Abstract
To explore the differences in mitogenome variation and phylogenetics among lineages of the Hemiptera superfamily Fulgoroidea, we sequenced four new mitogenomes of Caliscelidae: two species of the genus Bambusicaliscelis (Caliscelinae: Caliscelini), namely Bambusicaliscelis flavus and B. fanjingensis, and two species of the genus Youtuus (Ommatidiotinae: Augilini), namely Youtuus strigatus and Y. erythrus. The four mitogenomes were 15,922–16,640 bp (base pair) in length, with 37 mitochondrial genes and an AT-rich region. Gene content and arrangement were similar to those of most other sequenced hexapod mitogenomes. All protein-coding genes (PCGs) started with a canonical ATN or GTG and ended with TAA or an incomplete stop codon single T. Except for two transfer RNAs (tRNAs; trnS1 and trnV) lacking a dihydrouridine arm in the four species and trnC lacking a dihydrouridine stem in the Youtuus species, the remaining tRNAs could fold into canonical cloverleaf secondary structures. Phylogenetic analyses based on sequence data of 13 PCGs in the 28 Fulgoroidea species and two outgroups revealed that Delphacidae was monophyletic with strong support. Our data suggest that Fulgoridae is more ancient than Achilidae. Furthermore, Flatidae, Issidae, and Ricaniidae always cluster to form a sister group to Caliscelidae.
1. Introduction
Caliscelidae is a relatively small family within the large and diverse planthopper superfamily Fulgoroidea that comprises approximately 240 described species from 77 genera divided into five tribes (Caliscelini, Peltonotellini, Adenissini, Ommatidiotini, and Augilini) and two subfamilies (Ommatidiotinae and Caliscelinae) [1,2]. These species are extensively distributed worldwide. The genus Bambusicaliscelis was established by Chen and Zhang (2011) [3] and belongs to the tribe Caliscelini of the subfamily Caliscelinae. The genus Youtuus was established by Gong and Chen (2018) [4] and belongs to the tribe Augilini of the subfamily Ommatidiotinae. These two genera are restrictively distributed in southern China and primarily inhabit bamboo. They directly injure bamboo using their sucking and piercing mouthparts and indirectly damage bamboo by transmitting plant pathogens; this severely hampers bamboo production.
To date, a total of 13,774 species from 2465 genera in 33 families of Fulgoroidea have been recorded worldwide [2]. Despite their great diversity, no more than 30 complete or near-complete mitogenomes have been reported from six planthopper families. Further, the amount of data recorded about these families follow the order Delphacidae > Achilidae > Fulgoridae > Ricaniidae/Issidae > Flatidae [5]. Overall, the data distribution is extremely unbalanced, and mitogenomes have not been reported in some families, whereas for others, there are no recorded molecular data. At present, most studies still rely on morphology for identifying and describing Caliscelidae, and few studies have discussed the taxonomic status of Caliscelidae within the superfamily Fulgoroidea based on molecular fragments (i.e., 18S, 28S, histone 3, and wingless) [6,7]. However, there is no available information about the complete mitogenomes of caliscelids, hindering molecular phylogenetic analyses among the Caliscelidae genera and species using molecular data. Moreover, the phylogenetic relationship between Caliscelidae and other planthopper families is limited. Therefore, better knowledge of the phylogenetic relationships among the members of Caliscelidae may be obtained by sequencing their mitogenomes. Further, additional information about taxa and more data are required to obtain a more completed and supported phylogeny of the Fulgoroidea superfamily in the future.
The insect mitogenome is a compact, double-stranded, closed circular molecule [8] containing 37 genes encoding 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs; rrnS and rrnL), and a noncoding AT-rich region [8,9]. Due to the small size of mtDNA as well as maternal inheritance, conserved gene content, a relatively fast evolutionary rate, and evolutionarily low levels of recombination, mitogenome sequences are the most widely used molecular tools for insect species identification and are becoming increasingly important for phylogenetic and evolutionary analyses of insects at the family level [10,11,12,13,14,15,16].
In the present study, we sequenced and annotated the complete mitogenomes of four Caliscelidae species, two from Bambusicaliscelis and two from Youtuus: Bambusicaliscelis flavus, B. fanjingensis, Youtuus strigatus, and Y. erythrus. Further, we analyzed the characteristics of these mitogenomes, including nucleotide composition, tRNA secondary structure, codon usage, gene overlaps, and noncoding regions, and compared the genetic distances between Caliscelidae and six other reported planthopper families. Comparative mitogenomics of these new sequences will provide a preliminary understanding of the structures and basic characteristics of Caliscelidae mitogenomes. Moreover, phylogenetic analyses using these mitogenomes can reveal the phylogenetic position of Caliscelidae and shed light on its phylogenetic relationships with other families within Fulgoroidea. Overall, the molecular data obtained in this study can improve our understanding of the phylogeny and evolution of Fulgoroidea.
