Evaluation of Computationally Designed Peptides against TWEAK, a Cytokine of the Tumour Necrosis Factor Ligand Family
Abstract
1. Introduction
2. Results
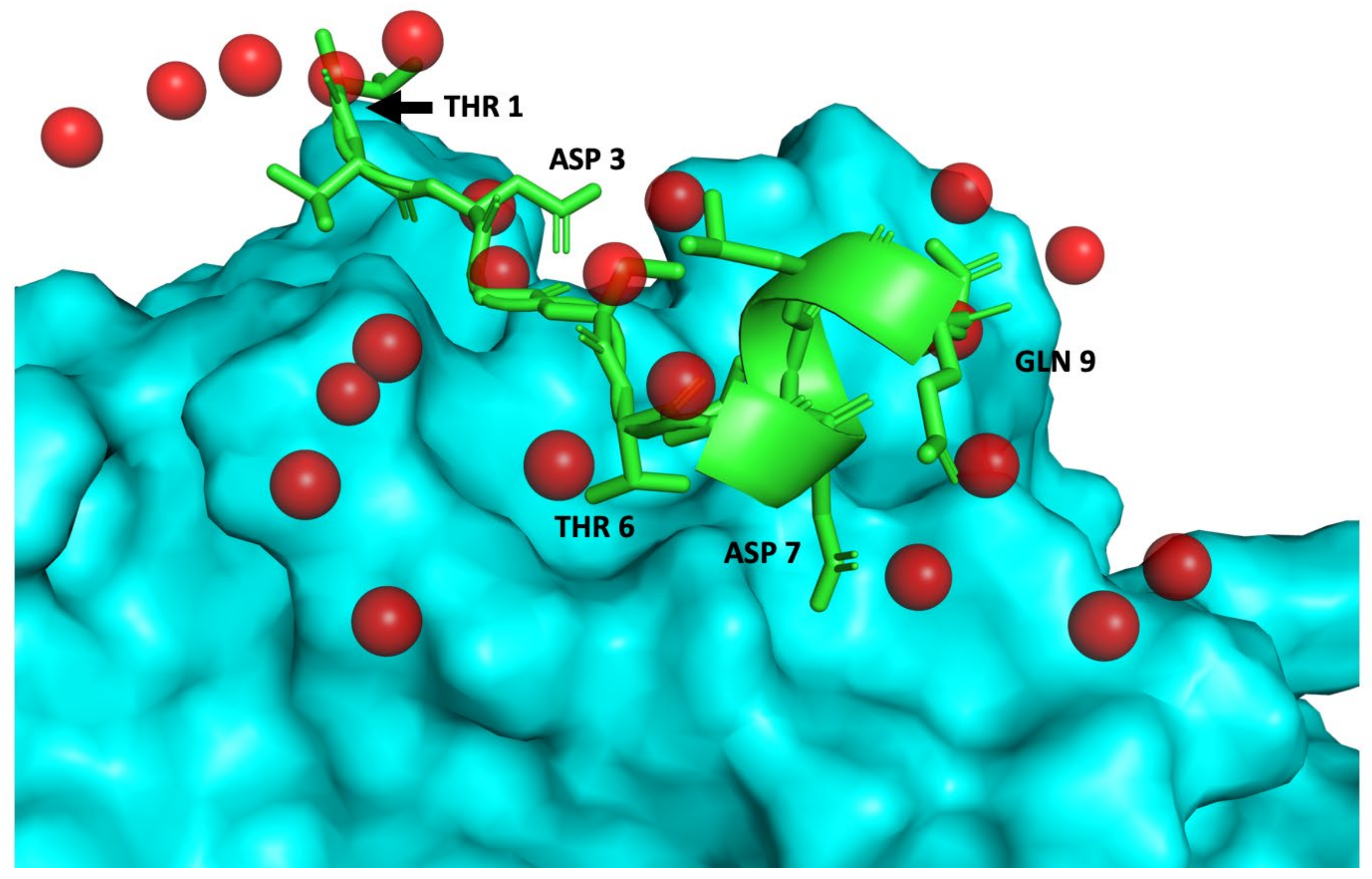
2.1. Computational Design of Peptides
2.2. Characterization of Brain Metastatic Cell Lines for Cell-Based Screening of Peptides
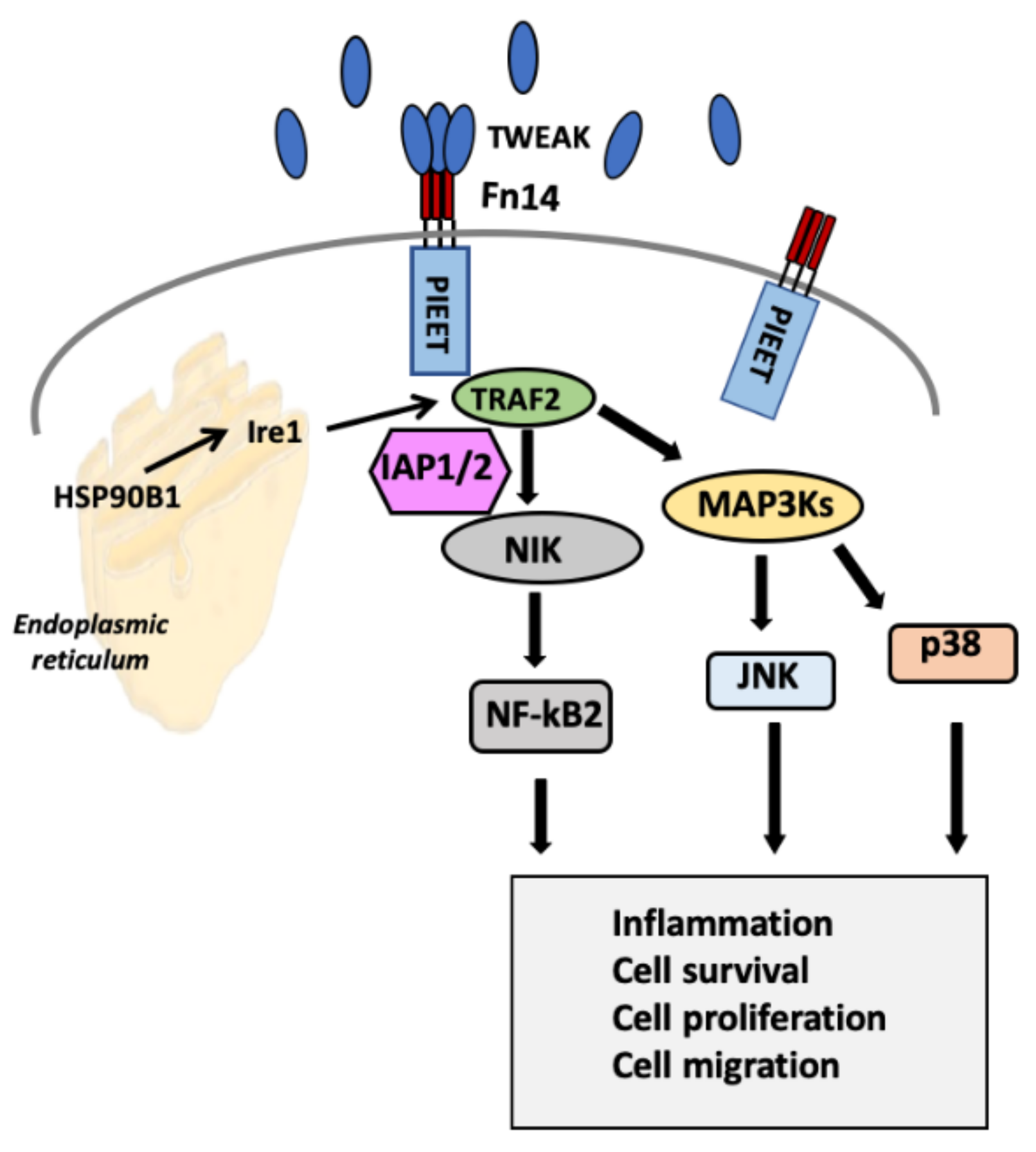
2.3. Selection of Genes for Cell-Based Screening of Peptides
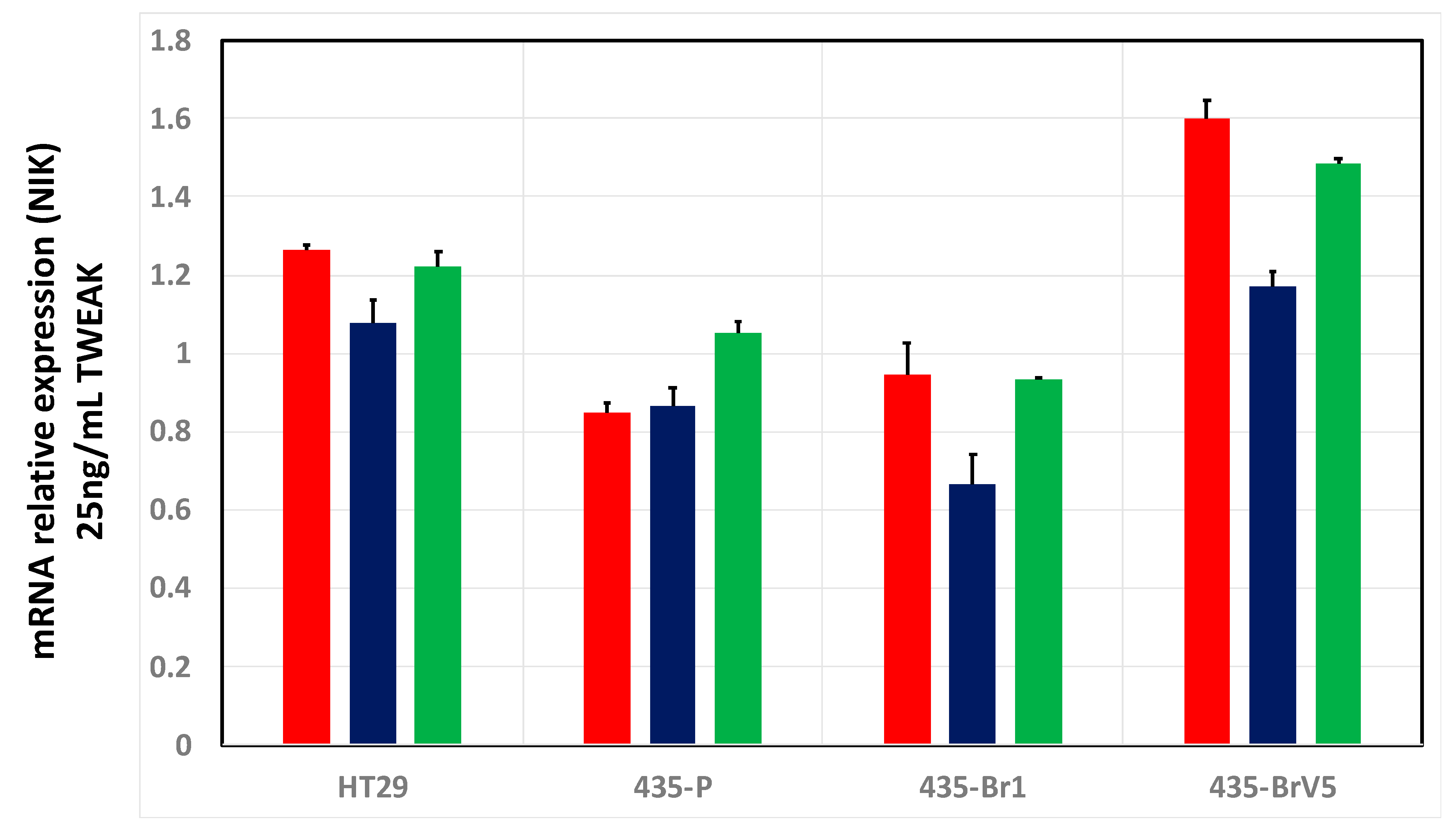
2.4. Cell-Based Screening of Pooled and Individual Peptides on Gene Expression Microarrays
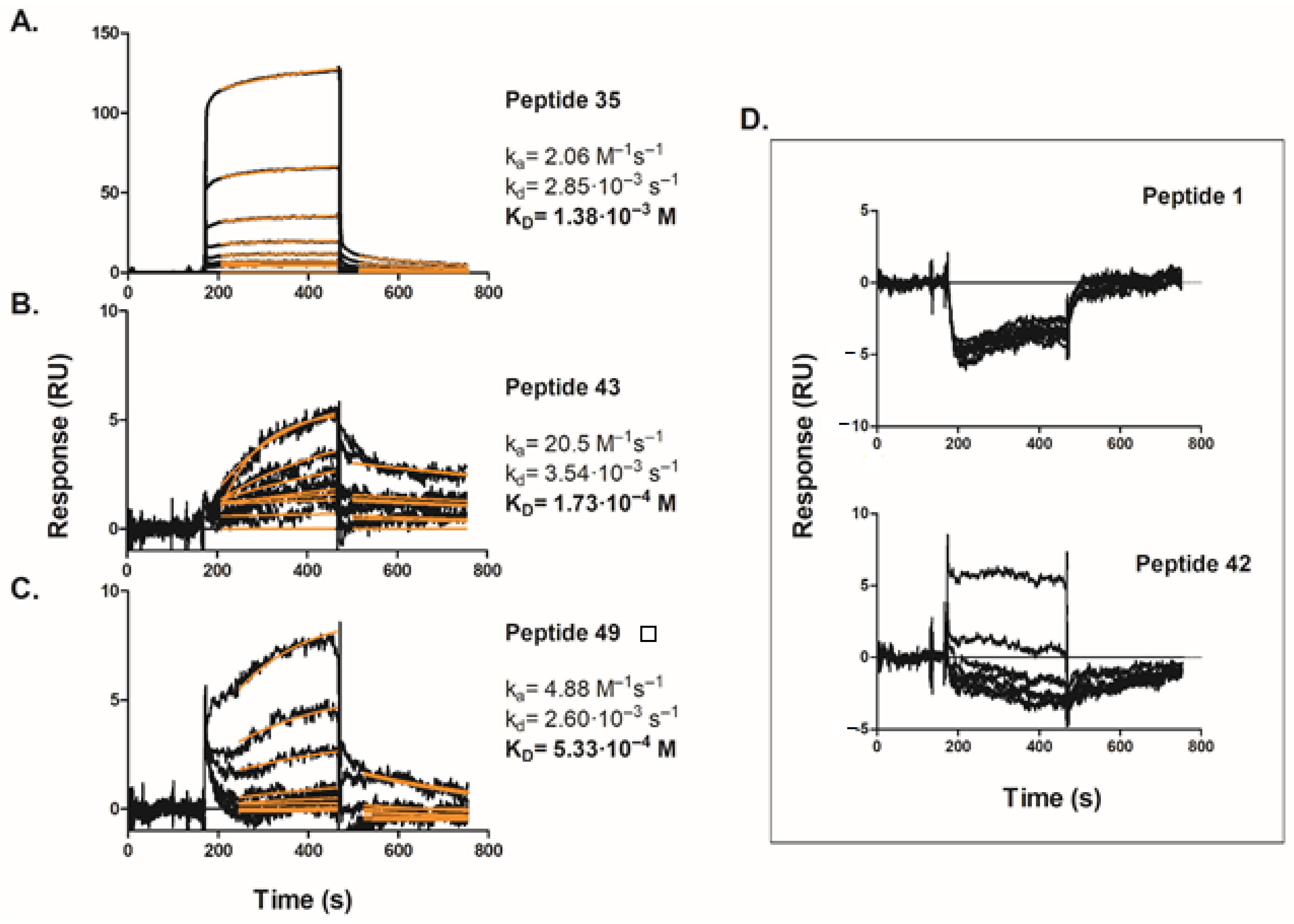
2.5. In Vitro Peptide Screening by Surface Plasmon Resonance
3. Discussion
4. Materials and Methods
4.1. Computational Design, Ranking and Selection of Peptides Mimicking the Interaction of a Neutralizing Antibody with TWEAK
4.2. Peptide Synthesis and Characterization
4.3. Cell Lines
4.4. Cell Culture Conditions and Treatments for Screening Peptide Activities
4.5. Transcriptomic Analysis
4.6. Surface Plasmon Resonance (SPR) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SPR | Surface plasmon resonance |
| TWEAK | Tumor necrosis factor-like WEAK inducer of apoptosis |
| Fn14 | Fibroblast growth factor-inducible 14 |
| MW | Molecular weight |
References
- Wells, J.A.; McClendon, C.L. Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nat. Cell Biol. 2007, 450, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Mullard, A. Protein–protein interaction inhibitors get into the groove. Nat. Rev. Drug Discov. 2012, 11, 173–175. [Google Scholar] [CrossRef] [PubMed]
