A Systematic Review on HOX Genes as Potential Biomarkers in Colorectal Cancer: An Emerging Role of HOXB9
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Eligibility Criteria and Study Selection
2.3. Data Extraction, Synthesis and Quality Assessment
3. Results
3.1. Study Selection
3.2. Clinicopathological Characteristics and Prognostic Significance of HOX Dysregulation in CRC
3.2.1. Study Characteristics
3.2.2. Findings
3.3. Functional Role of HOX Genes in CRC Progression
3.3.1. Study Characteristics
3.3.2. Findings
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Chow, F.C.L.; Chok, K.S.H. Colorectal liver metastases: An update on multidisciplinary approach. World J. Hepatol. 2019, 11, 150–172. [Google Scholar] [CrossRef]
- East, J.E.; Atkin, W.S.; Bateman, A.C.; Clark, S.K.; Dolwani, S.; Ket, S.N.; Leedham, S.J.; Phull, P.S.; Rutter, M.D.; Shepherd, N.A.; et al. British Society of Gastroenterology position statement on serrated polyps in the colon and rectum. Gut 2017, 66, 1181–1196. [Google Scholar] [CrossRef]
- Paschos, K.A.; Majeed, A.W.; Bird, N.C. Natural history of hepatic metastases from colorectal cancer—Pathobiological pathways with clinical significance. World J. Gastroenterol. 2014, 20, 3719–3737. [Google Scholar] [CrossRef]
- Li, B.; Huang, Q.; Wei, G.H. The role of hox transcription factors in cancer predisposition and progression. Cancers 2019, 11, 528. [Google Scholar] [CrossRef] [PubMed]
- Castelli-Gair Hombría, J.; Lovegrove, B. Beyond homeosis—HOX function in morphogenesis and organogenesis. Differentiation 2003, 71, 461–476. [Google Scholar] [CrossRef] [PubMed]
- Durston, A.; Wacker, S.; Bardine, N.; Jansen, H. Time Space Translation: A Hox Mechanism for Vertebrate A-P Patterning. Curr. Genom. 2012, 13, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Quinonez, S.C.; Innis, J.W. Human HOX gene disorders. Mol. Genet. Metab. 2014, 111, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. HOX genes and their role in the development of human cancers. J. Mol. Med. 2014, 92, 811–823. [Google Scholar] [CrossRef]
- Collins, C.T.; Hess, J.L. Role of HOXA9 in leukemia: Dysregulation, cofactors and essential targets. Oncogene 2016, 35, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.M.; Mouw, J.K.; Unger, M.A.; Lakins, J.N.; Gbegnon, M.K.; Clemmer, V.B.; Benezra, M.; Licht, J.D.; Boudreau, N.J.; Tsai, K.K.C.; et al. HOXA9 regulates BRCA1 expression to modulate human breast tumor phenotype. J. Clin. Investig. 2010, 120, 1535–1550. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, Q.; He, C.; Liang, D.; Yi, Q.; Shi, J.; Wan, B.; Yang, R.; Li, L.; Sha, S.; et al. HOXB9 inhibits proliferation in gastric carcinoma cells via suppression of phosphorylated-Akt and NF-κB-dependent Snail expression. Dig. Liver Dis. 2019, 51, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, T.; Takahashi, F.; Chiba, N.; Brachtel, E.; Takahashi, M.; Godin-Heymann, N.; Gross, K.W.; Vivanco, M.D.M.; Wijendran, V.; Shioda, T.; et al. HOXB9, a gene overexpressed in breast cancer, promotes tumorigenicity and lung metastasis. Proc. Natl. Acad. Sci. USA 2010, 107, 1100–1105. [Google Scholar] [CrossRef] [PubMed]
- Brotto, D.B.; Siena, Á.D.D.; de Barros, I.I.; Carvalho, S.; Muys, B.; Goedert, L.; Cardoso, C.; Plaça, J.R.; Ramão, A.; Squire, J.A.; et al. Contributions of HOX genes to cancer hallmarks: Enrichment pathway analysis and review. Tumor Biol. 2020, 42, 1010428320918050. [Google Scholar] [CrossRef] [PubMed]
- Haria, D.; Naora, H. Homeobox Gene Deregulation: Impact on the Hallmarks of Cancer. Cancer Hallm. 2014, 1, 67–76. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bhatlekar, S.; Czymmek, K.; Viswanathan, V.; Gonye, G.; Boman, B. Role of HOX genes in regulation of stem cell populations in normal and malignant colon tissue. Cancer Res. 2012, 72. [Google Scholar] [CrossRef]
