MEK6 Overexpression Exacerbates Fat Accumulation and Inflammatory Cytokines in High-Fat Diet-Induced Obesity
Abstract
:1. Introduction
2. Results
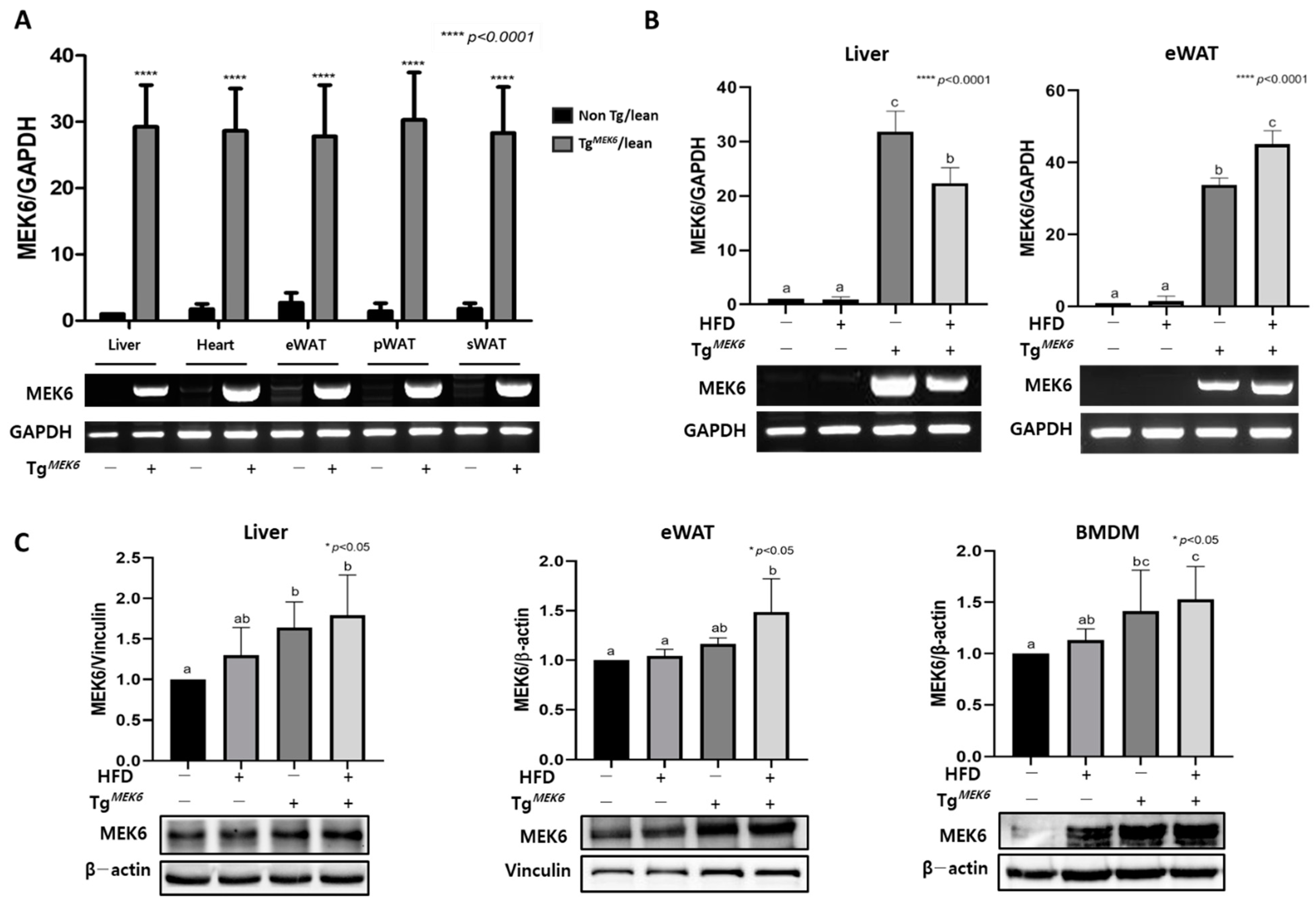
2.1. MEK6 Overexpression in Tissues of TgMEK6 Compared to Non-Tg
2.2. MEK6 Overexpression on Changes of Weight Gain and Dietary Intake
2.3. MEK6 Overexpression Alters Plasma Biochemistry
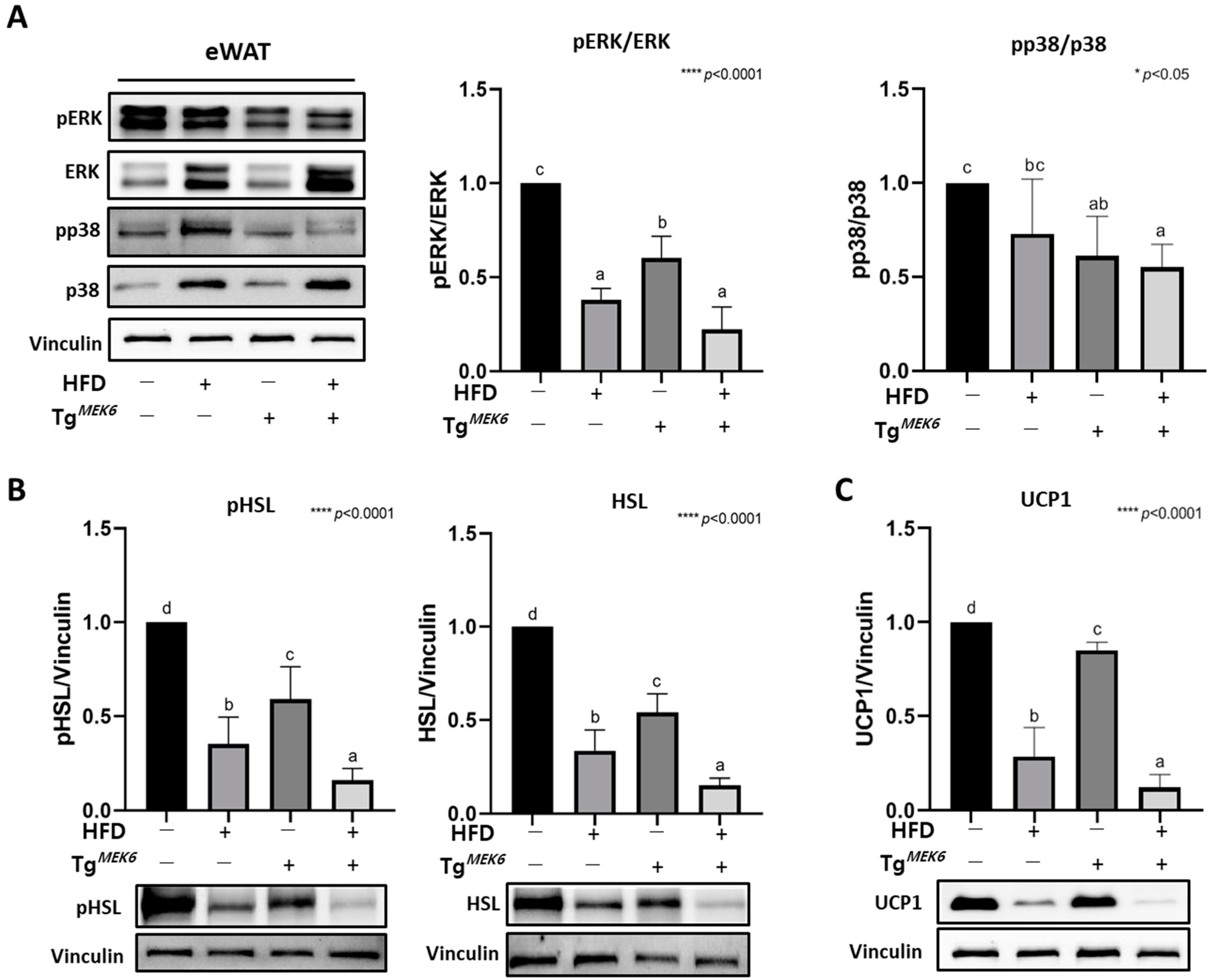
2.4. MEK6 Overexpression on Activation of p38/ERK Associated with Thermogenesis in WAT
2.5. MEK6 Overexpression Induces Adipocyte Hypertrophy and Adipogenesis in the WAT
2.6. MEK6 Overexpression Regulates PPARα, UCP2, and Activation of p38/ERK in the Liver
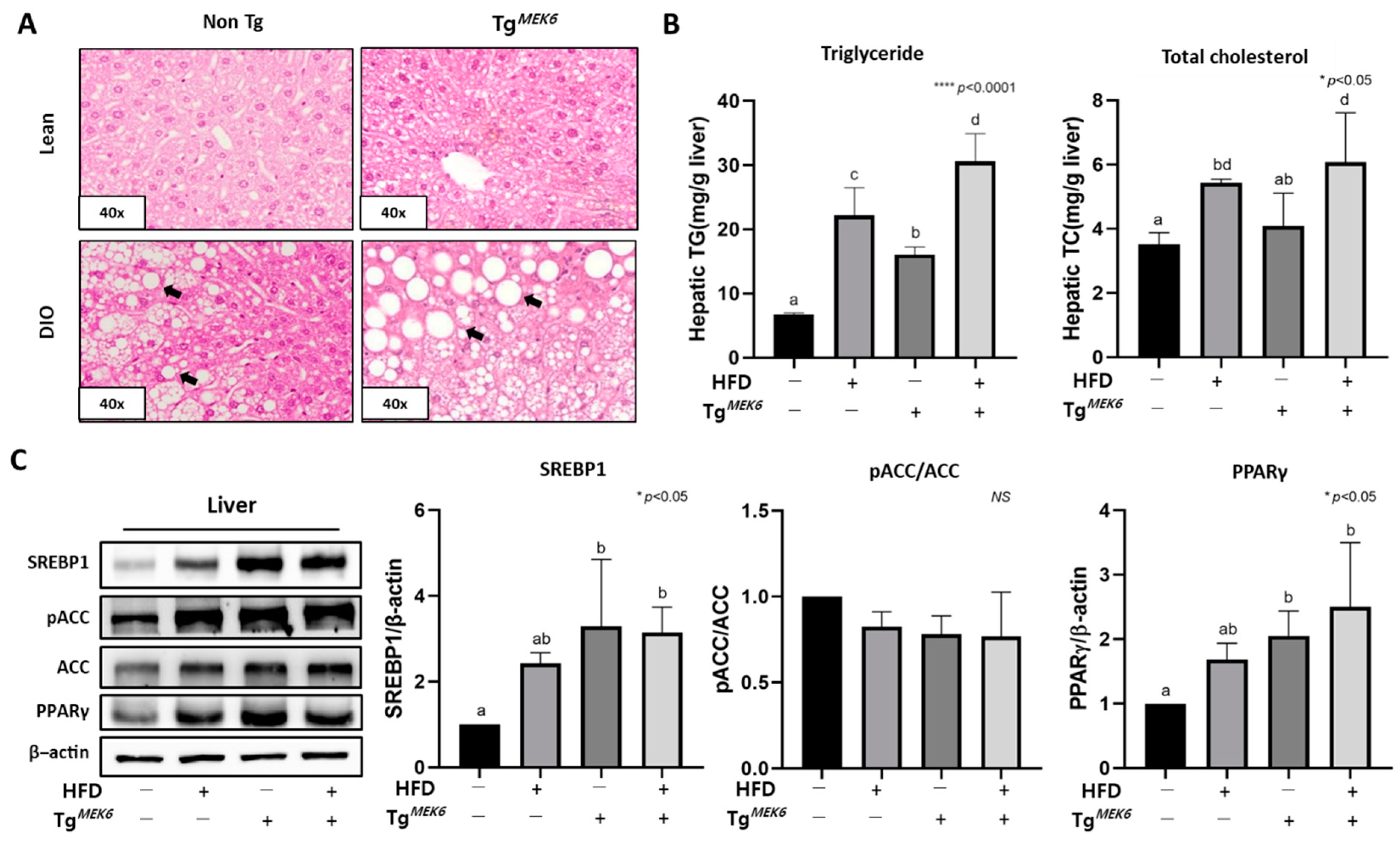
2.7. MEK6 Overexpression Regulates Fat Accumulation and Lipogenic Factors in the Liver
2.8. MEK6 Overexpression Regulates Adipokines in the WAT and Plasma
2.9. MEK6 Overexpression Regulates Cytokines in Bone-Marrow-Derived Macrophage
3. Discussion
4. Materials and Methods
4.1. Animal Design
4.2. MEK6 Overexpression and PCR Genotyping in C57BL/6N Mice
4.3. Histological Analysis of the Liver and Adipose Tissue
4.4. Bone-Marrow-Derived Macrophages (BMDM) Isolation and Differentiation
4.5. Biochemical Parameters
4.6. Preparation of Protein Extracts and Western Blot Analysis
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jehan, S.; Zizi, F.; Pandi-Perumal, S.R.; McFarlane, S.I.; Jean-Louis, G.; Myers, A.K. Energy imbalance: Obesity, associated comorbidities, prevention, management and public health implications. Adv. Obes. Weight. Manag. Control 2020, 10, 146–161. [Google Scholar] [CrossRef]
- Haslam, D.; James, W.P. Obesity. Lancet 2005, 366, 1197–1209. [Google Scholar] [CrossRef]
- Hill, J.O.; Wyatt, H.R.; Melanson, E.L. Genetic and environmental contributions to obesity. Med. Clin. N. Am. 2000, 84, 333–346. [Google Scholar] [CrossRef]
- WHO. Obesity: Preventing and Managing the Global Epidemic; World Health Organization technical report series; WHO: Geneva, Switzerland, 2000; Volume 894, pp. 1–253. [Google Scholar]
- Kivimäki, M.; Kuosma, E.; Ferrie, J.E.; Luukkonen, R.; Nyberg, S.T.; Alfredsson, L.; Batty, G.; Brunner, E.; Fransson, E.; Goldberg, M.; et al. Overweight, obesity, and risk of cardiometabolic multimorbidity: Pooled analysis of individual-level data for 120 813 adults from 16 cohort studies from the USA and Europe. Lancet Public Health 2017, 2, e277–e285. [Google Scholar] [CrossRef] [Green Version]
