Innate Immunity as an Executor of the Programmed Death of Individual Organisms for the Benefit of the Entire Population
Abstract
1. Introduction
2. Innate Immunity, Pathogen-Associated Molecular Patterns, and Pattern Recognition Receptors
3. Damage Associated Molecular Patterns (DAMPs) and Their Receptors
4. PAMP- and DAMP-Dependent Pathologies
4.1. Bacterial Infections
4.2. Viral Infections
4.3. COVID-19
4.4. Sepsis
4.5. Sterile Inflammation
4.6. Neuroinflammation and Neurodegeneration
5. Suppression of Inflammation by Targeting Mitochondria
6. Biomarkers of the All-Cause Death
7. Innate Immunity-Mediated Phenoptosis as a Common Cause of Human Mortality
8. Phenoptosis and Aging
9. Co-Evolution of Immunity and Phenoptosis
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Skulachev, V.P. Phenoptosis: Programmed death of an organism. Biochemistry 1999, 64, 1418–1426. [Google Scholar]
- Skulachev, V.P. Aging is a specific biological function rather than the result of a disorder in complex living systems: Biochemical evidence in support of Weismann’s hypothesis. Biochemistry 1997, 62, 1191–1195. [Google Scholar] [PubMed]
- Hamilton, W.D. The genetical evolution of social behaviour. I. J. Theor. Biol. 1964, 7, 17–52. [Google Scholar] [CrossRef]
- Skulachev, V.P.; Shilovsky, G.A.; Putyatina, T.S.; Popov, N.A.; Markov, A.V.; Skulachev, M.V.; Sadovnichii, V.A. Perspectives of Homo sapiens lifespan extension: Focus on external or internal resources. Aging 2020, 12, 5566–5584. [Google Scholar] [CrossRef]
- Jiang, W.; Wei, Y.; Long, Y.; Owen, A.; Wang, B.; Wu, X.; Luo, S.; Dang, Y.; Ma, D.K. A genetic program mediates cold-warming response and promotes stress-induced phenoptosis in C. elegans. eLife 2018, 7, e35037. [Google Scholar] [CrossRef] [PubMed]
- Lohr, J.N.; Galimov, E.R.; Gems, D. Does senescence promote fitness in Caenorhabditis elegans by causing death? Ageing Res. Rev. 2019, 50, 58–71. [Google Scholar] [CrossRef]
- Galimov, E.R.; Gems, D. Shorter life and reduced fecundity can increase colony fitness in virtual Caenorhabditis elegans. Aging Cell 2020, 19, e13141. [Google Scholar] [CrossRef] [PubMed]
- Lidsky, P.V.; Andino, R. Epidemics as an adaptive driving force determining lifespan setpoints. Proc. Natl. Acad. Sci. USA 2020, 117, 17937–17948. [Google Scholar] [CrossRef]
- Nirmala, J.G.; Lopus, M. Cell death mechanisms in eukaryotes. Cell Biol. Toxicol. 2020, 36, 145–164. [Google Scholar] [CrossRef] [PubMed]
- Janeway, C.A. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 1989, 54 Pt 1, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Barton, G.M.; Medzhitov, R. Toll-like receptors and their ligands. Curr. Top. Microbiol. Immunol. 2002, 270, 81–92. [Google Scholar]
- Janeway, C.A.; Medzhitov, R. Lipoproteins take their toll on the host. Curr. Biol. 1999, 9, R879–R882. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Palm, N.W.; Medzhitov, R. Pattern recognition receptors and control of adaptive immunity. Immunol. Rev. 2009, 227, 221–233. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Geijtenbeek, T.B.; Gringhuis, S.I. Signalling through C-type lectin receptors: Shaping immune responses. Nat. Rev. Immunol. 2009, 9, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Karijolich, J. Know thyself: RIG-I-like receptor sensing of DNA virus infection. J. Virol. 2019, 93, e01085-19. [Google Scholar] [CrossRef]
- Franchi, L.; Warner, N.; Viani, K.; Nuñez, G. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 2009, 227, 106–128. [Google Scholar] [CrossRef]
- Man, S.M.; Kanneganti, T.D. Regulation of inflammasome activation. Immunol. Rev. 2015, 265, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Platnich, J.M.; Muruve, D.A. NOD-like receptors and inflammasomes: A review of their canonical and non-canonical signaling pathways. Arch. Biochem. Biophys. 2019, 670, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Briard, B.; Place, D.E.; Kanneganti, T.D. DNA sensing in the innate immune response. Physiology 2020, 35, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Bai, X.C.; Chen, Z.J. Structures and Mechanisms in the cGAS-STING Innate Immunity Pathway. Immunity 2020, 53, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Matzinger, P. Tolerance, danger, and the extended family. Annu. Rev. Immunol. 1994, 12, 991–1045. [Google Scholar] [CrossRef] [PubMed]
- Seong, S.Y.; Matzinger, P. Hydrophobicity: An ancient damage-associated molecular pattern that initiates innate immune responses. Nat. Rev. Immunol. 2004, 4, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Han, Z.; Oppenheim, J.J. Alarmins and immunity. Immunol. Rev. 2017, 280, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Gong, T.; Liu, L.; Jiang, W.; Zhou, R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 2020, 20, 95–112. [Google Scholar] [CrossRef] [PubMed]
- Man, S.M.; Karki, R.; Kanneganti, T.D. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol. Rev. 2017, 277, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Thiam, H.R.; Wong, S.L.; Wagner, D.D.; Waterman, C.M. Cellular mechanisms of NETosis. Annu. Rev. Cell Dev. Biol. 2020, 36, 191–218. [Google Scholar] [CrossRef]
- Andersson, U.; Yang, H.; Harris, H. High-mobility group box 1 protein (HMGB1) operates as an alarmin outside as well as inside cells. Semin. Immunol. 2018, 38, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.; Brenner, M.; Wang, P. Extracellular CIRP (eCIRP) and inflammation. J. Leukoc. Biol. 2019, 106, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Roan, F.; Obata-Ninomiya, K.; Ziegler, S.F. Epithelial cell-derived cytokines: More than just signaling the alarm. J. Clin. Investig. 2019, 129, 1441–1451. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, S.; Zelenay, S.; Sancho, D.; Hanc, P.; Kjaer, S.; Feest, C.; Fletcher, G.; Durkin, C.; Postigo, A.; Skehel, M.; et al. F-actin Is an evolutionarily conserved damage-sssociated molecular pattern Rrecognized by DNGR-1, a receptor for dead cells. Immunity 2012, 36, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Bukrinsky, M. Extracellular cyclophilins in health and disease. Biochim. Biophys. Acta 2015, 1850, 2087–2095. [Google Scholar] [CrossRef]
