Genome-Wide Identification, Characterization and Expression Analysis of the CIPK Gene Family in Potato (Solanum tuberosum L.) and the Role of StCIPK10 in Response to Drought and Osmotic Stress
Abstract
:1. Introduction
2. Results
2.1. Identification, Gene Duplication and the Phylogenetic Tree of StCIPK Family Genes
2.2. Analysis of Gene Structure and Cis-Acting Elements
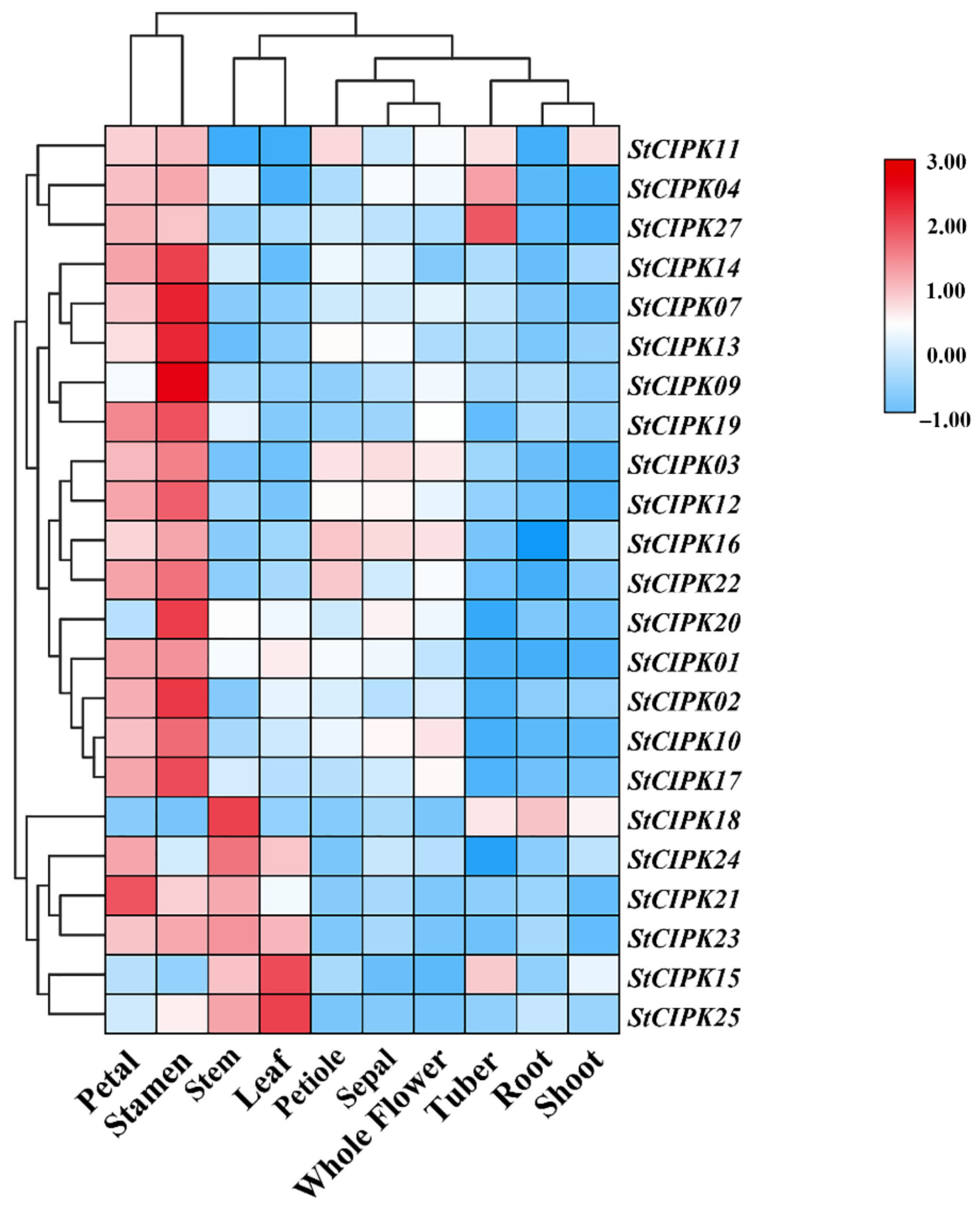
2.3. Expression Analysis of StCIPK Genes in Different Tissues
2.4. Expression Analysis of StCIPK Genes under Different Abiotic Stresses and ABA treatment
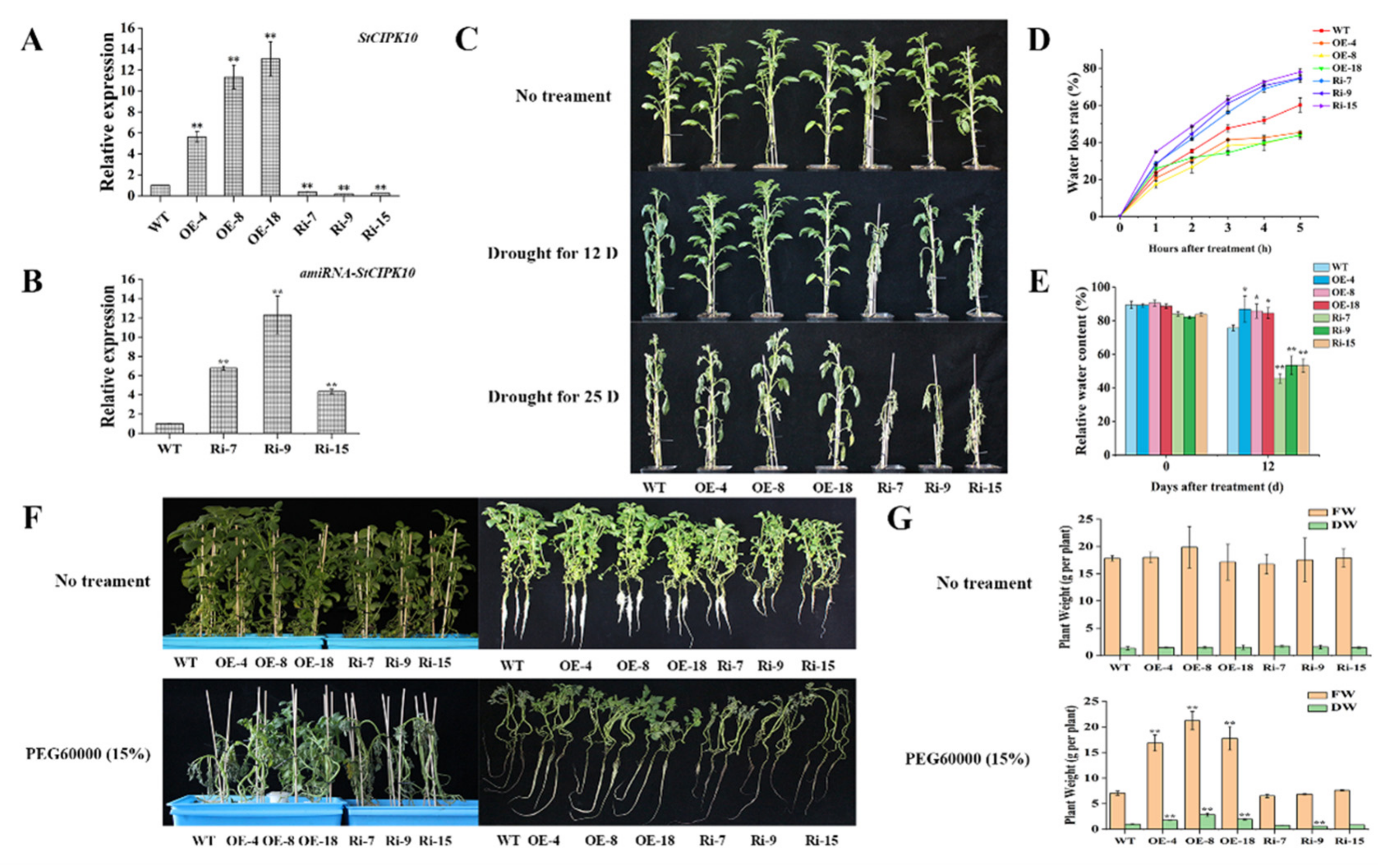
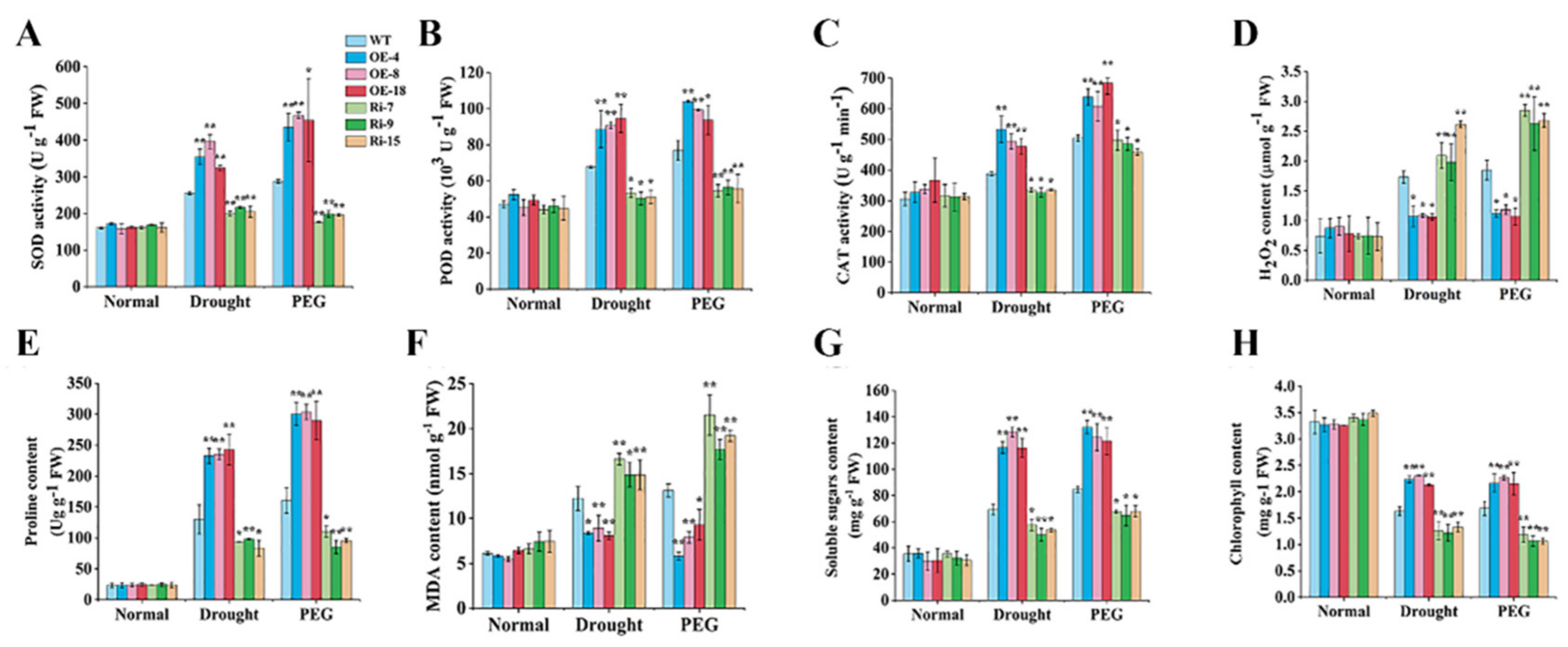
2.5. StCIPK10 Positively Modulates Responses to Drought and Osmotic Stress in Potato
2.6. Overexpression of StCIPK10 Increases ABA Sensitivity and Stomatal Closure in Transgenic Potato
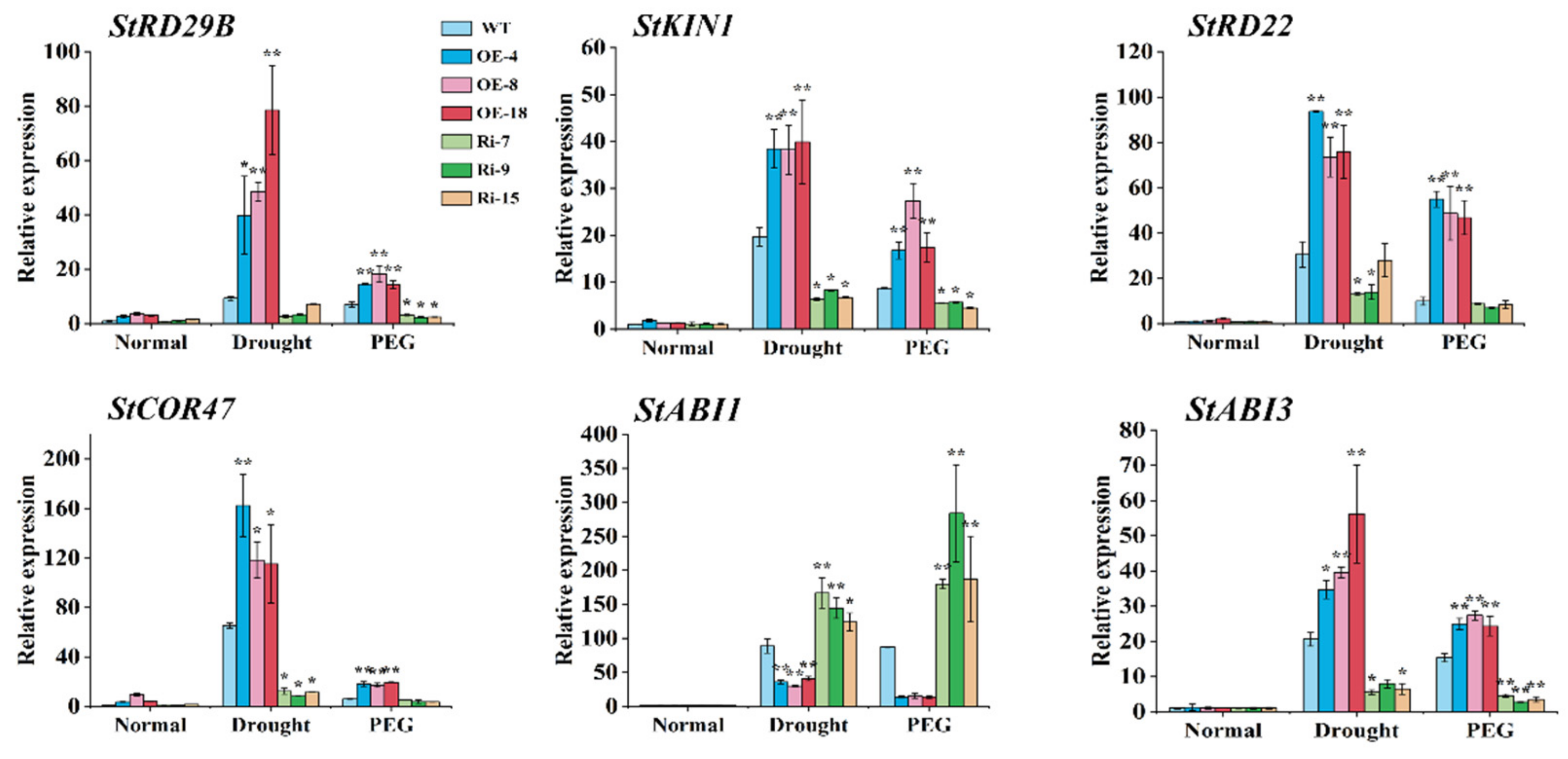
2.7. StCIPK10 Regulates Expression of Stress-Induced Genes in Transgenic Potato
2.8. Interaction between StCIPK10 and StCBL Proteins
3. Discussion
3.1. StCIPK Gene Expansion and Evolution in Potato
3.2. StCIPK10 is a Positive Regulator of Potato Tolerance to Drought and Osmotic Stress of Potato
3.3. StCIPK10 Is a Positive Regulator of Potato Tolerance to Drought and Osmotic Stress
3.4. StCIPK10 Is a Positive Regulator of ABA-Dependent Responses
3.5. Interactive Mechanism of StCIPK10 with Upstream Regulators
4. Materials and Methods
4.1. Identification of StCIPK Genes
4.2. Chromosomal Localization, Phylogenetic and Gene Duplication Analysis
4.3. Cis-Acting Element and Gene Structure Analysis
4.4. Plant Materials and Treatments
4.5. RNA Isolation and Gene Expression Analysis
4.6. Subcelluar Localization Analysis of StCIPK10
4.7. Protein Interaction Assay
4.8. Production of Transgenic Plants
4.9. Assays for Drought and Osmotic Stress Tolerance
4.10. Assays for Stomatal Movement and Phenotype under ABA Treatment
4.11. Validation of amiRNA Target Genes using RLM 5′-RACE
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Batistic, O.; Kudla, J. Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 2004, 219, 915–924. [Google Scholar] [CrossRef]
- Hetherington, A.M.; Brownlee, C. The generation of Ca2+ signals in plants. Annu. Rev. Plant Biol. 2004, 55, 401–427. [Google Scholar] [CrossRef] [Green Version]
- Mcainsh, M.R.; Pittman, J.K.J.N.P. Shaping the calcium signature. New Phytol. 2008, 181, 275–294. [Google Scholar] [CrossRef]
- Kudla, J.; Batistic, O.; Hashimoto, K. Calcium signals: The lead currency of plant information processing. Plant Cell 2010, 22, 541–563. [Google Scholar] [CrossRef]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef] [Green Version]
- Kudla, J.; Xu, Q.; Harter, K.; Gruissem, W.; Luan, S. Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. USA 1999, 96, 4718–4723. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Kim, K.N.; Ritz, O.; Albrecht, V.; Gupta, R.; Harter, K.; Luan, S.; Kudla, J. Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 1999, 11, 2393–2405. [Google Scholar] [CrossRef]
