Nuclear Reorganization in Hippocampal Granule Cell Neurons from a Mouse Model of Down Syndrome: Changes in Chromatin Configuration, Nucleoli and Cajal Bodies
Abstract
1. Introduction
2. Results
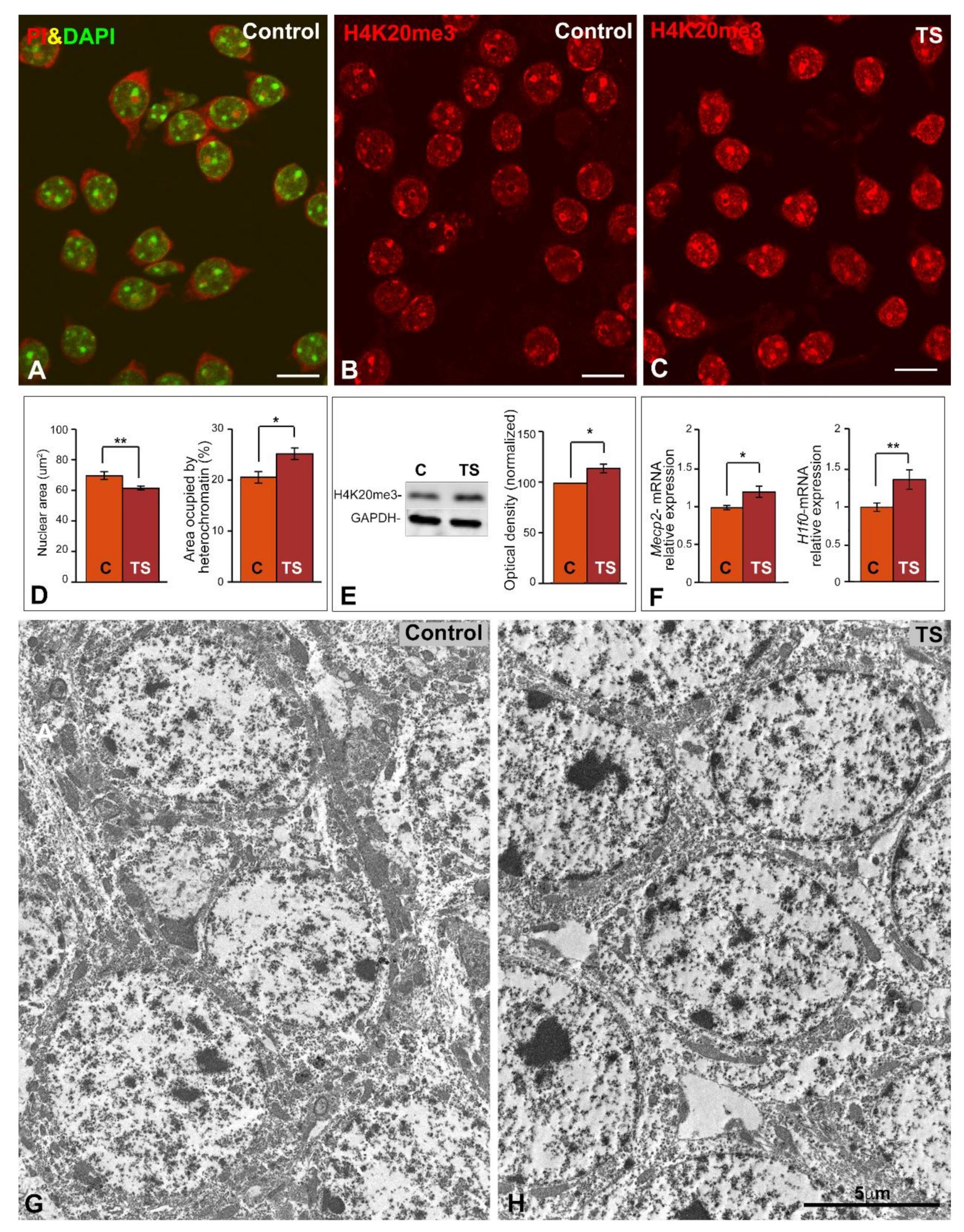
2.1. Reduced Nuclear Size and Increased Heterochromatinization Are Nuclear Features in TS GCs
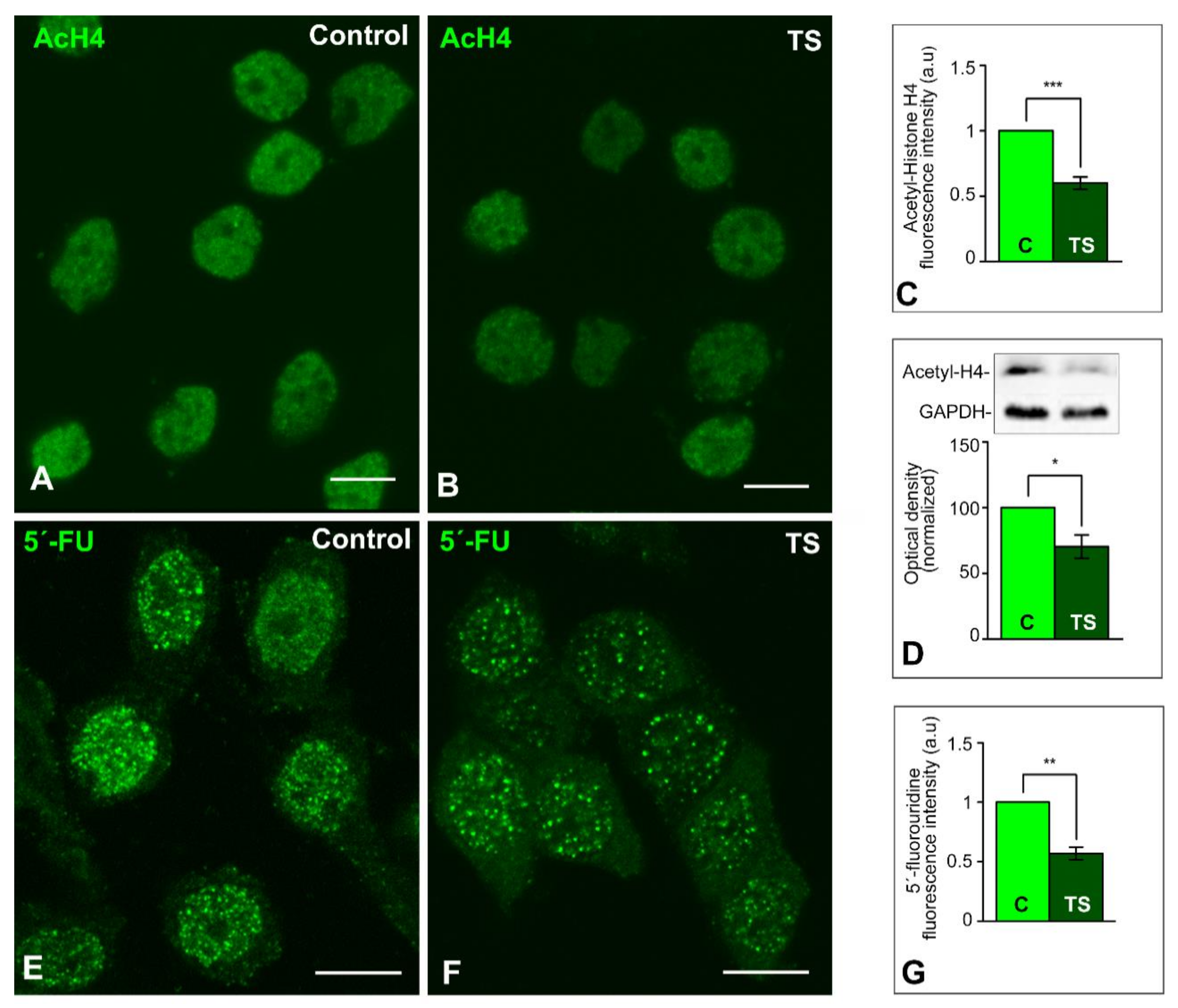
2.2. Chromatin Reorganization in TS GCs Induces a Decrease in Global Transcription Rate
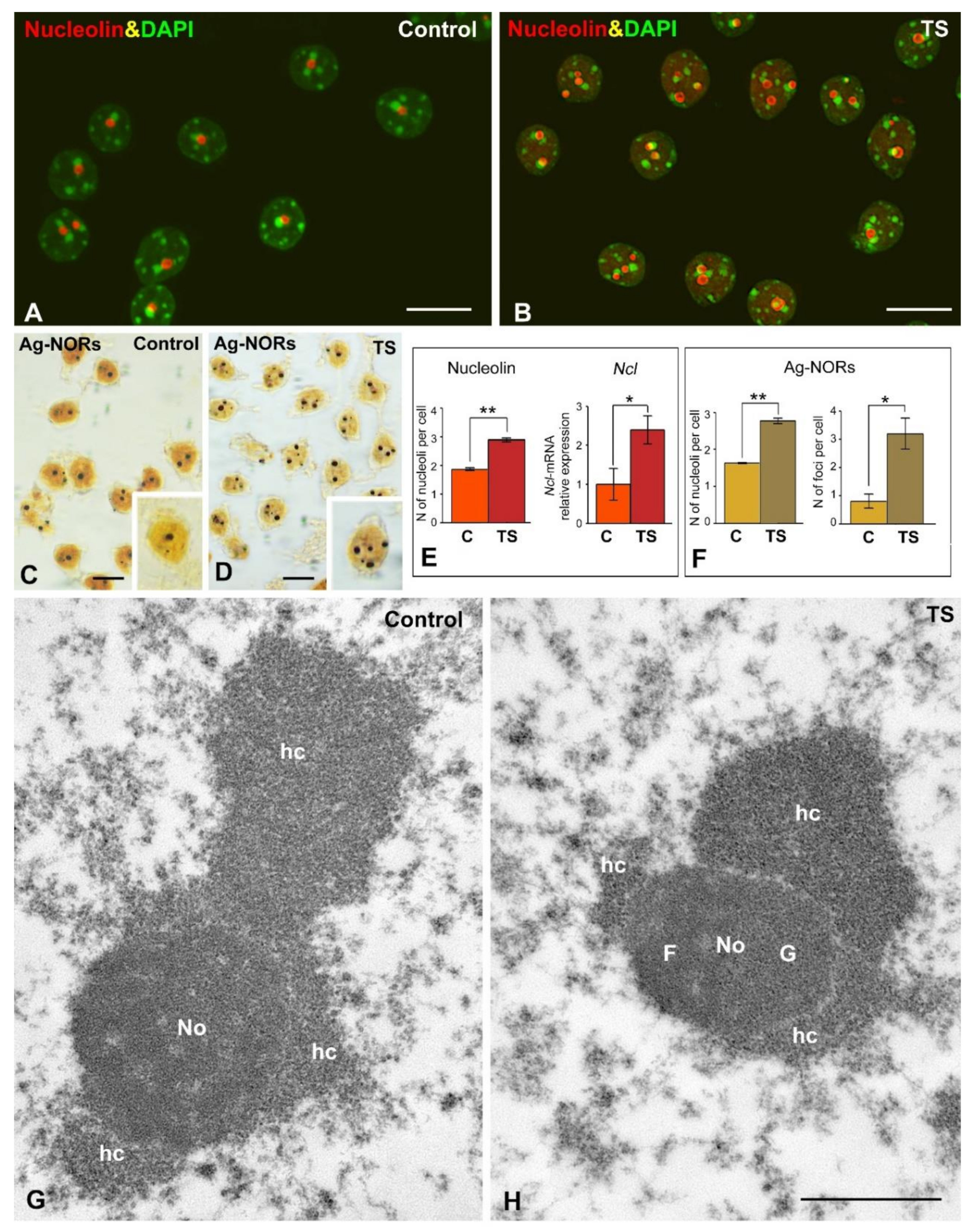
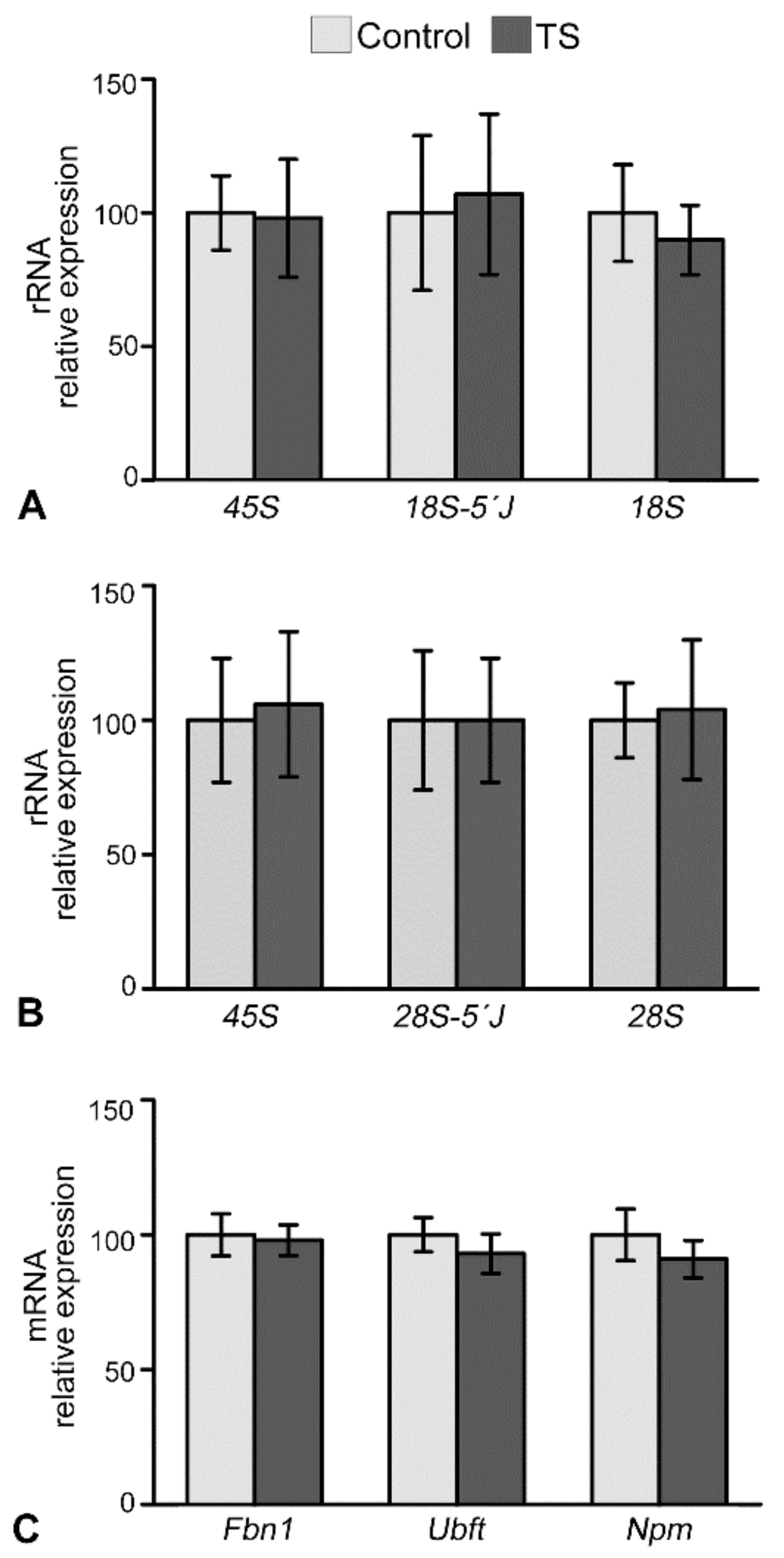
2.3. The Reorganization of Chromosomes in TS GCs Is Associated with Reduced Nucleolar Fusion, Resulting in a Higher Number of Nucleoli
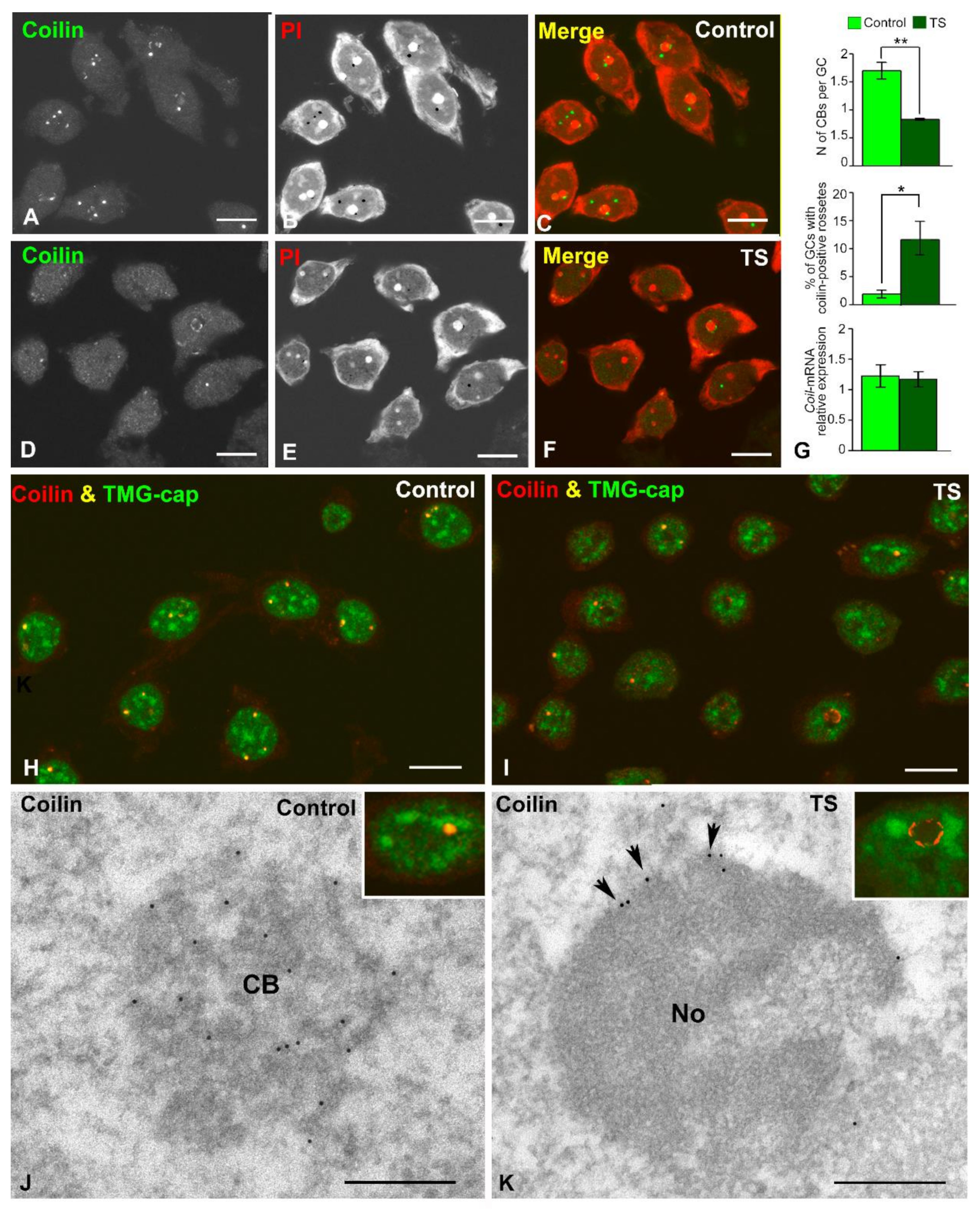
2.4. The Nuclear Reorganization in GCs of the TS Mouse Results in Reduced CB Number and Coilin Redistribution
3. Discussion
4. Conclusions
5. Materials and methods
5.1. Animals
5.2. Immunofluorescence and Confocal Microscopy
5.3. In Situ Transcription Assays with 5′-Fluorouridine
5.4. Conventional and Immunoelectron Microscopy
5.5. Ag-NOR Staining
5.6. Western Blotting
5.7. Real Time Quantitative PCR (qRT-PCR)
5.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shin, M.; Besser, L.M.; Kucik, J.E.; Lu, C.; Siffel, C.; Correa, A. The Congenital Anomaly Multistate Prevalence and Survival (CAMPS) Collaborative. Prevalence of Down Syndrome Among Children and Adolescents in 10 Regions of the United States. Pediatrics 2009, 124, 1565–1571. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Dhanasekaran, A.R.; Gardiner, K.J. Mouse models of Down syndrome: Gene content and consequences. Mamm. Genome 2016, 27, 538–555. [Google Scholar] [CrossRef] [PubMed]
- Rueda, N.; Flórez, J.; Martínez-Cué, C. Mouse Models of Down Syndrome as a Tool to Unravel the Causes of Mental Disabilities. Neural Plast. 2012, 2012, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Herault, Y.; Delabar, J.M.; Fisher, E.M.C.; Tybulewicz, V.L.J.; Yu, E.; Brault, V. Rodent models in Down syndrome research: Impact and future opportunities. Dis. Model. Mech. 2017, 10, 1165–1186. [Google Scholar] [CrossRef]
- Antonarakis, S.E.; Skotko, B.G.; Rafii, M.S.; Strydom, A.; Pape, S.E.; Bianchi, D.W.; Sherman, S.L.; Reeves, R.H. Down syndrome. Nat. Rev. Dis. Primers 2020, 6, 9. [Google Scholar] [CrossRef]
- Sturgeon, X.; Gardiner, K.J. Transcript catalogs of human chromosome 21 and orthologous chimpanzee and mouse regions. Mamm. Genome 2011, 22, 261–271. [Google Scholar] [CrossRef]
- Bartesaghi, R.; Guidi, S.; Ciani, E. Is it possible to improve neurodevelopmental abnormalities in Down syndrome? Rev. Neurosci. 2011, 22, 419–455. [Google Scholar] [CrossRef]
