Inhibitors of Nucleotide Excision Repair Decrease UVB-Induced Mutagenesis—An In Vitro Study
Abstract
1. Introduction
2. Results
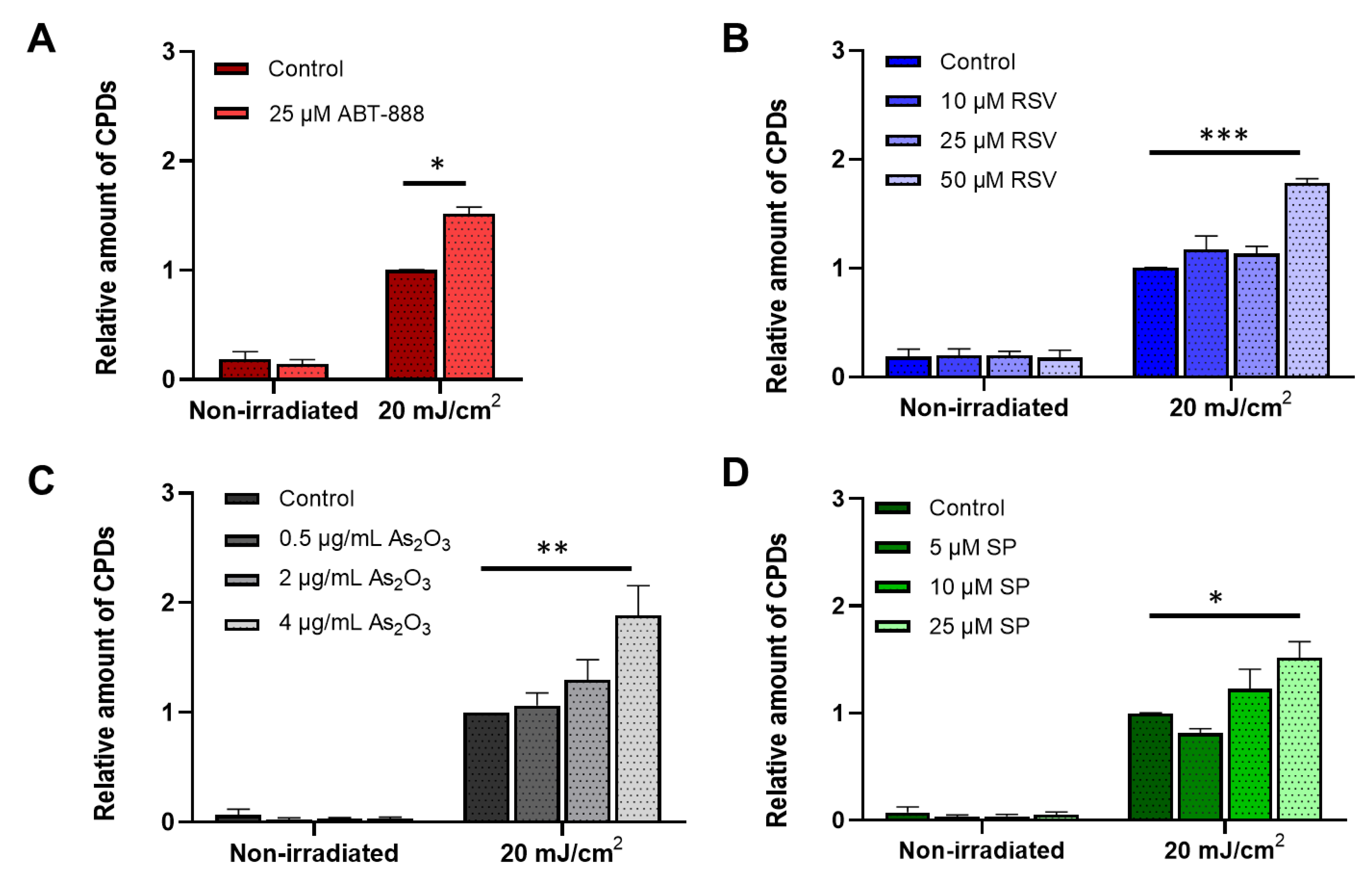
2.1. All Tested Chemicals Enhance CPD Formation after UVB Irradiation
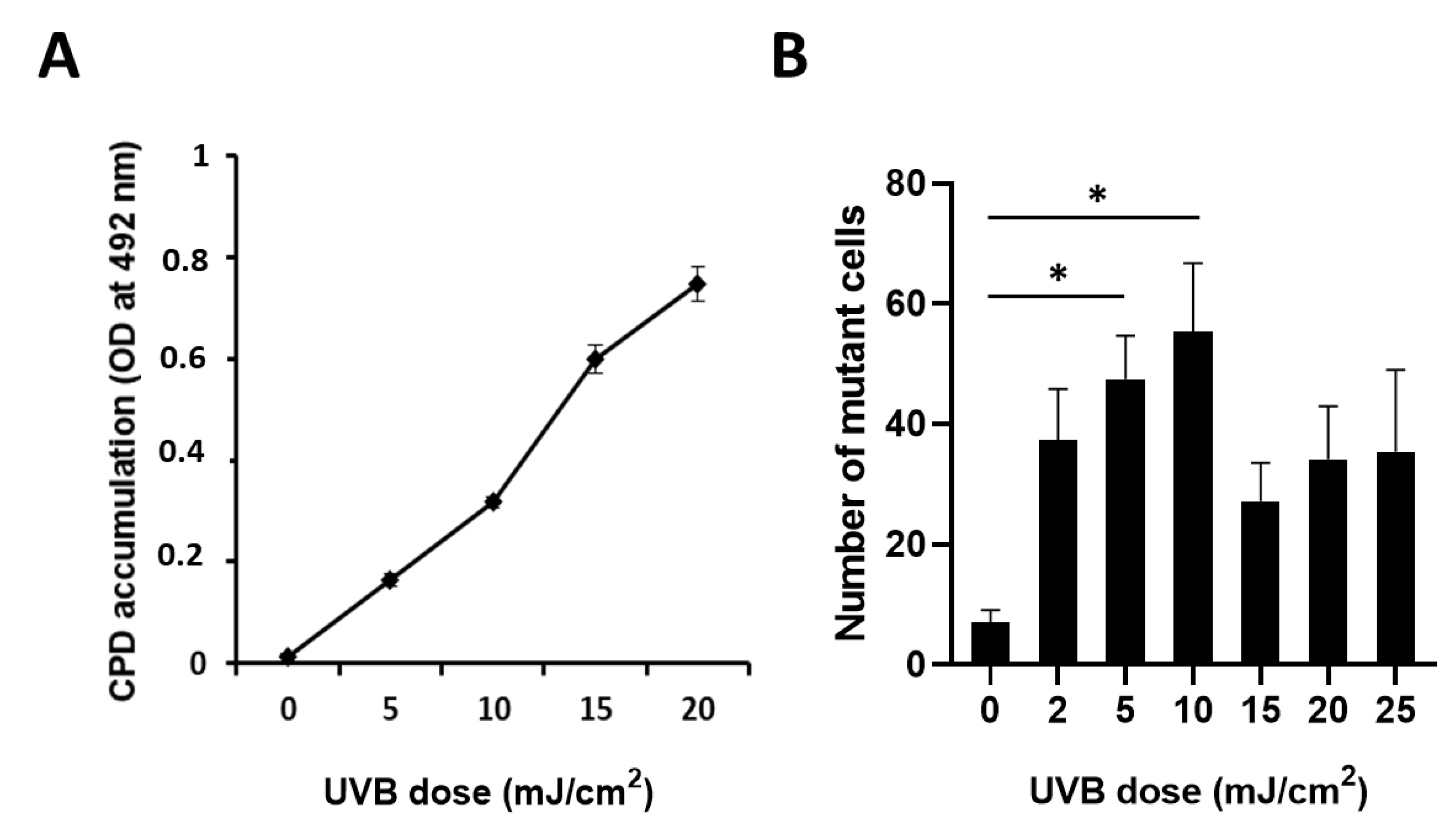
2.2. CPD Accumulation and UVB-Induced Mutagenesis Show a Nonlinear Relationship
2.3. Veliparib, Arsenic-Trioxide, and Spironolactone Treatments Prevent UVB-Induced Mutagenesis
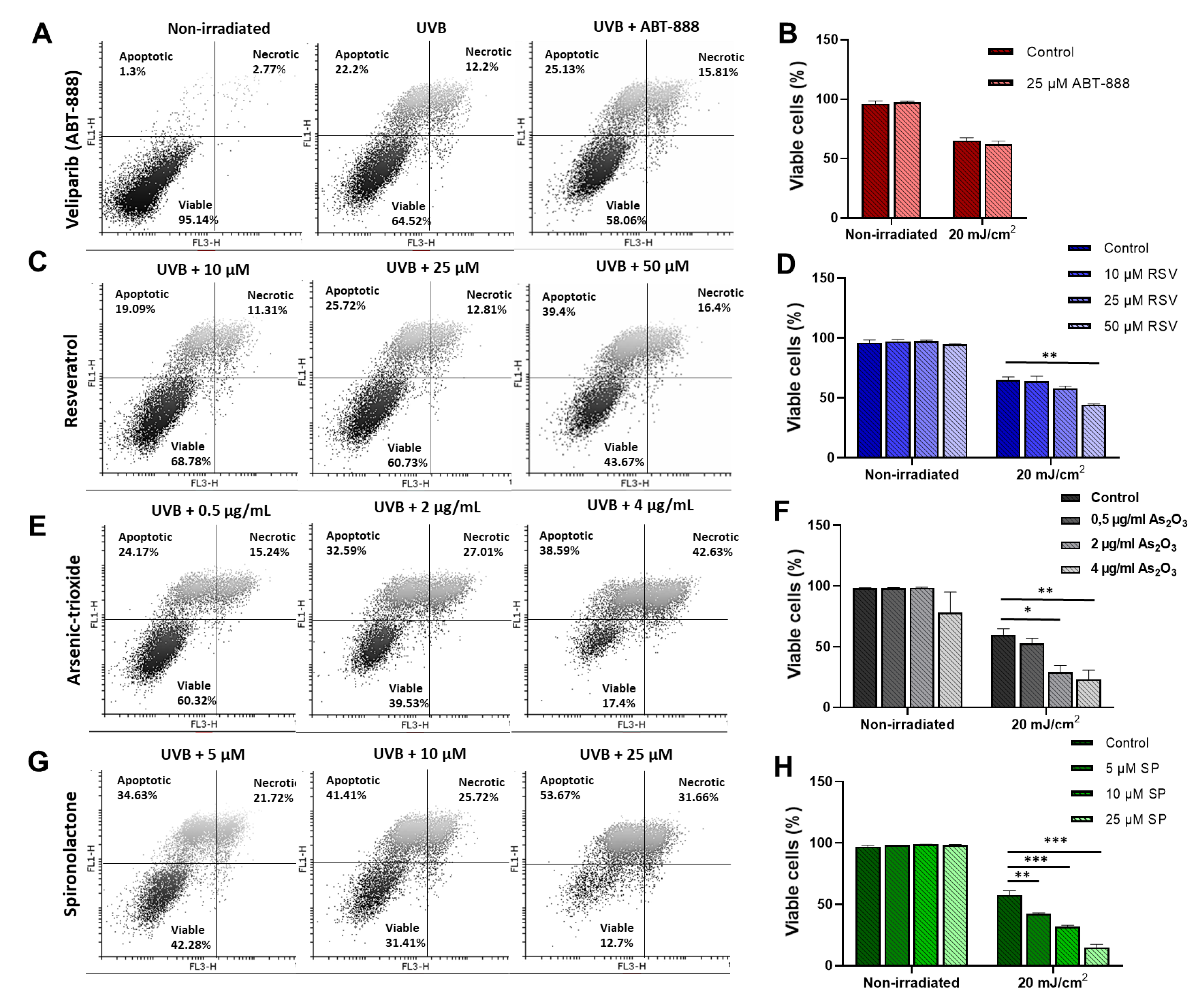
2.4. Resveratrol, Arsenic Trioxide, and Spironolactone Enhanced UVB-Induced Apoptosis
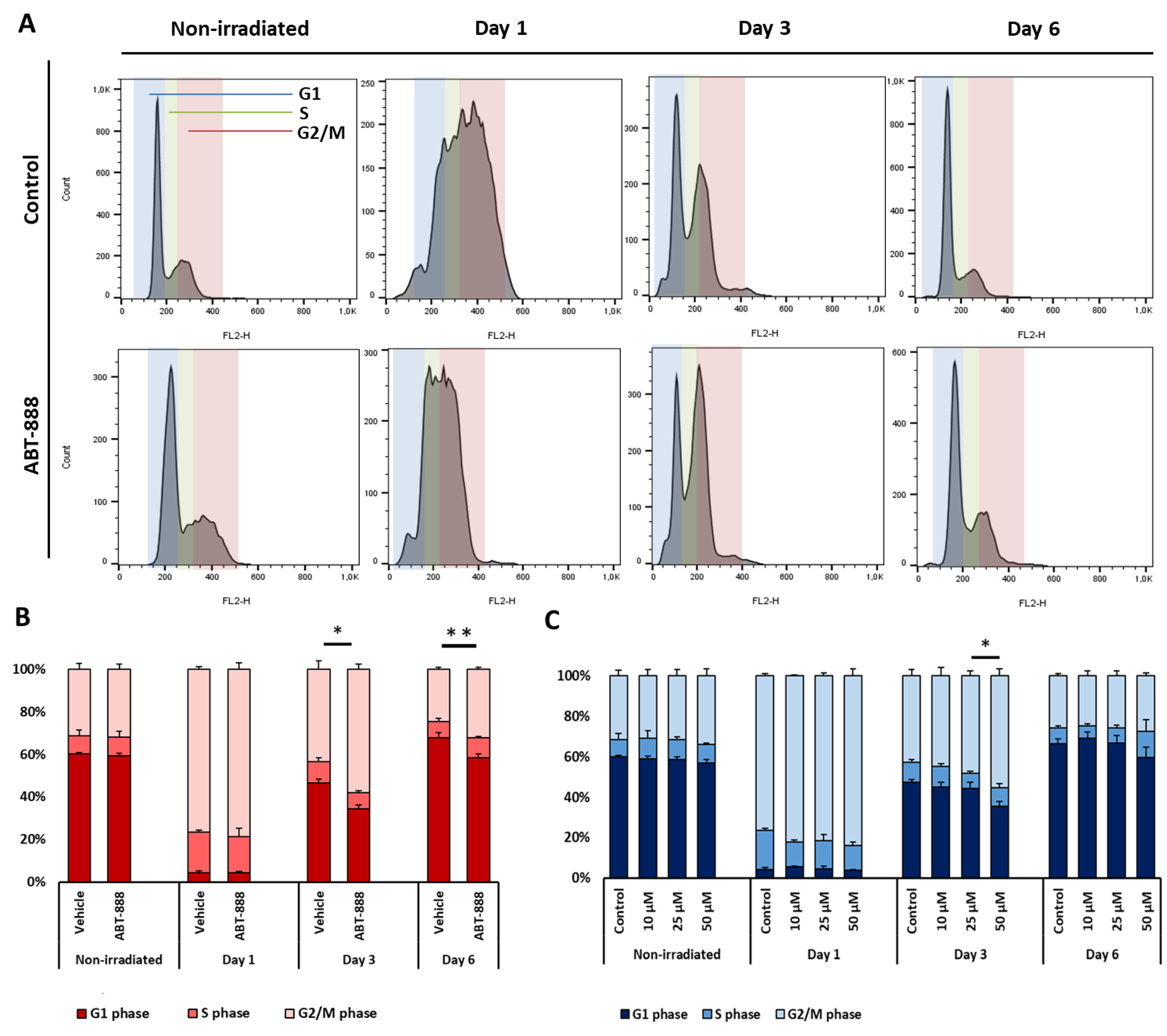
2.5. Veliparib and Resveratrol Augment UVB-Induced Cell Cycle Arrest
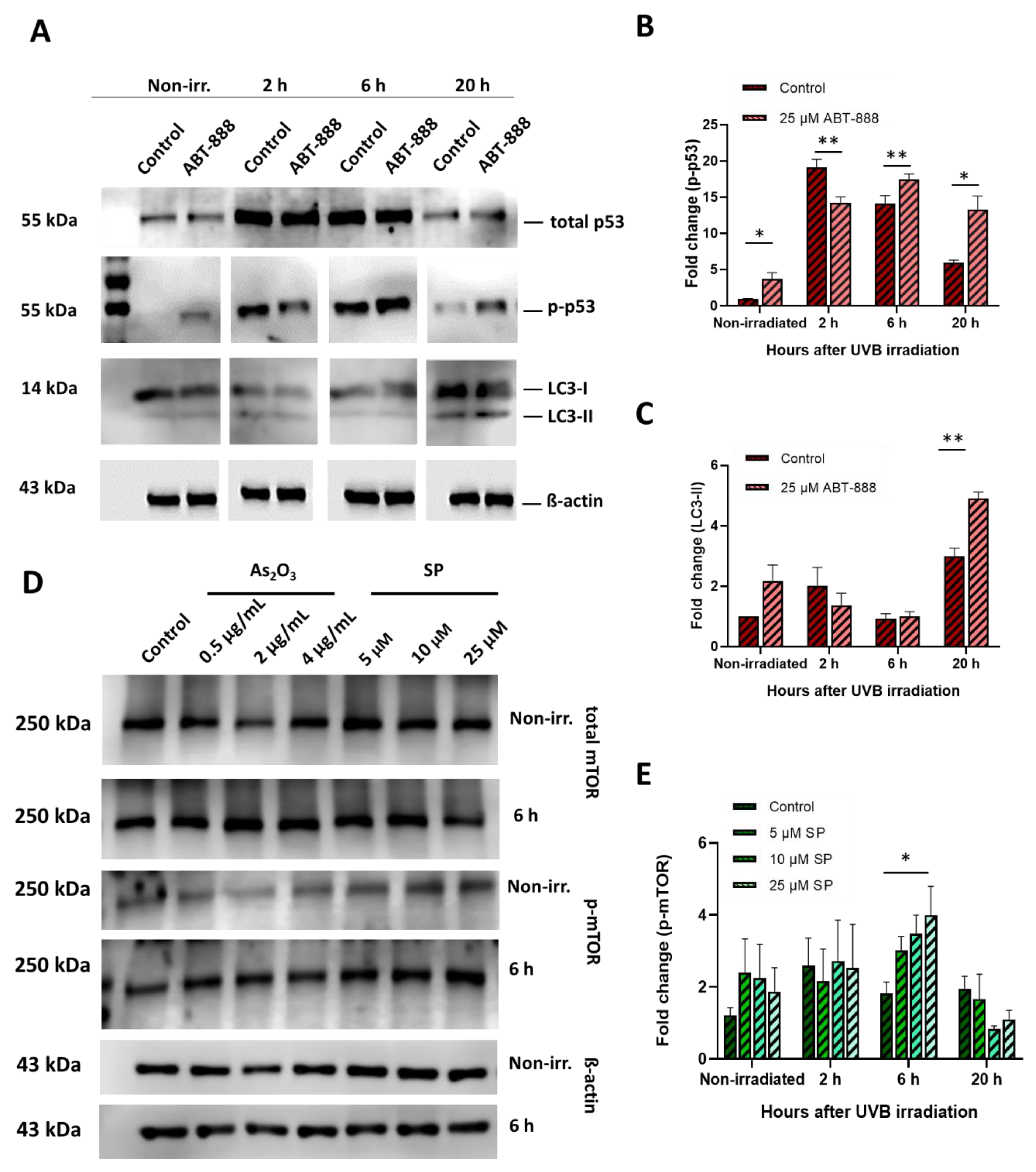
2.6. Altered Protein Expression in Diverse Stress–Response Pathways May Orchestrate UVB-Induced Mutagenesis
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Treatment
4.3. UVB Irradiation
4.4. CPD-Specific Enzyme-Linked Immunosorbent Assay (ELISA)
4.5. HPRT Gene Mutation Assay
4.6. Apoptosis Assay
4.7. Cell Cycle Analysis
4.8. Western Blot
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 6-4PP | Pyrimidine (6-4) pyrimidone photoproduct |
| ABT-888 | Veliparib |
| As2O3 | Arsenic trioxide |
| CHO | Chinese hamster ovary cell line |
| CPD | Cyclobutane–pyrimidine dimer |
| HPRT | Hypoxanthine phosphoribosyltransferase 1 |
| LC3 | Microtubule-associated protein 1A/1B-light chain 3 |
| mTOR | mammalian target of rapamycin |
| NER | Nucleotide excision repair |
| p53 | Cellular tumor antigen p53 |
| PARP1 | Poly [ADP-ribose] polymerase 1 |
| RSV | Resveratrol |
| SP | Spironolactone |
| XP | Xeroderma pigmentosum |
| UVB | Ultraviolet B radiation |
References
- Olsen, C.; Green, A.C.; Pandeya, N.; Whiteman, D. Trends in Melanoma Incidence Rates in Eight Susceptible Populations through 2015. J. Investig. Dermatol. 2019, 139, 1392–1395. [Google Scholar] [CrossRef]
- Glazer, A.M.; Winkelmann, R.R.; Farberg, A.S.; Rigel, D.S. Analysis of Trends in US Melanoma Incidence and Mortality. JAMA Dermatol. 2017, 153, 225–226. [Google Scholar] [CrossRef]
- Sacchetto, L.; Zanetti, R.; Comber, H.; Bouchardy, C.; Brewster, D.; Broganelli, P.; López, M.D.C.; Coza, D.; Galceran, J.; Gavin, A.; et al. Trends in incidence of thick, thin and in situ melanoma in Europe. Eur. J. Cancer 2018, 92, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Apalla, Z.; Lallas, A.; Sotiriou, E.; Lazaridou, E.; Ioannides, D. Epidemiological trends in skin cancer. Dermatol. Pract. Concept. 2017, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Griffin, L.L.; Ali, F.R.; Lear, J. Non-melanoma skin cancer. Clin. Med. 2016, 16, 62–65. [Google Scholar] [CrossRef]
- Boukamp, P. Non-melanoma skin cancer: What drives tumor development and progression? Carcinogenesis 2005, 26, 1657–1667. [Google Scholar] [CrossRef]