- Riddell, J.R.; Wang, X.Y.; Minderman, H.; Gollnick, S.O. Peroxiredoxin 1 stimulates secretion of proinflammatory cytokines by binding to TLR4. J. Immunol. 2010, 184, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Quintana, F.J.; Cohen, I.R. Heat shock proteins as endogenous adjuvants in sterile and septic inflammation. J. Immunol. 2005, 175, 2777–2782. [Google Scholar] [CrossRef]
- Bozza, M.T.; Jeney, V. Pro-inflammatory actions of heme and other hemoglobin-derived DAMPs. Front. Immunol. 2020, 11, 1323. [Google Scholar] [CrossRef] [PubMed]
- Fruitwala, S.; El-Naccache, D.W.; Chang, T.L. Multifaceted immune functions of human defensins and underlying mechanisms. Semin. Cell Dev. Biol. 2019, 88, 163–172. [Google Scholar] [CrossRef]
- Yang, D.; Chen, Q.; Su, S.B.; Zhang, P.; Kurosaka, K.; Caspi, R.R.; Michalek, S.M.; Rosenberg, H.F.; Zhang, N.; Oppenheim, J.J. Eosinophil-derived neurotoxin acts as an alarmin to activate the TLR2-MyD88 signal pathway in dendritic cells and enhances Th2 immune responses. J. Exp. Med. 2008, 205, 79–90. [Google Scholar] [CrossRef]
- Tewary, P.; Yang, D.; de la Rosa, G.; Li, Y.; Finn, M.W.; Krensky, A.M.; Clayberger, C.; Oppenheim, J.J. Granulysin activates antigen-presenting cells through TLR4 and acts as an immune alarmin. Blood 2010, 116, 3465–3474. [Google Scholar] [CrossRef]
- Idzko, M.; Ferrari, D.; Eltzschig, H.K. Nucleotide signalling during inflammation. Nature 2014, 509, 310–317. [Google Scholar] [CrossRef]
- Joosten, L.A.B.; Crişan, T.O.; Bjornstad, P.; Johnson, R.J. Asymptomatic hyperuricaemia: A silent activator of the innate immune system. Nat. Rev. Rheumatol. 2020, 16, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Frevert, C.W.; Felgenhauer, J.; Wygrecka, M.; Nastase, M.V.; Schaefer, L. Danger-associated molecular patterns derived from the extracellular matrix provide temporal control of innate immunity. J. Histochem. Cytochem. 2018, 66, 213–227. [Google Scholar] [CrossRef]
- Rodríguez-Nuevo, A.; Zorzano, A. The sensing of mitochondrial DAMPs by non-immune cells. Cell Stress 2019, 3, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, T.J.; Bajwa, E.; Klegeris, A. Cytochrome c can be released into extracellular space and modulate functions of human astrocytes in a toll-like receptor 4-dependent manner. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 129400. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Warren, S.; Adjemian, S.; Agostinis, P.; Martinez, A.B.; Chan, T.A.; Coukos, G.; Demaria, S.; Deutsch, E.; et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J. Immunother. Cancer 2020, 8, e000337. [Google Scholar] [CrossRef]
- Denning, N.L.; Aziz, M.; Gurien, S.D.; Wang, P. DAMPs and NETs in sepsis. Front. Immunol. 2019, 10, 2536. [Google Scholar] [CrossRef] [PubMed]
- Ekaney, M.L.; Otto, G.P.; Sossdorf, M.; Sponholz, C.; Boehringer, M.; Loesche, W.; Rittirsch, D.; Wilharm, A.; Kurzai, O.; Bauer, M.; et al. Impact of plasma histones in human sepsis and their contribution to cellular injury and inflammation. Crit. Care 2014, 18, 543. [Google Scholar] [CrossRef]
- Calvo-Rodriguez, M.; García-Rodríguez, C.; Villalobos, C.; Núñez, L. Role of Toll like receptor 4 in Alzheimer’s disease. Front. Immunol. 2020, 11, 1588. [Google Scholar] [CrossRef]
- Iyer, S.S.; He, Q.; Janczy, J.R.; Elliott, E.I.; Zhong, Z.; Olivier, A.K.; Sadler, J.J.; Knepper-Adrian, V.; Han, R.; Qiao, L. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity 2013, 39, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Tyurina, Y.Y.; Poloyac, S.M.; Tyurin, V.A.; Kapralov, A.A.; Jiang, J.; Anthonymuthu, T.S.; Kapralova, V.I.; Vikulina, A.S.; Jung, M.-Y.; Epperly, M.W. A mitochondrial pathway for biosynthesis of lipid mediators. Nat. Chem. 2014, 6, 542–552. [Google Scholar] [CrossRef]
- Rocha, D.; Caldas, A.; Oliveira, L.; Bressan, J.; Hermsdorff, H. Saturated fatty acids trigger TLR4-mediated inflammatory response. Atherosclerosis 2016, 244, 211–215. [Google Scholar] [CrossRef]
- Basu, S.; Binder, R.J.; Ramalingam, T.; Srivastava, P.K. CD91 is a common receptor for heat shock proteins gp96, hsp90, hsp70, and calreticulin. Immunity 2001, 14, 303–313. [Google Scholar] [CrossRef]
- Zhou, B.; Liu, J.; Zeng, L.; Zhu, S.; Wang, H.; Billiar, T.R.; Kroemer, G.; Klionsky, D.J.; Zeh, H.J.; Jiang, J.; et al. Extracellular SQSTM1 mediates bacterial septic death in mice through insulin receptor signalling. Nat. Microbiol. 2020, 5, 1576–1587. [Google Scholar] [CrossRef] [PubMed]
- Migeotte, I.; Communi, D.; Parmentier, M. Formyl peptide receptors: A promiscuous subfamily of G protein-coupled receptors controlling immune responses. Cytokine Growth Factor Rev. 2006, 17, 501–519. [Google Scholar] [CrossRef] [PubMed]
- Shimada, K.; Crother, T.R.; Karlin, J.; Dagvadorj, J.; Chiba, N.; Chen, S.; Ramanujan, V.K.; Wolf, A.J.; Vergnes, L.; Ojcius, D.M.; et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 2012, 36, 401–414. [Google Scholar] [CrossRef]
- Orozco, L.D.; Bennett, B.J.; Farber, C.R.; Ghazalpour, A.; Pan, C.; Che, N.; Wen, P.; Qi, H.X.; Mutukulu, A.; Siemers, N.; et al. Unraveling inflammatory responses using systems genetics and gene-environment interactions in macrophages. Cell 2012, 151, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Bourque, D.L.; Bhuiyan, T.R.; Genereux, D.P.; Rashu, R.; Ellis, C.N.; Chowdhury, F.; Khan, A.I.; Alam, N.H.; Paul, A.; Hossain, L.; et al. Analysis of the human mucosal response to cholera reveals sustained activation of innate immune signaling pathways. Infect. Immun. 2018, 86, e00594-17. [Google Scholar] [CrossRef] [PubMed]
- Satitsri, S.; Pongkorpsakol, P.; Srimanote, P.; Chatsudthipong, V.; Muanprasat, C. Pathophysiological mechanisms of diarrhea caused by the Vibrio cholerae O1 El Tor variant: An in vivo study in mice. Virulence 2016, 7, 789–805. [Google Scholar] [CrossRef] [PubMed]
- John, C.M.; Phillips, N.J.; Stein, D.C.; Jarvis, G.A. Innate immune response to lipooligosaccharide: Pivotal regulator of the pathobiology of invasive Neisseria meningitidis infections. Pathog. Dis. 2017, 75, ftx030. [Google Scholar] [CrossRef] [PubMed]