- Contestabile, A.; Fila, T.; Ceccarelli, C.; Bonasoni, P.; Bonapace, L.; Santini, D.; Bartesaghi, R.; Ciani, E. Cell cycle alteration and decreased cell proliferation in the hippocampal dentate gyrus and in the neocortical germinal matrix of fetuses with down syndrome and in Ts65Dn mice. Hippocampus 2007, 17, 665–678. [Google Scholar] [CrossRef]
- Llorens-Martín, M.; Rueda, N.; Tejeda, G.S.; Flórez, J.; Trejo, J.; Martínez-Cué, C. Effects of voluntary physical exercise on adult hippocampal neurogenesis and behavior of Ts65Dn mice, a model of Down syndrome. Neuroscience 2010, 171, 1228–1240. [Google Scholar] [CrossRef]
- Rueda, N.; Vidal, V.; García-Cerro, S.; Puente, A.; Campa, V.; Lantigua, S.; Narcís, O.; Bartesaghi, R.; Martínez-Cué, C. Prenatal, but not Postnatal, Curcumin Administration Rescues Neuromorphological and Cognitive Alterations in Ts65Dn Down Syndrome Mice. J. Nutr. 2020, 150, 2478–2489. [Google Scholar] [CrossRef]
- Haydar, T.F.; Reeves, R.H. Trisomy 21 and early brain development. Trends Neurosci. 2012, 35, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Kleschevnikov, A.M.; Yu, J.; Kim, J.; Lysenko, L.V.; Zeng, Z.; Yu, Y.E.; Mobley, W.C. Evidence that increased Kcnj6 gene dose is necessary for deficits in behavior and dentate gyrus synaptic plasticity in the Ts65Dn mouse model of Down syndrome. Neurobiol. Dis. 2017, 103, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Giacomini, A.; Stagni, F.; Emili, M.; Guidi, S.; Salvalai, M.E.; Grilli, M.; Vidal-Sanchez, V.; Martinez-Cué, C.; Bartesaghi, R. Treat-ment with corn oil improves neurogenesis and cognitive performance in the Ts65Dn mouse model of Down syndrome. Brain Res. Bull. 2018, 140, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Villarroya, O.; Ballestín, R.; López-Hidalgo, R.; Mulet, M.; Blasco-Ibáñez, J.M.; Crespo, C.; Nacher, J.; Gilabert-Juan, J.; Varea, E. Morphological alterations in the hippocampus of the Ts65Dn mouse model for Down Syndrome correlate with structural plasticity markers. Histol. Histopathol. 2018, 33, 101–115. [Google Scholar] [PubMed]
- Boisvert, F.-M.; Van Koningsbruggen, S.; Navascués, J.; Lamond, A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007, 8, 574–585. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, M. Chromosome Territories. Cold Spring Harb. Perspect. Biol. 2010, 2, a003889. [Google Scholar] [CrossRef]
- Mao, Y.S.; Zhang, B.; Spector, D.L. Biogenesis and function of nuclear bodies. Trends Genet. 2011, 27, 295–306. [Google Scholar] [CrossRef]
- Machyna, M.; Kehr, S.; Straube, K.; Kappei, D.; Buchholz, F.; Butter, F.; Ule, J.; Hertel, J.; Stadler, P.F.; Neugebauer, K.M. The coilin interactome identifies hundreds of small noncoding RNAs that traffic through Cajal bodies. Mol. Cell 2014, 56, 389–399. [Google Scholar] [CrossRef]
- Maass, P.G.; Barutcu, A.R.; Rinn, J.L. Interchromosomal interactions: A genomic love story of kissing chromosomes. J. Cell Biol. 2019, 218, 27–38. [Google Scholar] [CrossRef]