- Shariq, O.A.; McKenzie, T.J. Obesity-related hypertension: A review of pathophysiology, management, and the role of metabolic surgery. Gland. Surg. 2020, 9, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Mokdad, A.H.; Ford, E.S.; Bowman, B.A.; Dietz, W.H.; Vinicor, F.; Bales, V.S.; Marks, J.S. Prevalence of obesity, diabetes, and obesity-related health risk factors, 2001. JAMA 2003, 289, 76–79. [Google Scholar] [CrossRef]
- Calle, E.E.; Thun, M.J. Obesity and cancer. Oncogene 2004, 23, 6365–6378. [Google Scholar] [CrossRef] [PubMed]
- Stefan, N.; Birkenfeld, A.L.; Schulze, M.B.; Ludwig, D. Obesity and impaired metabolic health in patients with COVID-19. Nat. Rev. Endocrinol. 2020, 16, 341–342. [Google Scholar] [CrossRef] [Green Version]
- Dietz, W.; Santos-Burgoa, C.J.O. Obesity and its implications for COVID-19 mortality. Obesity (Silver Spring) 2020, 28, 1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, B.M.; Cheung, T.T.; Samaranayake, N.R. Safety of antiobesity drugs. Ther. Adv. Drug Saf. 2013, 4, 171–181. [Google Scholar] [CrossRef] [Green Version]
- Shang, A.; Gan, R.-Y.; Xu, X.-Y.; Mao, Q.-Q.; Zhang, P.-Z.; Li, H.-B. Effects and mechanisms of edible and medicinal plants on obesity: An updated review. Crit. Rev. Food Sci. Nutr. 2020, 61, 2061–2077. [Google Scholar] [CrossRef]
- Son, J.W.; Kim, S.J.D.; Journal, M. Comprehensive review of current and upcoming anti-obesity drugs. Diabetes Metab. J. 2020, 44, 802–818. [Google Scholar] [CrossRef]
- Lee, M.; Lee, Y.; Kang, I.; Shin, J.; Sorn, S.R. RMR-Related MAP2K6 Gene Variation on the Risk of Overweight/Obesity in Children: A 3-Year Panel Study. J. Pers. Med. 2021, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Roux, P.P.; Blenis, J. ERK and p38 MAPK-activated protein kinases: A family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 2004, 68, 320–344. [Google Scholar] [CrossRef] [Green Version]
- Rahman, S.M.T.; Zhou, W.; Deiters, A.; Haugh, J.M. Optical control of MAP kinase kinase 6 (MKK6) reveals that it has divergent roles in pro-apoptotic and anti-proliferative signaling. J. Biol. Chem. 2020, 295, 8494–8504. [Google Scholar] [CrossRef]
- Tanaka, N.; Kamanaka, M.; Enslen, H.; Dong, C.; Wysk, M.; Davis, R.J.; Flavell, R.A. Differential involvement of p38 mitogen-activated protein kinase kinases MKK3 and MKK6 in T-cell apoptosis. EMBO Rep. 2002, 3, 785–791. [Google Scholar] [CrossRef] [Green Version]