- de Zwaan, S.E.; Haass, N.K. Genetics of basal cell carcinoma. Australas J. Dermatol. 2010, 51, 81–92. [Google Scholar] [CrossRef]
- Emri, G.; Paragh, G.; Tósaki, Á.; Janka, E.; Kollár, S.; Hegedűs, C.; Gellén, E.; Horkay, I.; Koncz, G.; Remenyik, É. Ultraviolet radiation-mediated development of cutaneous melanoma: An update. J. Photochem. Photobiol. B Biol. 2018, 185, 169–175. [Google Scholar] [CrossRef] [PubMed]
- de Lima-Bessa, K.M.; Armelini, M.G.; Chiganças, V.; Jacysyn, J.F.; Amarante-Mendes, G.P.; Sarasin, A.; Menck, C.F.M. CPDs and 6-4PPs play different roles in UV-induced cell death in normal and NER-deficient human cells. DNA Repair 2008, 7, 303–312. [Google Scholar] [CrossRef]
- Pfeifer, G.P.; You, Y.-H.; Besaratinia, A. Mutations induced by ultraviolet light. Mutat. Res. 2005, 571, 19–31. [Google Scholar] [CrossRef]
- Brash, D.E. UV signature mutations. Photochem. Photobiol. 2015, 91, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.-L.; Nakajima, S.; Ma, L.; Walter, B.; Yasui, A.; Ethell, D.W.; Owen, L.B. Differential biologic effects of CPD and 6-4PP UV-induced DNA damage on the induction of apoptosis and cell-cycle arrest. BMC Cancer 2005, 5, 135. [Google Scholar] [CrossRef]
- Meador, J.A.; Walter, R.B.; Mitchell, D.L. Induction, Distribution and Repair of UV Photodamage in the Platyfish, Xiphophorus signum. Photochem. Photobiol. 2000, 72, 260–266. [Google Scholar] [CrossRef]
- Wei, L.; Christensen, S.R.; Fitzgerald, M.E.; Graham, J.; Hutson, N.D.; Zhang, C.; Huang, Z.; Hu, Q.; Zhan, F.; Xie, J.; et al. Ultradeep sequencing differentiates patterns of skin clonal mutations associated with sun-exposure status and skin cancer burden. Sci. Adv. 2021, 7, eabd7703. [Google Scholar] [CrossRef]
- You, Y.H.; Szabó, P.E.; Pfeifer, G.P. Cyclobutane pyrimidine dimers form preferentially at the major p53 mutational hotspot in UVB-induced mouse skin tumors. Carcinogenesis. 2000, 21, 2113–2117. [Google Scholar] [CrossRef] [PubMed]
- Scharer, O.D. Nucleotide excision repair in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013, 5, a012609. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, A.; Kobayashi, N.; Muramatsu, T.; Yamashina, Y.; Shirai, T.; Hashimoto, M.W.; Ikenaga, M.; Mori, T. Three-dimensional visualization of ultraviolet-induced DNA damage and its repair in human cell nuclei. J. Investig. Dermatol. 1998, 110, 143–148. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jans, J.; Schul, W.; Sert, Y.-G.; Rijksen, Y.; Rebel, H.; Eker, A.P.; Nakajima, S.; Van Steeg, H.; De Gruijl, F.R.; Yasui, A.; et al. Powerful skin cancer protection by a CPD-photolyase transgene. Curr. Biol. 2005, 15, 105–115. [Google Scholar] [CrossRef] [PubMed]