- Karpen, G.H.; Schaefer, J.E.; Laird, C.D. A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev. 1988, 2, 1745–1763. [Google Scholar] [CrossRef]
- McStay, B. Nucleolar organizer regions: Genomic ‘dark matter’ requiring illumination. Genes Dev. 2016, 30, 1598–1610. [Google Scholar] [CrossRef] [PubMed]
- Zatsepina, O.V. Cytogenetic instability of chromosomal nucleolar organizer regions (NORs) in cloned mouse L929 fibro-blasts. Chromosome Res. 2019, 27, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.J.; Roth, M.B. ncl-1 Is Required for the Regulation of Cell Size and Ribosomal RNA Synthesis in Caenorhabditis elegans. J. Cell Biol. 1998, 140, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Verdun, D.; Roussel, P.; Thiry, M.; Sirri, V.; Lafontaine, D.L. The nucleolus: Structure/function relationship in RNA matabolism. Wiley Interdisci. Rev. RNA 2010, 1, 415–431. [Google Scholar] [CrossRef]
- Baltanas, F.C.; Casafont, I.; Weruaga, E.; Alonso, J.R.; Berciano, M.T.; Lafarga, M. Nucleolar disruption and Cajal body disas-sembly are nuclear hallmarks of DNA damage-induced neurodegeneration in Purkinje cells. Brain Pathol. 2011, 21, 374–388. [Google Scholar] [CrossRef]
- Baltanas, F.C.; Berciano, M.T.; Tapia, O.; Narcis, J.O.; Lafarga, V.; Díaz, D.; Weruaga, E.; Santos, E.; Lafarga, M. Nucleolin reor-ganization and nucleolar stress in Purkinje cells of mutant PCD mice. Neurobiol. Dis. 2019, 127, 312–322. [Google Scholar] [CrossRef]
- Hernández-Ortega, K.; Garcia-Esparcia, P.; Gil, L.; Lucas, J.J.; Ferrer, I. Altered Machinery of Protein Synthesis in Alzheimer’s: From the Nucleolus to the Ribosome. Brain Pathol. 2016, 26, 593–605. [Google Scholar] [CrossRef]
- Parlato, R.; Kreiner, G. Nucleolar activity in neurodegenerative diseases: A missing piece of the puzzle? J. Mol. Med. 2012, 91, 541–547. [Google Scholar] [CrossRef]
- Tapia, O.; Narcís, J.O.; Riancho, J.; Tarabal, O.; Piedrafita, L.; Calderó, J.; Berciano, M.T.; Lafarga, M. Cellular bases of the RNA metab-olism dysfunction in motor neurons of a murine model of spinal muscular atrophy: Role of Cajal bodies and the nucleolus. Neurobiol. Dis. 2017, 108, 83–99. [Google Scholar] [CrossRef]
- Gall, J.G. The centennial of the Cajal body. Nat. Rev. Mol. Cell Biol. 2003, 4, 975–980. [Google Scholar] [CrossRef]
- Lafarga, M.; Casafont, I.; Bengoechea, R.; Tapia, O.; Berciano, M.T. Cajal’s contribution to the knowledge of the neuronal cell nucleus. Chromosoma 2009, 118, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Machyna, M.; Heyn, P.; Neugebauer, K.M. Cajal bodies: Where form meets function. Wiley Interdiscip. Rev. RNA 2013, 4, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Sawyer, I.A.; Sung, M.-H.; Sturgill, D.; Shevtsov, S.P.; Pegoraro, G.; Hakim, O.; Baek, S.; Hager, G.L.; Dundr, M. Cajal bodies are linked to genome conformation. Nat. Commun. 2016, 7, 10966. [Google Scholar] [CrossRef] [PubMed]