- Hrabak, E.M.; Chan, C.W.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Halfter, U.; Ishitani, M.; Zhu, J.K. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 2001, 13, 1383–1399. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Barrena, M.J.; Martinez-Ripoll, M.; Zhu, J.K.; Albert, A. The structure of the Arabidopsis thaliana SOS3: Molecular mechanism of sensing calcium for salt stress response. J. Mol. Biol. 2005, 345, 1253–1264. [Google Scholar] [CrossRef]
- Sanchez-Barrena, M.J.; Fujii, H.; Angulo, I.; Martinez-Ripoll, M.; Zhu, J.K.; Albert, A. The structure of the C-terminal domain of the protein kinase AtSOS2 bound to the calcium sensor AtSOS3. Mol. Cell 2007, 26, 427–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.; Du, W.; Yang, Y.; Schumaker, K.S.; Guo, Y. A calcium-independent activation of the Arabidopsis SOS2-like protein kinase24 by its interacting SOS3-like calcium binding protein1. Plant Physiol. 2014, 164, 2197–2206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, K.; Eckert, C.; Anschütz, U.; Scholz, M.; Held, K.; Waadt, R.; Reyer, A.; Hippler, M.; Becker, D.; Kudla, J. Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases (CIPKs) is required for full activity of CBL-CIPK complexes toward their target proteins. J. Biol. Chem. 2012, 287, 7956–7968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinl, S.; Kudla, J. The CBL-CIPK Ca2+-decoding signaling network: Function and perspectives. New Phytol. 2009, 184, 517–528. [Google Scholar] [CrossRef]
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef] [Green Version]
- Quan, R.; Lin, H.; Mendoza, I.; Zhang, Y.; Cao, W.; Yang, Y.; Shang, M.; Chen, S.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 2007, 19, 1415–1431. [Google Scholar] [CrossRef] [Green Version]
- Behera, S.; Long, Y.; Schmitz-Thom, I.; Wang, X.P.; Zhang, C.; Li, H.; Steinhorst, L.; Manishankar, P.; Ren, X.L.; Offenborn, J.N.; et al. Two spatially and temporally distinct Ca2+ signals convey Arabidopsis thaliana responses to K+ deficiency. New Phytol. 2017, 213, 739–750. [Google Scholar] [CrossRef] [Green Version]
- Nieves-Cordones, M.; Caballero, F.; Martinez, V.; Rubio, F. Disruption of the Arabidopsis thaliana inward-rectifier K+ channel AKT1 improves plant responses to water stress. Plant Cell Physiol. 2012, 53, 423–432. [Google Scholar] [CrossRef]
- D’Angelo, C.; Weinl, S.; Batistic, O.; Pandey, G.K.; Cheong, Y.H.; Schultke, S.; Albrecht, V.; Ehlert, B.; Schulz, B.; Harter, K.; et al. Alternative complex formation of the Ca-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J. 2006, 48, 857–872. [Google Scholar] [CrossRef]