- Javed, S.; Langley, S.E.M. Importance of HOX genes in normal prostate gland formation, prostate cancer development and its early detection. BJU Int. 2014, 113, 535–540. [Google Scholar] [CrossRef]
- Francis, J.C.; Gardiner, J.R.; Renaud, Y.; Chauhan, R.; Weinstein, Y.; Gomez-Sanchez, C.; Lefrançois-Martinez, A.M.; Bertherat, J.; Val, P.; Swain, A. HOX genes promote cell proliferation and are potential therapeutic targets in adrenocortical tumours. Br. J. Cancer 2020, 20, 805–816. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.A.; Carrano, F.M.; Agresta, F.; Alarçon, I.; Azran, C.; et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. J. Clin. Epidemiol. 2009, 62, e1–e34. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Zhang, Y.; Kwong, J.S.W.; Zhang, C.; Li, S.; Sun, F.; Niu, Y.; Du, L. The methodological quality assessment tools for preclinical and clinical studies, systematic review and meta-analysis, and clinical practice guideline: A systematic review. J. Evid. Based Med. 2015, 8, 2–10. [Google Scholar] [CrossRef]
- Hooijmans, C.R.; Rovers, M.M.; De Vries, R.B.M.; Leenaars, M.; Ritskes-Hoitinga, M.; Langendam, M.W. SYRCLE’s risk of bias tool for animal studies. BMC Med. Res. Methodol. 2014, 14, 43. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.T.; Altman, D.G.; Gøtzsche, P.C.; Jüni, P.; Moher, D.; Oxman, A.D.; Savović, J.; Schulz, K.F.; Weeks, L.; Sterne, J.A.C. The Cochrane Collaboration’s tool for assessing risk of bias in randomised trials. BMJ 2011, 343, d5928. [Google Scholar] [CrossRef]
- Liu, M.; Xiao, Y.; Tang, W.; Li, J.; Hong, L.; Dai, W.; Zhang, W.; Peng, Y.; Wu, X.; Wang, J.; et al. HOXD9 promote epithelial-mesenchymal transition and metastasis in colorectal carcinoma. Cancer Med. 2020, 9, 3932–3943. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, C.; Wang, Y.; Ma, S.; Cao, W.; Guan, F. HOXC11 functions as a novel oncogene in human colon adenocarcinoma and kidney renal clear cell carcinoma. Life Sci. 2020, 243, 117230. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.T.; Jiang, D.; Yuan, J.; Cui, Y.M.; Shi, X.W.; Chen, C.M.; Bian, X.W.; Deng, Y.J.; Ding, Y.Q. HOXB7 as a prognostic factor and mediator of colorectal cancer progression. Clin. Cancer Res. 2011, 17, 3569–3578. [Google Scholar] [CrossRef] [PubMed]
- Kanai, M.; Hamada, J.I.; Takada, M.; Asano, T.; Murakawa, K.; Takahashi, Y.; Murai, T.; Tada, M.; Miyamoto, M.; Kondo, S.; et al. Aberrant expressions of HOX genes in colorectal and hepatocellular carcinomas. Oncol. Rep. 2010, 23, 843–851. [Google Scholar]
- Ying, Y.; Wang, Y.; Huang, X.; Sun, Y.; Zhang, J.; Li, M.; Zeng, J.; Wang, M.; Xiao, W.; Zhong, L.; et al. Oncogenic HOXB8 is driven by MYC-regulated super-enhancer and potentiates colorectal cancer invasiveness via BACH1. Oncogene 2020, 39, 1004–1017. [Google Scholar] [CrossRef]
- Cantile, M.; Franco, R.; Tschan, A.; Baumhoer, D.; Zlobec, I.; Schiavo, G.; Forte, I.; Bihl, M.; Liguori, G.; Botti, G.; et al. HOX D13 expression across 79 tumor tissue types. Int. J. Cancer 2009, 125, 1532–1541. [Google Scholar] [CrossRef] [PubMed]
- Groene, J.; Mansmann, U.; Meister, R.; Staub, E.; Roepcke, S.; Heinze, M.; Klaman, I.; Bruemmendorf, T.; Hermann, K.; Loddenkemper, C.; et al. Transcriptional census of 36 microdissected colorectal cancers yields a gene signature to distinguish UICC II and III. Int. J. Cancer 2006, 119, 1829–1836. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Kim, R.S.; Zhang, H.; Lee, S.J.; Sheng, H.; Loehrer, P.J.; Gardner, T.A.; Jeng, M.H.; Kao, C. HOXB13 is downregulated in colorectal cancer to confer TCF4-mediated transactivation. Br. J. Cancer 2005, 92, 2233–2239. [Google Scholar] [CrossRef] [PubMed]