- Arias Escayola, D.; Neugebauer, K.M. Dynamics and function of nuclear bodies during embryogenesis. Biochemistry 2018, 57, 2462–2469. [Google Scholar] [CrossRef] [PubMed]
- Shevtsov, S.P.; Dundr, M. Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 2011, 13, 167–173. [Google Scholar] [CrossRef]
- Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121. [Google Scholar] [CrossRef]
- Pena, E.; Berciano, M.T.; Fernandez, R.; Ojeda, J.L.; Lafarga, M. Neuronal body size correlates with the number of nucleoli and Cajal bodies, and with the organization of the splicing machinery in rat trigeminal ganglion neurons. J. Comp. Neurol. 2001, 430, 250–263. [Google Scholar] [CrossRef]
- Berciano, M.T.; Novell, M.; Terán-Villagrá, N.; Casafont, I.; Bengoechea, R.; Val-Bernal, J.F.; Lafarga, M. Cajal body number and nucleolar size correlate with the cell body mass in human sensory ganglia neurons. J. Struct. Biol. 2007, 158, 410–420. [Google Scholar] [CrossRef]
- Narcís, J.O.; Tapia, O.; Tarabal, O.; Piedrafita, L.; Calderó, J.; Berciano, M.T.; Lafarga, M. Accumulation of poly(A) RNA in nuclear granules enriched in Sam68 in motor neurons from the SMNΔ7 mouse model of SMA. Sci. Rep. 2018, 8, 9646. [Google Scholar]
- Savino, T.M.; Gébrane-Younès, J.; De Mey, J.; Sibarita, J.-B.; Hernandez-Verdun, D. Nucleolar Assembly of the Rrna Processing Machinery in Living Cells. J. Cell Biol. 2001, 153, 1097–1110. [Google Scholar] [CrossRef]
- Ito, K.; Takizawa, T. Nuclear Architecture in the Nervous System: Development, Function, and Neurodevelopmental Diseases. Front. Genet. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Kemeny, S.; Tatout, C.; Salaun, G.; Pebrel-Richard, C.; Goumy, C.; Ollier, N.; Maurin, E.; Pereira, B.; Vago, P.; Gouas, L. Spatial organization of chromosome territories in the interphase nucleus of trisomy 21 cells. Chromosome 2018, 127, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Letourneau, A.; Santoni, F.A.; Bonilla, X.; Sailani, M.R.; Gonzalez, D.; Kind, J.; Chevalier, C.; Thurman, R.; Sandstrom, R.S.; Hibaoui, Y.; et al. Domains of ge-nome-wide gene expression dysregulation in Down’s syndrome. Nature 2014, 508, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Kahlem, P.; Sultan, M.; Herwig, R.; Steinfath, M.; Balzereit, D.; Eppens, B.; Saran, N.G.; Pletcher, M.T.; South, S.T.; Stetten, G.; et al. Transcript Level Alterations Reflect Gene Dosage Effects Across Multiple Tissues in a Mouse Model of Down Syndrome. Genome Res. 2004, 14, 1258–1267. [Google Scholar] [CrossRef] [PubMed]