- Dhillon, A.S.; Hagan, S.; Rath, O.; Kolch, W. MAP kinase signalling pathways in cancer. Oncogene 2007, 26, 3279–3290. [Google Scholar] [CrossRef] [Green Version]
- Wagner, E.F.; Nebreda, R. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer 2009, 9, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Enslen, H.; Brancho, D.M.; Davis, R.J. Molecular determinants that mediate selective activation of p38 MAP kinase isoforms. EMBO J. 2000, 19, 1301–1311. [Google Scholar] [CrossRef] [Green Version]
- Enslen, H.; Raingeaud, J.; Davis, R.J. Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by the MAP kinase kinases MKK3 and MKK6. J. Biol. Chem. 1998, 273, 1741–1748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Li, R.; Meng, Y.; Li, S.; Donelan, W.; Zhao, Y.; Qi, L.; Zhang, M.; Wang, X.; Cui, T.; et al. Irisin stimulates browning of white adipocytes through mitogen-activated protein kinase p38 MAP kinase and ERK MAP kinase signaling. Diabetes 2013, 63, 514–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matesanz, N.; Bernardo, E.; Acin-Perez, R.; Manieri, E.; Pérez-Sieira, S.; Cosido, L.H.; Montalvo-Romeral, V.; Mora, A.; Rodríguez, E.; Leiva-Vega, L.; et al. MKK6 controls T3-mediated browning of white adipose tissue. Nat. Commun. 2017, 8, 856. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Lee, M. Beiging Modulates Inflammatory Adipogenesis in Salt-Treated and MEK6–Transfected Adipocytes. Cells 2021, 10, 1106. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Eo, H.; Park, K.; Jin, M.; Park, E.-J.; Kim, S.-H.; Park, J.E.; Kim, S. A water-soluble extract from Cucurbita moschata shows anti-obesity effects by controlling lipid metabolism in a high fat diet-induced obesity mouse model. Biochem. Biophys. Res. Commun. 2007, 359, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Kopelman, P.G. Obesity as a medical problem. Nature 2000, 404, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Maury, E.; Brichard, S. Adipokine dysregulation, adipose tissue inflammation and metabolic syndrome. Mol. Cell. Endocrinol. 2010, 314, 1–16. [Google Scholar] [CrossRef]