- de Boer, J.; Hoeijmakers, J.H. Nucleotide excision repair and human syndromes. Carcinogenesis 2000, 21, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, G.; Garvey, D.R.; Singh, C.K.; Mintie, C.A.; Ahmad, N. Effects and Mechanism of Nicotinamide Against UVA- and/or UVB-mediated DNA Damages in Normal Melanocytes. Photochem. Photobiol. 2019, 95, 331–337. [Google Scholar] [CrossRef]
- Surjana, D.; Halliday, G.M.; Damian, D.L. Nicotinamide enhances repair of ultraviolet radiation-induced DNA damage in human keratinocytes and ex vivo skin. Carcinogenesis 2013, 34, 1144–1149. [Google Scholar] [CrossRef] [PubMed]
- Katiyar, S.K.; Vaid, M.; Van Steeg, H.; Meeran, S.M. Green tea polyphenols prevent UV-induced immunosuppression by rapid repair of DNA damage and enhancement of nucleotide excision repair genes. Cancer Prev. Res. 2010, 3, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Sheu, H.; Huang, Y.S.; Tsai, T.R.; Kuo, K. A novel function of emodin: Enhancement of the nucleotide excision repair of UV- and cisplatin-induced DNA damage in human cells. Biochem. Pharmacol. 1999, 58, 49–57. [Google Scholar] [CrossRef]
- Vaid, M.; Sharma, S.D.; Katiyar, S.K. Proanthocyanidins inhibit photocarcinogenesis through enhancement of DNA repair and xeroderma pigmentosum group A-dependent mechanism. Cancer Prev. Res. 2010, 3, 1621–1629. [Google Scholar] [CrossRef] [PubMed]
- Guillermo-Lagae, R.; Deep, G.; Ting, H.; Agarwal, C.; Agarwal, R. Silibinin enhances the repair of ultraviolet B-induced DNA damage by activating p53-dependent nucleotide excision repair mechanism in human dermal fibroblasts. Oncotarget 2015, 6, 39594–39606. [Google Scholar] [CrossRef]
- Keuser, B.; Khobta, A.; Galle, K.; Anderhub, S.; Schulz, I.; Pauly, K.; Epe, B. Influences of histone deacetylase inhibitors and resveratrol on DNA repair and chromatin compaction. Mutagenesis 2013, 28, 569–576. [Google Scholar] [CrossRef][Green Version]
- Okui, T.; Fujiwara, Y. Inhibition of human excision DNA repair by inorganic arsenic and the co-mutagenic effect in V79 Chinese hamster cells. Mutat. Res. 1986, 172, 69–76. [Google Scholar] [CrossRef]
- Holcomb, N.; Goswami, M.; Han, S.G.; Scott, T.; D’Orazio, J.; Orren, D.K.; Gairola, C.G.; Mellon, I. Inorganic arsenic inhibits the nucleotide excision repair pathway and reduces the expression of XPC. DNA Repair 2017, 52, 70–80. [Google Scholar] [CrossRef]
- Hegedűs, C.; Boros, G.; Fidrus, E.; Kis, G.N.; Antal, M.; Juhász, T.; Janka, E.A.; Jankó, L.; Paragh, G.; Emri, G.; et al. PARP1 Inhibition Augments UVB-Mediated Mitochondrial Changes-Implications for UV-Induced DNA Repair and Photocarcinogenesis. Cancers 2019, 12, 5. [Google Scholar] [CrossRef]
- Sergey, A.; Mériam, A.; Brino, L.; Marc-Egly, J.; Larsen, A.K.; Coin, F. A small molecule screen identifies an inhibitor of DNA repair inducing the degradation of TFIIH and the chemosensitization of tumor cells to platinum. Chem. Biol. 2014, 21, 398–407. [Google Scholar]