- Tolle, L.B.; Standiford, T.J. Danger-associated molecular patterns (DAMPs) in acute lung injury. J. Pathol. 2013, 229, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Raoof, M.; Chen, Y.; Sumi, Y.; Sursal, T.; Junger, W.; Brohi, K.; Itagaki, K.; Hauser, C.J. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 2010, 464, 104–107. [Google Scholar] [CrossRef]
- Potey, P.M.; Rossi, A.G.; Lucas, C.D.; Dorward, D.A. Neutrophils in the initiation and resolution of acute pulmonary inflammation: Understanding biological function and therapeutic potential. J. Pathol. 2019, 247, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Y.; Segovia, J.A.; Chang, T.H.; Morris, I.R.; Berton, M.T.; Tessier, P.A.; Tardif, M.R.; Cesaro, A.; Bose, S. DAMP molecule S100A9 acts as a molecular pattern to enhance inflammation during influenza A virus infection: Role of DDX21-TRIF-TLR4-MyD88 pathway. PLoS Pathog. 2014, 10, e1003848. [Google Scholar] [CrossRef]
- Nosaka, N.; Yashiro, M.; Yamada, M.; Fujii, Y.; Tsukahara, H.; Liu, K.; Nishibori, M.; Matsukawa, A.; Morishima, T. Anti-high mobility group box-1 monoclonal antibody treatment provides protection against influenza A virus (H1N1)-induced pneumonia in mice. Crit. Care 2015, 19, 249. [Google Scholar] [CrossRef]
- Imai, Y.; Kuba, K.; Neely, G.G.; Yaghubian-Malhami, R.; Perkmann, T.; van Loo, G.; Ermolaeva, M.; Veldhuizen, R.; Leung, Y.H.; Wang, H.; et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell 2008, 133, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Nhu, Q.M.; Shirey, K.; Teijaro, J.R.; Farber, D.L.; Netzel-Arnett, S.; Antalis, T.M.; Fasano, A.; Vogel, S.N. Novel signaling interactions between proteinase-activated receptor 2 and Toll-like receptors in vitro and in vivo. Mucosal Immunol. 2010, 3, 29–39. [Google Scholar] [CrossRef]
- Hosakote, Y.M.; Brasier, A.R.; Casola, A.; Garofalo, R.P.; Kurosky, A. Respiratory syncytial virus infection triggers epithelial HMGB1 release as a damage-associated molecular pattern promoting a monocytic inflammatory response. J. Virol. 2016, 90, 9618–9631. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.; Loh, Z.; Ullah, M.A.; Lynch, J.P.; Werder, R.B.; Collinson, N.; Zhang, V.; Dondelinger, Y.; Bertrand, M.J.M.; Everard, M.L.; et al. Respiratory syncytial virus infection promotes necroptosis and HMGB1 release by airway epithelial cells. Am. J. Respir. Crit. Care Med. 2020, 201, 1358–1371. [Google Scholar] [CrossRef] [PubMed]
- Dib, P.R.B.; Quirino-Teixeira, A.C.; Merij, L.B.; Pinheiro, M.B.M.; Rozini, S.V.; Andrade, F.B.; Hottz, E.D. Innate immune receptors in platelets and platelet-leukocyte interactions. J. Leukoc. Biol. 2020, 108, 1157–1182. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Chen, Y.; Xia, H.; Wang, C.; Tan, C.Y.; Cai, X.; Liu, Y.; Ji, F.; Xiong, P.; Liu, R.; et al. Transcriptional and proteomic insights into the host response in fatal COVID-19 cases. Proc. Natl. Acad. Sci. USA 2020, 117, 28336–28343. [Google Scholar] [CrossRef] [PubMed]
- Cain, D.W.; Cidlowski, J.A. After 62 years of regulating immunity, dexamethasone meets COVID-19. Nat. Rev. Immunol. 2020, 20, 587–588. [Google Scholar] [CrossRef]
- Chen, L.; Long, X.; Xu, Q.; Tan, J.; Wang, G.; Cao, Y.; Wei, J.; Luo, H.; Zhu, H.; Huang, L.; et al. Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients. Cell Mol. Immunol. 2020, 17, 992–994. [Google Scholar] [CrossRef]
- Chen, R.; Huang, Y.; Quan, J.; Liu, J.; Wang, H.; Billiar, T.R.; Lotze, M.T.; Zeh, H.J.; Kang, R.; Tang, D. HMGB1 as a potential biomarker and therapeutic target for severe COVID-19. Heliyon 2020, 6, e05672. [Google Scholar] [CrossRef]
- Silvin, A.; Chapuis, N.; Dunsmore, G.; Goubet, A.G.; Dubuisson, A.; Derosa, L.; Almire, C.; Hénon, C.; Kosmider, O.; Droin, N.; et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell 2020, 182, 1401.e18–1418.e18. [Google Scholar] [CrossRef]
- Guo, Q.; Zhao, Y.; Li, J.; Liu, J.; Yang, X.; Guo, X.; Kuang, M.; Xia, H.; Zhang, Z.; Cao, L.; et al. Induction of alarmin S100A8/A9 mediates activation of aberrant neutrophils in the pathogenesis of COVID-19. Cell Host Microbe 2021, 29, 222.e4–235.e4. [Google Scholar] [CrossRef] [PubMed]
- Andargie, T.E.; Tsuji, N.; Seifuddin, F.; Jang, M.K.; Yuen, P.S.; Kong, H.; Tunc, I.; Singh, K.; Charya, A.; Wilkins, K.; et al. Cell-free DNA maps COVID-19 tissue injury and risk of death and can cause tissue injury. JCI Insight 2021, 6, e147610. [Google Scholar] [CrossRef]
- Scozzi, D.; Cano, M.; Ma, L.; Zhou, D.; Zhu, J.H.; O’Halloran, J.A.; Goss, C.; Rauseo, A.M.; Liu, Z.; Sahu, S.K.; et al. Circulating mitochondrial DNA is an early indicator of severe illness and mortality from COVID-19. JCI Insight 2021, 6, e143299. [Google Scholar] [CrossRef] [PubMed]
- Zizzo, G.; Cohen, P.L. Imperfect storm: Is interleukin-33 the Achilles heel of COVID-19? Lancet Rheumatol. 2020, 2, e779–e790. [Google Scholar] [CrossRef]
- Sundén-Cullberg, J.; Norrby-Teglund, A.; Rouhiainen, A.; Rauvala, H.; Herman, G.; Tracey, K.J.; Lee, M.L.; Andersson, J.; Tokics, L.; Treutiger, C.J. Persistent elevation of high mobility group box-1 protein (HMGB1) in patients with severe sepsis and septic shock. Crit. Care Med. 2005, 33, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Dong, H.; Zhong, Y.; Huang, J.; Lv, J.; Li, J. The cold-inducible RNA-binding protein (CIRP) level in peripheral blood predicts sepsis outcome. PLoS ONE 2015, 10, e0137721. [Google Scholar] [CrossRef]
- Gelain, D.P.; de Bittencourt Pasquali, M.A.; Comim, C.M.; Grunwald, M.S.; Ritter, C.; Tomasi, C.D.; Alves, S.C.; Quevedo, J.; Dal-Pizzol, F.; Moreira, J.C. Serum heat shock protein 70 levels, oxidant status, and mortality in sepsis. Shock 2011, 35, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, D.J.; Toltl, L.J.; Swystun, L.L.; Pogue, J.; Liaw, K.L.; Weitz, J.I.; Cook, D.J.; Fox-Robichaud, A.E.; Liaw, P.C.; Canadian Critical Care Translational Biology Group. Prognostic utility and characterization of cell-free DNA in patients with severe sepsis. Crit. Care 2012, 16, R151. [Google Scholar] [CrossRef]
- Li, S.; Hu, Q.; Huang, J.; Wu, X.; Ren, J. Mitochondria-derived damage-associated molecular patterns in sepsis: From bench to bedside. Oxid. Med. Cell Longev. 2019, 2019, 6914849. [Google Scholar] [CrossRef] [PubMed]
