A Supramolecular Assembly of Hemoproteins Formed in a Star-Shaped Structure via Heme–Heme Pocket Interactions
Abstract
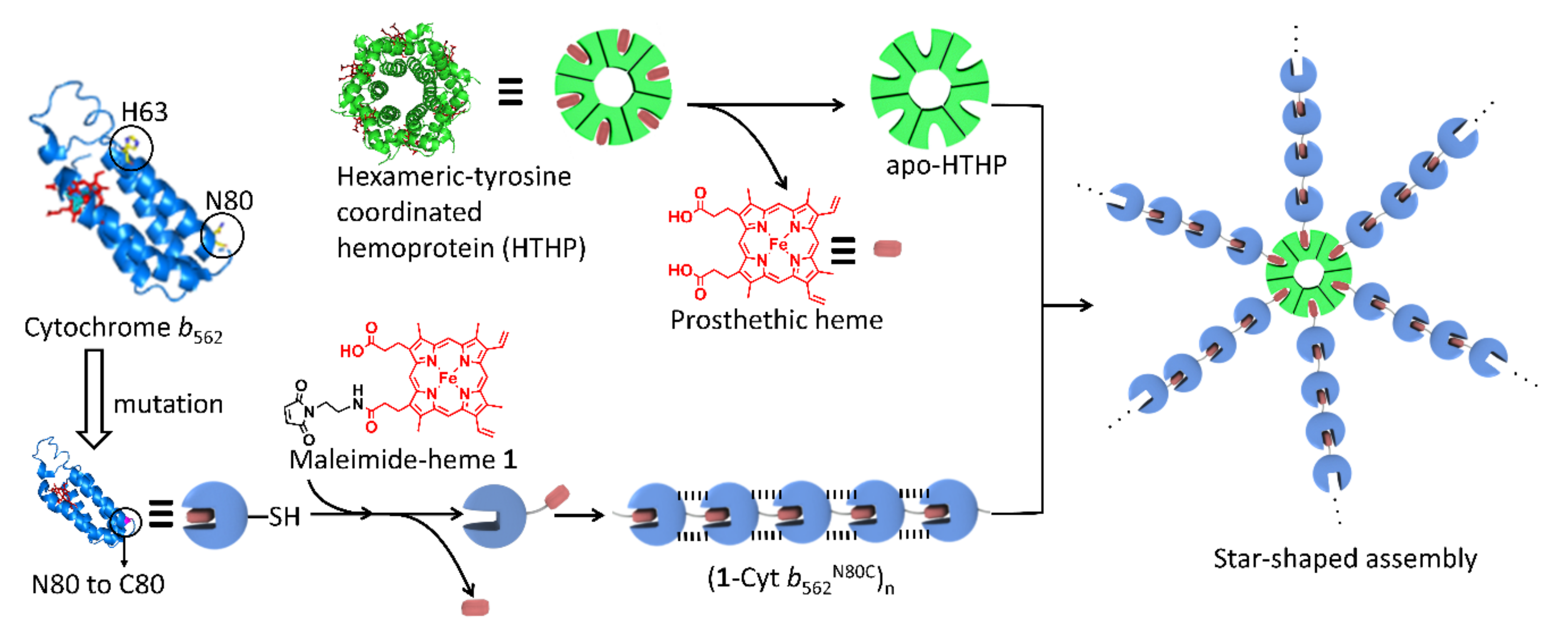
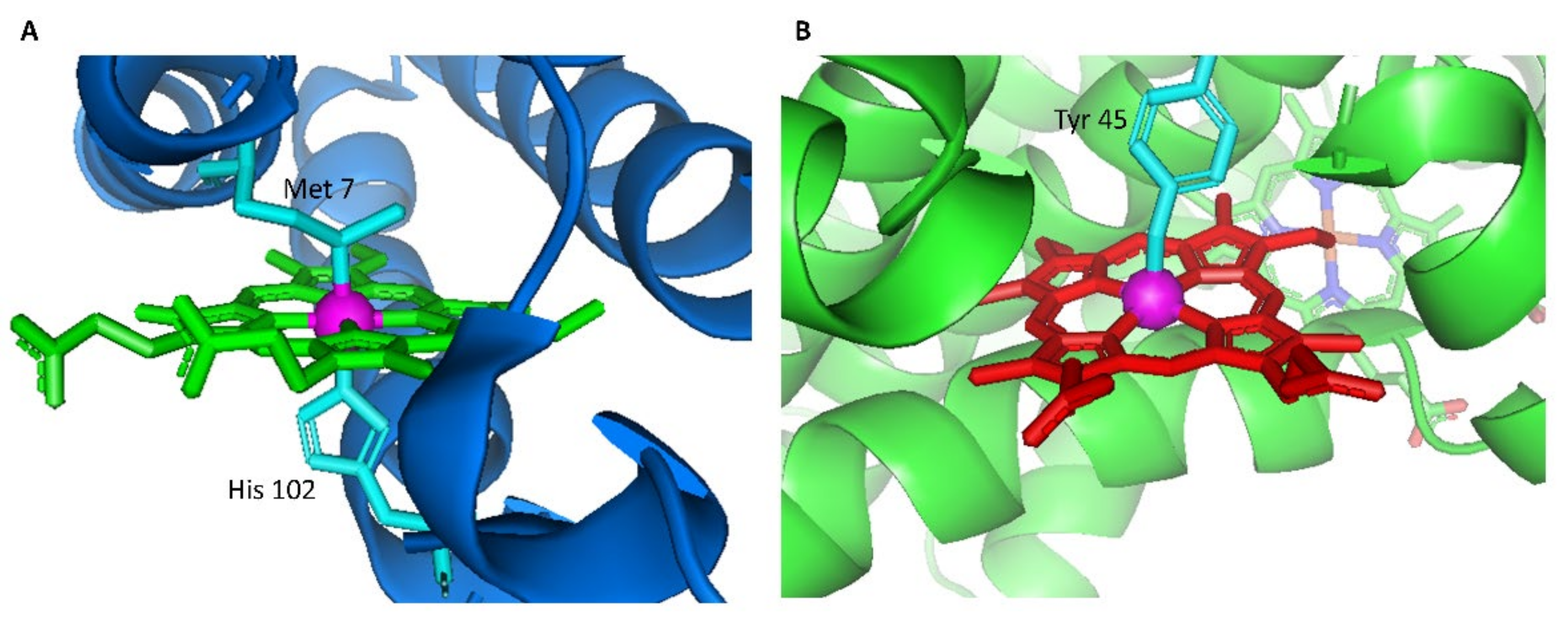
1. Introduction
2. Results and Discussion
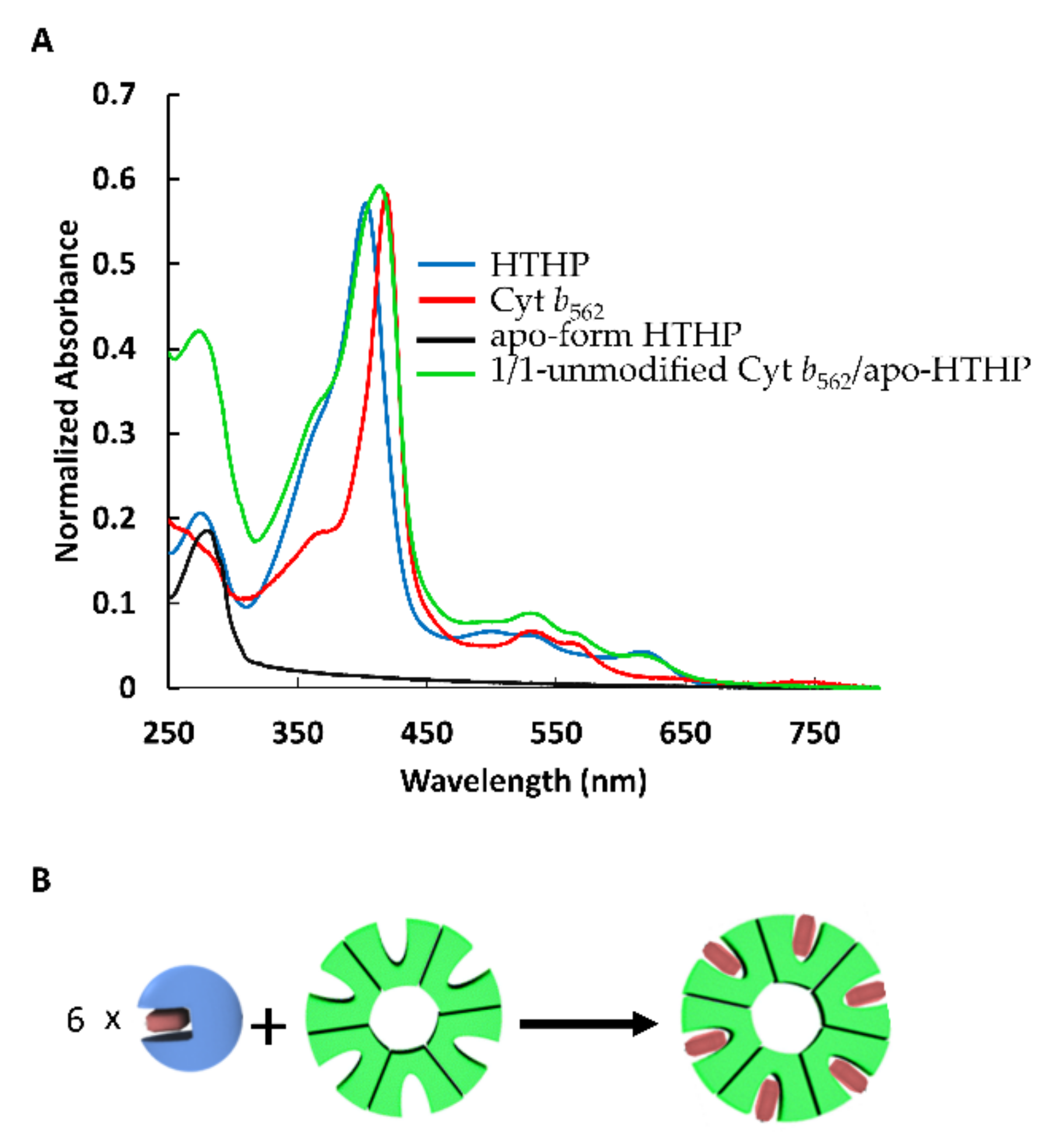
2.1. Heme Transfer from Unmodified Cytochrome b562 to apoHTHP
2.2. Assembly of Modified Cytochrome b562 with apo-form of HTHP
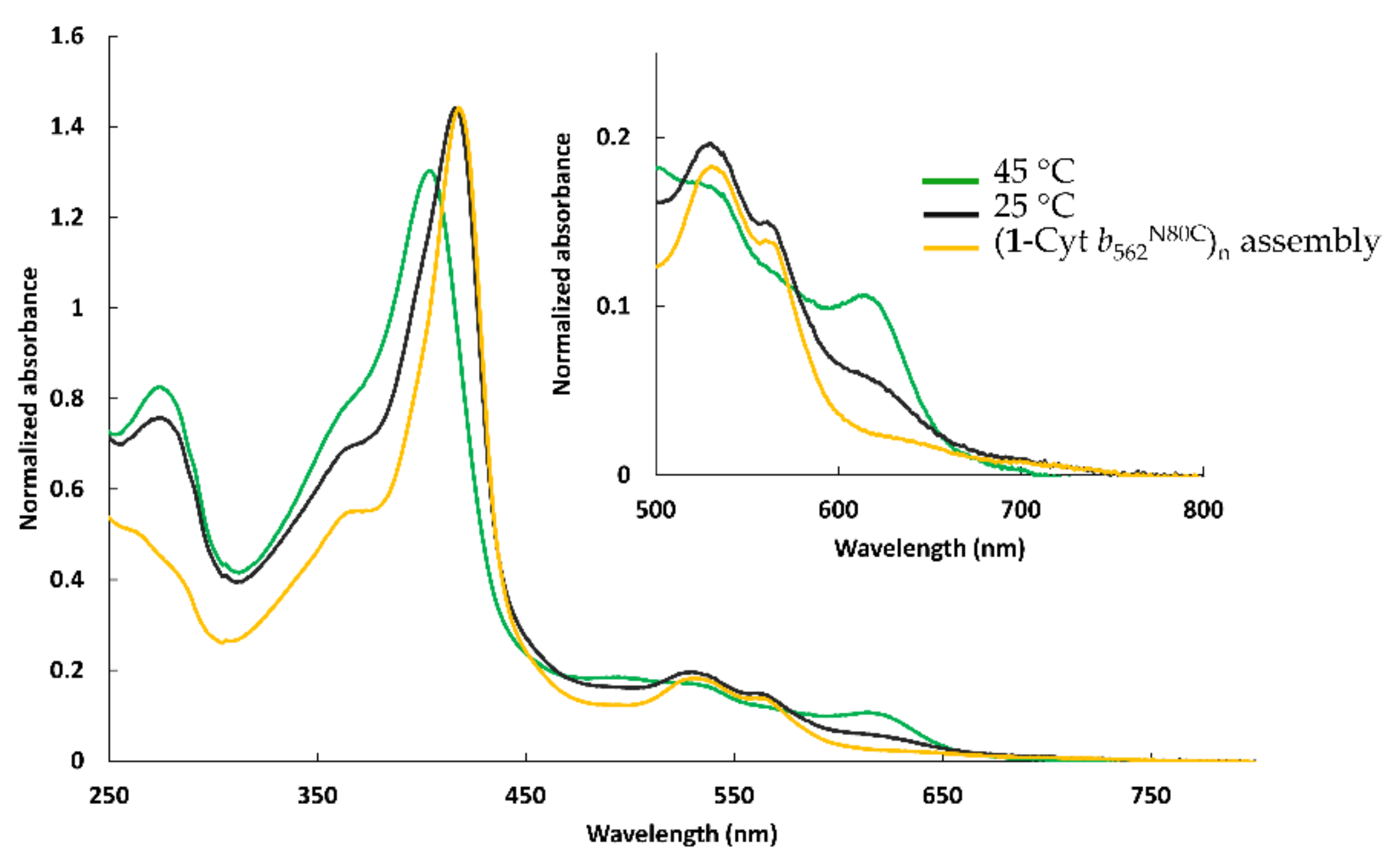
2.2.1. Assembly of (1-Cyt b562N80C)n with apoHTHP
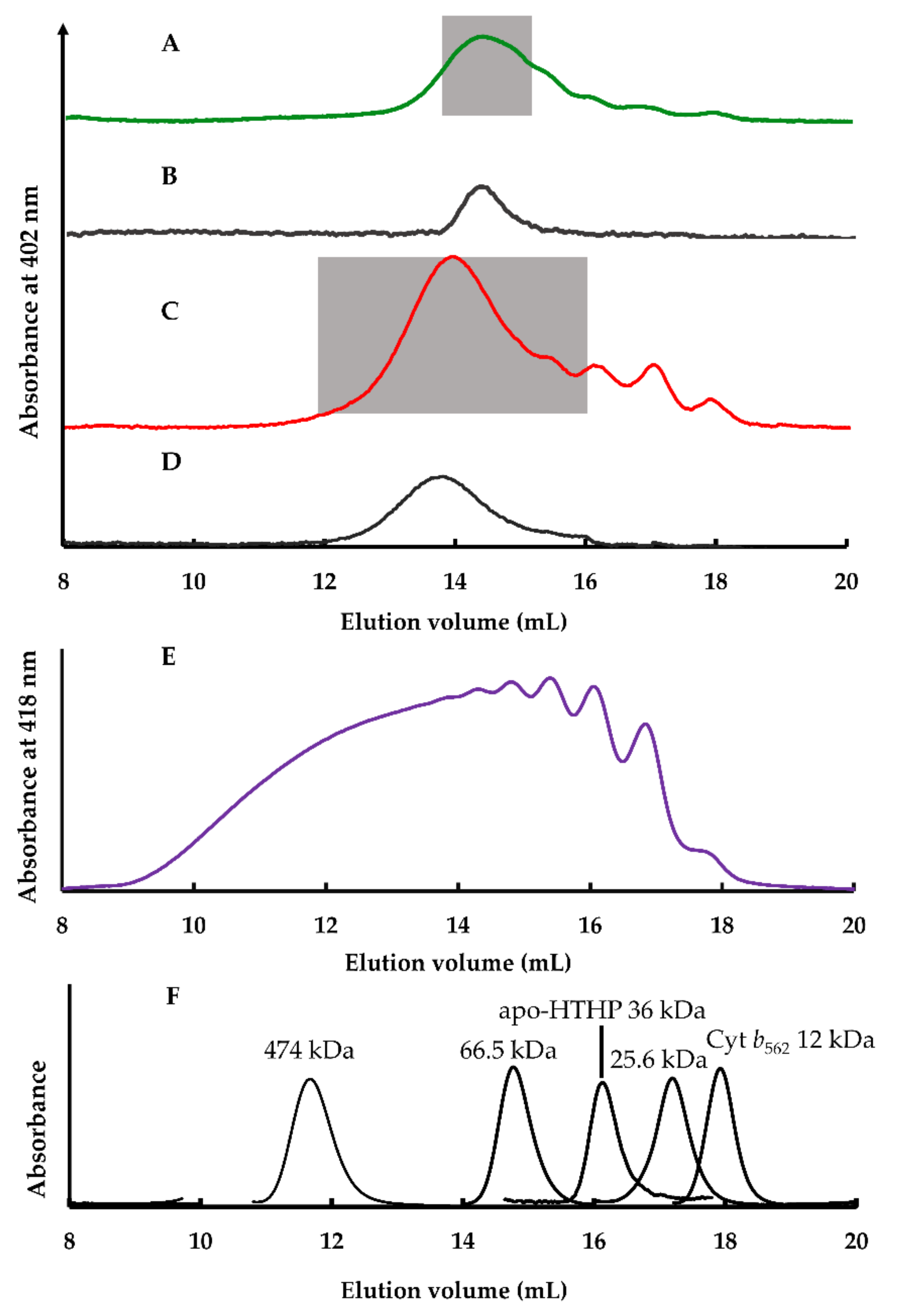
2.2.2. Size Exclusion Chromatography Analysis
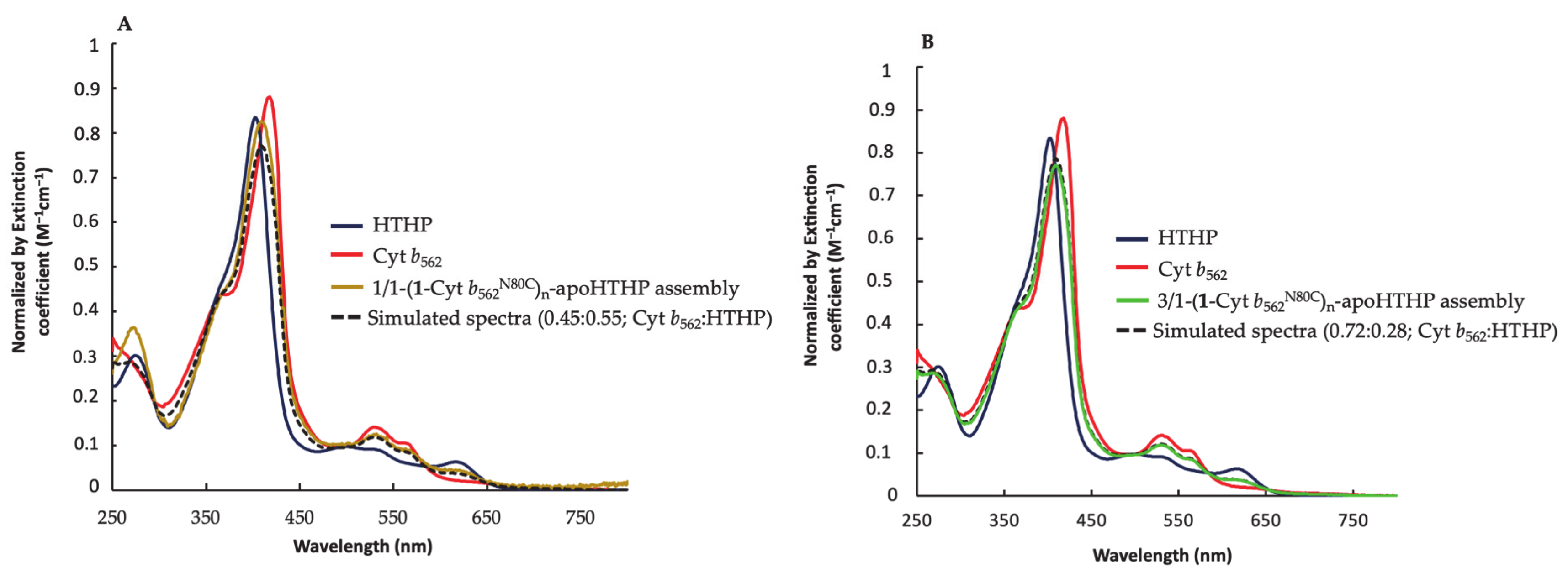
2.2.3. UV-Vis Spectra of Fractionated Assemblies
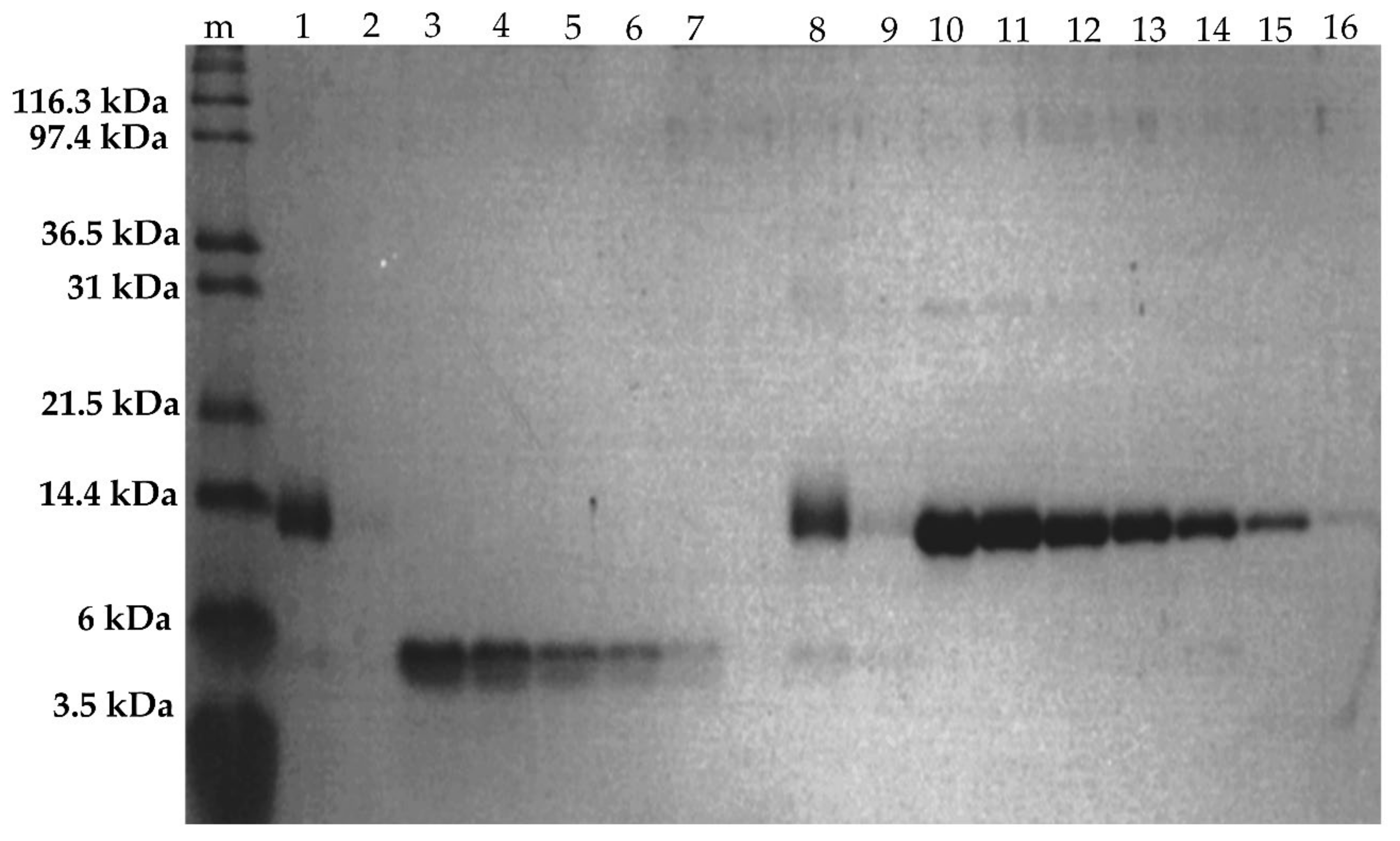
2.2.4. SDS-PAGE of Fractionated Assemblies
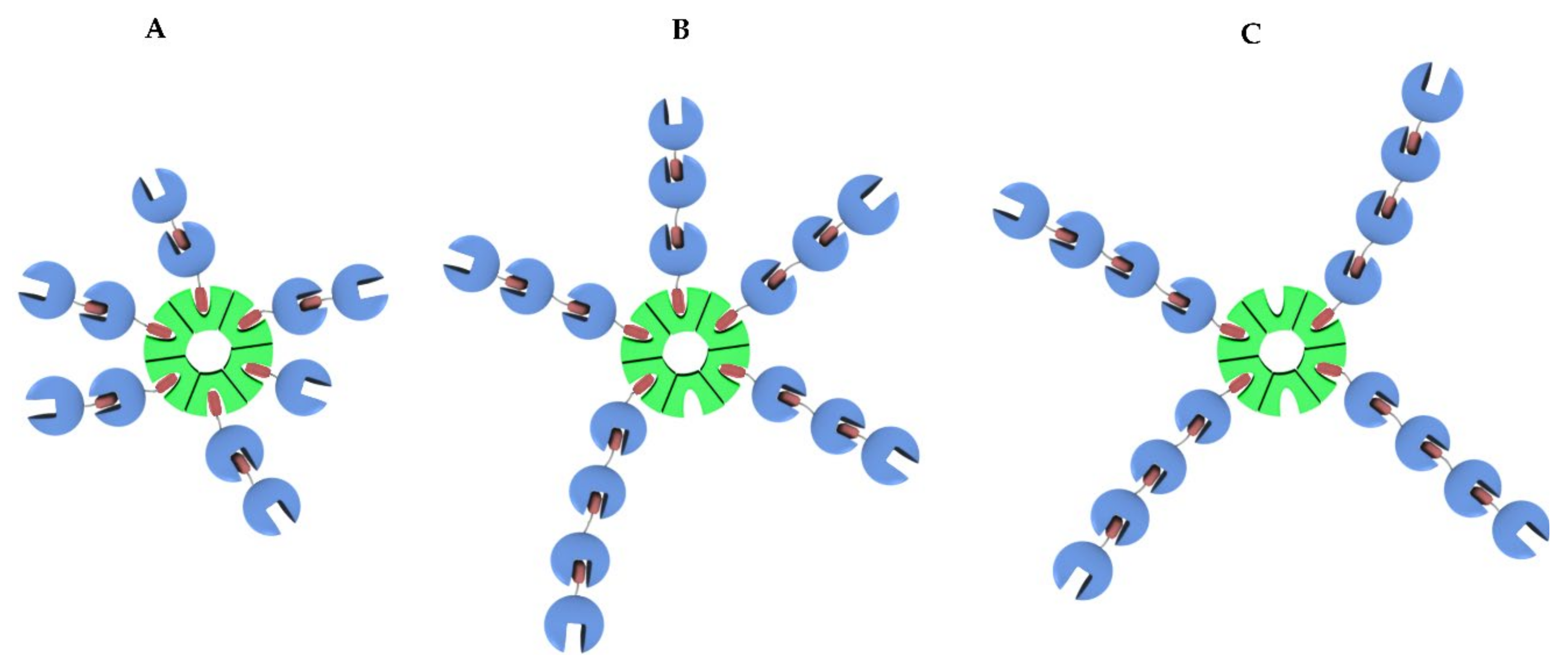
2.2.5. Estimation of Apparent Structures of (1-Cyt b562N80C)n-apoHTHP Assemblies
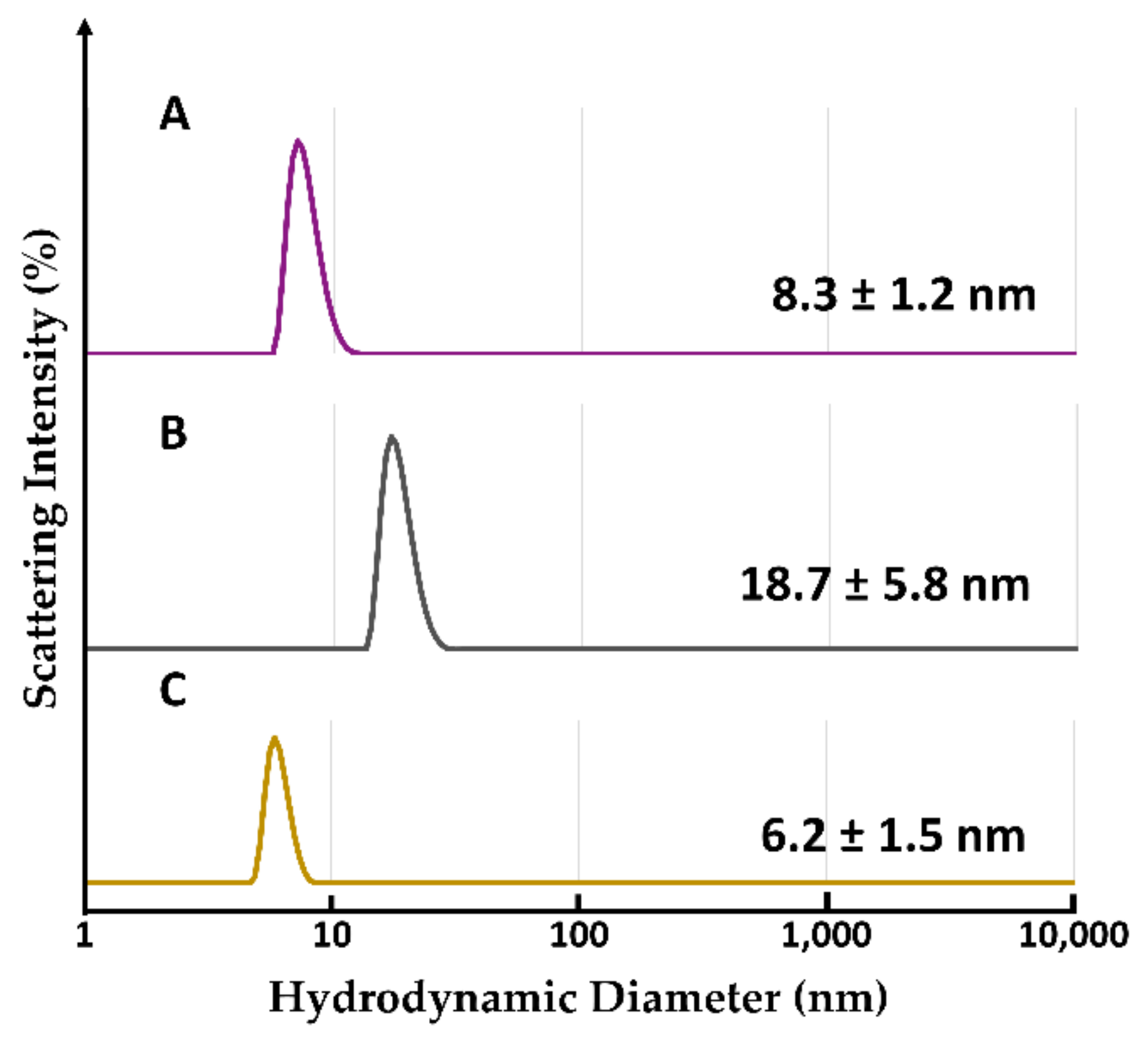
2.2.6. Evaluation of Hydrodynamic Diameter by Dynamic Light Scattering Analysis
3. Materials and Methods
3.1. Instruments and Materials
3.2. Preparation of Unmodified Cyt b562 with apoHTHP
3.3. Preparation of Assembly by apoHTHP and Equimolar (1-Cyt b562N80C)n