- Lyle, R.; Gehrig, C.; Neergaard-Henrichsen, C.; Deutsch, S.; Antonarakis, S.E. Gene expression from the aneuploidy chro-mosome in a trisomy mouse model of Down syndrome. Genome Res. 2004, 14, 1268–1274. [Google Scholar] [CrossRef] [PubMed]
- Rachidi, M.; Lopes, C. Mental retardation and associated neurological dysfunctions in Down syndrome: A consequence of dysregulation in critical chromosome 21 genes and associated molecular pathways. Eur. J. Paediatr. Neurol. 2008, 12, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Do, C.; Xing, Z.; Yu, Y.E.; Tycko, B. Catherine Trans-acting epigenetic effects of chromosomal aneuploidies: Lessons from Down syndrome and mouse models. Epigenomics 2017, 9, 189–207. [Google Scholar] [CrossRef] [PubMed]
- Alldred, M.J.; Lee, S.H.; Petkova, E.; Ginsberg, S.D. Expression profile analysis of vulnerable CA1 pyramidal neurons in young-Middle-Aged Ts65Dn mice. J. Comp. Neurol. 2014, 523, 61–74. [Google Scholar] [CrossRef]
- Kelley, C.M.; Ginsberg, S.D.; Alldred, M.J.; Strupp, B.J.; Mufson, E.J. Maternal Choline Supplementation Alters Basal Forebrain Cholinergic Neuron Gene Expression in the Ts65Dn Mouse Model of Down Syndrome. Dev. Neurobiol. 2019, 79, 664–683. [Google Scholar] [CrossRef]
- Mentis, A.-F.A. Epigenomic engineering for Down syndrome. Neurosci. Biobehav. Rev. 2016, 71, 323–327. [Google Scholar] [CrossRef]
- Nelson, D.M.; Jaber-Hijazi, F.; Cole, J.J.; Robertson, N.A.; Pawlikowski, J.S.; Norris, K.T.; Criscione, S.W.; Pchelintsev, N.A.; Piscitello, D.; Stong, N.; et al. Mapping H4K20me3 onto the chromatin landscape of senescent cells indicates a function in control of cell senescence and tumor suppression through preservation of genetic and epigenetic stability. Genome Biol. 2016, 17, 158. [Google Scholar] [CrossRef] [PubMed]
- Karachentsev, D.; Sarma, K.; Reinberg DSteward, R. PR-Ser7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev. 2005, 19, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, M.; Walter, D.; Gillespie, P.J.; Izard, F.; Fahrenkrog, B.; Lleres, D.; Lerdrup, M.; Johansen, J.V.; Hansen, K.; Julien, E.; et al. Histone H4K20 methylation mediated chromatin compaction threshold ensures genome integrity by limiting DNA replication licensing. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Webster, M.; Witkin, K.L.; Cohen-Fix, O. Sizing up the nucleus: Nuclear shape, size and nuclear-envelope assembly. J. Cell Sci. 2009, 122, 1477–1486. [Google Scholar] [CrossRef]
- Castillo-Iglesias, M.S.; Berciano, M.T.; Narcis, J.O.; Val-Bernal, J.F.; Rodriguez-Rey, J.C.; Tapia, O.; Lafarga, M. Reorganization of the nuclear compartments involved in transcription and RNA processing in myonuclei of type I spinal muscular atrophy. Histochem. Cell Biol. 2019, 152, 227–237. [Google Scholar] [CrossRef]
