NnABI4-Mediated ABA Regulation of Starch Biosynthesis in Lotus (Nelumbo nucifera Gaertn)
Abstract
1. Introduction
2. Results
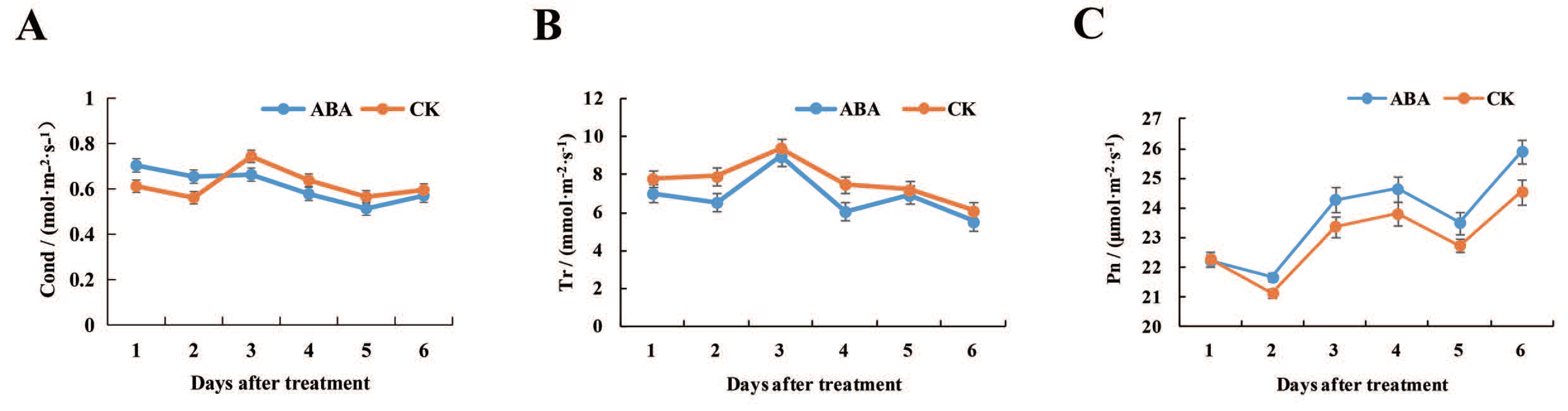
2.1. Effects of Exogenous ABA on Photosynthetic Characteristics of Lotus Leaves
2.2. Effect of Exogenous ABA Treatment on the Total Starch Content of Lotus
2.3. Expression Profile of Key Genes in Starch Pathway after ABA Treatment
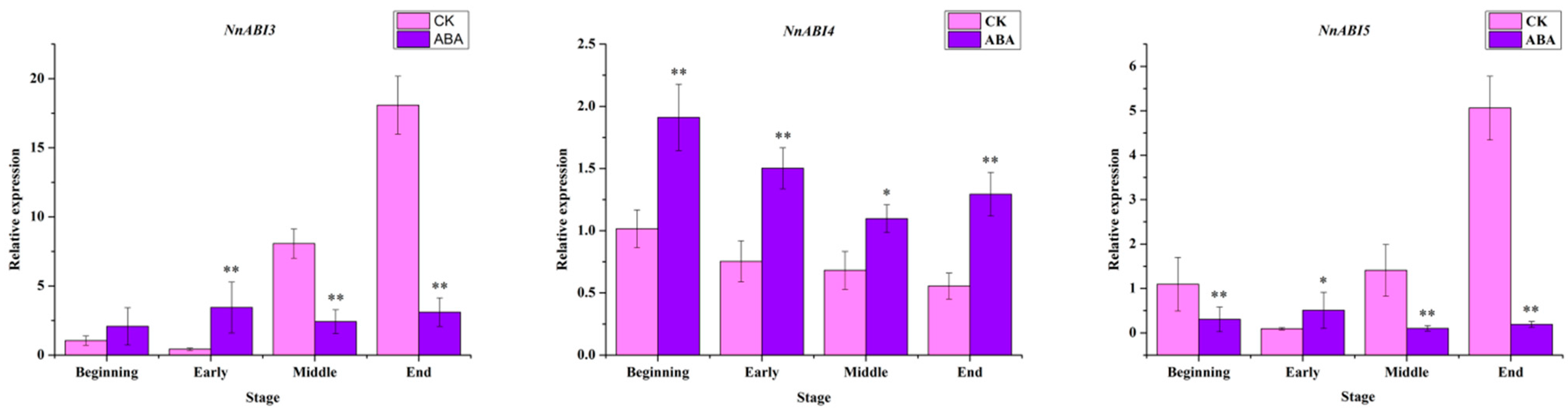
2.4. Expression Profile of Key Genes in ABA Signaling Pathway after ABA Treatment
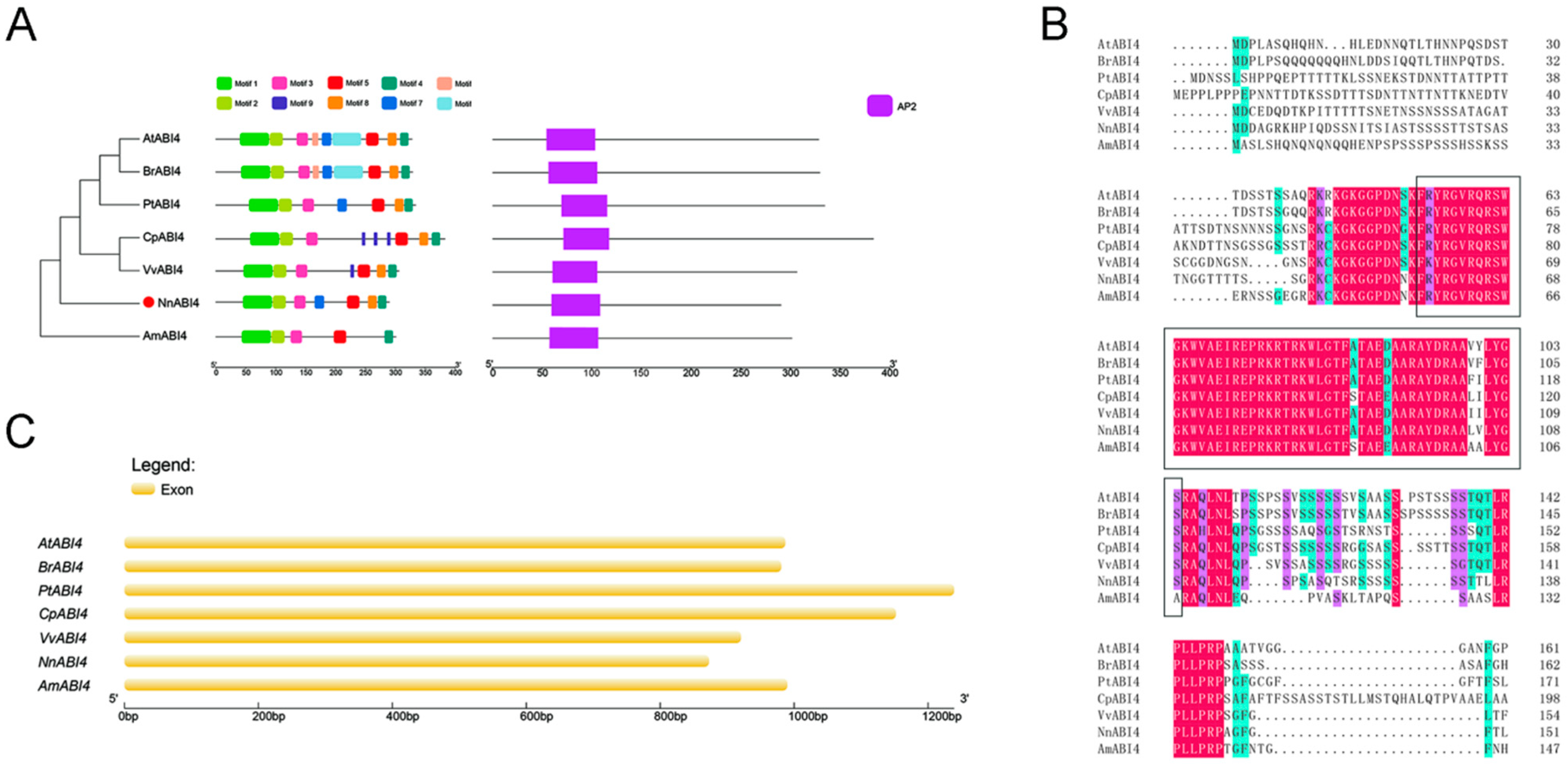
2.5. Sequence Analysis of ABI4
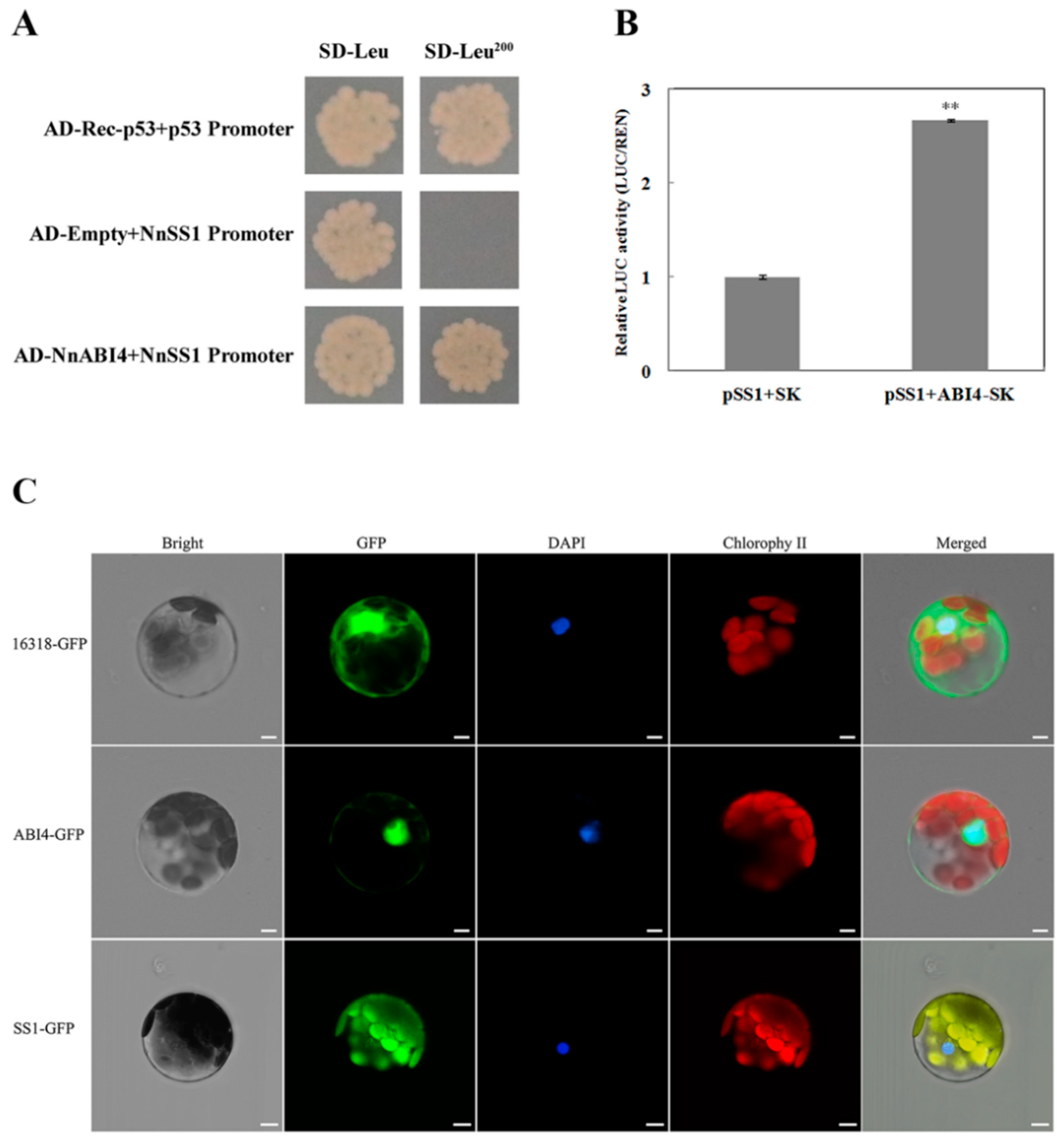
2.6. NnAB14 Directly Promotes the Expression of NnSS1
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Sample Preparation
4.3. Determination of Photosynthetic Parameters of Leaves
4.4. Determination of Starch Content
4.5. Quantitative Real-Time PCR Analysis
4.6. Y1H Assays
4.7. Transient Dual-Luciferase Detection
4.8. Subcellular Localization
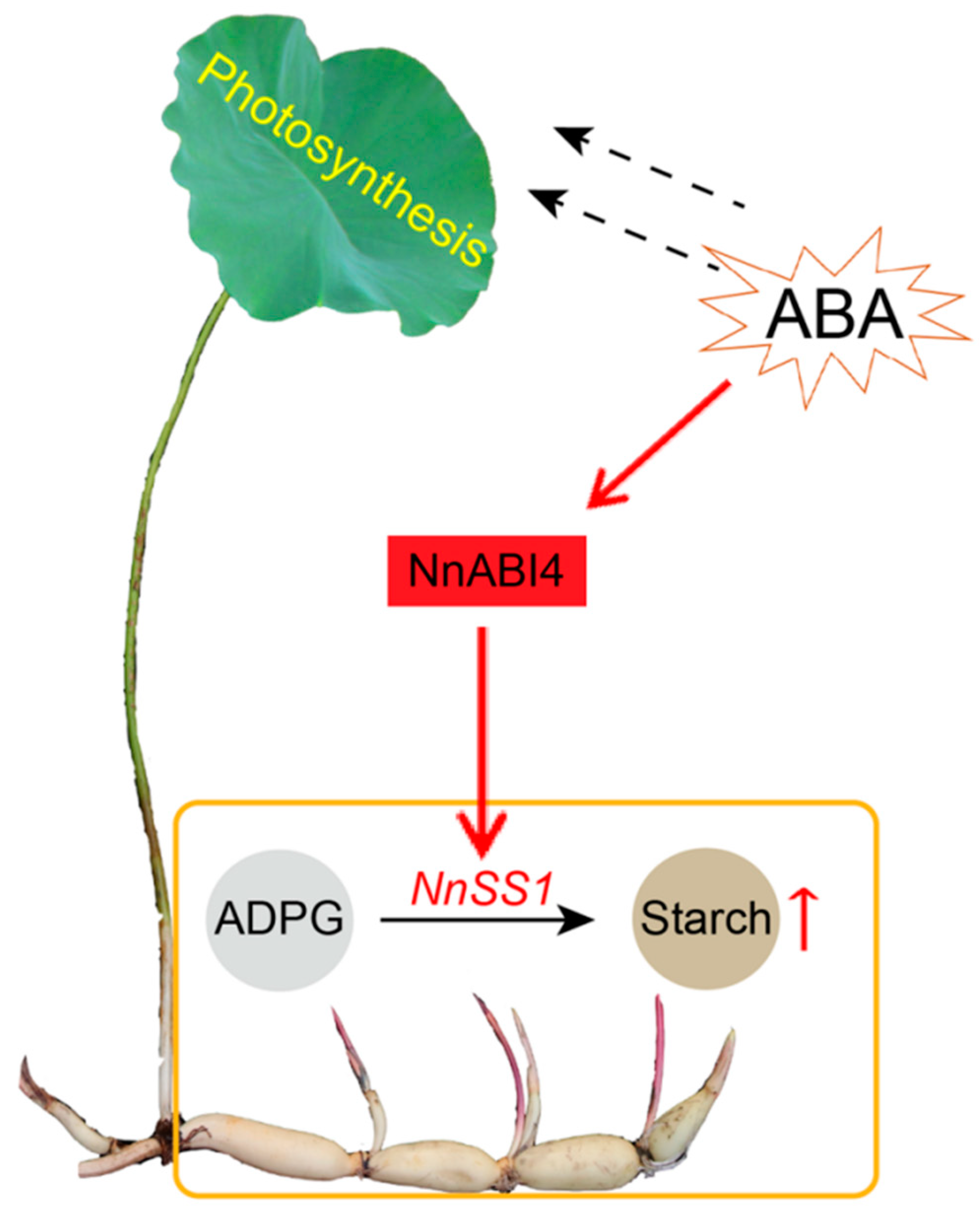
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kötting, O.; Kossmann, J.; Zeeman, S.C.; Lloyd, J.R. Regulation of starch metabolism: The age of enlightenment? Curr. Opin. Plant Biol. 2010, 13, 320–328. [Google Scholar] [CrossRef]
- Thitisaksakul, M.; Jiménez, R.C.; Arias, M.C.; Beckles, D.M. Effects of environmental factors on cereal starch biosynthesis and composition. J. Cereal Sci. 2012, 56, 67–80. [Google Scholar] [CrossRef]
- Njoku, D.N.; Mbah, E.U. Assessment of yield components of some cassava (Manihot esculenta Crantz) genotypes using multivariate analysis such as path coefficients. Open Agric. 2020, 5, 516–528. [Google Scholar] [CrossRef]
- Tian, Z.; Qian, Q.; Liu, Q.; Yan, M.; Liu, X.; Yan, C.; Liu, G.; Gao, Z.; Tang, S.; Zeng, D.; et al. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Acad. Sci. USA 2009, 106, 21760–21765. [Google Scholar] [CrossRef]