- Valkov, E.; Sharpe, T.; Marsh, M.; Greive, S.; Hyvönen, M. Targeting Protein–Protein Interactions and Fragment-Based Drug Discovery. Topics Curr. Chem. 2011, 317, 145–179. [Google Scholar] [CrossRef]
- Cunningham, A.D.; Qvit, N.; Mochly-Rosen, D. Peptides and peptidomimetics as regulators of protein–protein interactions. Curr. Opin. Struct. Biol. 2017, 44, 59–66. [Google Scholar] [CrossRef]
- Lau, J.L.; Dunn, M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef]
- Hu, G.; Zeng, W.; Xia, Y. TWEAK/Fn14 signaling in tumors. Tumor Biol. 2017, 39, 1010428317714624. [Google Scholar] [CrossRef]
- Chicheportiche, Y.; Bourdon, P.R.; Xu, H.; Hsu, Y.-M.; Scott, H.S.; Hession, C.; Garcia, I.; Browning, J.L. TWEAK, a New Secreted Ligand in the Tumor Necrosis Factor Family That Weakly Induces Apoptosis. J. Biol. Chem. 1997, 272, 32401–32410. [Google Scholar] [CrossRef]
- Wiley, S.R.; Cassiano, L.; Lofton, T.; Davis-Smith, T.; Winkles, J.A.; Lindner, V.; Liu, H.; Daniel, T.O.; Smith, C.A.; Fanslow, W.C. A Novel TNF Receptor Family Member Binds TWEAK and Is Implicated in Angiogenesis. Immunity 2001, 15, 837–846. [Google Scholar] [CrossRef]
- Winkles, J.A. The TWEAK–Fn14 cytokine–receptor axis: Discovery, biology and therapeutic targeting. Nat. Rev. Drug Discov. 2008, 7, 411–425. [Google Scholar] [CrossRef]
- Martínez-Aranda, A.; Hernández, V.; Moreno, F.; Baixeras, N.; Cuadras, D.; Urruticoechea, A.; Gil-Gil, M.; Vidal, N.; Andreu, X.; Seguí, M.A.; et al. Predictive and Prognostic Brain Metastases Assessment in Luminal Breast Cancer Patients: FN14 and GRP94 from Diagnosis to Prophylaxis. Front. Oncol. 2017, 7, 283. [Google Scholar] [CrossRef]