3.4. Preparation of Assembly Produced by apoHTHP and the Three Equivalent (1-Cyt b562N80C)n
3.5. SEC Analyses, Sample Preparations and SEC Fractionation
3.6. SDS-PAGE
3.7. Protein Quantification by Image Analysis of SDS-PAGE
3.8. Hydrodynamic Diameter by DLS Measurement
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| PDB | Protein Data Bank |
| TEM | Transmission electron microscopy |
| AFM | Atomic force microscopy |
References
- Marsh, J.A.; Hernandez, H.; Hall, Z.; Anhert, S.E.; Perica, T.; Robinson, C.V.; Teichmann, S.A. Protein Complexes Are Under Evolutionary Selection to Assemble via Ordered Pathways. Cell 2013, 153, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K. Synthetic approaches to construct viral capsid-like spherical nanomaterials. Chem. Commun. 2018, 54, 8944–8959. [Google Scholar] [CrossRef] [PubMed]
- Inaba, H.; Matsuura, K. Peptide Nanomaterials Designed from Natural Supramolecular Systems. Chem. Rec. 2019, 19, 843–858. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Luo, Q.; Hou, C.; Liu, J. Nanostructures based on protein self-assembly: From hierarchical construction to bioinspired materials. Nano Today 2017, 14, 16–41. [Google Scholar] [CrossRef]