- Drerup, M.M.; Schlücking, K.; Hashimoto, K.; Manishankar, P.; Steinhorst, L.; Kuchitsu, K.; Kudla, J. The Calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol. Plant 2013, 6, 559–569. [Google Scholar] [CrossRef] [Green Version]
- Pandey, G.K.; Grant, J.J.; Cheong, Y.H.; Kim, B.G.; Li, L.G.; Luan, S. Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol. Plant 2008, 1, 238–248. [Google Scholar] [CrossRef]
- Tsou, P.L.; Lee, S.Y.; Allen, N.S.; Winter-Sederoff, H.; Robertson, D. An ER-targeted calcium-binding peptide confers salt and drought tolerance mediated by CIPK6 in Arabidopsis. Planta 2012, 235, 539–552. [Google Scholar] [CrossRef]
- Zhao, J.; Sun, Z.; Zheng, J.; Guo, X.; Dong, Z.; Huai, J.; Gou, M.; He, J.; Jin, Y.; Wang, J.; et al. Cloning and characterization of a novel CBL-interacting protein kinase from maize. Plant Mol. Biol. 2009, 69, 661–674. [Google Scholar] [CrossRef]
- Xiang, Y.; Huang, Y.; Xiong, L. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 2007, 144, 1416–1428. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, T.; John, S.J.; Chen, M.; Chang, J.; Yang, G.; He, G.A. CBL-interacting protein kinase TaCIPK27 confers drought tolerance and exogenous ABA sensitivity in transgenic Arabidopsis. Plant Physiol. Biochem. 2018, 123, 103–113. [Google Scholar] [CrossRef]
- Wang, R.K.; Li, L.L.; Cao, Z.H.; Zhao, Q.; Li, M.; Zhang, L.Y.; Hao, Y.J. Molecular cloning and functional characterization of a novel apple MdCIPK6L gene reveals its involvement in multiple abiotic stress tolerance in transgenic plants. Plant Mol. Biol. 2012, 79, 123–135. [Google Scholar] [CrossRef]
- Meng, D.; Dong, B.; Niu, L.; Song, Z.; Wang, L.; Amin, R.; Cao, H.; Li, H.; Yang, Q.; Fu, Y. The pigeon pea CcCIPK14-CcCBL1 pair positively modulates drought tolerance by enhancing flavonoid biosynthesis. Plant J. 2021, 106, 1278–1297. [Google Scholar] [CrossRef]
- Kolukisaoglu, U.; Weinl, S.; Blazevic, D.; Batistic, O.; Kudla, J. Calcium sensors and their interacting protein kinases: Genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004, 134, 43–58. [Google Scholar] [CrossRef] [Green Version]
- Sun, T.; Wang, Y.; Wang, M.; Li, T.; Zhou, Y.; Wang, X.; Wei, S.; He, G.; Yang, G. Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 269. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Jiang, M.M.; Ren, L.; Liu, Y.; Chen, H.Y. Identification and characterization of CBL and CIPK gene families in eggplant (Solanum melongena L.). Mol. Genet. Genom. 2016, 291, 1769–1781. [Google Scholar] [CrossRef]
- Xi, Y.; Liu, J.; Dong, C.; Cheng, Z.M. The CBL and CIPK gene family in grapevine (Vitis vinifera): Genome-wide analysis and expression profiles in response to various abiotic stresses. Front. Plant Sci. 2017, 8, 978. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Gai, W.X.; Qiao, Y.M.; Ali, M.; Wei, A.M.; Luo, D.X.; Li, Q.H.; Gong, Z.H. Identification of CBL and CIPK gene families and functional characterization of CaCIPK1 under Phytophthora capsici in pepper (Capsicum annuum L.). BMC Genom. 2019, 20, 775. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, B.; Liu, W.Z.; Li, H.; Wang, L.; Wang, B.; Deng, M.; Liang, W.; Deyholos, M.K.; Jiang, Y.Q. Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol. 2014, 14, 8. [Google Scholar] [CrossRef] [Green Version]