- Kemp, M.G.; Krishnamurthy, S.; Kent, M.N.; Schumacher, D.L.; Sharma, P.; Excoffon, K.J.; Travers, J.B. Spironolactone Depletes the XPB Protein and Inhibits DNA Damage Responses in UVB-Irradiated Human Skin. J. Investig. Dermatol. 2019, 139, 448–454. [Google Scholar] [CrossRef]
- Ueda, M.; Matsuura, K.; Kawai, H.; Wakasugi, M.; Matsunaga, T. Spironolactone-induced XPB degradation depends on CDK7 kinase and SCF(FBXL18) E3 ligase. Genes Cells 2019, 24, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, A.E.W.; Menefee, M.E.; Hartmann, L.C.; Long, H.J.; Northfelt, D.W.; Reid, J.M.; Boakye-Agyeman, F.; Kayode, O.; Flatten, K.S.; Harrell, M.I.; et al. A Phase I Clinical Trial of the Poly(ADP-ribose) Polymerase Inhibitor Veliparib and Weekly Topotecan in Patients with Solid Tumors. Clin. Cancer Res. 2018, 24, 744–752. [Google Scholar] [CrossRef]
- Kummar, S.; Ji, J.; Morgan, R.; Lenz, H.-J.; Puhalla, S.L.; Belani, C.P.; Gandara, D.R.; Allen, D.; Kiesel, B.; Beumer, J.H.; et al. A phase I study of veliparib in combination with metronomic cyclophosphamide in adults with refractory solid tumors and lymphomas. Clin. Cancer Res. 2012, 18, 1726–1734. [Google Scholar] [CrossRef] [PubMed]
- Lowery, M.A.; Kelsen, D.P.; Capanu, M.; Smith, S.C.; Lee, J.W.; Stadler, Z.K.; Moore, M.J.; Kindler, H.L.; Golan, T.; Segal, A.; et al. Phase II trial of veliparib in patients with previously treated BRCA-mutated pancreas ductal adenocarcinoma. Eur. J. Cancer 2018, 89, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.S.; Blais, N.; Mazieres, J.; Martin Reck, C.; Jones, M.; Juhasz, E.; Urban, L.; Orlov, S.; Barlesi, F.; Kio, E.; et al. Randomized, Placebo-Controlled, Phase II Study of Veliparib in Combination with Carboplatin and Paclitaxel for Advanced/Metastatic Non-Small Cell Lung Cancer. Clin. Cancer Res. 2017, 23, 1937–1944. [Google Scholar] [CrossRef] [PubMed]
- Struthers, A.D.; Krum, H.; Williams, G.H. A comparison of the aldosterone-blocking agents eplerenone and spironolactone. Clin. Cardiol. 2008, 31, 153–158. [Google Scholar] [CrossRef]
- Lainscak, M.; Pelliccia, F.; Rosano, G.; Vitale, C.; Schiariti, M.; Greco, C.; Speziale, G.; Gaudio, C. Safety profile of mineralocorticoid receptor antagonists: Spironolactone and eplerenone. Int. J. Cardiol. 2015, 200, 25–29. [Google Scholar] [CrossRef]
- Soignet, S.L.; Frankel, S.R.; Douer, D.; Tallman, M.S.; Kantarjian, H.; Calleja, E.; Stone, R.M.; Kalaycio, M.; Scheinberg, D.A.; Steinherz, P.; et al. United States multicenter study of arsenic trioxide in relapsed acute promyelocytic leukemia. J. Clin. Oncol. 2001, 19, 3852–3860. [Google Scholar] [CrossRef] [PubMed]
- Cicconi, L.; Fenaux, P.; Kantarjian, H.; Tallman, M.; Sanz, M.A.; Lo-Coco, F. Molecular remission as a therapeutic objective in acute promyelocytic leukemia. Leukemia 2018, 32, 1671–1678. [Google Scholar] [CrossRef]