- Yeates, T.O.; Padilla, J.E. Designing supramolecular protein assemblies. Curr. Opin. Struct. Biol. 2002, 12, 464–470. [Google Scholar] [CrossRef]
- Malay, A.D.; Miyazaki, N.; Biela, A.; Chakraborti, S.; Majsterkiewicz, K.; Stupka, I.; Kaplan, C.S.; Kowalczyk, A.; Piette, B.M.A.G.; Hochberg, G.K.A.; et al. An ultra-stable gold- coordinated protein cage displaying reversible assembly. Nature 2019, 569, 438–442. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Dang, D.T.; van Dongen, J.L.J.; Brunsvel, L. Protein Dimerization Induced by Supramolecular Interactions with Cucurbit[8]uril. Angew. Che. Int. Ed. 2010, 49, 895–898. [Google Scholar] [CrossRef]
- Carlson, J.C.T.; Jena, S.S.; Flenniken, M.; Chou, T.; Siegel, R.A.; Wagner, C.R. Chemically Controlled Self-Assembly of Protein Nanorings. J. Am. Chem. Soc. 2006, 128, 7630–7638. [Google Scholar] [CrossRef]
- Brodin, J.D.; Ambroggio, X.I.; Tang, C.; Parent, K.N.; Baker, T.S.; Tezcan, F.A. Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays. Nat. Chem. 2012, 4, 375–382. [Google Scholar] [CrossRef]