- Mullins, E.; Milbourne, D.; Petti, C.; Doyle-Prestwich, B.M.; Meade, C. Potato in the age of biotechnology. Trends Plant Sci. 2006, 11, 254–260. [Google Scholar] [CrossRef] [Green Version]
- Anithakumari, A.M.; Nataraja, K.N.; Visser, R.G.; van der Linden, C.G. Genetic dissection of drought tolerance and recovery potential by quantitative trait locus mapping of a diploid potato population. Mol. Breed. 2012, 30, 1413–1429. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, A.; Huynh, H.D.; Endo, T.; Watanabe, K. Review of recent transgenic studies on abiotic stress tolerance and future molecular breeding in potato. Breed Sci. 2015, 65, 85–102. [Google Scholar] [CrossRef] [Green Version]
- Slater, A.T.; Cogan, N.O.; Hayes, B.J.; Schultz, L.; Dale, M.F.; Bryan, G.J.; Forster, J.W. Improving breeding efficiency in potato using molecular and quantitative genetics. Theor. Appl. Genet. 2014, 127, 2279–2292. [Google Scholar] [CrossRef]
- Na, J.K.; Kim, K.H.; Seong, E.S.; Kim, B.G.; Cho, K.S.; Cho, J.H. Overexpression of nuclear factor y subunit StNF-YA7 enhances drought tolerance in potato. Horitic. Environ. Biotechnol. 2017, 58, 170–177. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Y.; Li, D.; Feng, S.; Yang, J.; Zhang, J.; Zhang, J.; Wang, D.; Gan, Y. Improving salt tolerance in potato through overexpression of AtHKT1 gene. BMC Plant Biol. 2019, 19, 357. [Google Scholar] [CrossRef]
- Lee, H.E.; Shin, D.; Park, S.R.; Han, S.E.; Jeong, M.J.; Kwon, T.R.; Lee, S.K.; Park, S.C.; Yi, B.Y.; Kwon, H.B.; et al. Ethylene responsive element binding protein 1 (StEREBP1) from Solanum tuberosum increases tolerance to abiotic stress in transgenic potato plants. Biochem. Biophys. Res. Commun. 2007, 353, 863–868. [Google Scholar] [CrossRef]
- Huang, Z.; Duan, W.; Song, X.; Tang, J.; Wu, P.; Zhang, B.; Hou, X. Retention, molecular evolution, and expression divergence of the auxin/indole acetic acid and auxin response factor gene families in Brassica rapa shed light on their evolution patterns in plants. Genome Biol. Evol. 2015, 8, 302–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albrecht, V.; Ritz, O.; Linder, S.; Harter, K.; Kudla, J. The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 2001, 20, 1051–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaves-Sanjuan, A.; Sanchez-Barrena, M.J.; Gonzalez-Rubio, J.M.; Moreno, M.; Ragel, P.; Jimenez, M.; Pardo, J.M.; Martinez-Ripoll, M.; Quintero, F.J.; Albert, A. Structural basis of the regulatory mechanism of the plant CIPK family of protein kinases controlling ion homeostasis and abiotic stress. Proc. Natl. Acad. Sci. USA 2014, 111, E4532–E4541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000, 3, 217–223. [Google Scholar] [CrossRef]