- Savio, L.E.B.; de Andrade Mello, P.; da Silva, C.G.; Coutinho-Silva, R. The P2X7 Receptor in Inflammatory Diseases: Angel or Demon? Front. Pharmacol. 2018, 9, 52. [Google Scholar] [CrossRef]
- Timmermans, K.; Kox, M.; Scheffer, G.J.; Pickkers, P. Danger in the intensive care unit: DAMPs in critically ill patients. Shock 2016, 45, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V. Toll-like receptors in the pathogenesis of neuroinflammation. J. Neuroimmunol. 2019, 332, 16–30. [Google Scholar] [CrossRef] [PubMed]
- Venegas, C.; Heneka, M.T. Danger-associated molecular patterns in Alzheimer’s disease. J. Leukoc. Biol. 2017, 101, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Alam, A.; Hana, Z.; Jin, Z.; Suen, K.C.; Ma, D. Surgery, neuroinflammation and cognitive impairment. EBioMedicine 2018, 37, 547–556. [Google Scholar] [CrossRef]
- Boncyk, C.S.; Rengel, K.F.; Pandharipande, P.P.; Hughes, C.G. In the ICU—delirium post cardiac arrest. Curr. Opin. Crit. Care 2019, 25, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Ferro, J.M.; Oliveira, S. Neurologic manifestations of gastrointestinal and liver diseases. Curr. Neurol. Neurosci. Rep. 2014, 14, 487. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Kiernan, M.C.; Murray, A.; Rosner, M.H.; Ronco, C. Kidney-brain crosstalk in the acute and chronic setting. Nat. Rev. Nephrol. 2015, 11, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, M.E.; Christie, J.D.; Lanken, P.N.; Biester, R.C.; Thompson, B.T.; Bellamy, S.L.; Localio, A.R.; Demissie, E.; Hopkins, R.O.; Angus, D.C. The adult respiratory distress syndrome cognitive outcomes study: Long-term neuropsychological function in survivors of acute lung injury. Am. J. Respir. Crit. Care Med. 2012, 185, 1307–1315. [Google Scholar] [CrossRef] [PubMed]
- Mazeraud, A.; Righy, C.; Bouchereau, E.; Benghanem, S.; Bozza, F.A.; Sharshar, T. Septic-associated encephalopathy: A Comprehensive review. Neurotherapeutics 2020, 17, 392–403. [Google Scholar] [CrossRef] [PubMed]
- Lund-Sørensen, H.; Benros, M.E.; Madsen, T.; Sørensen, H.J.; Eaton, W.W.; Postolache, T.T.; Nordentoft, M.; Erlangsen, A. A nationwide cohort study of the association between hospitalization with infection and risk of death by suicide. JAMA Psychiatry 2016, 73, 912–919. [Google Scholar] [CrossRef]
- Hoogland, I.C.; Houbolt, C.; van Westerloo, D.J.; van Gool, W.A.; van de Beek, D. Systemic inflammation and microglial activation: Systematic review of animal experiments. J. Neuroinflamm. 2015, 12, 114. [Google Scholar] [CrossRef]
- Zrzavy, T.; Höftberger, R.; Berger, T.; Rauschka, H.; Butovsky, O.; Weiner, H.; Lassmann, H. Pro-inflammatory activation of microglia in the brain of patients with sepsis. Neuropathol. Appl. Neurobiol. 2019, 45, 278–290. [Google Scholar] [CrossRef]
- Arumugam, T.V.; Okun, E.; Tang, S.C.; Thundyil, J.; Taylor, S.M.; Woodruff, T.M. Toll-like receptors in ischemia-reperfusion injury. Shock 2009, 32, 4–16. [Google Scholar] [CrossRef]
- Zhang, J.; Takahashi, H.K.; Liu, K.; Wake, H.; Liu, R.; Maruo, T.; Date, I.; Yoshino, T.; Ohtsuka, A.; Mori, S.; et al. Anti-high mobility group box-1 monoclonal antibody protects the blood-brain barrier from ischemia-induced disruption in rats. Stroke 2011, 42, 1420–1428. [Google Scholar] [CrossRef]
- Helms, J.; Kremer, S.; Merdji, H.; Clere-Jehl, R.; Schenck, M.; Kummerlen, C.; Collange, O.; Boulay, C.; Fafi-Kremer, S.; Ohana, M.; et al. Neurologic features in severe SARS-CoV-2 infection. N. Engl. J. Med. 2020, 382, 2268–2270. [Google Scholar] [CrossRef]
- Mehta, M.M.; Weinberg, S.E.; Chandel, N.S. Mitochondrial control of immunity: Beyond ATP. Nat. Rev. Immunol. 2017, 17, 608–620. [Google Scholar] [CrossRef]
- Mills, E.L.; Kelly, B.; O’Neill, L.A.J. Mitochondria are the powerhouses of immunity. Nat. Immunol. 2017, 18, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Pinegin, B.; Vorobjeva, N.; Pashenkov, M.; Chernyak, B. The role of mitochondrial ROS in antibacterial immunity. J. Cell Physiol. 2018, 233, 3745–3754. [Google Scholar] [CrossRef]
- Vorobjeva, N.; Galkin, I.; Pletjushkina, O.; Golyshev, S.; Zinovkin, R.; Prikhodko, A.; Pinegin, V.; Kondratenko, I.; Pinegin, B.; Chernyak, B. Mitochondrial permeability transition pore is involved in oxidative burst and NETosis of human neutrophils. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165664. [Google Scholar] [CrossRef]
- West, A.P.; Shadel, G.S.; Ghosh, S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 2011, 11, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Skulachev, V.P.; Anisimov, V.N.; Antonenko, Y.N.; Bakeeva, L.E.; Chernyak, B.V.; Erichev, V.P.; Filenko, O.F.; Kalinina, N.I.; Kapelko, V.I.; Kolosova, N.G. An attempt to prevent senescence: A mitochondrial approach. Biochim. Biophys. Acta (BBA)-Bioenerg. 2009, 1787, 437–461. [Google Scholar] [CrossRef]
- Ji, J.; Kline, A.E.; Amoscato, A.; Samhan-Arias, A.K.; Sparvero, L.J.; Tyurin, V.A.; Tyurina, Y.Y.; Fink, B.; Manole, M.D.; Puccio, A.M. Lipidomics identifies cardiolipin oxidation as a mitochondrial target for redox therapy of brain injury. Nat. Neurosci. 2012, 15, 1407. [Google Scholar] [CrossRef] [PubMed]
- Lokhmatikov, A.V.; Voskoboynikova, N.; Cherepanov, D.A.; Skulachev, M.V.; Steinhoff, H.-J.; Skulachev, V.P.; Mulkidjanian, A.Y. Impact of antioxidants on cardiolipin oxidation in liposomes: Why mitochondrial cardiolipin serves as an apoptotic signal? Oxidative Med. Cell. Longev. 2016, 2016, 8679469. [Google Scholar] [CrossRef] [PubMed]
- Mulkidjanian, A.; Shalaeva, D.; Lyamzaev, K.; Chernyak, B. Does oxidation of mitochondrial cardiolipin trigger a chain of antiapoptotic reactions? Biochemistry 2018, 83, 1263–1278. [Google Scholar] [CrossRef] [PubMed]
- Lyamzaev, K.G.; Panteleeva, A.A.; Karpukhina, A.A.; Galkin, I.I.; Popova, E.N.; Pletjushkina, O.Y.; Rieger, B.; Busch, K.B.; Mulkidjanian, A.Y.; Chernyak, B.V. Novel fluorescent mitochondria-targeted probe MitoCLox reports lipid peroxidation in response to oxidative stress in vivo. Oxidative Med. Cell. Longev. 2020, 2020, 3631272. [Google Scholar] [CrossRef]