- Culp, P.A.; Choi, D.; Zhang, Y.; Yin, J.; Seto, P.; Ybarra, S.E.; Su, M.; Sho, M.; Steinle, R.; Wong, M.H.; et al. Antibodies to TWEAK Receptor Inhibit Human Tumor Growth through Dual Mechanisms. Clin. Cancer Res. 2010, 16, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.; Pang, F.-M.; Li, D.; Zhang, L.; Wu, J.; Liu, Z.-Q.; Li, X.; Yan, H. Overexpression of Fn14 in gliomas: Tumor progression and poor prognosis. Futur. Oncol. 2018, 14, 1273–1284. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, F.; Wang, C.; Wang, C.; Tang, Y.; Jiang, Z. Src Promotes Metastasis of Human Non-Small Cell Lung Cancer Cells through Fn14-Mediated NF-κB Signaling. Med Sci. Monit. 2018, 24, 1282–1294. [Google Scholar] [CrossRef] [PubMed]
- Whitsett, T.G.; Mathews, I.T.; Cardone, M.H.; Lena, R.J.; Pierceall, W.E.; Bittner, M.; Sima, C.; LoBello, J.; Weiss, G.J.; Tran, N.L. Mcl-1 mediates TWEAK/Fn14-induced non-small cell lung cancer survival and therapeutic response. Mol. Cancer Res. 2014, 12, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Cheng, C.; Hu, G.; Wang, X.; Liu, J.; Yan, Z.; Zeng, W.; Xia, Y. TWEAK Promotes the Proliferation of Squamous Cell Carcinoma Cells Through Activating cIAP1 Signals. Front. Oncol. 2020, 10, 439. [Google Scholar] [CrossRef] [PubMed]
- Ebner, N.; Von Haehling, S. Unlocking the wasting enigma: Highlights from the 8th Cachexia Conference. J. Cachex. Sarcopenia Muscle 2016, 7, 90–94. [Google Scholar] [CrossRef]
- Johnston, A.J.; Murphy, K.T.; Jenkinson, L.; Laine, D.; Emmrich, K.; Faou, P.; Weston, R.; Jayatilleke, K.M.; Schloegel, J.; Talbo, G.H.; et al. Targeting of Fn14 Prevents Cancer-Induced Cachexia and Prolongs Survival. Cell 2015, 162, 1365–1378. [Google Scholar] [CrossRef]