2. Materials and Methods
2.1. Species Collection and Taxonomic Identification
Specimens of Y. strigatus, Y. erythrus, and B. fanjingensis were collected from Guizhou Province in July 2018, and those of B. flavus were collected from Yunnan Province in August 2018. All fresh specimens were immediately preserved in absolute ethanol and then stored at −20 °C at the Institute of Entomology, Guizhou University, Guiyang, China. These four species were identified based on their morphological characteristics [3,4,17]; species identification was performed by Nian Gong.
2.2. DNA Extraction and Sequencing
Total genomic DNA was extracted from the muscle tissue of each Caliscelidae species using the Takara Genome DNA Extraction Kit (Sangon Biotech, Shanghai, China) according to the manufacturer’s instructions. Extracted DNA samples were stored at −20 °C. Mitogenomes were sequenced using a next-generation sequencing platform with Illumina Hiseq 2500 at OriGene (Beijing, China). Following quantification of the total DNA that was extracted, an Illumina TruSeq library with single species was generated from the pooled genomic DNA, with an average insert size of 480 bp. This library was sequenced on a full run of Illumina Hiseq 2500 with 500 cycles and paired-end sequencing (250 bp reads). Raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in the BIG Data Center, Chinese Academy of Sciences, under accession codes CRA003721.
2.3. Genome Assembly, Annotation, and Analysis
The quality of the raw sequences was evaluated using FastQC v0.11.4 (www.bioinformatics.babraham.ac.uk/projects/fastqc). Putative mitogenome reads with an average quality value of <Q30 were removed before assembly. Then, the clean sequences were assembled using MitoZ v2.4 software [18] with default parameters and the mitogenome of Ricania speculum (Fulgoroidea: Ricaniidae; NC_031369) as a reference.
The four mitogenomes were initially annotated using the MITOS web server (http://mitos.bioinf.uni-leipzig.de/index.py) [19] with invertebrate genetic codes. PCGs were predicted by determining theirs open reading frames using the invertebrate mitochondrial genetic codon. rRNA genes and the AT-rich regions were determined by comparisons with the homologous sequences of other planthoppers from GenBank (GenBank accession numbers: MH324927–MH324931). The locations and secondary structures of tRNA genes were predicted using the tRNAscan-SE search server (http://lowelab.ucsc. edu/tRNAscan-SE) and the MITOS web server (http://mitos.bioinf.uni-leipzig.de/index.py) with invertebrate codon predictors [20]. Mitogenomic circular maps were depicted and annotated using Geneious R9 [21].
Nucleotide composition and relative synonymous codon usage (RSCU) were calculated using PhyloSuite v1.2.2 [22], and RSCU figures were drawn using the fmsb package [23] of R 3.6.1 [24]. Strand asymmetry was calculated according to the following formulas [25]: AT skew = (A − T)/(A + T) and GC skew = (G − C)/(G + C). Tandem repeats in the AT-rich region were searched using the Tandem Repeats Finder program (http://tandem.bu.edu/trf/trf.basic.submit.html) [26]. The overlapping regions and intergenic spacers between genes were manually counted. The sequence data of the four insect mitogenomes have been deposited to GenBank under the accession numbers MW281858–MW281861 for B. flavus, B. fanjingensis, Y. strigatus, and Y. erythrus, respectively.
2.4. Phylogenetic Analyses
In addition to the four mitogenomes obtained in this study, the complete mitogenomes of 24 planthopper species were downloaded from GenBank for phylogenetic analyses; the mitogenomes of Maiestas dorsalis and Yanocephalus yanonis were used as outgroups. Detailed information and accession numbers of these mitogenomes are listed in Table S1.
The nucleotide sequences of 13 PCGs were aligned using MEGA v7.0.26 [27] with the Muscle [28]. Individual gene alignments were concatenated into a dataset using SequenceMatrix v1.7 [29]: nucleotide sequences of the 13 PCGs. The best partition schemes and substitution models for maximum likelihood (ML) and Bayesian inference (BI) analyses of the dataset were estimated using PartitionFinder v2.1.1 [30] with the greedy algorithm. Details of the best-fit partitioning schemes for ML and BI analyses are listed in Table S2. ML analyses were performed using IQ-Tree v1.6.3 [31], with 1000 replicates of ultrafast-likelihood bootstraping [32] to obtain node support values. BI analysis was performed using MrBayes v3.2.6 [33] under the following conditions: independent substitution model for each partition, four Markov chains, two independent runs each for 1,000,000 generations, sampling every 1000th generation, and the first 25% discarded as burn-in. When the average standard deviation of split frequencies fell below 0.01 and remained stable, stationarity was considered to have reached. Furthermore, an effective sample size value of more than 200 was used as a convergence diagnostic. When the bootstrap percentage (BP) was >75% or the Bayesian posterior probability (BPP) was >0.9, the results were considered credible. Consensus trees were viewed and edited using FigTree v1.4.2 [34].