- Bastings, M.M.C.; de Greef, T.F.A.; van Dongen, J.L.J.; Merkx, M.; Meijer, E.W. Macrocyclization of enzyme-based supramolecular polymers. Chem. Sci. 2010, 1, 79–88. [Google Scholar] [CrossRef]
- Ringler, P.; Schulz, G.E. Self-Assembly of Proteins into Designed Networks. Science 2003, 302, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.K.; Negishi, H.; Abe, S.; Ueno, T. Construction of supramolecular nanotubes from protein crystals. Chem. Sci. 2019, 10, 1046–1051. [Google Scholar] [CrossRef] [PubMed]
- Kitagishi, H.; Oohora, K.; Yamaguchi, H.; Sato, H.; Matsuo, T.; Harada, A.; Hayashi, T. Supramolecular Hemoprotein Linear Assembly by Successive Interprotein Heme-Heme Pocket Interactions. J. Am. Chem. Soc. 2007, 129, 10326–10327. [Google Scholar] [CrossRef] [PubMed]
- Onoda, A.; Takahashi, A.; Oohora, K.; Onuma, Y.; Hayashi, T. Fibrous Supramolecular Hemoprotein Assemblies Connected with Synthetic Heme Dimer and Apohemoprotein Dimer. Chem. Biodivers. 2012, 9, 1684–1692. [Google Scholar] [CrossRef] [PubMed]
- Kitagishi, H.; Kakikura, Y.; Yamaguchi, H.; Oohora, K.; Harada, A.; Hayashi, T. Self-Assembly of One- and Two-Dimensional Hemoprotein Systems by Polymerization through Heme-Heme Pocket Interactions. Angew. Chem. Int. Ed. 2009, 48, 1271–1274. [Google Scholar] [CrossRef] [PubMed]