- Boulamery, A.; Desplat-Jégo, S. Regulation of Neuroinflammation: What Role for the Tumor Necrosis Factor-Like Weak Inducer of Apoptosis/Fn14 Pathway? Front. Immunol. 2017, 8, 1534. [Google Scholar] [CrossRef]
- Roos, A.; Dhruv, H.D.; Mathews, I.T.; Inge, L.J.; Tuncali, S.; Hartman, L.K.; Chow, D.; Millard, N.; Yin, H.H.; Kloss, J.; et al. Identification of aurintricarboxylic acid as a selective inhibitor of the TWEAK-Fn14 signaling pathway in glioblastoma cells. Oncotarget 2017, 8, 12234–12246. [Google Scholar] [CrossRef]
- Zhang, X.; Winkles, J.A.; Gongora, M.C.; Polavarapu, R.; Michaelson, J.S.; Hahm, K.; Burkly, L.; Friedman, M.; Li, X.-J.; Yepes, M. TWEAK—Fn14 Pathway Inhibition Protects the Integrity of the Neurovascular Unit during Cerebral Ischemia. Br. J. Pharmacol. 2006, 27, 534–544. [Google Scholar] [CrossRef]
- Chopra, M.; Brandl, A.; Siegmund, D.; Mottok, A.; Schäfer, V.; Biehl, M.; Kraus, S.; Bäuerlein, C.A.; Ritz, M.; Mattenheimer, K.; et al. Blocking TWEAK-Fn14 interaction inhibits hematopoietic stem cell transplantation-induced intestinal cell death and reduces GVHD. Blood 2015, 126, 437–444. [Google Scholar] [CrossRef]
- Martínez-Aranda, A.; Hernández, V.; Guney, E.; Muixí, L.; Foj, R.; Baixeras, N.; Cuadras-Pallejà, D.; Moreno, V.; Urruticoechea, A.; Gil, M.; et al. FN14 and GRP94 expression are prognostic/predictive biomarkers of brain metastasis outcome that open up new therapeutic strategies. Oncotarget 2015, 6, 44254–44273. [Google Scholar] [CrossRef]