3. Results
3.1. Genome Organization and Composition
The complete mitogenomes of B. flavus (GenBank No.: MW281858; length: 16,053 bp), B. fanjingensis (GenBank No.: MW281859; length: 15,979 bp), Y. strigatus (GenBank No.: MW281860; length: 16,640 bp), and Y. erythrus (GenBank No.: MW281861; length: 15,922 bp) were identified as circular double-stranded molecules (Figure 1). They were of medium size compared to those of other species of the Fulgoroidea superfamily. Each newly sequenced mitogenome contained 37 typical mitochondrial genes (13 PCGs, 2 rRNAs, and 22 tRNAs) and an AT-rich region (Figure 1; Table 1). All four caliscelid planthoppers displayed an identical gene arrangement that was shared with the putative ancestral gene order [35,36]. Of the typical genes, 23, including 9 PCGs and 14 tRNAs, were encoded on the major strand (J-strand), whereas the remaining 14 genes, including 4 PCGs, 8 tRNAs, and 2 rRNAs, were encoded on the minor strand (N-strand).
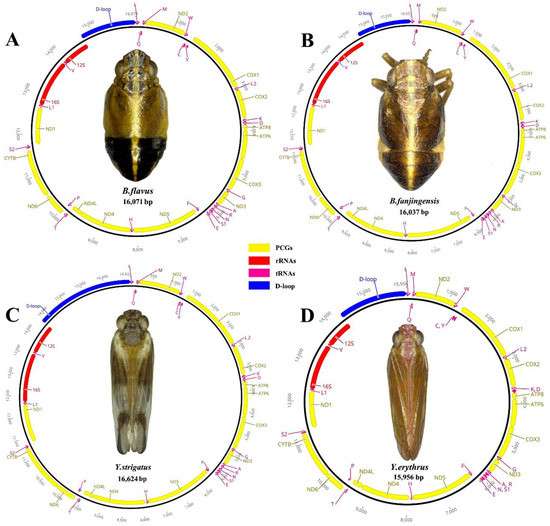
Figure 1.
Circular maps of the mitogenomes of Bambusicaliscelis flavus (A), B. fanjingensis (B), Youtuus strigatus (C), and Y. erythrus (D). Protein-coding, ribosomal, and transfer RNA genes are shown with standard abbreviations. Gene orientations are indicated by arrow directions. Protein-coding genes are denoted in yellow, transfer RNA genes in aubergine, the AT-rich region in blue, and the two ribosomal RNA genes in red. The PCGs and rRNAs are the standard abbreviations. Each tRNA is denoted as a one-letter symbol according to the IUPAC-IUB single-letter amino acid codes.

Table 1.
List of total size and intergenic nucleotides for mitochondrial genes of B. flavus, B. fanjingensis, Y. strigatus, and Y. erythrus, with lengths of genes, anticodons of tRNAs, and start/stop codons of protein-coding genes.
The nucleotide compositions and skewness levels of the newly obtained mitogenomes are provided in Table 2. The mean AT nucleotide content of the four mitogenomes was similar: 79.5% in B. flavus, 80.4% in B. fanjingensis, 79.8% in Y. strigatus, and 79.7% in Y. erythrus; these results indicate considerable AT bias. The AT-rich region had the highest AT content (82.3–89.1%), whereas PCGs had the lowest AT content (78.4–79.1%); these results are consistent with those of all previously sequenced planthopper mitogenomes [5]. Composition analysis of the new mitogenomes revealed that the whole mitogenomes exhibited positive AT skew (0.164–0.197), indicating that the presence of A was more than that of T, and negative GC skew (−0.293 to −0.238), indicating that the presence of C was more than that of G; these results correspond well with the AT bias observed in other Fulgoroidea mitogenomes.

Table 2.
Base composition of four mitochondrial whole genomes, 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and AT-rich regions.
3.2. PCGs and Codon Usage
The mitogenomes of all four planthoppers contained 13 PCGs. The concatenated lengths of the 13 PCGs in B. flavus, B. fanjingensis, Y. strigatus, and Y. erythrus were 11,039, 10,942, 10,933, and 10,901 bp, respectively; their respective average AT content was 78.4%, 78.8%, 79.1%, and 79%, respectively. The AT (−0.135 to −0.11) and GC (−0.062 to −0.042) skewness values of PCGs were similar among the four planthoppers (Table 2). Among the 13 PCGs, the nad5 gene was the largest and the atp8 gene was the smallest. Only four genes (nad1, nad4, nad4L, and nad5) were present on the minor strand, whereas the other nine PCGs (cox1, cox2, cox3, atp6, atp8, nad2, nad3, nad6, and cytb) were present on the major strand (Table 1; Figure 1).