- Fan, X.Y.; Guo, M.; Li, R.D.; Yang, Y.H.; Liu, M.; Zhu, Q.; Tang, S.Z.; Gu, M.H.; Xu, R.G.; Yan, C.J. Allelic variations in the soluble starch synthase II gene family result in changes of grain quality and starch properties in rice (Oryza sativa L.). J. Agric. Sci. 2016, 155, 129–140. [Google Scholar] [CrossRef]
- Umemoto, T.; Horibata, T.; Aoki, N.; Hiratsuka, M.; Yano, M.; Inouchi, N. Effects of variations in starch synthase on starch properties and eating quality of rice. Plant Prod. Sci. 2008, 11, 472–480. [Google Scholar] [CrossRef]
- Lin, L.; Cai, C.; Gilbert, R.G.; Li, E.; Wang, J.; Wei, C. Relationships between amylopectin molecular structures andfunctional properties of different-sized fractions of normal andhigh-amylose maize starches. Food Hydrocoll. 2016, 52, 359–368. [Google Scholar] [CrossRef]
- Syahariza, Z.A.; Sar, S.; Hasjim, J.; Tizzotti, M.J.; Gilbert, R.G. The importance of amylose and amylopectin fine structures for starch digestibility in cooked rice grains. Food Chem. 2013, 136, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wei, X.; Jiao, G.; Chen, W.; Wu, Y.; Sheng, Z.; Hu, S.; Xie, L.; Wang, J.; Tang, S.; et al. GBSS-BINDING PROTEIN, encoding a CBM48 domain-containing protein, affects rice quality and yield. J. Integr. Plant Biol. 2020, 62, 948–966. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Jiao, G.; Lin, H.; Sheng, Z.; Shao, G.; Xie, L.; Tang, S.; Xu, Q.; Hu, P. GRAIN INCOMPLETE FILLING 2 regulates grain filling and starch synthesis during rice caryopsis development. J. Integr. Plant Biol. 2017, 59, 134–153. [Google Scholar] [CrossRef]