- Gill, H.; Yim, R.; Lee, H.K.K.; Mak, V.; Lin, S.-Y.; Kho, B.; Yip, S.-F.; Lau, J.S.M.; Li, W.; Ip, H.-W.; et al. Long-term outcome of relapsed acute promyelocytic leukemia treated with oral arsenic trioxide-based reinduction and maintenance regimens: A 15-year prospective study. Cancer 2018, 124, 2316–2326. [Google Scholar] [CrossRef] [PubMed]
- Sirerol, J.A.; Feddi, F.; Mena, S.; Rodriguez, M.L.; Sirera, P.; Aupí, M.; Pérez, S.; Asensi, M.; Ortega, A.; Estrela, J.M. Topical treatment with pterostilbene, a natural phytoalexin, effectively protects hairless mice against UVB radiation-induced skin damage and carcinogenesis. Free Radic. Biol Med. 2015, 85, 1–11. [Google Scholar] [CrossRef]
- Fujimura, A.T.; Martinez, R.M.; Pinho-Ribeiro, F.A.; Silva, A.M.; Baracat, M.M.; Georgetti, S.R.; Verri, W.A.; Chorilli, M.; Casagrande, R. Resveratrol-Loaded Liquid-Crystalline System Inhibits UVB-Induced Skin Inflammation and Oxidative Stress in Mice. J. Nat. Prod. 2016, 79, 1329–1338. [Google Scholar] [CrossRef] [PubMed]
- Adhami, V.M.; Afaq, F.; Ahmad, N. Suppression of ultraviolet B exposure-mediated activation of NF-kappaB in normal human keratinocytes by resveratrol. Neoplasia 2003, 5, 74–82. [Google Scholar] [CrossRef]
- Subedi, L.; Lee, T.H.; Wahedi, H.M.; Baek, S.; Kim, S.Y. Corrigendum to “Resveratrol-Enriched Rice Attenuates UVB-ROS-Induced Skin Aging via Downregulation of Inflammatory Cascades”. Oxid. Med. Cell. Longev. 2018, 2018, 6052623. [Google Scholar] [CrossRef] [PubMed]
- Afaq, F.; Adhami, V.M.; Ahmad, N. Prevention of short-term ultraviolet B radiation-mediated damages by resveratrol in SKH-1 hairless mice. Toxicol. Appl. Pharmacol. 2003, 186, 28–37. [Google Scholar] [CrossRef]
- Vitale, N.; Kisslinger, A.; Paladino, S.; Procaccini, C.; Matarese, G.; Pierantoni, G.M.; Mancini, F.P.; Tramontano, D. Resveratrol couples apoptosis with autophagy in UVB-irradiated HaCaT cells. PLoS ONE 2013, 8, e80728. [Google Scholar] [CrossRef]
- Aziz, M.H.; Afaq, F.; Ahmad, N. Prevention of ultraviolet-B radiation damage by resveratrol in mouse skin is mediated via modulation in survivin. Photochem. Photobiol. 2005, 81, 25–31. [Google Scholar] [CrossRef]
- Back, J.H.; Zhu, Y.; Calabro, A.; Queenan, C.; Kim, A.S.; Arbesman, J.; Kim, A.L. Resveratrol-mediated downregulation of Rictor attenuates autophagic process and suppresses UV-induced skin carcinogenesis. Photochem. Photobiol. 2012, 88, 1165–1172. [Google Scholar] [CrossRef]
- Nollen, M.; Ebert, F.; Moser, J.; Mullenders, L.H.F.; Hartwig, A.; Schwerdtle, T. Impact of arsenic on nucleotide excision repair: XPC function, protein level, and gene expression. Mol. Nutr. Food Res. 2009, 53, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Boros, G. Investigation of UVB-Induced Cellular Mechanisms in Human Keratinocytes using a Novel Approach of Delivering In Vitro Synthesized mRNA Encoding Cyclobutane Pyrimidine Dimer-Specific Photolyase. Ph.D. Thesis, University of Debrecen, Debrecen, Hungary, 2015. [Google Scholar]