- Lyamzaev, K.G.; Tokarchuk, A.V.; Panteleeva, A.A.; Mulkidjanian, A.Y.; Skulachev, V.P.; Chernyak, B.V. Induction of autophagy by depolarization of mitochondria. Autophagy 2018, 14, 921–924. [Google Scholar] [CrossRef] [PubMed]
- Prola, A.; Blondelle, J.; Vandestienne, A.; Piquereau, J.; Denis, R.G.P.; Guyot, S.; Chauvin, H.; Mourier, A.; Maurer, M.; Henry, C.; et al. Cardiolipin content controls mitochondrial coupling and energetic efficiency in muscle. Sci. Adv. 2021, 7, eabd6322. [Google Scholar] [CrossRef] [PubMed]
- Kagan, V.E.; Tyurin, V.A.; Jiang, J.; Tyurina, Y.Y.; Ritov, V.B.; Amoscato, A.A.; Osipov, A.N.; Belikova, N.A.; Kapralov, A.A.; Kini, V. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat. Chem. Biol. 2005, 1, 223–232. [Google Scholar] [CrossRef]
- Kagan, V.E.; Bayır, H.A.; Belikova, N.A.; Kapralov, O.; Tyurina, Y.Y.; Tyurin, V.A.; Jiang, J.; Stoyanovsky, D.A.; Wipf, P.; Kochanek, P.M. Cytochrome c/cardiolipin relations in mitochondria: A kiss of death. Free Radic. Biol. Med. 2009, 46, 1439–1453. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, K.; Raundhal, M.; Chen, B.B.; Morse, C.; Tyurina, Y.Y.; Khare, A.; Oriss, T.B.; Huff, R.; Lee, J.S.; Croix, C.M.S. The mito-DAMP cardiolipin blocks IL-10 production causing persistent inflammation during bacterial pneumonia. Nat. Commun. 2017, 8, 13944. [Google Scholar] [CrossRef]
- Zhou, R.; Yazdi, A.S.; Menu, P.; Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011, 469, 221–225. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A. Cardiolipin and the Nlrp3 inflammasome. Cell Metab. 2013, 18, 610–612. [Google Scholar] [CrossRef] [PubMed]
- Bozza, F.A.; D’Avila, J.C.; Ritter, C.; Sonneville, R.; Sharshar, T.; Dal-Pizzol, F. Bioenergetics, mitochondrial dysfunction, and oxidative stress in the pathophysiology of septic encephalopathy. Shock 2013, 39 (Suppl. S1), 10–16. [Google Scholar] [CrossRef] [PubMed]
- Veloso, C.D.; Belew, G.D.; Ferreira, L.L.; Grilo, L.F.; Jones, J.G.; Portincasa, P.; Sardão, V.A.; Oliveira, P.J. A mitochondrial approach to cardiovascular risk and disease. Curr. Pharm. Des. 2019, 25, 3175–3194. [Google Scholar] [CrossRef] [PubMed]
- Bader, V.; Winklhofer, K.F. Mitochondria at the interface between neurodegeneration and neuroinflammation. Semin. Cell Dev. Biol. 2020, 99, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.H.; Griffiths, H.R. The dual role of Reactive Oxygen Species in autoimmune and inflammatory diseases: Evidence from preclinical models. Free Radic. Biol. Med. 2018, 125, 62–71. [Google Scholar] [CrossRef]
- Prauchner, C.A. Oxidative stress in sepsis: Pathophysiological implications justifying antioxidant co-therapy. Burns 2017, 43, 471–485. [Google Scholar] [CrossRef]
- Duran-Bedolla, J.; Montes de Oca-Sandoval, M.A.; Saldana-Navor, V.; Villalobos-Silva, J.A.; Rodriguez, M.C.; Rivas-Arancibia, S. Sepsis, mitochondrial failure and multiple organ dysfunction. Clin. Investig. Med. 2014, 37, E58–E69. [Google Scholar] [CrossRef] [PubMed]
- Fock, E.M.; Parnova, R.G. Protective effect of mitochondria-targeted antioxidants against inflammatory response to lipopolysaccharide challenge: A review. Pharmaceutics 2021, 13, 144. [Google Scholar] [CrossRef] [PubMed]
- Lowes, D.A.; Thottakam, B.M.; Webster, N.R.; Murphy, M.P.; Galley, H.F. The mitochondria-targeted antioxidant MitoQ protects against organ damage in a lipopolysaccharide-peptidoglycan model of sepsis. Free Radic. Biol. Med. 2008, 45, 1559–1565. [Google Scholar] [CrossRef] [PubMed]
- Supinski, G.S.; Murphy, M.P.; Callahan, L.A. MitoQ administration prevents endotoxin-induced cardiac dysfunction. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 297, R1095–R1102. [Google Scholar] [CrossRef]
- Bakeeva, L.E.; Barskov, I.V.; Egorov, M.V.; Isaev, N.K.; Kapelko, V.I.; Kazachenko, A.V.; Kirpatovsky, V.I.; Kozlovsky, S.V.; Lakomkin, V.L.; Levina, S.B.; et al. Mitochondria-targeted plastoquinone derivatives as tools to interrupt execution of the aging program. 2. Treatment of some ROS- and age-related diseases (heart arrhythmia, heart infarctions, kidney ischemia, and stroke). Biochemistry 2008, 73, 1288–1299. [Google Scholar] [CrossRef]
- McManus, M.J.; Murphy, M.P.; Franklin, J.L. The mitochondria-targeted antioxidant MitoQ prevents loss of spatial memory retention and early neuropathology in a transgenic mouse model of Alzheimer’s disease. J. Neurosci. 2011, 31, 15703–15715. [Google Scholar] [CrossRef]
- Kapay, N.A.; Popova, O.V.; Isaev, N.K.; Stelmashook, E.V.; Kondratenko, R.V.; Zorov, D.B.; Skrebitsky, V.G.; Skulachev, V.P. Mitochondria-targeted plastoquinone antioxidant SkQ1 prevents amyloid-β-induced impairment of long-term potentiation in rat hippocampal slices. J. Alzheimers Dis. 2013, 36, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Fink, M.P.; Macias, C.A.; Xiao, J.; Tyurina, Y.Y.; Delude, R.L.; Greenberger, J.S.; Kagan, V.E.; Wipf, P. Hemigramicidin-TEMPO conjugates: Novel mitochondria-targeted antioxidants. Crit. Care Med. 2007, 35 (Suppl. S9), S461–S467. [Google Scholar] [CrossRef]
- Weidinger, A.; Mullebner, A.; Paier-Pourani, J.; Banerjee, A.; Miller, I.; Lauterbock, L.; Duvigneau, J.C.; Skulachev, V.P.; Redl, H.; Kozlov, A.V. Vicious inducible nitric oxide synthase-mitochondrial reactive oxygen species cycle accelerates inflammatory response and causes liver injury in rats. Antioxid. Redox Signal. 2015, 22, 572–586. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Du, M.; Zhu, M.J. Quercetin suppresses NLRP3 inflammasome activation in epithelial cells triggered by Escherichia coli O157:H7. Free Radic. Biol. Med. 2017, 108, 760–769. [Google Scholar] [CrossRef]
- Yan, Y.; Jiang, W.; Spinetti, T.; Tardivel, A.; Castillo, R.; Bourquin, C.; Guarda, G.; Tian, Z.; Tschopp, J.; Zhou, R. Omega-3 fatty acids prevent inflammation and metabolic disorder through inhibition of NLRP3 inflammasome activation. Immunity 2013, 38, 1154–1163. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.H.; Ting, H.C.; Chen, W.W.; Chan, J.F.; Hsu, Y.H. Omega-3 and omega-6 fatty acid differentially impact cardiolipin remodeling in activated macrophage. Lipids Health Dis. 2018, 17, 201. [Google Scholar] [CrossRef]