- Kim, K.N.; Cheong, Y.H.; Grant, J.J.; Pandey, G.K.; Luan, S. CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 2003, 15, 411–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilmour, S.J.; Artus, N.N.; Thomashow, M.F. cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana. Plant Mol. Biol. 1992, 18, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, S15–S45. [Google Scholar] [CrossRef] [Green Version]
- Cai, C.C.; Wang, W.L.; Zhang, J.; Deng, M.S.; Huang, T.; Yu, L.P.; Li, L.Q.; Wang, X.Y. Identification and sequence analysis of CBL family genes in potato. Mol. Plant Breed. 2019, 17, 2778–2784. [Google Scholar] [CrossRef]
- Liu, J.; Chen, N.; Chen, F.; Cai, B.; Dal Santo, S.; Tornielli, G.B.; Pezzotti, M.; Cheng, Z.M. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genom. 2014, 15, 281. [Google Scholar] [CrossRef] [Green Version]
- Gorlova, O.; Fedorov, A.; Logothetis, C.; Amos, C.; Gorlov, I. Genes with a large intronic burden show greater evolutionary conservation on the protein level. BMC Evol. Biol. 2014, 14, 50. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Gu, Z.; Xin, D.; Hao, L.; Liu, C.; Huang, J.; Ma, B.; Zhang, H. Identification and characterization of putative CIPK genes in maize. J. Genet. Genom. 2011, 38, 77–87. [Google Scholar] [CrossRef]
- Flagel, L.E.; Wendel, J.F. Gene duplication and evolutionary novelty in plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef]
- Zhu, K.; Chen, F.; Liu, J.; Chen, X.; Hewezi, T.; Cheng, Z.M. Evolution of an intron-poor cluster of the CIPK gene family and expression in response to drought stress in soybean. Sci. Rep. 2016, 6, 28216–28225. [Google Scholar] [CrossRef] [Green Version]
- Ohta, M.; Guo, Y.; Halfter, U.; Zhu, J.K. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. USA 2003, 100, 11771–11776. [Google Scholar] [CrossRef] [Green Version]
- Yin, X.; Wang, Q.; Chen, Q.; Xiang, N.; Yang, Y.; Yang, Y. Genome-wide identification and functional analysis of the calcineurin B-like protein and calcineurin B-like protein-interacting protein knase gene families in turnip (Brassica rapa var. rapa). Front. Plant Sci. 2017, 8, 1191–1210. [Google Scholar] [CrossRef]
- Han, J.P.; Köster, P.; Drerup, M.M.; Scholz, M.; Li, S.; Edel, K.H.; Hashimoto, K.; Kuchitsu, K.; Hippler, M.; Kudla, J. Fine-tuning of RBOHF activity is achieved by differential phosphorylation and Ca2+ binding. New Phytol. 2019, 221, 1935–1949. [Google Scholar] [CrossRef]
- Kanwar, P.; Sanyal, S.K.; Tokas, I.; Yadav, A.K.; Pandey, A.; Kapoor, S.; Pandey, G.K. Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice. Cell Calcium 2014, 56, 81–95. [Google Scholar] [CrossRef]