- Okita, T.W. Is there an alternative pathway for starch synthesis? Plant Physiol. 1992, 100, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Hanashiro, I. Granule-bound starch synthase I is responsible for biosynthesis of extra-long unit chains of amylopectin in rice. Plant Cell Physiol. 2008, 49, 925–933. [Google Scholar] [CrossRef]
- Ball, S.G.; Morell, M.K. From bacterial glycogen to starch: Understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 2003, 54, 207. [Google Scholar] [CrossRef]
- Nicolas, S.; Paula, R.; Sandy, R.; Lucas, M.M.; Isaac, R.; Manuel, M.; Francisco, J.M.; Miroslav, O.; Abdellatif, B.; Véronique, P.; et al. Starch granule initiation in arabidopsis requires the presence of either class iv or class iii starch synthases. Plant Cell 2009, 21, 2443–2457. [Google Scholar]
- Naoko, F.; Mayumi, Y.; Noriko, A.; Takashi, O.; Akio, M.; Hirohiko, H.; Yasunori, N. Function and characterization of starch synthase i using mutants in rice. Plant Physiol. 2006, 140, 1070. [Google Scholar]
- Zhang, X.; Nicolas, S.; David, D.; Christophe, D.H.; Martha, G.J.; Alan, M.M. Overlapping functions of the starch synthases SSII and SSIII in amylopectin biosynthesis in Arabidopsis. BMC Plant Biol. 2008, 8, 96. [Google Scholar] [CrossRef]
- Hanashiro, I.; Tagawa, M.; Shibahara, S.; Iwata, K.; Takeda, Y. Examination of molar-based distribution of A, B and C chains of amylopectin by fluorescent labeling with 2-aminopyridine. Carbohydr. Res. 2002, 337, 1211–1215. [Google Scholar] [CrossRef]
- Li, Q.; Liu, X.; Zhang, C.; Jiang, L.; Jiang, M.; Zhong, M.; Fan, X.; Gu, M.; Liu, Q. Rice soluble starch synthase I: Allelic variation, expression, function, and interaction with waxy. Front. Plant Sci. 2018, 9, 1591. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Zhang, H.; Zhai, H.; Liu, Q.; He, S. A soluble starch synthase I gene, IbSSI, alters the content, composition, granule size and structure of starch in transgenic sweet potato. Sci. Rep. 2017, 7, 2315. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Smith, A.M. Starch biosynthesis. Plant Cell 1995, 7, 971. [Google Scholar]