- Oohora, K.; Hayashi, T. Hemoprotein-based supramolecular assembling systems. Curr. Opin. Chem. Biol. 2014, 19, 154–161. [Google Scholar] [CrossRef]
- Oohora, K.; Kajihara, R.; Fujimaki, N.; Uchihashi, T.; Hayashi, T. A ring-shaped hemoprotein trimer thermodynamically controlled by the supramolecular heme-heme pocket interaction. Chem. Commun. 2019, 55, 1544–1547. [Google Scholar] [CrossRef]
- Oohora, K.; Fujimaki, N.; Kajihara, R.; Watanabe, H.; Uchihashi, T.; Hayashi, T. Supramolecular Hemoprotein Assembly with a Periodic Structure Showing Heme-Heme Exciton Coupling. J. Am. Chem. Soc. 2018, 140, 10145–10148. [Google Scholar] [CrossRef]
- Hirayama, S.; Oohora, K.; Uchihashi, T.; Hayashi, T. Thermoresponsive Micellar Assembly Constructed from a Hexameric Hemoprotein Modified with Poly(N-isopropylacrylamide) toward an Artificial Light-Harvesting System. J. Am. Chem. Soc. 2020, 142, 1822–1831. [Google Scholar] [CrossRef]
- Oohora, K.; Hirayama, S.; Uchihashi, T.; Hayashi, T. Construction of a Hexameric Hemoprotein Sheet and Direct Observation of Dynamic Process of its Formation. Chem. Lett. 2020, 49, 186–190. [Google Scholar] [CrossRef]