- Sanyal, S.K.; Rao, S.; Mishra, L.K.; Sharma, M.; Pandey, G.K. Plant stress responses mediated by CBL-CIPK phosphorylation network. In The Enzymes; Lin, C., Luan, S., Eds.; Academic Press: Cambridge, MA, USA, 2016; Volume 40, pp. 31–64. [Google Scholar] [CrossRef]
- Bajji, M.; Kinet, J.-M.; Lutts, S. The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regul. 2002, 36, 61–70. [Google Scholar] [CrossRef]
- Cha-Um, S.; Takabe, T.; Kirdmanee, C. Ion contents, relative electrolyte leakage, proline accumulation, photosynthetic abilities and growth characters of oil palm seedlings in response to salt stress. Pak. J. Bot. 2010, 42, 2020–2191. [Google Scholar] [CrossRef]
- Rajashekar, C.B.; Panda, M. Water stress is a component of cold acclimation process essential for inducing full freezing tolerance in strawberry. Sci. Hortic. 2014, 174, 54–59. [Google Scholar] [CrossRef] [Green Version]
- Bose, J.; Rodrigo-Moreno, A.; Shabala, S. ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 2014, 65, 1241–1257. [Google Scholar] [CrossRef]
- Mottley, C.; Robinson, R.E.; Mason, R.P. Free radical formation in the oxidation of malondialdehyde and acetylacetone by peroxidase enzymes. Arch. Biochem. Biophys. 1991, 289, 153–160. [Google Scholar] [CrossRef]
- Al Hassan, M.; Chaura, J.; Donat-Torres, M.P.; Boscaiu, M.; Vicente, O. Antioxidant responses under salinity and drought in three closely related wild monocots with different ecological optima. AoB Plants 2017, 9, plx009. [Google Scholar] [CrossRef]
- Armengaud, P.; Thiery, L.; Buhot, N.; Grenier-De March, G.; Savouré, A. Transcriptional regulation of proline biosynthesis in Medicago truncatula reveals developmental and environmental specific features. Physiol. Plant. 2004, 120, 442–450. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, X.; Jing, R.; Chang, X.; Xie, H. Characterization of a common wheat (Triticum aestivum L.) TaSnRK2.7 gene involved in abiotic stress responses. J. Exp. Bot. 2011, 62, 975–988. [Google Scholar] [CrossRef] [Green Version]
- Cruz, T.M.D.; Carvalho, R.F.; Richardson, D.N.; Duque, P. Abscisic acid (ABA) regulation of Arabidopsis SR protein gene expression. Int. J. Mol. Sci. 2014, 15, 17541–17564. [Google Scholar] [CrossRef] [Green Version]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Yang, X.; Sun, W.; Miao, Y.; He, L.; Zhang, X. The calcium sensor CBL2 and its interacting kinase CIPK6 are involved in plant sugar homeostasis via interacting with tonoplast sugar transporter TST2. Plant Physiol. 2020, 183, 236–249. [Google Scholar] [CrossRef]