- Borzenkova, R.A.; Borovkova, M.P. Developmental patterns of phytohormone content in the cortex and pith of potato tubers as related to their growth and starch content. Russ. J. Plant Physiol. 2003, 50, 119–124. [Google Scholar] [CrossRef]
- Seiler, C.; Harshavardhan, V.T.; Rajesh, K.; Reddy, P.S.; Strickert, M.; Rolletschek, H.; Scholz, U.; Wobus, U.; Sreenivasulu, N. ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J. Exp. Bot. 2011, 62, 2615–2632. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Ye, N.; Yang, J.; Peng, X.; Zhang, J. Regulation of expression of starch synthesis genes by ethylene and ABA in relation to the development of rice inferior and superior spikelets. J. Exp. Bot. 2011, 62, 3907–3916. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Cai, T.; Yin, Y.; Yang, W.; Ni, Y.; Yang, D.; Wang, Z. Exogenous application of abscisic acid or gibberellin acid has different effects on starch granule size distribution in grains of wheat-sciencedirect. J. Integr. Agric. 2013, 12, 1551–1559. [Google Scholar] [CrossRef]
- Yang, D.; Luo, Y.; Ni, Y.; Yin, Y.; Yang, W.; Peng, D.; Cui, Z.; Wang, Z. Effects of exogenous ABA application on post-anthesis dry matter redistribution and grain starch accumulation of winter wheat with different staygreen characteristics. Crop J. 2014, 2, 144–153. [Google Scholar] [CrossRef]
- Sebastián, R.; Ximena, N.; Francisco, J.P. ABA promotes starch synthesis and storage metabolism in dormant grapevine buds. J. Plant Physiol. 2019, 234, 1–8. [Google Scholar]
- Ramon, M.; Rolland, F.; Thevelein, J.M.; Dijck, P.V.; Leyman, B. ABI4 mediates the effects of exogenous trehalose on Arabidopsis growth and starch breakdown. Plant Mol. Biol. 2007, 63, 195–206. [Google Scholar] [CrossRef]