The start and stop codons of all PCGs are listed in Table 1. In the four newly sequenced mitogenomes, most of the PCGs started with the typical ATN (ATA/T/G/C) codon, except for nad5 of B. flavus, B. fanjingensis, and Y. strigatus, which started with GTG. The complete termination codon TAA was more frequently used than TAG, whereas a single T was also used as the stop codon. The presence of an incomplete stop codon is common in insects [37], with reports suggesting that such a stop codon can be completed by post-transcriptional polyadenylation [10,38,39].
We determined the codon usage pattern, RSCU, and number in the PCGs of the four planthopper mitogenomes (Figure 2 and Figure 3; Table S3). The results demonstrated that codon usage was similar among the four species. The four most prevalent codons were Phe (UUU), Ile (AUU), Leu2 (UUA), and Met (AUA), accounting for more than half of the total number of amino acids. They all solely contained A or U, indicating high AT content in PCGs. The RSCU values of the PCGs indicated a pattern toward more A and T than G and C. In addition, codon usage showed an extremely high AT bias, which plays a key role in the AT bias of the whole mitogenome. All AT skew and GC skew of these PCGs were negative.
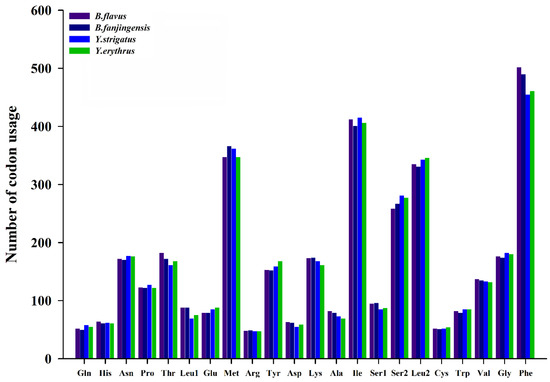
Figure 2.
Number of codon usages in the protein-coding genes of Bambusicaliscelis flavus, B. fanjingensis, Youtuus strigatus, and Y. erythrus mitogenomes. Each amino acid is denoted as a three-letter symbol according to the IUPAC-IUB three-letter amino acid codes.
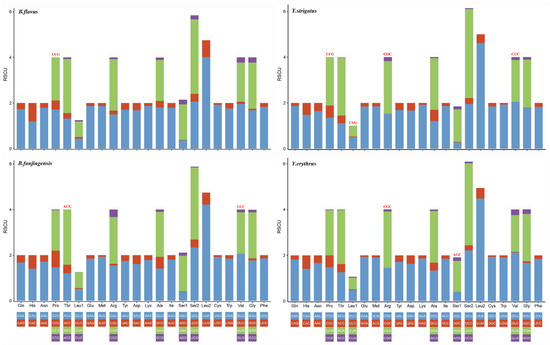
Figure 3.
Relative synonymous codon usage in the protein-coding genes of Bambusicaliscelis flavus, B. fanjingensis, Youtuus strigatus, and Y. erythrus mitogenomes. Codon families are indicated below the X-axis. The color of the codon family below the X-axis corresponds to the color above the X-axis. The stop codon is not given. Codons absent in mitochondrial genomes are shown at the top of columns.
3.3. Ribosomal RNA and Transfer RNA Genes
In the complete mitogenomes of the four species, two rRNA genes (rrnL and rrnS) were encoded on the minor strand, with the second-highest AT content (>80%), positive GC skew (0.278−0.328), and negative AT skew (−0.203 to −0.157) (Table 2). The lengths of rrnL ranged from 1221 bp (B. fanjingensis) to 1227 bp (B. flavus), whereas those of rrnS ranged from 717 bp (B. flavus) to 738 bp (B. fanjingensis). Similar to other sequenced planthopper mitogenomes, rrnL was located between trnL2 and trnV, whereas rrnS was located between trnV and the AT-rich region.
All the 22 tRNA genes of the four mitogenomes were identified, with their total length ranging from 1406 to 1478 bp. The length of each tRNA was 56–74 bp and interspersed throughout the entire mitogenome. Among the 22 tRNA genes, 14 were encoded by the major strand and the remaining 8 were encoded by the minor strand, with a high AT content (>79%) and positive AT and GC skew. Except for trnS1 and trnV in the four species and trnC in the Youtuus species, which lacked a stable dihydrouridine (DHU) stem and was replaced by a simple loop, the remaining tRNA genes in the four new mitogenomes could be folded into the canonical cloverleaf secondary structure (Figures S1−S4). All the anticodons of the 22 tRNA genes are known to be identical across Hemiptera [5]. According to the predicted secondary structure, we identified a total of six mismatched base pairs, namely G-U, A-A, A-G, A-C, U-C, and U-U, as well as an extra A nucleotide. The total number of mismatched base pairs detected in the four planthoppers was 31 in B. flavus, 28 in B. fanjingensis, 32 in Y. strigatus, and 35 in Y. erythrus.