- Oohora, K.; Hirayama, S.; Mashima, T.; Hayashi, T. Supramolecular dimerization of a hexameric hemoprotein via multiple pyrene-pyrene interactions. J. Porphyrins Phthalocyanine 2020, 24, 259–267. [Google Scholar] [CrossRef]
- Mashima, T.; Oohora, K.; Hayashi, T. Successive Energy Transfer within Multiple Photosensitizers Assembled in a Hexameric Hemoprotein Scaffold. Phys. Chem. Chem. Phys. 2018, 20, 3200–3209. [Google Scholar] [CrossRef] [PubMed]
- Oohora, K.; Mashima, T.; Ohkubo, K.; Fukuzumi, S.; Hayashi, T. Energy migration within hexameric hemoprotein reconstituted with Zn porphyrinoid molecules. Chem. Commun. 2015, 51, 11138–11140. [Google Scholar] [CrossRef] [PubMed]
- Itagaki, E.; Hager, L.P. Studies on Cytochrome b562 of Escherichia coli. J. Biol. Chem. 1966, 241, 3687–3695. [Google Scholar] [CrossRef]
- Jeoung, J.H.; Pippig, D.A.; Martins, B.M.; Wagener, N.; Dobbek, H. HTHP: A novel class of hexameric, tyrosine-coordinated heme proteins. J. Mol. Biol. 2007, 368, 1122–1131. [Google Scholar] [CrossRef]