- Hu, Y.; Li, Y.; Zhang, J.; Liu, H.; Tian, M.; Huang, Y. Binding of ABI4 to a CACCG motif mediates the ABA-induced expression of the ZmSSI gene in maize (Zea mays L.) endosperm. J. Exp. Bot. 2012, 63, 5979–5989. [Google Scholar] [CrossRef]
- Rook, F.; Corke, F.; Card, R.; Munz, G.; Smith, C.; Bevan, M.W. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 2010, 26, 421–433. [Google Scholar] [CrossRef]
- Ming, R.; VanBuren, R.; Liu, Y.; Yang, M.; Han, Y.; Li, L.; Zhang, Q.; Kim, M.J.; Schatz, M.C.; Campbell, M.; et al. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol. 2013, 14, R41. [Google Scholar] [CrossRef]
- Shen-Miller, J. Sacred lotus, the long-living fruits of China Antique. Seed Sci. Res. 2002, 12, 131–143. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, M.; Wang, S. Processing characteristics and flavour of full lotus root powder beverage. J. Sci. Food Agric. 2010, 90, 2482–2489. [Google Scholar] [CrossRef]
- Huang, L.; Li, M.; Cao, D.; Yang, P. Genetic dissection of rhizome yield-related traits in Nelumbo nucifera through genetic linkage map construction and QTL mapping. Plant Physiol. Biochem. 2021, 160, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Shen, Q.; Hu, L.; Hu, Y.; Ye, X.; Liu, D.; Chen, J. Physicochemical properties, structure and invitro digestibility on complex of starch with lotus (Nelumbo nucifera Gaertn.) leaf flavonoids. Food Hydrocoll. 2018, 81, 191–199. [Google Scholar] [CrossRef]
- Li, L.; Zhang, X.D.; Pan, E.C.; Sun, L.; Xie, K.; Li, G.U.; Cao, B.S. Relationship of Starch Synthesis with it’s related enzymes’ activities during rhizome development of lotus(Nelumbo nucifera Gaertn). Sci. Agric. Sin. 2006, 11, 2307–2312. [Google Scholar]