- Oliva, B.; Fernandez-Fuentes, N. Knowledge-based modeling of peptides at protein interfaces: PiPreD. Bioinformatics 2014, 31, 1405–1410. [Google Scholar] [CrossRef][Green Version]
- Lammens, A.; Baehner, M.; Kohnert, U.; Niewoehner, J.; Von Proff, L.; Schraeml, M.; Lammens, K.; Hopfner, K.-P. Crystal Structure of Human TWEAK in Complex with the Fab Fragment of a Neutralizing Antibody Reveals Insights into Receptor Binding. PLoS ONE 2013, 8, e62697. [Google Scholar] [CrossRef] [PubMed]
- Assi, S.A.; Tanaka, T.; Rabbitts, T.H.; Fernandez-Fuentes, N. PCRPi: Presaging Critical Residues in Protein interfaces, a new computational tool to chart hot spots in protein interfaces. Nucleic Acids Res. 2009, 38, e86. [Google Scholar] [CrossRef] [PubMed]
- Vucic, D. The Role of Ubiquitination in TWEAK-Stimulated Signaling. Front. Immunol. 2013, 4, 472. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef] [PubMed]
- Raju, R.; Nanjappa, V.; Balakrishnan, L.; Radhakrishnan, A.; Thomas, J.K.; Sharma, J.; Tian, M.; Palapetta, S.M.; Subbannayya, T.; Sekhar, N.R.; et al. NetSlim: High-confidence curated signaling maps. Database 2011, 2011, bar032. [Google Scholar] [CrossRef]
- Bhattacharjee, M.; Raju, R.; Radhakrishnan, A.; Nanjappa, V.; Muthusamy, B.; Singh, K.; Kuppusamy, D.; Lingala, B.T.; Pan, A.; Mathur, P.P.; et al. A Bioinformatics Resource for TWEAK-Fn14 Signaling Pathway. J. Signal Transduct. 2012, 2012, 1–10. [Google Scholar] [CrossRef]
- Kimple, A.J.; Muller, R.E.; Siderovski, D.P.; Willard, F.S. A Capture Coupling Method for the Covalent Immobilization of Hexahistidine Tagged Proteins for Surface Plasmon Resonance. Comput. Biol. 2010, 627, 91–100. [Google Scholar] [CrossRef]