- Budden, T.; Davey, R.J.; Vilain, R.E.; Ashton, K.A.; Braye, S.G.; Beveridge, N.J.; Bowden, N.A. Repair of UVB-induced DNA damage is reduced in melanoma due to low XPC and global genome repair. Oncotarget 2016, 7, 60940–60953. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.E. Mammalian cell HPRT gene mutation assay: Test methods. Methods Mol. Biol. 2012, 817, 55–67. [Google Scholar]
- Greinert, R.; Boguhn, O.; Harder, D.; Breitbart, E.W.; Mitchell, D.L.; Volkmer, B. The dose dependence of cyclobutane dimer induction and repair in UVB-irradiated human keratinocytes. Photochem. Photobiol. 2000, 72, 701. [Google Scholar] [CrossRef]
- Ikehata, H.; Mori, T.; Douki, T.; Cadet, J.; Yamamoto, M. Quantitative analysis of UV photolesions suggests that cyclobutane pyrimidine dimers produced in mouse skin by UVB are more mutagenic than those produced by UVC. Photochem. Photobiol. Sci. 2018, 17, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Liu, J.; Jin, J.; Ni, W.; Xu, W. Arsenic trioxide induces not only apoptosis but also autophagic cell death in leukemia cell lines via up-regulation of Beclin-1. Leuk. Res. 2007, 31, 329–339. [Google Scholar] [CrossRef]
- Kumar, S.; Yedjou, C.; Tchounwou, P.B. Arsenic trioxide induces oxidative stress, DNA damage, and mitochondrial pathway of apoptosis in human leukemia (HL-60) cells. J. Exp. Clin. Cancer Res. 2014, 33, 42. [Google Scholar] [CrossRef]
- Lee, C.-H.; Yu, C.-L.; Liao, W.-T.; Kao, Y.-H.; Chai, C.-Y.; Chen, G.-S.; Yu, H.-S. Effects and interactions of low doses of arsenic and UVB on keratinocyte apoptosis. Chem. Res. Toxicol. 2004, 17, 1199–1205. [Google Scholar] [CrossRef]
- Zhang, W.; Hanks, A.N.; Boucher, K.M.; Florell, S.R.; Allen, S.M.; Alexander, A.; Brash, D.E.; Grossman, U. UVB-induced apoptosis drives clonal expansion during skin tumor development. Carcinogenesis 2004, 26, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Eastman, A. Cell cycle checkpoints and their impact on anticancer therapeutic strategies. J. Cell. Biochem. 2004, 91, 223–231. [Google Scholar] [CrossRef]
- Paulovich, A.G.; Toczyski, D.P.; Hartwell, L.H. When Checkpoints Fail. Cell 1997, 88, 315–321. [Google Scholar] [CrossRef]
- Pavey, S.; Russell, T.; Gabrielli, B. G2 phase cell cycle arrest in human skin following UV irradiation. Oncogene 2001, 20, 6103–6110. [Google Scholar] [CrossRef]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 and Autophagy. Methods Mol. Biol. 2008, 445, 77–88. [Google Scholar]