- Cheng, L.; Liu, X.; Yin, J.; Yang, J.; Li, Y.; Hui, L.; Li, S.; Li, L. Activity and expression of ADP-glucose pyrophosphorylase during rhizome formation in lotus (Nelumbo nucifera Gaertn.). Bot. Stud. 2016, 57, 26. [Google Scholar] [CrossRef][Green Version]
- Lu, Y.; Li, L.; Zhou, Y.; Gao, Q.; Liang, G.; Chen, X.; Qi, X. Cloning and characterization of the wx gene encoding a granule-bound starch synthase in lotus (Nelumbo nucifera Gaertn). Plant Mol. Biol. Rep. 2012, 30, 1210–1217. [Google Scholar] [CrossRef]
- Jenner, C.F.; Hawker, J.S.; Siwek, K. The Synthesis of [14C]Starch From [14C]Sucrose in Isolated wheat grains is dependent upon the activity of soluble starch synthase. Plant Physiol. 1993, 20, 329–335. [Google Scholar]
- Li, L.; Shao, T.; Yang, H.; Chen, M.; Gao, X.; Long, X.; Shao, H.; Liu, Z.; Rengel, Z. The endogenous plant hormones and ratios regulate sugar and dry matter accumulation in Jerusalem artichoke in salt-soil. Sci. Total Environ. 2017, 578, 40–46. [Google Scholar] [CrossRef]
- Kobashi, K.; Gemma, H.; Iwahori, S. Sugar accumulation in peach fruit as affected by abscisic acid (ABA) treatment in relation to some sugar metabolizing enzymes. J. Jpn. Soc. Hortic. Sci. 1999, 68, 465–470. [Google Scholar] [CrossRef]
- Josef, B. Effect of abscisic acid on sorbitol uptake in growing apple fruits. J. Exp. Bot. 1983, 34, 737–743. [Google Scholar]
- Ma, Q.; Sun, M.; Liu, Y.; Lu, J.; Hu, D.; Hao, Y. Molecular cloning and functional characterization of the apple sucrose transporter gene MdSUT2. Plant Physiol. Biochem. 2016, 109, 442–451. [Google Scholar] [CrossRef]
- Wang, G.Q.; Chen, M.X.; Zhang, J.H.; Gao, B.; Liu, Y.G. Regulation gene expression in the remobilization of carbon reserves in rice stems during grain filling. Plant Cell Physiol. 2017, 58, 1391–1404. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, W.; Ju, C.; Li, Y.; Yang, J.; Zhang, J. Involvement of abscisic acid in fructan hydrolysis and starch biosynthesis in wheat under soil drying. Plant Growth Regul. 2016, 80, 265–279. [Google Scholar] [CrossRef]