- Elam, M.B.; Cowan, G.S.; Rooney, R.J.; Hiler, M.L.; Yellaturu, C.R.; Deng, X.; Howell, G.E.; Park, E.A.; Gerling, I.C.; Patel, D.; et al. Hepatic gene expression in morbidly obese women: Implications for disease susceptibility. Obesity 2009, 17, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Fujishiro, M.; Gotoh, Y.; Katagiri, H.; Sakoda, H.; Ogihara, T.; Anai, M.; Onishi, Y.; Ono, H.; Abe, M.; Shojima, N.; et al. Three mitogen-activated protein kinases inhibit insulin signaling by different mechanisms in 3T3-L1 adipocytes. Mol. Endocrinol. 2003, 17, 487–497. [Google Scholar] [CrossRef]
- Engelman, J.A.; Berg, A.H.; Lewis, R.Y.; Lin, A.; Lisanti, M.P.; Scherer, P.E. Constitutively active mitogen-activated protein kinase kinase 6 (MKK6) or salicylate induces spontaneous 3T3-L1 adipogenesis. J. Biol. Chem. 1999, 274, 35630–35638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozer, J.; Ratner, M.; Shaw, M.; Bailey, W.; Schomaker, S. The current state of serum biomarkers of hepatotoxicity. Toxicology 2008, 245, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.-S.; Lee, B.-S.; Ma, J.Y. Ethanol extract of Mylabris phalerata inhibits M2 polarization induced by recombinant IL-4 and IL-13 in murine macrophages. Evid.-Based Complement. Altern. Med. 2017, 2017, 4218468. [Google Scholar] [CrossRef] [Green Version]
- Thevkar-Nagesh, P.; Rawal, S.; Zahr, T.; Fisher, E.A.; Garabedian, M. Transcriptional regulation of ACSL1 by CHREBP and NF-kappa B in macrophages during hyperglycemia and inflammation. bioRxiv 2020. [Google Scholar] [CrossRef]
- Boone, L.; Meyer, D.; Cusick, P.; Ennulat, D.; Bolliger, A.P.; Everds, N.; Meador, V.; Elliott, G.; Honor, D.; Bounous, D.; et al. Selection and interpretation of clinical pathology indicators of hepatic injury in preclinical studies. Veter-Clin. Pathol. 2005, 34, 182–188. [Google Scholar] [CrossRef]
- Galvin, Z.; McDonough, A.; Ryan, J.; Stewart, S. Blood alanine aminotransferase levels> 1,000 IU/l–causes and outcomes. Clin. Med. 2015, 15, 244. [Google Scholar] [CrossRef] [Green Version]
- Longo, M.; Zatterale, F.; Naderi, J.; Parrillo, L.; Formisano, P.; Raciti, G.A.; Beguinot, F.; Miele, C. Adipose tissue dysfunction as determinant of obesity-associated metabolic complications. Int. J. Mol. Sci. 2019, 20, 2358. [Google Scholar] [CrossRef] [Green Version]
- Spalding, K.L.; Arner, E.; Westermark, P.O.; Bernard, S.; Buchholz, B.A.; Bergmann, O.; Blomqvist, L.; Hoffstedt, J.; Näslund, E.; Britton, T.; et al. Dynamics of fat cell turnover in humans. Nature 2008, 453, 783–787. [Google Scholar] [CrossRef]
