2021). Nanopore Sequencing Resolves Elusive Long Tandem-Repeat Regions in Mitochondrial Genomes. International Journal of Molecular Sciences, 22(4), 1811. https://doi.org/10.3390/ijms22041811