- Quintero, F.J.; Ohta, M.; Shi, H.; Zhu, J.K.; Pardo, J.M. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA 2002, 99, 9061–9066. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Copley, R.R.; Schmidt, S.; Ciccarelli, F.D.; Doerks, T.; Schultz, J.; Ponting, C.P.; Bork, P. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 2004, 32, D142–D144. [Google Scholar] [CrossRef] [Green Version]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Vannozzi, A.; Wang, G.; Zhong, Y.; Corso, M.; Cavallini, E.; Cheng, Z.M. A comprehensive survey of the grapevine VQ gene family and its transcriptional correlation with WRKY proteins. Front. Plant Sci. 2015, 6, 417–433. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△CT method. Methods 2001, 25, 402–410. [Google Scholar] [CrossRef]
- Pieczynski, M.; Marczewski, W.; Hennig, J.; Dolata, J.; Bielewicz, D.; Piontek, P.; Wyrzykowska, A.; Krusiewicz, D.; Strzelczyk-Zyta, D.; Konopka-Postupolska, D.; et al. Down-regulation of CBP80 gene expression as a strategy to engineer a drought-tolerant potato. Plant Biotechnol. J. 2013, 11, 459–469. [Google Scholar] [CrossRef]
- Si, H.J.; Xie, C.H.; Liu, J. An efficient protocol for Agrobacterium-mediated transformation with microtuber and the introduction of an antisense class I patatin gene into potato. Acta Agron. Sin. 2003, 29, 801–805. [Google Scholar] [CrossRef]
- Bouchabke, O.; Chang, F.; Simon, M.; Voisin, R.; Pelletier, G.; Durand-Tardif, M. Natural variation in Arabidopsis thaliana as a tool for highlighting differential drought responses. PLoS ONE 2008, 27, e1705. [Google Scholar] [CrossRef]
- Luo, Q.; Wei, Q.; Wang, R.; Zhang, Y.; Zhang, F.; He, Y.; Zhou, S.; Feng, J.; Yang, G.; He, G. BdCIPK31, a calcineurin B-like protein-interacting protein kinase, regulates plant response to drought and salt stress. Front. Plant Sci. 2017, 8, 1184. [Google Scholar] [CrossRef] [Green Version]




| Gene Name | Gene ID | Chr. 1 | Genomic Location (bp) | CDS Length (bp) 2 | No. of Exons | Protein Length (aa) 3 | MW (kDa) 4 | pI 5 |
|---|---|---|---|---|---|---|---|---|
| StCIPK01 | Soltu.DM.01G005660.2 | 1 | 5,154,274–5,159,709 | 1317 | 14 | 438 | 50.23 | 6.72 |
| StCIPK02 | Soltu.DM.02G002430.1 | 2 | 11,576,701–11,584,119 | 1368 | 14 | 455 | 51.10 | 8.92 |
| StCIPK03 | Soltu.DM.02G015750.1 | 2 | 31,880,731–31,885,458 | 1437 | 1 | 478 | 53.76 | 8.60 |
| StCIPK04 | Soltu.DM.02G015770.1 | 2 | 31,899,161–31,901,152 | 1377 | 2 | 458 | 52.14 | 9.05 |
| StCIPK05 | Soltu.DM.03G000420.1 | 3 | 383,299–385,208 | 1302 | 1 | 433 | 48.63 | 9.26 |
| StCIPK06 | Soltu.DM.03G001400.1 | 3 | 1,308,274–3,10,351 | 1113 | 2 | 370 | 42.44 | 8.59 |
| StCIPK07 | Soltu.DM.04G031510.2 | 4 | 65,585,586–65,591,402 | 1344 | 14 | 447 | 50.63 | 6.38 |
| StCIPK08 | Soltu.DM.05G019250.1 | 5 | 43,311,668–43,315,613 | 1485 | 12 | 494 | 56.43 | 7.64 |
| StCIPK09 | Soltu.DM.05G022320.1 | 5 | 47,506,266–47,510,051 | 1338 | 1 | 445 | 50.20 | 9.07 |
| StCIPK10 | Soltu.DM.05G023210.1 | 5 | 48,492,654–48,497,748 | 1371 | 12 | 456 | 51.44 | 6.45 |
| StCIPK11 | Soltu.DM.06G002800.1 | 6 | 2,846,349–2,848,922 | 1341 | 2 | 446 | 50.96 | 8.47 |
| StCIPK12 | Soltu.DM.06G002810.1 | 6 | 2,851,745–2,855,743 | 1413 | 1 | 470 | 53.82 | 8.27 |
| StCIPK13 | Soltu.DM.06G010870.1 | 6 | 31,710,868–31,712,193 | 1326 | 1 | 441 | 49.74 | 9.10 |