- Singleton, M.K.; Gonzalez, M.L.; Leung, K.N.; Yasui, D.H.; Schroeder, D.I.; Dunaway, K.; LaSalle, J.M. MeCP2 is required for global heterochromatic and nucleolar changes during activity-dependent neuronal maturation. Neurobiol. Dis. 2011, 43, 190–200. [Google Scholar] [CrossRef]
- Flanagan, T.W.; Brown, D.T. Molecular dynamics of histone H1. Biochim. Biophys. Acta 2016, 1859, 468–475. [Google Scholar] [CrossRef]
- Zhang, T.; Cooper, S.; Brockdorff, N. The interplay of histone modifications—Writers that read. EMBO Rep. 2015, 16, 1467–1481. [Google Scholar] [CrossRef]
- Rieder, D.; Trajanoski, Z.; McNally, J.G. Transcription factories. Front. Genet. 2012, 3, 221. [Google Scholar] [CrossRef]
- Casafont, I.; Palanca, A.R.; Lafarga, V.; Mata-Garrido, J.; Berciano, M.T.; Lafarga, M. Dynamic Behavior of the RNA Polymerase II and the Ubiquitin Proteasome System During the Neuronal DNA Damage Response to Ionizing Radiation. Mol. Neurobiol. 2015, 53, 6799–6808. [Google Scholar] [CrossRef]
- Caudron-Herger, M.; Pankert, T.; Seiler, J.; Nemeth, A.; Voit, R.; Grummt, I.; Rippe, K. Alu element-containing RNAs maintain nuclear structure and function. EMBO J. 2015, 34, 2758–2774. [Google Scholar] [CrossRef] [PubMed]
- Carmo-Fonseca, M. Assembly of the nucleolus: In need of revision. EMBO J. 2015, 34, 2731–2732. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mayer, C.; Bierhoff, H.; Grummt, I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 2005, 19, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Sheltzer, J.M.; Torres, E.M.; Dunham, M.J.; Amon, A. Transcriptional consequences of aneuploidy. Proc. Natl. Acad. Sci. USA 2012, 109, 12644–12649. [Google Scholar] [CrossRef]
- Mennel, H.D.; Moller, I. Morphometric investigation on nuclear and nucleolar arrangement and AgNOR content in the rat hippocampus under normal and ischemic conditions. Exp. Toxic Pathol. 1994, 46, 491–501. [Google Scholar] [CrossRef]
- Yazdani, M.; Deogracias, R.; Guy, J.; Poot, R.A.; Bird, A.; Barde, Y.-A. Disease Modeling Using Embryonic Stem Cells: MeCP2 Regulates Nuclear Size and RNA Synthesis in Neurons. Stem. Cells 2012, 30, 2128–2139. [Google Scholar] [CrossRef]
- Hansen, J.C.; Ghosh, R.P.; Woodcock, C.L. Binding of the Rett syndrome protein, MeCP2, to methylated and unmethylated DNA and chromatin. IUBMB Life 2010, 62, 732–738. [Google Scholar] [CrossRef]
- García-Cerro, S.; Rueda, N.; Vidal, V.; Lantigua, S.; Martínez-Cué, C. Normalizing the gene dosage of Dyrk1A in a mouse model of Down syndrome rescues several Alzheimer’s disease phenotypes. Neurobiol. Dis. 2017, 106, 76–88. [Google Scholar] [CrossRef]
- Vihervaara, A.; Duarte, F.M.; Lis, J.T. Molecular mechanisms driving transcriptional stress responses. Nat. Rev. Genet. 2018, 19, 385–397. [Google Scholar] [CrossRef]