- Takashi, A.; Kouichi, M.; Tatsuhito, F. Gene expression of ADP-glucose pyrophosphorylase and starch contents in rice cultured cells are cooperatively regulated by sucrose and ABA. Plant Cell Physiol. 2005, 46, 937–946. [Google Scholar]
- Akihiro, T.; Umezawa, T.; Ueki, C.; Lobna, B.M.; Mizuno, K.; Ohta, M.; Fujimura, T. Genome wide cDNA-AFLP analysis of genes rapidly induced by combined sucrose and ABA treatment in rice cultured cells. FEBS Lett. 2006, 580, 5947–5952. [Google Scholar] [CrossRef][Green Version]
- Chen, J.; Huang, B.; Li, Y.; Du, H.; Gu, Y.G.; Liu, H.; Zhang, J.; Huang, Y. Synergistic influence of sucrose and abscisic acid on the genes involved in starch synthesis in maize endosperm-ScienceDirect. Carbohydr. Res. 2011, 346, 1684–1691. [Google Scholar] [CrossRef] [PubMed]
- Bossi, F.; Cordoba, E.; Dupré, P.; Mendoza, M.S.; Román, C.S.; León, P. The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J. 2009, 59, 359–374. [Google Scholar] [CrossRef]
- Kong, Y.; Chen, S.; Yang, Y.; An, C. ABA-insensitive (ABI) 4 and ABI5 synergistically regulate DGAT1 expression in Arabidopsis seedlings under stress. FEBS Lett. 2013, 587, 3076–3082. [Google Scholar] [CrossRef] [PubMed]
- Wind, J.J.; Peviani, A.; Snel, B.; Hanson, J.; Smeekens, S.C. ABI4: Versatile activator and repressor. Trends Plant Sci. 2013, 18, 125–132. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Niu, X.; Helentjaris, T.; Bate, N.J. Maize ABI4 binds coupling element1 in abscisic acid and sugar response genes. Plant Cell. 2002, 14, 2565–2575. [Google Scholar] [CrossRef]