- Darlington, G.J.; Ross, S.E.; MacDougald, O. The role of C/EBP genes in adipocyte differentiation. J. Biol. Chem. 1998, 273, 30057–30060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siersbæk, R.; Nielsen, R.; Mandrup, S. PPARγ in adipocyte differentiation and metabolism–novel insights from genome-wide studies. FEBS Lett. 2010, 584, 3242–3249. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.S.; Subramaniam, S.; Dramane, G.; Khelifi, D.; Khan, N.A. ERK1 and ERK2 activation modulates diet-induced obesity in mice. Biochimie 2017, 137, 78–87. [Google Scholar] [CrossRef]
- Jiao, P.; Feng, B.; Li, Y.; He, Q.; Xu, H. Hepatic ERK activity plays a role in energy metabolism. Mol. Cell. Endocrinol. 2013, 375, 157–166. [Google Scholar] [CrossRef] [Green Version]
- Pheiffer, C.; Jacobs, C.; Patel, O.; Ghoor, S.; Muller, C.; Louw, J. Expression of UCP2 in Wistar rats varies according to age and the severity of obesity. J. Physiol. Biochem. 2015, 72, 25–32. [Google Scholar] [CrossRef]
- Gao, W.; Xu, W.; Li, H. The expression and role of peroxisome proliferator-activated receptor α in atherosclerosis. Mol. Med. Rep. 2013, 7, 862–868. [Google Scholar] [CrossRef]
- Horimoto, M.; Fülöp, P.; Derdak, Z.; Wands, J.R.; Baffy, G. Uncoupling protein-2 deficiency promotes oxidant stress and delays liver regeneration in mice. Hepatology 2004, 39, 386–392. [Google Scholar] [CrossRef]
- Ricquier, D.; Bouillaud, F. The uncoupling protein homologues: UCP1, UCP2, UCP3, StUCP and AtUCP. Biochem. J. 2000, 345, 161–179. [Google Scholar] [CrossRef] [PubMed]
- Emre, Y.; Hurtaud, C.; Nübel, T.; Criscuolo, F.; Ricquier, D.; Cassard-Doulcier, A.-M. Mitochondria contribute to LPS-induced MAPK activation via uncoupling protein UCP2 in macrophages. Biochem. J. 2007, 402, 271–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senese, R.; Cioffi, F.; De Matteis, R.; Petito, G.; De Lange, P.; Silvestri, E.; Lombardi, A.; Moreno, M.; Goglia, F.; Lanni, A. 3,5 Diiodo-L-Thyronine (T2) promotes the browning of white adipose tissue in high-fat diet-induced overweight male rats housed at thermoneutrality. Cells 2019, 8, 256. [Google Scholar] [CrossRef] [Green Version]
- Matesanz, N.; Nikolic, I.; Leiva, M.; Pulgarin-Alfaro, M.; Santamans, A.M.; Bernardo, E.; Mora, A.; Herrera-Melle, L.; Rodriguez, E.; Beiroa, D. p38α blocks brown adipose tissue thermogenesis through p38δ inhibition. PLoS Biol. 2018, 16, e2004455. [Google Scholar] [CrossRef]