- Mashima, T.; Oohora, K.; Hayashi, T. Substitution of amino acid residue axially coordinating to the heme molecule in hexameric tyrosine-coordinated hemoprotein to enhance peroxidase activity. J. Porphyrins Phthalocyanine 2017, 21, 823–831. [Google Scholar] [CrossRef]
- Robinson, C.R.; Liu, Y.; Thomson, J.A.; Sturtevant, J.M.; Sligar, S.G. Energetics of Heme Binding to Native and Denatured States of Cytochrome b562. Biochemistry 1997, 36, 16141–16146. [Google Scholar] [CrossRef]
- Hargrove, M.S.; Barrick, D.; Olson, J.S. The Association Rate Constant for Heme Binding to Globin Is Independent of Protein Structure. Biochemistry 1996, 35, 11293–11299. [Google Scholar] [CrossRef]
- Oohora, K.; Onuma, Y.; Tanaka, Y.; Onoda, A.; Hayashi, T. A supramolecular assembly based on an engineered hemoprotein exhibiting a thermal stimulus-driven conversion to a new distinct supramolecular structure. Chem. Commun. 2017, 53, 6879–6882. [Google Scholar] [CrossRef]
| Assembly | Components | Concentration as a Monomer (μM) |
|---|---|---|
| 1/1-(1-Cyt b562N80C)n-apoHTHP assembly | 1-Cyt b562N80C | 4.7 ± 0.19 |
| apoHTHP | 2.6 ± 0.44 | |
| 3/1-(1-Cyt b562N80C)n-apoHTHP assembly | 1-Cyt b562N80C | 8.3 ± 1.2 |