- Day, E.S.; Cote, S.M.; Whitty, A. Binding Efficiency of Protein–Protein Complexes. Biochemistry 2012, 51, 9124–9136. [Google Scholar] [CrossRef] [PubMed]
- Preusser, M.; Winkler, F.; Valiente, M.; Manegold, C.; Moyal, E.; Widhalm, G.; Tonn, J.-C.; Zielinski, C. Recent advances in the biology and treatment of brain metastases of non-small cell lung cancer: Summary of a multidisciplinary roundtable discussion. ESMO Open 2018, 3, e000262. [Google Scholar] [CrossRef]
- Sood, V.D.; Baker, D. Recapitulation and Design of Protein Binding Peptide Structures and Sequences. J. Mol. Biol. 2006, 357, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Kortemme, T.; Baker, D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl. Acad. Sci. USA 2002, 99, 14116–14121. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Peinado, C.; Defaus, S.; Sans-Comerma, L.; Valle, J.; Andreu, D. Decoding the human serum interactome of snake-derived antimicrobial peptide Ctn[15-34]: Toward an explanation for unusually long half-life. J. Proteom. 2019, 204, 103372. [Google Scholar] [CrossRef]
- Martínez-Aranda, A.; Hernández, V.; Picón, C.; Modolell, I.; Sierra, A. Development of a Preclinical Therapeutic Model of Human Brain Metastasis with Chemoradiotherapy. Int. J. Mol. Sci. 2013, 14, 8306–8327. [Google Scholar] [CrossRef]
- Perez, J.G.; Tran, N.L.; Rosenblum, M.G.; Schneider, C.S.; Connolly, N.P.; Kim, A.J.; Woodworth, G.F.; Winkles, J.A. The TWEAK receptor Fn14 is a potential cell surface portal for targeted delivery of glioblastoma therapeutics. Oncogene 2016, 35, 2145–2155. [Google Scholar] [CrossRef]