- Cao, W.; Daniel, K.W.; Robidoux, J.; Puigserver, P.; Medvedev, A.V.; Bai, X.; Floering, L.M.; Spiegelman, B.M.; Collins, S. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell. Biol. 2004, 24, 3057–3067. [Google Scholar] [CrossRef] [Green Version]
- Holm, C. Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Biochem. Soc. Trans. 2003, 31, 1120–1124. [Google Scholar] [CrossRef]
- Rosenwald, M.; Perdikari, A.; Rülicke, T.; Wolfrum, C. Bi-directional interconversion of brite and white adipocytes. Nature 2013, 15, 659–667. [Google Scholar] [CrossRef]
- Granneman, J.G.; Burnazi, M.; Zhu, Z.; Schwamb, L.A. White adipose tissue contributes to UCP1-independent thermogenesis. Am. J. Physiol. Metab. 2003, 285, E1230–E1236. [Google Scholar] [CrossRef] [Green Version]
- Weisberg, S.P.; McCann, D.; Desai, M.; Rosenbaum, M.; Leibel, R.L.; Ferrante, A.W., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Investig. 2003, 112, 1796–1808. [Google Scholar] [CrossRef]
- Cinti, S.; Mitchell, G.; Barbatelli, G.; Murano, I.; Ceresi, E.; Faloia, E.; Wang, S.; Fortier, M.; Greenberg, A.S.; Obin, M.S. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J. Lipid Res. 2005, 46, 2347–2355. [Google Scholar] [CrossRef] [Green Version]
- Frederich, R.C.; Hamann, A.; Anderson, S.; Löllmann, B.; Lowell, B.B.; Flier, J.S. Leptin levels reflect body lipid content in mice: Evidence for diet-induced resistance to leptin action. Nat. Med. 1995, 1, 1311–1314. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, K.-Y.; Diaz, P.A.; Heo, M.; Leibel, R.L. Determinants of leptin gene expression in fat depots of lean mice. Am. J. Physiol. Integr. Comp. Physiol. 2002, 282, R226–R234. [Google Scholar] [CrossRef] [Green Version]
- Van Harmelen, V.; Reynisdottir, S.; Eriksson, P.; Thorne, A.; Hoffstedt, J.; Lonnqvist, F.; Arner, P. Leptin secretion from subcutaneous and visceral adipose tissue in women. Diabetes 1998, 47, 913–917. [Google Scholar] [CrossRef]
- Arita, Y.; Kihara, S.; Ouchi, N.; Takahashi, M.; Maeda, K.; Miyagawa, J.-I.; Hotta, K.; Shimomura, I.; Nakamura, T.; Miyaoka, K.J.B.; et al. Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem. Biophys. Res. Commun. 1999, 257, 79–83. [Google Scholar] [CrossRef]
- Matsuzawa, Y. The metabolic syndrome and adipocytokines. FEBS Lett. 2006, 580, 2917–2921. [Google Scholar] [CrossRef] [Green Version]