3.4. Overlapping and Intergenic Spacer Regions
We found 13–22 intergenic spacers in the four mitogenomes, which ranged in size from 1 to 58 bp. The longest (58 bp) intergenic spacer was identified in B. fanjingensis, between the trnF and nad5 genes. The four planthoppers had 4–10 overlapping genes, with overlaps ranging in size from 1 to 8 bp (Table 1). Two gene overlaps were conserved among the four newly sequenced mitogenomes: trnW–trnC (8 bp: AAGCCTTA) and atp8–atp6 (7 bp: ATGATAA (in Youtuus) or ATAATAA (in Bambusicaliscelis. A 7 bp overlapping region (TTAACAT (in Bambusicaliscelis) or TTAGCAT (in Youtuus)) located between nad4 and nad4L was detected in three of the four mitogenomes (excluding Y. erythrus). Further, an 8 bp overlapping region (ATGTTTAA) located between nad6 and cytb was detected in both the Youtuus species but not in either of the Bambusicaliscelis species.
3.5. AT-Rich Region
The AT-rich region is believed to be involved in regulating the replication and transcription of mtDNA in insects; in the four planthoppers, it was located between rrnS and trnI. Among the four sequenced mitogenomes of Caliscelidae, the lengths of the AT-rich regions considerably differed in the four mitogenomes, ranging from 1230 bp in B. fanjingensis to 2288 bp in Y. strigatus. Nevertheless, length differences among the mitogenomes were consistent with differences in their total length. The AT-rich region had the highest AT content, ranging from 82.3% in Y. strigatus to 89.1% in B. fanjingensis, with positive AT skew (0.011−0.068) and negative GC skew (−0.405 to −0.169) (Table 2). The structural organization of the AT-rich regions in the four planthopper mitogenomes is illustrated in Figures S5−S8. Four repeat regions were present in B. flavus, three were present in B. fanjingensis and Y. strigatus, and two were present in Y. erythrus. The largest repeat unit was 217 bp, with two repeats in B. flavus.
3.6. Phylogenetic Relationships
Phylogenetic relationships among the 28 Fulgoroidea species (including the four newly sequenced mitogenomes) and two outgroup species were determined by concatenating the nucleotide sequences of the 13 PCGs using BI and ML analyses. A total of two phylogenetic trees (BI-PCG, and ML-PCG) were reconstructed using BI and ML analyses (Figure 4). In two trees, 10 Delphacidae species formed a steadily monophyletic group with high support (BPP = 1/BP = 100) and were located at the basal position of the phylogenetic tree. This finding is consistent with that of previous studies [5,40,41,42,43,44]. In all trees, four Fulgoridae species formed a cluster that was located near the clade of Delphacidae with high support (BPP = 1/BP = 100), whereas four families (Caliscelidae, Ricaniidae, Flatidae, and Issidae) gathered to form a sister group to Achilidae. Moreover, three families (Ricaniidae, Flatidae, and Issidae) assembled to form a sister group to Caliscelidae. The four Caliscelidae species always clustered with high support (BPP = 1/BP = 100).
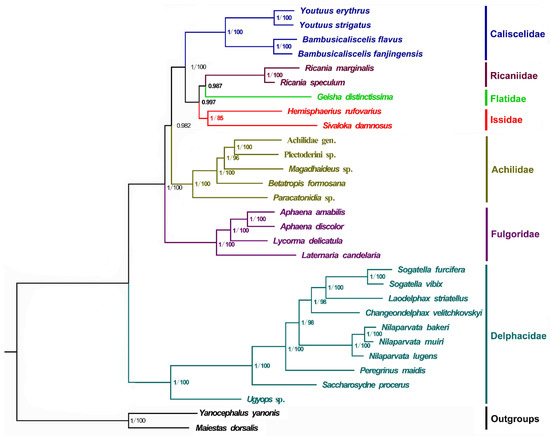
Figure 4.
Phylogenetic trees of Fulgoroidea inferred using MrBayes (Bayesian inference) and maximum likelihood (ML) analysis based on the nucleotide sequences of the 13 protein-coding genes. Bayesian posterior probabilities (BPPs) and bootstrap percentage (BP) are indicated on branches; a BP below 75 is not shown.
4. Discussion
In the present study, we successfully sequenced and analyzed the complete mitogenomes of four Caliscelidae species. Among the four mitogenomes, the length ranged from 15,922 bp in Y. erythrus to 16,640 bp in Y. strigatus. The difference in length is mainly attributed to the difference in the size of the intergenic spacer regions and the length of the AT-rich region. The intergenic spacer regions of Bambusicaliscelis are longer than those of Youtuus, whereas the AT-rich region of Youtuus is longer than that of Bambusicaliscelis. Except for atp8 (37 bp), nad5 (59 bp), and nad4 (58 bp), the length differences of each PCG in the four newly sequenced mitogenomes did not exceed 23 bp; this result suggests that the PCGs have relatively conserved characteristics among species. However, the length variation of tRNAs and rRNAs among the different species was limited, which is mainly due to the stability of their secondary structures.