- Pestinger, V.; Wijeratne, S.S.K.; Rodriguez-Melendez, R.; Zempleni, J. Novel histone biotinylation marks are enriched in repeat regions and participate in repression of transcriptionally competent genes. J. Nutr. Biochem. 2011, 22, 328–333. [Google Scholar] [CrossRef]
- Singh, M.P.; Wijeratne, S.S.K.; Zempleni, J. Biotinylation of lysine 16 in histone H4 contributes toward nucleosome condensation. Arch. Biochem. Biophys. 2013, 529, 105–111. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Canzonetta, C.; Mulligan, C.; Deutsch, S.; Ruf, S.; O’Doherty, A.; Lyle, R.; Borel, C.; Lin-Marq, N.; Delom, F.; Groet, J.; et al. DYRK1A-dosage imbalance perturbs NRSF/REST levels, deregulating pluripotency and em-bryonic stem cell fate in Down syndrome. Am. J. Hum. Genet. 2008, 83, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, R.; Hassan, M.Q.; Pratap, J.; Lian, J.B.; Montecino, M.A.; van Wijnen, A.J.; Stein, J.L.; Imbalzano, A.N.; Stein, G.S. The human SWI/SNF complex associates with RUNX1 to control transcription of hematopoietic target genes. J. Cell Physiol. 2010, 225, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Antonarakis, S.E.; Lyle, R.; Dermitzakis, E.T.; Reymond, A.; Deutsch, S. Chromosome 21 and Down syndrome: From genomics to pathophysiology. Nat. Rev. Genet. 2004, 5, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Zidovska, A.; Weitz, D.A.; Mitchison, T.J. Micron-scale coherence in interphase chromatindynamics. Proc. Natl. Acad. Sci. USA 2013, 110, 15555–15560. [Google Scholar] [CrossRef]
- Caragine, C.M.; Haley, S.C.; Zidovska, A. Nucleolar dynamics and interactions with nucleoplasm in living cells. eLife 2019, 8. [Google Scholar] [CrossRef]
- Falahati, H.; Pelham-Webb, B.; Blythe, S.; Wieschaus, E. Nucleation by rRNA Dictates the Precision of Nucleolus Assembly. Curr. Biol. 2016, 26, 277–285. [Google Scholar] [CrossRef]
- Das, S.; Cong, R.; Shandilya, J.; Senepati, P.; Moindrot, B.; Monier, K.; Delage, H.; Mongelard, F.; Kumar, S.; Kundu, T.K.; et al. Characterization of nucleolin K88 acetylation defines a new pool of nucleolin colocalizing with pre-mRNA splicing factors. FEBS Lett. 2013, 587, 417–424. [Google Scholar] [CrossRef]
- Palanca, A.R.; Casafont, I.; Berciano, M.T.; Lafarga, M. Reactive nucleolar and Cajal body responses to proteasome inhibition in sensory ganglion neurons. Biochim. Biophys. Acta 2014, 1842, 848–859. [Google Scholar] [CrossRef]
- Lafarga, V.; Tapia, O.; Sharma, S.; Bengoechea, R.; Stoecklin, G.; Lafarga, M.; Berciano, M.T. CBP-mediated SMN acetylation modulates Cajal body biogenesis and the cytoplasmic targeting of SMN. Cell. Mol. Life Sci. 2017, 75, 527–546. [Google Scholar] [CrossRef]