- Mandal, P.; Pratt, B.T.; Barnes, M.; McMullen, M.R.; Nagy, L.E. Molecular mechanism for adiponectin-dependent M2 macrophage polarization: Link between the metabolic and innate immune activity of full-length adiponectin. J. Biol. Chem. 2011, 286, 13460–13469. [Google Scholar] [CrossRef] [Green Version]
- Ouchi, N.; Parker, J.L.; Lugus, J.J.; Walsh, K. Adipokines in inflammation and metabolic disease. Nat. Rev. Immunol. 2011, 11, 85–97. [Google Scholar] [CrossRef]
- Hui, X.; Gu, P.; Zhang, J.; Nie, T.; Pan, Y.; Wu, D.; Feng, T.; Zhong, C.; Wang, Y.; Lam, K.S.; et al. Adiponectin enhances cold-induced browning of subcutaneous adipose tissue via promoting M2 macrophage proliferation. Cell Metab. 2015, 22, 279–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acedo, S.C.; Gambero, S.; Cunha, F.G.P.; Lorand-Metze, I.; Gambero, A. Participation of leptin in the determination of the macrophage phenotype: An additional role in adipocyte and macrophage crosstalk. Vitr. Cell. Dev. Biol.-Anim. 2013, 49, 473–478. [Google Scholar] [CrossRef]
- Unamuno, X.; Gómez-Ambrosi, J.; Rodríguez, A.; Becerril, S.; Frühbeck, G.; Catalán, V. Adipokine dysregulation and adipose tissue inflammation in human obesity. Eur. J. Clin. Investig. 2018, 48, e12997. [Google Scholar] [CrossRef] [Green Version]
- Folch, J.; Lees, M.; Stanley, G.S. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]





| Group | Non_Tg | TgMEK6 | p-Value | |||
|---|---|---|---|---|---|---|
| Lean (n = 10) | DIO (n = 10) | Lean (n = 10) | DIO (n = 10) | |||
| Body Weight (g) | Initial | 19.82 ± 0.82 a | 22.34 ± 0.73 b | 23.12 ± 1.23 b | 23.26 ± 1.00 b | <0.0001 **** |
| Final | 30.75 ± 1.54 a | 49.72 ± 1.76 c | 33.56 ± 2.27 b | 52.44 ± 1.72 d | <0.0001 **** | |
| Body weight gain (g/day) | 0.11 ± 0.02 a | 0.28 ± 0.02 b | 0.11 ± 0.02 a | 0.30 ± 0.09 c | <0.0001 **** | |
| Total body weight gain (g) | 10.92 ± 1.83 a | 27.39 ± 1.29 b | 10.44 ± 1.47 a | 29.18 ± 1.92 c | <0.0001 **** | |
| Dietary intake (g/day body weight) | 0.10 ± 0.01 b | 0.05 ± 0.01 a | 0.11 ± 0.01 c | 0.05 ± 0.01 a | <0.0001 **** | |
| Total dietary intake (g/body weight) | 10.52 ± 0.65 b | 5.25 ± 0.29 a | 11.88 ± 0.70 c | 5.72 ± 0.30 a | <0.0001 **** | |
| Food efficiency ratio | 1.05 ± 0.23 a | 5.23 ± 0.44 b | 0.89 ± 0.16 a | 5.12 ± 0.55 b | <0.0001 **** | |
| Tissue weight (g/100g bodyweight) | ||||||