Among the secondary structures of the 22 putative tRNAs, trnS1 and trnV lacked a DHU stem and formed a simple loop in the four species. The DHU arm of trnS1 formed a simple loop, which has been recognized in many metazoans [45,46,47]; however, we are the first to report the lack of a DHU stem in trnV in Caliscelidae. We found that trnC also lacks a DHU stem and forms a simple loop in the two Youtuus species; this finding is in contrast to the normal structure observed in the two Bambusicaliscelis species and may be related to the evolution of these species. The genera Bambusicaliscelis and Youtuus are part of the tribes Caliscelini and Augilini, respectively, and the species in these two tribes differ considerably based on appearance. For instance, the species occurring in Augilini have hind wings, whereas those occurring in Caliscelini exhibit hind-wing degeneration. These are species-specific differences.
In many Fulgoroidea species, an 8 bp overlapping AAGCCTTA region between trnW and trnC has been detected [5]; the four newly sequenced mitogenomes of Caliscelidae also show this characteristic. Further, a 7 bp overlap between atp8 and atp6 was detected in other planthoppers. In the present study, analysis showed that the 7 bp overlapping region in Bambusicaliscelis is coded as ATAATAA and that in Youtuus is coded as ATGATAA. Perhaps, owing to differences between the genera, there is a A/G difference. An 8 bp overlapping region (ATGTTTAA) was located between nad6 and cytb in both the Youtuus species but not in either of the Bambusicaliscelis species; these are species-specific differences.
In the phylogenetic analysis, Delphacidae species formed a steady monophyletic group with high support (BPP = 1/BP = 100) and were located at the basal position of the phylogenetic tree, consistent with the findings of previous studies [5,40,41,42,43,44]. In contrast to the study of Xu et al. [5], who found Achilidae to be more ancient than Fulgoridae, our analysis indicated that Fulgoridae was more ancient than Achilidae; this discrepancy may be associated with the number of selected species and the different taxa used in each study. However, our result is consistent with the divergence time estimation of Fulgoroidea analyzed by Song and Liang, wherein the chronogram of Fulgoroidea showed that the lineage Achilidae could be traced back to the early Palaeocene era (46 Mya), whereas Fulgoridae diverged in the early Cretaceous era (approximately 78 Mya) [48]. In addition, we found that Flatidae, Issidae, and Ricaniidae always gathered to form a sister group to Caliscelidae. The placement of the Ricaniidae sister to Flatidae and Issidae was consistently supported in both analyses, with strong nodal supports (BPP > 0.98), consistent with the findings of morphological, nuclear genes and mitochondrial genes [6,40,41]. Because Issidae had only two effective mitogenome sequences, the paraphyletic of Issidae observed in other molecular phylogenetic analyses was not supported in this study [43]. The four Caliscelidae species always clustered with high support (BPP = 1/BP = 100). Although the representative mitogenomes from Fulgoroidea are still limited, the findings of phylogenetic analyses will be more reliable and convincing because information about more mitogenomes and genomes of Fulgoroidea species is available in databases.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/22/3/1348/s1: Figure S1. Secondary structures of transfer RNAs (tRNAs) in the mitogenome of Bambusicaliscelis flavus. Dashes indicate Watson–Crick base pairing, and dots indicate GU base pairing; Figure S2. Secondary structures of transfer RNAs (tRNAs) in the mitogenome of Bambusicaliscelis fanjingensis. Dashes indicate Watson–Crick base pairing, and dots indicate GU base pairing; Figure S3. Secondary structures of transfer RNAs (tRNAs) in the mitogenome of Youtuus strigatus. Dashes indicate Watson–Crick base pairing, and dots indicate GU base pairing; Figure S4. Secondary structures of transfer RNAs (tRNAs) in the mitogenome of Youtuus erythrus. Dashes indicate Watson–Crick base pairing, and dots indicate GU base pairing; Figure S5. Features present in the AT-rich region of the Bambusicaliscelis flavus mitogenome. The four tandem repeats are shaded in yellow, gray, purple-red, and blue, respectively. * indicates a mismatch;—indicates an insertion or deletion; Figure S6. Features present in the AT-rich region of the Bambusicaliscelis fanjingensis mitogenome. The three tandem repeats are shaded in yellow, gray, and blue, respectively. * indicates a mismatch;—indicates an insertion or deletion; Figure S7. Features present in the AT-rich region of the Youtuus strigatus mitogenome. The three tandem repeats are shaded in yellow, gray, and blue, respectively. * indicates a mismatch;—indicates an insertion or deletion; Figure S8. Features present in the AT-rich region of the Youtuus erythrus mitogenome. The two tandem repeats are shaded in yellow and gray, respectively. * indicates a mismatch;—indicates an insertion or deletion; Table S1. Mitogenomes used in this study; Table S2. The partition schemes and best substitution models used in the phylogenetic analysis; Table S3. Codon number and relative synonymous codon usage (RSCU) in the four mitochondrial PCGs.