| apoHTHP | 3.2 ± 0.51 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soon, J.W.; Oohora, K.; Hirayama, S.; Hayashi, T. A Supramolecular Assembly of Hemoproteins Formed in a Star-Shaped Structure via Heme–Heme Pocket Interactions. Int. J. Mol. Sci. 2021, 22, 1012. https://doi.org/10.3390/ijms22031012
Soon JW, Oohora K, Hirayama S, Hayashi T. A Supramolecular Assembly of Hemoproteins Formed in a Star-Shaped Structure via Heme–Heme Pocket Interactions. International Journal of Molecular Sciences. 2021; 22(3):1012. https://doi.org/10.3390/ijms22031012
Chicago/Turabian StyleSoon, Julian Wong, Koji Oohora, Shota Hirayama, and Takashi Hayashi. 2021. "A Supramolecular Assembly of Hemoproteins Formed in a Star-Shaped Structure via Heme–Heme Pocket Interactions" International Journal of Molecular Sciences 22, no. 3: 1012. https://doi.org/10.3390/ijms22031012
APA StyleSoon, J. W., Oohora, K., Hirayama, S., & Hayashi, T. (2021). A Supramolecular Assembly of Hemoproteins Formed in a Star-Shaped Structure via Heme–Heme Pocket Interactions. International Journal of Molecular Sciences, 22(3), 1012. https://doi.org/10.3390/ijms22031012