| StCIPK14 | Soltu.DM.06G010880.1 | 6 | 31,745,058–31,746,792 | 1332 | 1 | 443 | 50.07 | 8.90 |
| StCIPK15 | Soltu.DM.06G024260.1 | 6 | 50,088,724–50,090,478 | 1353 | 1 | 450 | 51.02 | 8.92 |
| StCIPK16 | Soltu.DM.06G032750.1 | 6 | 57,566,917–57,568,428 | 1260 | 2 | 419 | 48.00 | 8.27 |
| StCIPK17 | Soltu.DM.07G000510.1 | 7 | 636,765–638,572 | 1275 | 1 | 424 | 47.66 | 9.08 |
| StCIPK18 | Soltu.DM.09G010080.1 | 7 | 18,714,464–18,716,381 | 1413 | 1 | 470 | 53.04 | 8.59 |
| StCIPK19 | Soltu.DM.08G015880.1 | 8 | 40,832,281–40,834,116 | 1371 | 1 | 456 | 52.35 | 8.63 |
| StCIPK20 | Soltu.DM.09G010290.1 | 9 | 14,851,369–14,853,146 | 1353 | 1 | 450 | 50.95 | 8.54 |
| StCIPK21 | Soltu.DM.09G025570.1 | 9 | 55,141,943–55,143,603 | 1317 | 2 | 437 | 49.44 | 8.90 |
| StCIPK22 | Soltu.DM.10G022340.1 | 10 | 53,624,978–53,626,957 | 1311 | 1 | 436 | 49.23 | 9.36 |
| StCIPK23 | Soltu.DM.11G018660.1 | 11 | 35,353,412–35,361,571 | 1398 | 13 | 465 | 53.07 | 7.24 |
| StCIPK24 | Soltu.DM.12G027440.1 | 12 | 2,229,648–2,31,821 | 1299 | 1 | 432 | 48.60 | 9.00 |
| StCIPK25 | Soltu.DM.12G026670.4 | 12 | 2,830,602–2,838,182 | 1341 | 14 | 446 | 50.63 | 8.87 |
| StCIPK26 | Soltu.DM.12G018730.1 | 12 | 18,210,942–18,212,405 | 1464 | 1 | 487 | 54.77 | 9.18 |
| StCIPK27 | Soltu.DM.12G002020.1 | 12 | 59,751,790–59,759,517 | 1596 | 11 | 531 | 60.01 | 5.97 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, R.; Liu, W.; Li, S.; Zhu, X.; Yang, J.; Zhang, N.; Si, H. Genome-Wide Identification, Characterization and Expression Analysis of the CIPK Gene Family in Potato (Solanum tuberosum L.) and the Role of StCIPK10 in Response to Drought and Osmotic Stress. Int. J. Mol. Sci. 2021, 22, 13535. https://doi.org/10.3390/ijms222413535
Ma R, Liu W, Li S, Zhu X, Yang J, Zhang N, Si H. Genome-Wide Identification, Characterization and Expression Analysis of the CIPK Gene Family in Potato (Solanum tuberosum L.) and the Role of StCIPK10 in Response to Drought and Osmotic Stress. International Journal of Molecular Sciences. 2021; 22(24):13535. https://doi.org/10.3390/ijms222413535
Chicago/Turabian StyleMa, Rui, Weigang Liu, Shigui Li, Xi Zhu, Jiangwei Yang, Ning Zhang, and Huaijun Si. 2021. "Genome-Wide Identification, Characterization and Expression Analysis of the CIPK Gene Family in Potato (Solanum tuberosum L.) and the Role of StCIPK10 in Response to Drought and Osmotic Stress" International Journal of Molecular Sciences 22, no. 24: 13535. https://doi.org/10.3390/ijms222413535
APA StyleMa, R., Liu, W., Li, S., Zhu, X., Yang, J., Zhang, N., & Si, H. (2021). Genome-Wide Identification, Characterization and Expression Analysis of the CIPK Gene Family in Potato (Solanum tuberosum L.) and the Role of StCIPK10 in Response to Drought and Osmotic Stress. International Journal of Molecular Sciences, 22(24), 13535. https://doi.org/10.3390/ijms222413535