Author Contributions
Investigation, formal analysis, and writing—original draft: N.G.; resources, investigation, and formal analysis: L.Y.; and validation, project administration, and funding acquisition: X.-S.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (grant nos. 32060343 and 31472033); the Science and Technology Support Program of Guizhou Province (grant no. 20201Y129); and the Program of Excellent Innovation Talents, Guizhou Province (grant no. 20154021).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant nos. 32060343 and 31472033); the Science and Technology Support Program of Guizhou Province (grant no. 20201Y129); and the Program of Excellent Innovation Talents, Guizhou Province (grant no. 20154021).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gnezdilov, V.M. A Modern System of the Family Caliscelidae Amyot et Serville (Homoptera, Fulgoroidea). Зоологический Журнал/Entomol. Rev. 2013, 92, 1309–1311. [Google Scholar] [CrossRef]
- Bourgoin, T. FLOW (Fulgoromorpha Lists on The Web): A World Knowledge Base Dedicated to Fulgoromorpha. Version 8 Updated 1 October 2020. Available online: http://hemiptera-databases.org/flow/ (accessed on 17 October 2020).
- Chen, X.-S.; Zhang, Z.-G. Bambusicaliscelis, a New Bamboo-Feeding Planthopper Genus of Caliscelini (Hemiptera: Fulgoroidea: Caliscelidae: Caliscelinae), with Descriptions of Two New Species and Their Fifth-Instar Nymphs from Southwestern China. Ann. Entomol. Soc. Am. 2011, 104, 95–104. [Google Scholar] [CrossRef]
- Gong, N.; Yang, L.; Chen, X.-S. Youtuus, a new bamboo-feeding genus of the tribe Augilini with two new species from China (Hemiptera, Fulgoromorpha, Caliscelidae). ZooKeys 2018, 783, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.-Y.; Long, J.-K.; Chen, X.-S. Comparative analysis of the complete mitochondrial genomes of five Achilidae species (Hemiptera: Fulgoroidea) and other Fulgoroidea reveals conserved mitochondrial genome organization. PeerJ 2019, 7, e6659. [Google Scholar] [CrossRef] [PubMed]
- Urban, J.M.; Cryan, J.R. Evolution of the planthoppers (Insecta: Hemiptera: Fulgoroidea). Mol. Phylogenet. Evol. 2007, 42, 556–572. [Google Scholar] [CrossRef]
- Gwiazdowski, R.A.; Foottit, R.G.; Maw, H.E.L.; Hebert, P.D.N. The Hemiptera (Insecta) of Canada: Constructing a Reference Library of DNA Barcodes. PLoS ONE 2015, 10, e0125635. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Wolstenholme, D.R. Animal Mitochondrial DNA: Structure and Evolution. Adv. Clin. Chem. 1992, 141, 173–216. [Google Scholar] [CrossRef]
- Cao, S.-Y.; Wu, X.-B.; Yan, P.; Hu, Y.-L.; Su, X.; Jiang, Z. Complete nucleotide sequences and gene organization of mitochondrial genome of Bufo gargarizans. Mitochondrion 2006, 6, 186–193. [Google Scholar] [CrossRef]
- E Cook, C.; Yue, Q.; Akam, M. Mitochondrial genomes suggest that hexapods and crustaceans are mutually paraphyletic. Proc. R. Soc. B Boil. Sci. 2005, 272, 1295–1304. [Google Scholar] [CrossRef]
- Ma, C.; Yang, P.; Jiang, F.; Chapuis, M.; Shali, Y.; Sword, G.A.; Kang, L. Mitochondrial genomes reveal the global phylogeography and dispersal routes of the migratory locust. Mol. Ecol. 2012, 21, 4344–4358. [Google Scholar] [CrossRef] [PubMed]
- Nardi, F.; Spinsanti, G.; Boore, J.L.; Carapelli, A.; Dallai, R.; Frati, F. Hexapod origins: Monophyletic or paraphyletic. Science 2003, 299, 1887–1889. [Google Scholar] [CrossRef] [PubMed]
- Saccone, C.; de Giorgi, C.; Gissi, C.; Pesole, G.; Reyes, A. Evolutionary genomics in Metazoa: The mitochondrial DNA as a model system. Gene 1999, 238, 195–209. [Google Scholar] [CrossRef]
- Timmermans, M.J.T.N.; Lees, D.C.; Simonsen, T.J. Towards a mitogenomic phylogeny of Lepidoptera. Mol. Phylogenet. Evol. 2014, 79, 169–178. [Google Scholar] [CrossRef]
- Zhang, P.; Zhou, H.; Liang, D.; Liu, Y.-F.; Chen, Y.-Q.; Qu, L.-H. The complete mitochondrial genome of a tree frog, Polypedates megacephalus (Amphibia: Anura: Rhacophoridae), and a novel gene organization in living amphibians. Gene 2005, 346, 133–143. [Google Scholar] [CrossRef]