| Liver | 3.74 ± 0.50 a | 4.45 ± 0.69 bc | 3.99 ± 0.17 ab | 4.67 ± 1.00 c | <0.05 * | |
| Heart | 0.36 ± 0.04 b | 0.25 ± 0.02 a | 0.43 ± 0.04 c | 0.26 ± 0.03 a | <0.0001 **** | |
| Kidney | 1.19 ± 0.14 c | 0.77 ± 0.07 a | 1.27 ± 0.09 c | 0.87 ± 0.10 b | <0.0001 **** | |
| Total fat | 2.92 ± 1.11 a | 7.77 ± 0.99 b | 3.42 ± 1.57 a | 7.50 ± 0.91 b | <0.0001 **** | |
| Perirenal fat (pWAT) | 0.48 ± 0.27 a | 2.25 ± 0.58 b | 0.88 ± 0.36 a | 2.67 ± 0.39 b | <0.0001 **** | |
| Visceral fat (vWAT) | 0.85 ± 0.46 a | 3.05 ± 0.61 d | 0.66 ± 0.30 a | 2.43 ± 0.59 c | <0.05 * | |
| Epididymal fat (eWAT) | 1.59 ± 0.73 a | 2.46 ± 0.32 b | 1.88 ± 1.05 ab | 2.41 ± 0.76 b | <0.0001 **** | |
| Group | Non Tg | TgMEK6 | p-Value | ||
|---|---|---|---|---|---|
| Lean (n = 10) | DIO (n = 10) | Lean (n = 10) | DIO (n = 10) | ||
| Blood glucose (mg/dL) | 119.1 ± 25.02 a | 185.71 ± 36.09 b | 123.22 ± 29.75 a | 199.94 ± 48.57 b | <0.0001 **** |
| AST (IU/L) | 47.48 ± 1.62 a | 63.83 ± 1.14 b | 59.29 ± 3.76 c | 76.46 ± 1.64 d | <0.001 *** |
| ALT (IU/L) | 8.62 ± 0.40 a | 48.95 ± 1.78 b | 15.38 ± 0.70 c | 71.74 ± 5.30 d | <0.001 *** |
| AST/ALT ratio | 5.51 ± 0.25 c | 1.30 ± 0.04 a | 3.86 ± 0.15 b | 1.07 ± 0.06 a | <0.001 *** |
| TG (mg/dL) | 60.50 ± 2.74 a | 77.61 ± 3.46 c | 70.25 ± 4.13 b | 89.47 ± 3.86 d | <0.01 ** |
| TC (mg/dL) | 146.08 ± 3.71 a | 243.92 ± 9.32 c | 187.92 ± 7.13 b | 285.00 ± 2.83 d | <0.0001 **** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Lee, M. MEK6 Overexpression Exacerbates Fat Accumulation and Inflammatory Cytokines in High-Fat Diet-Induced Obesity. Int. J. Mol. Sci. 2021, 22, 13559. https://doi.org/10.3390/ijms222413559
Lee S, Lee M. MEK6 Overexpression Exacerbates Fat Accumulation and Inflammatory Cytokines in High-Fat Diet-Induced Obesity. International Journal of Molecular Sciences. 2021; 22(24):13559. https://doi.org/10.3390/ijms222413559
Chicago/Turabian StyleLee, Suyeon, and Myoungsook Lee. 2021. "MEK6 Overexpression Exacerbates Fat Accumulation and Inflammatory Cytokines in High-Fat Diet-Induced Obesity" International Journal of Molecular Sciences 22, no. 24: 13559. https://doi.org/10.3390/ijms222413559
APA StyleLee, S., & Lee, M. (2021). MEK6 Overexpression Exacerbates Fat Accumulation and Inflammatory Cytokines in High-Fat Diet-Induced Obesity. International Journal of Molecular Sciences, 22(24), 13559. https://doi.org/10.3390/ijms222413559