- Simopoulos, A.P. Omega-3 fatty acids in inflammation and autoimmune diseases. J. Am. Coll. Nutr. 2002, 21, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Marine omega-3 fatty acids and inflammatory processes: Effects, mechanisms and clinical relevance. Biochim. Biophys. Acta 2015, 1851, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.; Kettunen, J.; Würtz, P.; Haller, T.; Havulinna, A.S.; Kangas, A.J.; Soininen, P.; Esko, T.; Tammesoo, M.-L.; Mägi, R. Biomarker profiling by nuclear magnetic resonance spectroscopy for the prediction of all-cause mortality: An observational study of 17,345 persons. PLoS Med. 2014, 11, e1001606. [Google Scholar] [CrossRef]
- Langley, R.J.; Tsalik, E.L.; van Velkinburgh, J.C.; Glickman, S.W.; Rice, B.J.; Wang, C.; Chen, B.; Carin, L.; Suarez, A.; Mohney, R.P. An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci. Transl. Med. 2013, 5, ra95–ra195. [Google Scholar] [CrossRef]
- Ryan, D.G.; O’Neill, L.A. Krebs cycle reborn in macrophage immunometabolism. Annu. Rev. Immunol. 2020, 38, 289–313. [Google Scholar] [CrossRef] [PubMed]
- Williams, N.C.; O’Neill, L.A. A role for the Krebs cycle intermediate citrate in metabolic reprogramming in innate immunity and inflammation. Front. Immunol. 2018, 9, 141. [Google Scholar] [CrossRef]
- Cantu, D.; Schaack, J.; Patel, M. Oxidative inactivation of mitochondrial aconitase results in iron and H2O2-mediated neurotoxicity in rat primary mesencephalic cultures. PLoS ONE 2009, 4, e7095. [Google Scholar] [CrossRef] [PubMed]
- Deelen, J.; Kettunen, J.; Fischer, K.; van der Spek, A.; Trompet, S.; Kastenmüller, G.; Boyd, A.; Zierer, J.; van den Akker, E.B.; Ala-Korpela, M. A metabolic profile of all-cause mortality risk identified in an observational study of 44,168 individuals. Nat. Commun. 2019, 10, 3346. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Omega-3 fatty acids and inflammatory processes. Nutrients 2010, 2, 355–374. [Google Scholar] [CrossRef]
- Mori, T.A.; Beilin, L.J. Omega-3 fatty acids and inflammation. Curr. Atheroscler. Rep. 2004, 6, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Omega-3 fatty acids and inflammatory processes: From molecules to man. Biochem. Soc. Trans. 2017, 45, 1105–1115. [Google Scholar] [CrossRef]
- Di Nicolantonio, J.J.; O’Keefe, J.H. Importance of maintaining a low omega–6/omega–3 ratio for reducing inflammation. Open Heart 2018, 5, e000946. [Google Scholar] [CrossRef]
- Carracedo, M.; Artiach, G.; Arnardottir, H.; Bäck, M. The Resolution of Inflammation through Omega-3 Fatty Acids in Atherosclerosis, Intimal Hyperplasia, and Vascular Calcification. In Seminars in Immunopathology; Springer: Berlin/Heidelberg, Germany, 2019; pp. 757–766. [Google Scholar]
- Giacobbe, J.; Benoiton, B.; Zunszain, P.; Pariante, C.M.; Borsini, A. The anti-inflammatory role of omega-3 polyunsaturated fatty acids metabolites in pre-clinical models of psychiatric, neurodegenerative and neurological disorders. Front. Psychiatry 2020, 11, 122. [Google Scholar] [CrossRef]
- Harris, W.S.; Tintle, N.L.; Imamura, F.; Qian, F.; Korat, A.V.A.; Marklund, M.; Djousse, L.; Bassett, J.K.; Carmichael, P.H.; Chen, Y.Y.; et al. Blood n-3 fatty acid levels and total and cause-specific mortality from 17 prospective studies. Nat. Commun. 2021, 12, 2329. [Google Scholar] [CrossRef] [PubMed]
- Sommer, S.S. Does cancer kill the individual and save the species? Hum. Mutat. 1994, 3, 166–169. [Google Scholar] [CrossRef]
- He, Q.; Fu, Y.; Tian, D.; Yan, W. The contrasting roles of inflammasomes in cancer. Am. J. Cancer Res. 2018, 8, 566–583. [Google Scholar] [PubMed]
- Hernandez, C.; Huebener, P.; Schwabe, R.F. Damage-associated molecular patterns in cancer: A double-edged sword. Oncogene 2016, 35, 5931–5941. [Google Scholar] [CrossRef]
- McCall, K.D.; Muccioli, M.; Benencia, F. Toll-Like Receptors Signaling in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1223, 81–97. [Google Scholar] [PubMed]
- Wattenberg, M.M.; Beatty, G.L. Overcoming immunotherapeutic resistance by targeting the cancer inflammation cycle. Semin. Cancer Biol. 2020, 65, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Zuo, M.; Yalavarthi, S.; Gockman, K.; Madison, J.A.; Shi, H.; Woodard, W.; Lezak, S.P.; Lugogo, N.L.; Knight, J.S.; et al. Neutrophil extracellular traps and thrombosis in COVID-19. J. Thromb Thrombolysis 2021, 51, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Bonaventura, A.; Vecchie, A.; Dagna, L.; Martinod, K.; Dixon, D.L.; van Tassell, B.W.; Dentali, F.; Montecucco, F.; Massberg, S.; Levi, M.; et al. Endothelial dysfunction and immunothrombosis as key pathogenic mechanisms in COVID-19. Nat. Rev. Immunol. 2021, 21, 319–329. [Google Scholar] [CrossRef]
- Plotnikov, E.Y.; Morosanova, M.A.; Pevzner, I.B.; Zorova, L.D.; Manskikh, V.N.; Pulkova, N.V.; Galkina, S.I.; Skulachev, V.P.; Zorov, D.B. Protective effect of mitochondria-targeted antioxidants in an acute bacterial infection. Proc. Natl. Acad. Sci. USA 2013, 110, E3100–E3108. [Google Scholar] [CrossRef] [PubMed]
- Plotnikov, E.Y.; Pevzner, I.B.; Zorova, L.D.; Chernikov, V.P.; Prusov, A.N.; Kireev, I.I.; Silachev, D.N.; Skulachev, V.P.; Zorov, D.B. Mitochondrial damage and mitochondria-targeted antioxidant protection in LPS-induced acute kidney injury. Antioxidants 2019, 8, 176. [Google Scholar] [CrossRef]
- Zakharova, V.V.; Pletjushkina, O.Y.; Galkin, I.I.; Zinovkin, R.A.; Chernyak, B.V.; Krysko, D.V.; Bachert, C.; Krysko, O.; Skulachev, V.P.; Popova, E.N. Low concentration of uncouplers of oxidative phosphorylation decreases the TNF-induced endothelial permeability and lethality in mice. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 968–977. [Google Scholar] [CrossRef] [PubMed]
- Asher, A.; Tintle, N.L.; Myers, M.; Lockshon, L.; Bacareza, H.; Harris, W.S. Blood omega-3 fatty acids and death from COVID-19: A pilot study. Prostaglandins Leukot Essent Fatty Acids 2021, 166, 102250. [Google Scholar] [CrossRef] [PubMed]