- Toiyama, Y.; Mizoguchi, A.; Kimura, K.; Araki, T.; Yoshiyama, S.; Sakaguchi, K.; Miki, C.; Kusunoki, M. Persistence of gene expression changes in noninflamed and inflamed colonic mucosa in ulcerative colitis and their presence in colonic carcinoma. World J. Gastroenterol. 2005, 11, 5151–5155. [Google Scholar] [PubMed]
- Vider, B.Z.; Zimber, A.; Hirsch, D.; Estlein, D.; Chastre, E.; Prevot, S.; Gespach, C.; Yaniv, A.; Gazit, A. Human colorectal carcinogenesis is associated with deregulation of homeobox gene expression. Biochem. Biophys. Res. Commun. 1997, 232, 742–748. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wu, F.; Jiang, Z. Effect of HOXA6 on the proliferation, apoptosis, migration and invasion of colorectal cancer cells. Int. J. Oncol. 2018, 52, 2093–2100. [Google Scholar] [CrossRef]
- Yuan, Y.; Sun, S.; Jiao, N.; Shu, Y.; Zhang, Y. Upregulation of HOXA10 Protein Expression Predicts Poor Prognosis for Colorectal Cancer. Genet. Test. Mol. Biomarkers 2018, 22, 1–8. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, G.; Ding, L.; Jiang, T.; Shao, S.; Gao, Y.; Lu, Y. HOXA3 promotes tumor growth of human colon cancer through activating EGFR/Ras/Raf/MEK/ERK signaling pathway. J. Cell. Biochem. 2018, 119, 2864–2874. [Google Scholar] [CrossRef]
- Tatangelo, F.; Di Mauro, A.; Scognamiglio, G.; Aquino, G.; Lettiero, A.; Delrio, P.; Avallone, A.; Cantile, M.; Botti, G. Posterior HOX genes and HOTAIR expression in the proximal and distal colon cancer pathogenesis. J. Transl. Med. 2018, 16, 350. [Google Scholar] [CrossRef]
- Song, J.; Wang, T.; Xu, W.; Wang, P.; Wan, J.; Wang, Y.; Zhan, J.; Zhang, H. HOXB9 acetylation at K27 is responsible for its suppression of colon cancer progression. Cancer Lett. 2018, 426, 63–72. [Google Scholar] [CrossRef]
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. Role of HOX genes in stem cell differentiation and cancer. Stem Cells Int. 2018, 2018, 1–15. [Google Scholar] [CrossRef]
- Watanabe, Y.; Saito, M.; Saito, K.; Matsumoto, Y.; Kanke, Y.; Onozawa, H.; Hayase, S.; Sakamoto, W.; Ishigame, T.; Momma, T.; et al. Upregulated HOXA9 expression is associated with lymph node metastasis in colorectal cancer. Oncol. Lett. 2018, 15, 2756–2762. [Google Scholar] [CrossRef] [PubMed]
- Mansour, M.A.; Senga, T. HOXD8 exerts a tumor-suppressing role in colorectal cancer as an apoptotic inducer. Int. J. Biochem. Cell Biol. 2017, 88, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Carbone, C.; Piro, G.; Simionato, F.; Ligorio, F.; Cremolini, C.; Loupakis, F.; Alè, G.; Rossini, D.; Merz, V.; Santoro, R.; et al. Homeobox B9 mediates resistance to anti-VEGF therapy in colorectal cancer patients. Clin. Cancer Res. 2017, 23, 4312–4322. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Feng, Q.; He, G.; Yang, L.; Tang, W.; Lao, X.; Zhu, D.; Lin, Q.; Xu, P.; Wei, Y.; et al. Silencing homeobox C6 inhibits colorectal cancer cell proliferation. Oncotarget 2016, 7, 29216–29227. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.; Zhao, X.; Zuo, X.; Peng, Z. MiR-10b promotes invasion by targeting HOXD10 in colorectal cancer. Oncol. Lett. 2016, 12, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Pan, J.; Lu, X.; Chi, P. Role of miR-196 and its target gene HoxB8 in the development and proliferation of human colorectal cancer and the impact of neoadjuvant chemotherapy with FOLFOX4 on their expression. Oncol. Lett. 2016, 12, 4041–4047. [Google Scholar] [CrossRef]
- Hoshino, Y.; Hayashida, T.; Hirata, A.; Takahashi, H.; Chiba, N.; Ohmura, M.; Wakui, M.; Jinno, H.; Hasegawa, H.; Maheswaran, S.; et al. Bevacizumab terminates homeobox B9-induced tumor proliferation by silencing microenvironmental communication. Mol. Cancer 2014, 13, 102. [Google Scholar] [CrossRef]