- Schackert, G.; Price, J.E.; Bucana, C.D.; Fidler, I.J. Unique patterns of brain metastasis produced by different human carcinomas in athymic nude mice. Int. J. Cancer 1989, 44, 892–897. [Google Scholar] [CrossRef]
- Chambers, A.F. MDA-MB-435 and M14 Cell Lines: Identical but not M14 Melanoma? Cancer Res. 2009, 69, 5292–5293. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badia-Villanueva, M.; Defaus, S.; Foj, R.; Andreu, D.; Oliva, B.; Sierra, A.; Fernandez-Fuentes, N. Evaluation of Computationally Designed Peptides against TWEAK, a Cytokine of the Tumour Necrosis Factor Ligand Family. Int. J. Mol. Sci. 2021, 22, 1066. https://doi.org/10.3390/ijms22031066
Badia-Villanueva M, Defaus S, Foj R, Andreu D, Oliva B, Sierra A, Fernandez-Fuentes N. Evaluation of Computationally Designed Peptides against TWEAK, a Cytokine of the Tumour Necrosis Factor Ligand Family. International Journal of Molecular Sciences. 2021; 22(3):1066. https://doi.org/10.3390/ijms22031066
Chicago/Turabian StyleBadia-Villanueva, Miriam, Sira Defaus, Ruben Foj, David Andreu, Baldo Oliva, Angels Sierra, and Narcis Fernandez-Fuentes. 2021. "Evaluation of Computationally Designed Peptides against TWEAK, a Cytokine of the Tumour Necrosis Factor Ligand Family" International Journal of Molecular Sciences 22, no. 3: 1066. https://doi.org/10.3390/ijms22031066
APA StyleBadia-Villanueva, M., Defaus, S., Foj, R., Andreu, D., Oliva, B., Sierra, A., & Fernandez-Fuentes, N. (2021). Evaluation of Computationally Designed Peptides against TWEAK, a Cytokine of the Tumour Necrosis Factor Ligand Family. International Journal of Molecular Sciences, 22(3), 1066. https://doi.org/10.3390/ijms22031066