- Acevedo-Hernández, G.J.; León, P.; Herrera-Estrella, L.R. Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. Plant J. 2005, 43, 506–519. [Google Scholar] [CrossRef]
- Koussevitzky, S.; Nott, A.; Mockler, T.C.; Hong, F.; Sachetto-Martins, G.; Surpin, M.; Lim, J.; Mittler, R.; Chory, J. Signals from chloroplasts converge to regulate nuclear gene expression. Science 2007, 316, 715–719. [Google Scholar] [CrossRef]
- Rook, F.; Hadingham, S.A.; Li, Y.; Bevan, M.W. Sugar and ABA response pathways and the control of gene expression. Plant Cell Environ. 2010, 29, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Xie, S.X.; Xiao, Q.; Wei, B.; Zheng, L.; Wang, Y.; Cao, Y.; Zhang, X.; Long, T.; Li, Y.; et al. Sucrose and ABA regulate starch biosynthesis in maize through a novel transcription factor, ZmEREB156. Sci. Rep. 2016, 6, 27590. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, P.; Liu, A.; Zhang, Y.; Feng, K.; Zhao, S.; Li, L. NnABI4-Mediated ABA Regulation of Starch Biosynthesis in Lotus (Nelumbo nucifera Gaertn). Int. J. Mol. Sci. 2021, 22, 13506. https://doi.org/10.3390/ijms222413506
Wu P, Liu A, Zhang Y, Feng K, Zhao S, Li L. NnABI4-Mediated ABA Regulation of Starch Biosynthesis in Lotus (Nelumbo nucifera Gaertn). International Journal of Molecular Sciences. 2021; 22(24):13506. https://doi.org/10.3390/ijms222413506
Chicago/Turabian StyleWu, Peng, Ailian Liu, Yongyan Zhang, Kai Feng, Shuping Zhao, and Liangjun Li. 2021. "NnABI4-Mediated ABA Regulation of Starch Biosynthesis in Lotus (Nelumbo nucifera Gaertn)" International Journal of Molecular Sciences 22, no. 24: 13506. https://doi.org/10.3390/ijms222413506
APA StyleWu, P., Liu, A., Zhang, Y., Feng, K., Zhao, S., & Li, L. (2021). NnABI4-Mediated ABA Regulation of Starch Biosynthesis in Lotus (Nelumbo nucifera Gaertn). International Journal of Molecular Sciences, 22(24), 13506. https://doi.org/10.3390/ijms222413506