- Toldo, S.; Bussani, R.; Nuzzi, V.; Bonaventura, A.; Mauro, A.G.; Cannata, A.; Pillappa, R.; Sinagra, G.; Nana-Sinkam, P.; Sime, P.; et al. Inflammasome formation in the lungs of patients with fatal COVID-19. Inflamm. Res. 2021, 70, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Relja, B.; Land, W.G. Damage-associated molecular patterns in trauma. Eur. J. Trauma Emerg. Surg. 2020, 46, 751–775. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Yalavarthi, S.; Shi, H.; Gockman, K.; Zuo, M.; Madison, J.A.; Blair, C.; Weber, A.; Barnes, B.J.; Egeblad, M.; et al. Neutrophil extracellular traps in COVID-19. JCI Insight 2020, 5, e138999. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Suarez, J.S.; Minaai, M.; Li, S.; Gaudino, G.; Pass, H.I.; Carbone, M.; Yang, H. HMGB1 as a therapeutic target in disease. J. Cell Physiol. 2021, 236, 3406–3419. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Huang, Y.; Mao, P.; Zhang, J.; Li, Y. Sepsis and ARDS: The dark side of histones. Mediators Inflamm. 2015, 2015, 205054. [Google Scholar] [CrossRef] [PubMed]
- Palm, N.W.; Medzhitov, R. Not so fast: Adaptive suppression of innate immunity. Nat. Med. 2007, 13, 1142–1144. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.D.; Zhao, J.; Auh, S.; Yang, X.; Du, P.; Tang, H.; Fu, Y.-X. Adaptive immune cells temper initial innate responses. Nat. Med. 2007, 13, 1248–1252. [Google Scholar] [PubMed]
- Remolina, S.C.; Chang, P.L.; Leips, J.; Nuzhdin, S.V.; Hughes, K.A. Genomic basis of aging and life-history evolution in Drosophila melanogaster. Evolution 2012, 66, 3390–3403. [Google Scholar] [CrossRef]
- Carnes, M.U.; Campbell, T.; Huang, W.; Butler, D.G.; Carbone, M.A.; Duncan, L.H.; Harbajan, S.V.; King, E.M.; Peterson, K.R.; Weitzel, A.; et al. The genomic basis of postponed senescence in Drosophila melanogaster. PLoS ONE 2015, 10, e0138569. [Google Scholar] [CrossRef] [PubMed]
- Fabian, D.K.; Garschall, K.; Klepsatel, P.; Santos-Matos, G.; Sucena, É.; Kapun, M.; Lemaitre, B.; Schlötterer, C.; Arking, R.; Flatt, T. Evolution of longevity improves immunity in. Evol. Lett. 2018, 2, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Kounatidis, I.; Chtarbanova, S.; Cao, Y.; Hayne, M.; Jayanth, D.; Ganetzky, B.; Ligoxygakis, P. NF-κB immunity in the brain determines fly lifespan in healthy aging and age-related neurodegeneration. Cell Rep. 2017, 19, 836–848. [Google Scholar] [CrossRef]
- Lin, Y.R.; Parikh, H.; Park, Y. Stress resistance and lifespan enhanced by downregulation of antimicrobial peptide genes in the Imd pathway. Aging 2018, 10, 622–631. [Google Scholar] [CrossRef] [PubMed]
- Gorbunova, V.; Seluanov, A.; Kennedy, B.K. The world goes bats: Living longer and tolerating viruses. Cell Metab. 2020, 32, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Irving, A.T.; Ahn, M.; Goh, G.; Anderson, D.E.; Wang, L.F. Lessons from the host defences of bats, a unique viral reservoir. Nature 2021, 589, 363–370. [Google Scholar] [CrossRef]
- Banerjee, A.; Baker, M.L.; Kulcsar, K.; Misra, V.; Plowright, R.; Mossman, K. Novel insights into immune systems of bats. Front. Immunol. 2020, 11, 26. [Google Scholar] [CrossRef]
- Goh, G.; Ahn, M.; Zhu, F.; Lee, L.B.; Luo, D.; Irving, A.T.; Wang, L.F. Complementary regulation of caspase-1 and IL-1beta reveals additional mechanisms of dampened inflammation in bats. Proc. Natl. Acad. Sci. USA 2020, 117, 28939–28949. [Google Scholar] [CrossRef] [PubMed]
- Morrison, C.E.; Hogg, C.J.; Gales, R.; Johnson, R.N.; Grueber, C.E. Low innate immune-gene diversity in the critically endangered orange-bellied parrot (Neophema chrysogaster). Emu—Austral Ornithol. 2020, 120, 56–64. [Google Scholar] [CrossRef]
- Lin, T.; Buffenstein, R. The Unusual Immune System of the Naked Mole-Rat. Adv. Exp. Med. Biol. 2021, 1319, 315–327. [Google Scholar]
- Hilton, H.G.; Rubinstein, N.D.; Janki, P.; Ireland, A.T.; Bernstein, N.; Fong, N.L.; Wright, K.M.; Smith, M.; Finkle, D.; Martin-McNulty, B.; et al. Single-cell transcriptomics of the naked mole-rat reveals unexpected features of mammalian immunity. PLoS Biol. 2019, 17, e3000528. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, S.A.; Wasserman, S.A. Conventional and non-conventional Drosophila Toll signaling. Dev. Comp. Immunol. 2014, 42, 16–24. [Google Scholar] [CrossRef]
- Higashitsuji, H.; Fujita, T.; Higashitsuji, H.; Fujita, J. Mammalian cold-inducible RNA-binding protein facilitates wound healing through activation of AMP-activated protein kinase. Biochem. Biophys. Res. Commun. 2020, 533, 1191–1197. [Google Scholar] [CrossRef] [PubMed]
- Bolk, L. The Problem of Human Development; Gustav Fischer: Jena, Germany, 1926. [Google Scholar]
- Skulachev, V.P.; Holtze, S.; Vyssokikh, M.Y.; Bakeeva, L.E.; Skulachev, M.V.; Markov, A.V.; Hildebrandt, T.B.; Sadovnichii, V.A. Neoteny, Prolongation of Youth: From Naked Mole Rats to “Naked Apes” (Humans). Physiol. Rev. 2017, 97, 699–720. [Google Scholar] [CrossRef] [PubMed]
- Singh, T.; Newman, A.B. Inflammatory markers in population studies of aging. Ageing Res. Rev. 2011, 10, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.C.; Goldstein, D.R.; Montgomery, R.R. Age-dependent dysregulation of innate immunity. Nat. Rev. Immunol. 2013, 13, 875–887. [Google Scholar] [CrossRef]
- Sanada, F.; Taniyama, Y.; Muratsu, J.; Otsu, R.; Shimizu, H.; Rakugi, H.; Morishita, R. Source of chronic inflammation in aging. Front. Cardiovasc. Med. 2018, 5, 12. [Google Scholar] [CrossRef]
- Franceschi, C.; Capri, M.; Monti, D.; Giunta, S.; Olivieri, F.; Sevini, F.; Panourgia, M.P.; Invidia, L.; Celani, L.; Scurti, M.; et al. Inflammaging and anti-inflammaging: A systemic perspective on aging and longevity emerged from studies in humans. Mech. Ageing Dev. 2007, 128, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Salvioli, S.; Capri, M.; Valensin, S.; Tieri, P.; Monti, D.; Ottaviani, E.; Franceschi, C. Inflamm-aging, cytokines and aging: State of the art, new hypotheses on the role of mitochondria and new perspectives from systems biology. Curr. Pharm. Des. 2006, 12, 3161–3171. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, C.; Garagnani, P.; Parini, P.; Giuliani, C.; Santoro, A. Inflammaging: A new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 2018, 14, 576–590. [Google Scholar] [CrossRef] [PubMed]