- Ryan, K.M. p53 and autophagy in cancer: Guardian of the genome meets guardian of the proteome. Eur. J. Cancer 2011, 47, 44–50. [Google Scholar] [CrossRef]
- Strozyk, E.; Kulms, D. The role of AKT/mTOR pathway in stress response to UV-irradiation: Implication in skin carcinogenesis by regulation of apoptosis, autophagy and senescence. Int. J. Mol. Sci. 2013, 14, 15260–15285. [Google Scholar] [CrossRef]
- Thomas-Schoemann, A.; Batteux, F.; Mongaret, C.; Nicco, C.; Chéreau, C.; Annereau, M.; Dauphin, A.; Goldwasser, F.; Weill, B.; Lemare, F.; et al. Arsenic trioxide exerts antitumor activity through regulatory T cell depletion mediated by oxidative stress in a murine model of colon cancer. J. Immunol. 2012, 189, 5171–5177. [Google Scholar] [CrossRef]
- Carone, L.; Oxberry, S.G.; Twycross, R.; Charlesworth, S.; Mihalyo, M.; Wilcock, A. Spironolactone. J. Pain Symptom Manag. 2017, 53, 288–292. [Google Scholar] [CrossRef]
- Li, S.; Topatana, W.; Juengpanich, S.; Cao, J.; Hu, J.; Zhang, B.; Ma, D.; Cai, X.; Chen, M. Development of synthetic lethality in cancer: Molecular and cellular classification. Signal Transduct. Target Ther. 2020, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Helleday, T. The underlying mechanism for the PARP and BRCA synthetic lethality: Clearing up the misunderstandings. Mol. Oncol. 2011, 5, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.J.; Tutt, A.N.; Ashworth, A. Synthetic Lethality and Cancer Therapy: Lessons Learned from the Development of PARP Inhibitors. Annu. Rev. Med. 2015, 66, 455–470. [Google Scholar] [CrossRef]
- Chaudhuri, A.R.; Nussenzweig, A.R.C.A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Pines, A.; Vrouwe, M.G.; Marteijn, J.A.; Typas, D.; Luijsterburg, M.S.; Cansoy, M.; Mullenders, L. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J. Cell Biol. 2012, 199, 235–249. [Google Scholar] [CrossRef]
- Robu, M.; Shah, R.G.; Purohit, N.K.; Zhou, P.; Naegeli, H.; Shah, G.M. Poly(ADP-ribose) polymerase 1 escorts XPC to UV-induced DNA lesions during nucleotide excision repair. Proc. Natl. Acad. Sci. USA 2017, 114, E6847–E6856. [Google Scholar] [CrossRef]
- Thorslund, T.; Von Kobbe, C.; Harrigan, J.A.; Indig, F.E.; Christiansen, M.; Stevnsner, T.; Bohr, V.A. Cooperation of the Cockayne syndrome group B protein and poly(ADP-ribose) polymerase 1 in the response to oxidative stress. Mol. Cell. Biol. 2005, 25, 7625–7636. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.; Ho, Y.-S.; Wang, Y.-J. Arsenic trioxide induces autophagy and apoptosis in human glioma cells in vitro and in vivo through downregulation of survivin. J. Mol. Med. 2011, 89, 927–941. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Gheorghiu, L.; Drumm, M.; Clayman, R.; Eidelman, A.; Wszolek, M.F.; Olumi, A.; Feldman, A.; Wang, M.; Marcar, L.; et al. PARP-1 inhibition with or without ionizing radiation confers reactive oxygen species-mediated cytotoxicity preferentially to cancer cells with mutant TP53. Oncogene 2018, 37, 2793–2805. [Google Scholar] [CrossRef] [PubMed]