- Cioce, M.; Boulon, S.; Matera, A.G.; Lamond, A.I. UV-induced fragmentation of Cajal bodies. J. Cell Biol. 2006, 175, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Gilder, A.S.; Do, P.M.; I Carrero, Z.; Cosman, A.M.; Broome, H.J.; Velma, V.; Martinez, L.A.; Hebert, M.D. Coilin participates in the suppression of RNA polymerase I in response to cisplatin-induced DNA damage. Mol. Biol. Cell 2011, 22, 1070–1079. [Google Scholar] [CrossRef] [PubMed]
- Hebert, M.D.; Poole, A.R. Towards an understanding of regulating Cajal body activity by protein modification. RNA Biol. 2017, 14, 761–778. [Google Scholar] [CrossRef] [PubMed]
- Hebert, M.D.; Shpargel, K.B.; Ospina, J.K.; Tucker, K.E.; Matera, A.G. Coilin Methylation Regulates Nuclear Body Formation. Dev. Cell 2002, 3, 329–337. [Google Scholar] [CrossRef]
- Navascués, J.; Bengoechea, R.; Tapia, O.; Casafont, I.; Berciano, M.; Lafarga, M. SUMO-1 transiently localizes to Cajal bodies in mammalian neurons. J. Struct. Biol. 2008, 163, 137–146. [Google Scholar] [CrossRef]
- Hetman, M.; Pietrzak, M. Emerging roles of the neuronal nucleolus. Trends Neurosci. 2012, 35, 305–314. [Google Scholar] [CrossRef]
- Hainmueller, T.; Bartos, M. Dentate gyrus circuits for encoding, retrieval and discrimination of episodic memories. Nat. Rev. Neurosci. 2020, 21, 153–168. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrouh, N.H.; Farr, A.L.; Randall, R.J. Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puente-Bedia, A.; Berciano, M.T.; Tapia, O.; Martínez-Cué, C.; Lafarga, M.; Rueda, N. Nuclear Reorganization in Hippocampal Granule Cell Neurons from a Mouse Model of Down Syndrome: Changes in Chromatin Configuration, Nucleoli and Cajal Bodies. Int. J. Mol. Sci. 2021, 22, 1259. https://doi.org/10.3390/ijms22031259
Puente-Bedia A, Berciano MT, Tapia O, Martínez-Cué C, Lafarga M, Rueda N. Nuclear Reorganization in Hippocampal Granule Cell Neurons from a Mouse Model of Down Syndrome: Changes in Chromatin Configuration, Nucleoli and Cajal Bodies. International Journal of Molecular Sciences. 2021; 22(3):1259. https://doi.org/10.3390/ijms22031259
Chicago/Turabian StylePuente-Bedia, Alba, María T. Berciano, Olga Tapia, Carmen Martínez-Cué, Miguel Lafarga, and Noemí Rueda. 2021. "Nuclear Reorganization in Hippocampal Granule Cell Neurons from a Mouse Model of Down Syndrome: Changes in Chromatin Configuration, Nucleoli and Cajal Bodies" International Journal of Molecular Sciences 22, no. 3: 1259. https://doi.org/10.3390/ijms22031259
APA StylePuente-Bedia, A., Berciano, M. T., Tapia, O., Martínez-Cué, C., Lafarga, M., & Rueda, N. (2021). Nuclear Reorganization in Hippocampal Granule Cell Neurons from a Mouse Model of Down Syndrome: Changes in Chromatin Configuration, Nucleoli and Cajal Bodies. International Journal of Molecular Sciences, 22(3), 1259. https://doi.org/10.3390/ijms22031259