- Gong, N.; Yang, L.; Chen, X.-S. Two new species of the bamboo-feeding genus Bambusicaliscelis Chen & Zhang, 2011 from China (Hemiptera, Fulgoromorpha, Caliscelidae). ZooKeys 2018, 776, 81–89. [Google Scholar] [CrossRef]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Gärtner, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Li, W.-X.; Jakovlić, I.; Zou, H.; Zhang, J.; Wang, G.-T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. bioRxiv 2018, 489088. [Google Scholar] [CrossRef]
- Bivand, R.; Anselin, L.; Berke, O.; Bernat, A.; Carvalho, M.; Chun, Y.; Dormann, C.F.; Dray, S.; Halbersma, R.; Lewin-Koh, N. spdep: Spatial dependence: Weighting schemes, statistics and models. GeoHealth 2019, 4, e2020GH000276. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.A.; Österman, J.; Wahlberg, N.; Nesme, X.; Lavire, C.; Vial, L.; Paulin, L.; de Lajudie, P.; Lindström, K. Phy-logeny of the Rhizobium-Allorhizobium-Agrobacterium clade supports the delineation of Neorhizobium gen. nov. Sys-Temat. Appl. Microbiol. 2014, 37, 208–215. [Google Scholar]
- Dai, L.-S.; Zhu, B.-J.; Qian, C.; Zhang, C.-F.; Li, J.; Wang, L.; Wei, G.-Q.; Liu, C. The complete mitochondrial genome of the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Mitochondrial DNA 2016, 27, 1512–1513. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.-S.; Zhu, B.-J.; Zhao, Y.; Zhang, C.-F.; Liu, C.-L. Comparative Mitochondrial Genome Analysis of Eligma narcissus and other Lepidopteran Insects Reveals Conserved Mitochondrial Genome Organization and Phylogenetic Relationships. Sci. Rep. 2016, 6, 26387. [Google Scholar] [CrossRef]
- Kim, I.; Lee, E.M.; Seol, K.Y.; Yun, E.Y.; Lee, Y.B.; Hwang, J.S.; Jin, B.R. The mitochondrial genome of the Korean hairstreak, Coreana raphaelis (Lepidoptera: Lycaenidae). Insect Mol. Biol. 2006, 15, 217–225. [Google Scholar] [CrossRef]
- Boore, J.L. Complete Mitochondrial Genome Sequence of the Polychaete Annelid Platynereis dumerilii. Mol. Biol. Evol. 2001, 18, 1413–1416. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nat. Cell Biol. 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Muir, F. XLVIII—On the classification of the Fulgoroidea. J. Nat. Hist. 1930, 6, 461–478. [Google Scholar] [CrossRef]
- Asche, M. Preliminary thoughts on the phylogeny of Fulgoromorpha (Homoptera: Auchenorrhyncha). In Proceedings of the 6th Auchenorrhyncha, Turin, Italy, 7–11 September 1987; pp. 47–53. [Google Scholar]
- Bourgoin, T.; Steffen-Campbell, J.; Campbell, B. Molecular Phylogeny of Fulgoromorpha (Insecta, Hemiptera, Archaeorrhyncha). The Enigmatic Tettigometridae: Evolutionary Affiliations and Historical Biogeography. Cladistics 1997, 13, 207–224. [Google Scholar] [CrossRef]
- Yeh, W.B.; Yang, C.T.; Hui, C.F. A molecular phylogeny of planthoppers (Hemiptera: Fulgoroidea) inferred from mito-chondrial 16S rDNA sequences. Zool. Stud. 2005, 44, 519–535. [Google Scholar]
- Gnezdilov, V.M. Notes on the phylogenetic relationships of planthoppers of the family issidae (Hemiptera, Fulgoroidea) of the Western Palaearctic fauna, with descriptions of two new genera. Entomol. Rev. 2016, 96, 332–347. [Google Scholar] [CrossRef]
- Sheffield, N.C.; Song, H.; Cameron, S.L.; Whiting, M.F. A Comparative Analysis of Mitochondrial Genomes in Coleoptera (Arthropoda: Insecta) and Genome Descriptions of Six New Beetles. Mol. Biol. Evol. 2008, 25, 2499–2509. [Google Scholar] [CrossRef]
- Cao, S.-S.; Du, Y.-Z. Characterization of the complete mitochondrial genome of Chilo auricilius and comparison with three other rice stem borers. Gene 2014, 548, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.-J.; Zhu, W.-C.; Rong, X.; Liu, J.; Ding, X.-L.; Hong, X.-Y. The complete mitochondrial genome sequence of Sogatella furcifera (Horváth) and a comparative mitogenomic analysis of three predominant rice planthoppers. Gene 2014, 533, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Liang, A.-P. A Preliminary Molecular Phylogeny of Planthoppers (Hemiptera: Fulgoroidea) Based on Nuclear and Mitochondrial DNA Sequences. PLoS ONE 2013, 8, e58400. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).