- Vance, S.M.; Liu, E.; Zhao, L.; Parsels, J.D.; Parsels, L.A.; Brown, J.L.; Maybaum, J.; Lawrence, T.S.; Morgan, M.A. Selective radiosensitization of p53 mutant pancreatic cancer cells by combined inhibition of Chk1 and PARP1. Cell Cycle 2011, 10, 4321–4329. [Google Scholar] [CrossRef]
- Valenzuela, M.T.; Guerrero, R.; Nuñez, M.I.; De Almodóvar, J.M.R.; Sarker, M.; De Murcia, G.; Oliver, F.J. PARP-1 modifies the effectiveness of p53-mediated DNA damage response. Oncogene 2002, 21, 1108–1116. [Google Scholar] [CrossRef]
- Kubota, E.; Williamson, C.T.; Ye, R.; Elegbede, A.; Peterson, L.; Lees-Miller, S.P.; Bebb, D.G. Low ATM protein expression and depletion of p53 correlates with olaparib sensitivity in gastric cancer cell lines. Cell Cycle 2014, 13, 2129–2137. [Google Scholar] [CrossRef]
- Kanai, M.; Hanashiro, K.; Kim, S.-H.; Hanai, S.; Boulares, A.H.; Miwa, M.; Fukasawa, K. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat. Cell Biol. 2007, 9, 1175–1183. [Google Scholar] [CrossRef]
- Lorin, S.; Hamaï, A.; Mehrpour, M.; Codogno, P. Autophagy regulation and its role in cancer. Semin. Cancer Biol. 2013, 23, 361–379. [Google Scholar] [CrossRef]
- Boros, G.; Miko, E.; Muramatsu, H.; Weissman, D.; Emri, E.; Rózsa, D.; Nagy, G.; Juhász, A.; Juhász, I.; Van Der Horst, G.; et al. Transfection of pseudouridine-modified mRNA encoding CPD-photolyase leads to repair of DNA damage in human keratinocytes: A new approach with future therapeutic potential. J. Photochem. Photobiol. B Biol. 2013, 129, 93–99. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fidrus, E.; Hegedűs, C.; Janka, E.A.; Paragh, G.; Emri, G.; Remenyik, É. Inhibitors of Nucleotide Excision Repair Decrease UVB-Induced Mutagenesis—An In Vitro Study. Int. J. Mol. Sci. 2021, 22, 1638. https://doi.org/10.3390/ijms22041638
Fidrus E, Hegedűs C, Janka EA, Paragh G, Emri G, Remenyik É. Inhibitors of Nucleotide Excision Repair Decrease UVB-Induced Mutagenesis—An In Vitro Study. International Journal of Molecular Sciences. 2021; 22(4):1638. https://doi.org/10.3390/ijms22041638
Chicago/Turabian StyleFidrus, Eszter, Csaba Hegedűs, Eszter Anna Janka, György Paragh, Gabriella Emri, and Éva Remenyik. 2021. "Inhibitors of Nucleotide Excision Repair Decrease UVB-Induced Mutagenesis—An In Vitro Study" International Journal of Molecular Sciences 22, no. 4: 1638. https://doi.org/10.3390/ijms22041638
APA StyleFidrus, E., Hegedűs, C., Janka, E. A., Paragh, G., Emri, G., & Remenyik, É. (2021). Inhibitors of Nucleotide Excision Repair Decrease UVB-Induced Mutagenesis—An In Vitro Study. International Journal of Molecular Sciences, 22(4), 1638. https://doi.org/10.3390/ijms22041638