- Skulachev, M.V.; Antonenko, Y.N.; Anisimov, V.N.; Chernyak, B.V.; Cherepanov, D.A.; Chistyakov, V.A.; Egorov, M.V.; Kolosova, N.G.; Korshunova, G.A.; Lyamzaev, K.G.; et al. Mitochondrial-targeted plastoquinone derivatives. Effect on senescence and acute age-related pathologies. Curr. Drug Targets 2011, 12, 800–826. [Google Scholar] [CrossRef] [PubMed]
- Refardt, D.; Kümmerli, R. Defying bacteriophages: Contrasting altruistic with individual-based resistance mechanisms in Escherichia coli. Commun. Integr. Biol. 2013, 6, e25159. [Google Scholar] [CrossRef]
- Koonin, E.V.; Zhang, F. Coupling immunity and programmed cell suicide in prokaryotes: Life-or-death choices. Bioessays 2017, 39, 1–9. [Google Scholar] [CrossRef]
- Fan, Y.; Hoshino, T.; Nakamura, A. Identification of a VapBC toxin-antitoxin system in a thermophilic bacterium Thermus thermophilus HB27. Extremophiles 2017, 21, 153–161. [Google Scholar] [CrossRef][Green Version]
- Li, Z.; Song, Q.; Wang, Y.; Xiao, X.; Xu, J. Identification of a functional toxin-antitoxin system located in the genomic island PYG1 of piezophilic hyperthermophilic archaeon Pyrococcus yayanosii. Extremophiles 2018, 22, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Débarre, F.; Lion, S.; van Baalen, M.; Gandon, S. Evolution of host life-history traits in a spatially structured host-parasite system. Am. Nat. 2012, 179, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S.; Wolf, Y.I. Evolutionary Genomics of Defense Systems in Archaea and Bacteria. Annu. Rev. Microbiol. 2017, 71, 233–261. [Google Scholar] [CrossRef]
- Westra, E.R.; van Houte, S.; Gandon, S.; Whitaker, R. The ecology and evolution of microbial CRISPR-Cas adaptive immune systems. Philos. Trans. R Soc. Lond. B Biol. Sci. 2019, 374, 20190101. [Google Scholar] [CrossRef]
- Puigbo, P.; Makarova, K.S.; Kristensen, D.M.; Wolf, Y.I.; Koonin, E.V. Reconstruction of the evolution of microbial defense systems. BMC Evol. Biol. 2017, 17, 94. [Google Scholar] [CrossRef]
- Brennan, J.J.; Gilmore, T.D. Evolutionary origins of Toll-like receptor signaling. Mol. Biol. Evol. 2018, 35, 1576–1587. [Google Scholar] [CrossRef]
- Arandjelovic, S.; Ravichandran, K.S. Phagocytosis of apoptotic cells in homeostasis. Nat. Immunol. 2015, 16, 907–917. [Google Scholar] [CrossRef]
- Bamboat, Z.M.; Ocuin, L.M.; Balachandran, V.P.; Obaid, H.; Plitas, G.; DeMatteo, R.P. Conventional DCs reduce liver ischemia/reperfusion injury in mice via IL-10 secretion. J. Clin. Investig. 2010, 120, 559–569. [Google Scholar] [CrossRef]
- Kimbrell, D.A.; Beutler, B. The evolution and genetics of innate immunity. Nat. Rev. Genet 2001, 2, 256–267. [Google Scholar] [CrossRef]
- Stein, C.; Caccamo, M.; Laird, G.; Leptin, M. Conservation and divergence of gene families encoding components of innate immune response systems in zebrafish. Genome Biol. 2007, 8, R251. [Google Scholar] [CrossRef] [PubMed]
- Boyle, J.P.; Mayle, S.; Parkhouse, R.; Monie, T.P. Comparative Genomic and Sequence Analysis Provides Insight into the Molecular Functionality of NOD1 and NOD2. Front. Immunol. 2013, 4, 317. [Google Scholar] [CrossRef] [PubMed]
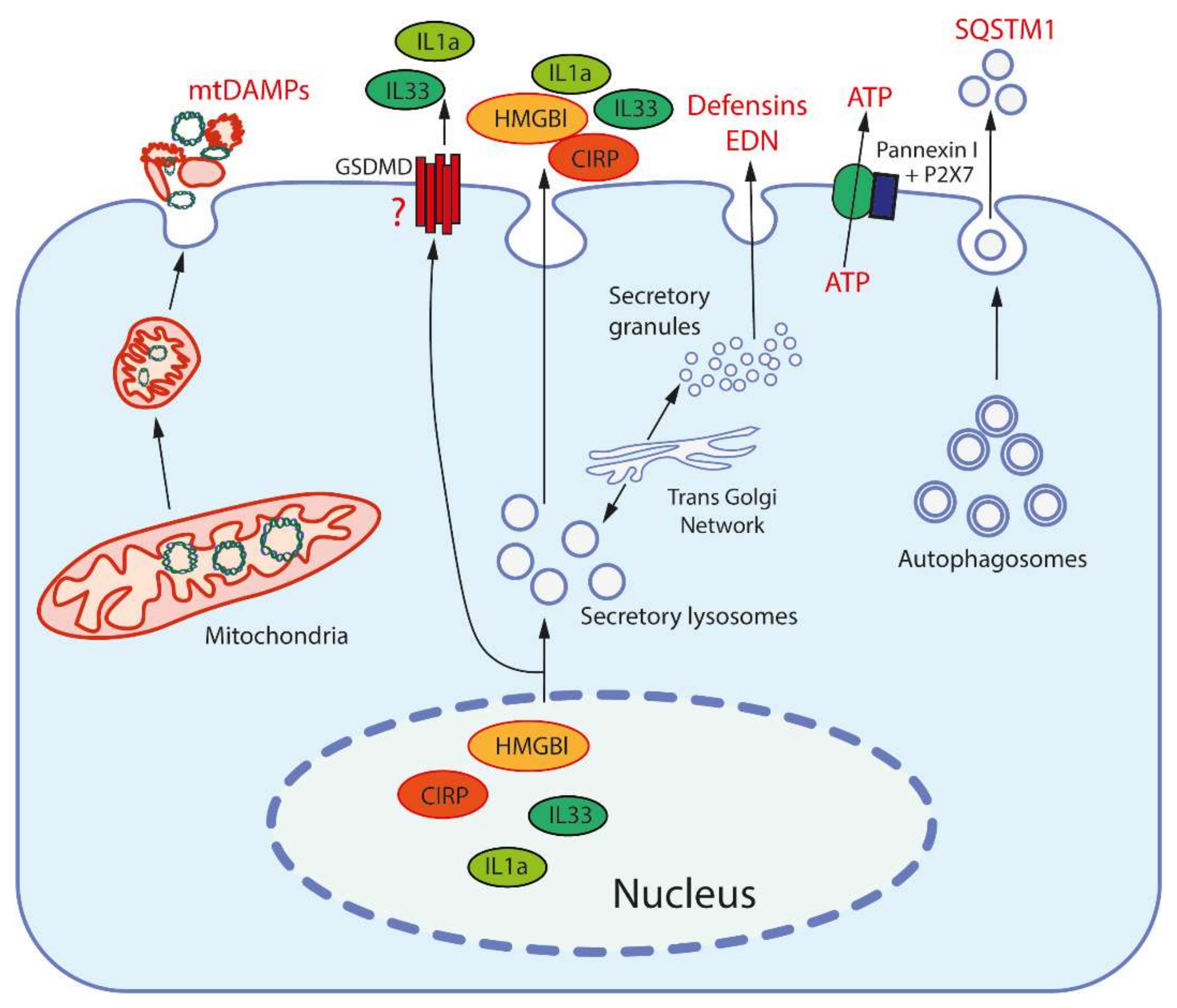
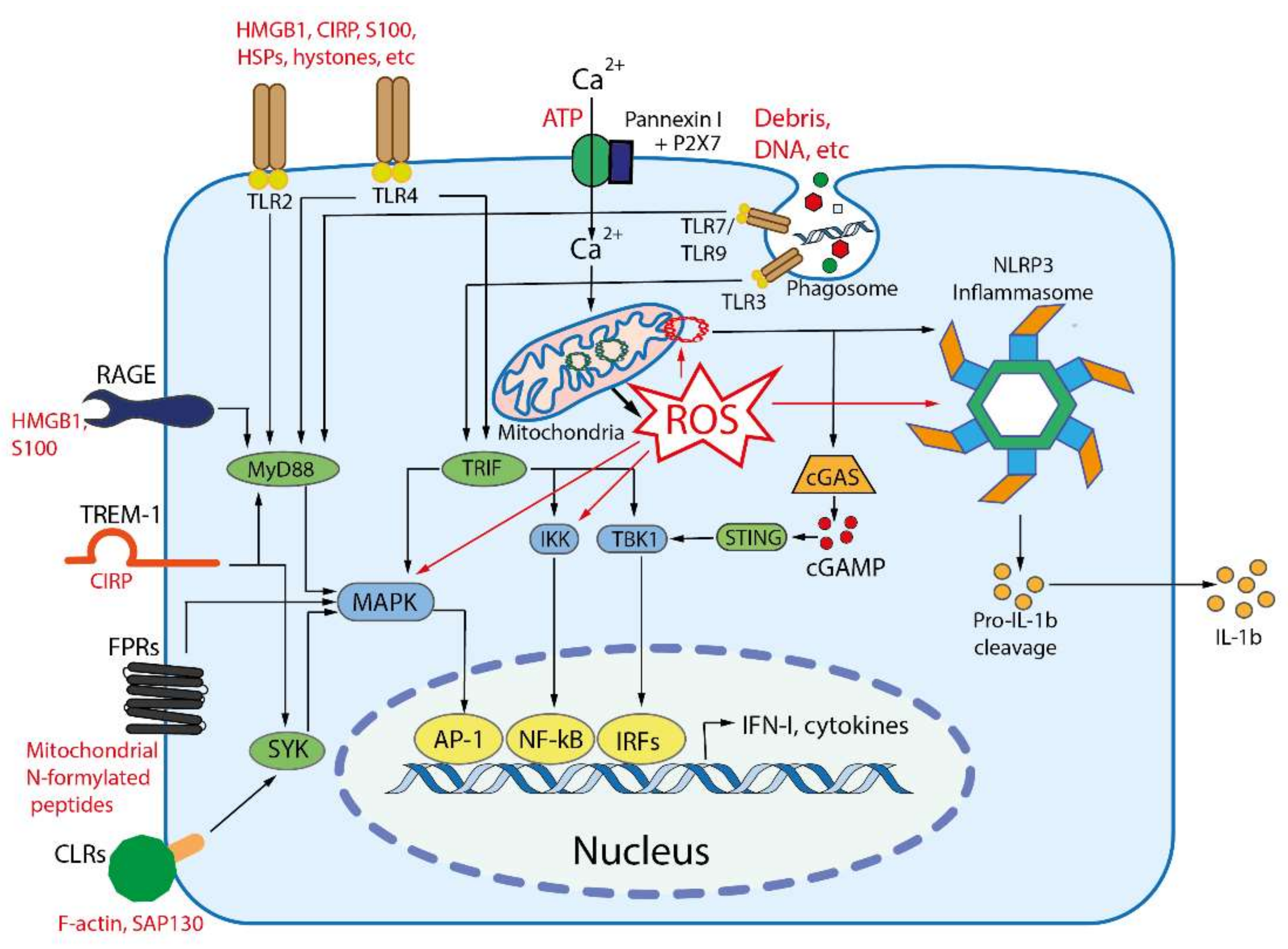
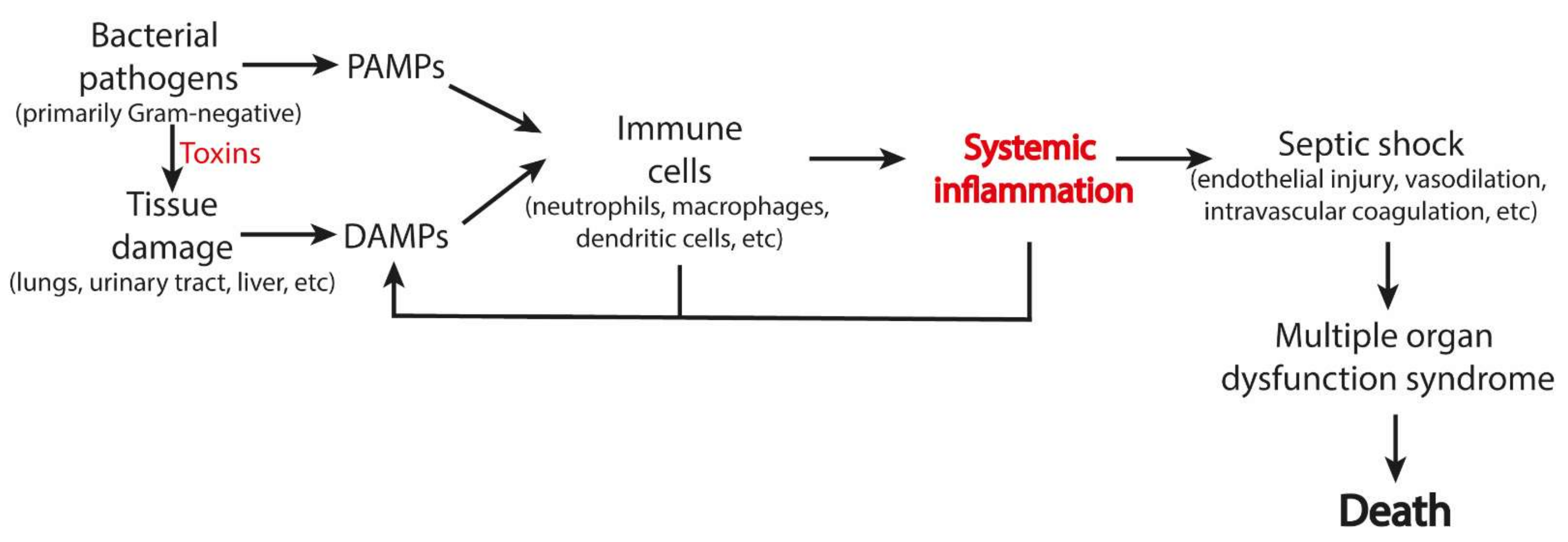
| # | Origin | DAMPs | Receptors | Ref |
|---|---|---|---|---|
| 1 | Nucleus | HMGB1 | TLR2, TLR4, RAGE | [29] |
| CIRP | TLR4, TREM-1 | [30] | ||
| Histones | TLR2, TLR4 | [47] | ||
| SAP130 | Mincle | [26] | ||
| IL-1α | IL1R1 | [31] | ||
| IL-33 | ST2 | [31] | ||
| DNA | cGAS, AIM2, RAGE, IFI16 | [21] | ||
| 2 | Cytosol | S100 proteins | TLR2, TLR4, RAGE | [31] |
| HSPs | TLR2, TLR4, CD91 | [35] | ||
| F-Actin | DNGR-1, TREM1 | [32] | ||
| Cyclophilin A | CD147 | [33] | ||
| Peroxiredoxin 1 | TLR4 | [34] | ||
| Oxidized hemoglobin, heme | TLR4 | [36] | ||
| Amyloid β | TLR4 | [48] | ||
| ATP, ADP | P2X7R, P2Y2R, P2Y12R, | [40] | ||
| Uric acid | TREM-1, TLR2, TLR4, P2X7, NLRP3 | [26] | ||
| mRNA | TLR3 | [26] | ||
| microRNAs | TLR7 | [26] | ||
| SNAPIN | TLR2 | [26] | ||
| AGEs | RAGE | [26] | ||
| 3 | Mitochondria | Formyl peptides | FPR1 | [43] |
| mtDNA | TLR9, NLRP3 | [43] | ||
| Cardiolipin | NLRP3, TREM2 | [49] | ||
| Cytochrome c | TLR4 | [44] | ||
| Oxygenated mitochondrial fatty acids | TRL4 | [50,51] | ||
| TFAM | RAGE | [43] | ||
| 4 | ER, secretory granules, autophagosomes | Defensins | TLR4 | [37] |
| Cathelicidins | P2X7, FPR2 | [37] | ||
| Eosinophil-derived neurotoxin | TLR2 | [38] | ||
| Granulisin | TLR4 | [39] | ||
| Calreticulin | CD91 | [52] | ||
| Gp96 | TLR2, TLR4, CD91 | [52] | ||
| Sequestosome-1 (SQSTM1 or p62) | INSR | [53] | ||
| 5 | Extracellular matrix | Heparan sulphate, versican, aggrecan | TLR4 | [42] |
| Proteoglycans (biglycan, decorin, etc.) | TLR2, TLR4, CD14, NLRP3 | [42] | ||
| Tenascin-C | TLR4 | [42] | ||
| Fibrinogen | TLR4 | [42] | ||
| Fibronectin | TLR2, TLR4 | [42] | ||
| Low molecular weight hyaluronan | TLR2, TLR4, NLRP3 | [42] | ||
| 6 | Tumor cells | Annexin A1 | FPR1 | [45] |
| PAUF | TLR4 | [45] | ||
| API5 | TLR4 | [45] | ||
| Rps-3 | TLR4 | [45] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chernyak, B.V.; Lyamzaev, K.G.; Mulkidjanian, A.Y. Innate Immunity as an Executor of the Programmed Death of Individual Organisms for the Benefit of the Entire Population. Int. J. Mol. Sci. 2021, 22, 13480. https://doi.org/10.3390/ijms222413480
Chernyak BV, Lyamzaev KG, Mulkidjanian AY. Innate Immunity as an Executor of the Programmed Death of Individual Organisms for the Benefit of the Entire Population. International Journal of Molecular Sciences. 2021; 22(24):13480. https://doi.org/10.3390/ijms222413480
Chicago/Turabian StyleChernyak, Boris V., Konstantin G. Lyamzaev, and Armen Y. Mulkidjanian. 2021. "Innate Immunity as an Executor of the Programmed Death of Individual Organisms for the Benefit of the Entire Population" International Journal of Molecular Sciences 22, no. 24: 13480. https://doi.org/10.3390/ijms222413480
APA StyleChernyak, B. V., Lyamzaev, K. G., & Mulkidjanian, A. Y. (2021). Innate Immunity as an Executor of the Programmed Death of Individual Organisms for the Benefit of the Entire Population. International Journal of Molecular Sciences, 22(24), 13480. https://doi.org/10.3390/ijms222413480