- Zhan, J.; Niu, M.; Wang, P.; Zhu, X.; Li, S.; Song, J.; He, H.; Wang, Y.; Xue, L.; Fang, W.; et al. Elevated HOXB9 expression promotes differentiation and predicts a favourable outcome in colon adenocarcinoma patients. Br. J. Cancer 2014, 111, 883–893. [Google Scholar] [CrossRef]
- Huang, K.; Yuan, R.; Wang, K.; Hu, J.; Huang, Z.; Yan, C.; Shen, W.; Shao, J. Overexpression of HOXB9 promotes metastasis and indicates poor prognosis in colon cancer. Chin. J. Cancer Res. 2014, 26, 72–80. [Google Scholar]
- Li, H.; Zhu, G.; Xing, Y.; Zhu, Y.; Piao, D. miR-4324 functions as a tumor suppressor in colorectal cancer by targeting HOXB2. J. Int. Med. Res. 2019, 48, 300060519883731. [Google Scholar] [CrossRef]
- Li, H.; Li, J.; Yang, T.; Lin, S. MicroRNA-433 represses proliferation and invasion of colon cancer cells by targeting homeobox A1. Oncol. Res. 2018, 26, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Bhatlekar, S.; Viswanathan, V.; Fields, J.Z.; Boman, B.M. Overexpression of HOXA4 and HOXA9 genes promotes self-renewal and contributes to colon cancer stem cell overpopulation. J. Cell. Physiol. 2018, 233, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhao, Q.; Zhou, J.; Shi, R. miR-429 mediates tumor growth and metastasis in colorectal cancer. Am. J. Cancer Res. 2017, 7, 218–233. [Google Scholar] [PubMed]
- Chen, F.; Sun, G.; Peng, J. RNAi-mediated HOXD3 knockdown inhibits growth in human RKO cells. Oncol. Rep. 2016, 36, 1793–1798. [Google Scholar] [CrossRef]
- Kasiri, S.; Ansari, K.I.; Hussain, I.; Bhan, A.; Mandal, S.S. Antisense oligonucleotide mediated knockdown of HOXC13 affects cell growth and induces apoptosis in tumor cells and over expression of HOXC13 induces 3D-colony formation. RSC Adv. 2013, 3, 3260–3269. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Su, C.; Zhu, Y.; Li, H.; Liu, N.; Xu, T.; Sun, C.; Lv, Y. MicroRNA-544a Regulates Migration and Invasion in Colorectal Cancer Cells via Regulation of Homeobox A10. Dig. Dis. Sci. 2016, 61, 2535–2544. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, K.; Motiwala, T.; Claus, R.; Yan, P.; Kutay, H.; Datta, J.; Majumder, S.; Bai, S.; Majumder, A.; Huang, T.; et al. HOXB13, a target of DNMT3B, is methylated at an upstream CpG island, and functions as a tumor suppressor in primary colorectal tumors. PLoS ONE 2010, 5, e10338. [Google Scholar] [CrossRef] [PubMed]
- El Din, K.S.; Loree, J.; Sayre, E.C.; Gill, S.; Brown, C.; Dau, H.; De Vera, M.A. Trends in the epidemiology of young-onset colorectal cancer: A worldwide systematic review. BMC Cancer 2020, 20, 288. [Google Scholar] [CrossRef]
- Jin, X.; Dai, L.; Ma, Y.; Wang, J.; Yan, H.; Jin, Y.; Zhu, X.; Liu, Z. Homeobox proteins are potential biomarkers and therapeutic targets in gastric cancer: A systematic review and meta-analysis. BMC Cancer 2020, 20, 866. [Google Scholar] [CrossRef] [PubMed]
- Kachgal, S.; Mace, K.A.; Boudreau, N.J. The dual roles of homeobox genes in vascularization and wound healing. Cell Adhes. Migr. 2012, 6, 457–470. [Google Scholar] [CrossRef]
- Yu, M.; Zhan, J.; Zhang, H. HOX family transcription factors: Related signaling pathways and post-translational modifications in cancer. Cell Signal. 2020, 66, 109469. [Google Scholar] [CrossRef]
- Wan, J.; Xu, W.; Zhan, J.; Ma, J.; Li, X.; Xie, Y.; Wang, J.; Zhu, W.G.; Luo, J.; Zhang, H. PCAF-mediated acetylation of transcriptional factor HOXB9 suppresses lung adenocarcinoma progression by targeting oncogenic protein JMJD6. Nucleic Acids Res. 2016, 44, 10662–10675. [Google Scholar] [CrossRef] [PubMed]
- Ladam, F.; Sagerström, C.G. Hox regulation of transcription: More complex(es). Dev. Dyn. 2014, 243, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Contarelli, S.; Fedele, V.; Melisi, D. HOX genes family and cancer: A novel role for homeobox B9 in the resistance to anti-angiogenic therapies. Cancers 2020, 21, 3299. [Google Scholar] [CrossRef]
- Morgan, R.; El-Tanani, M.; Hunter, K.D.; Harrington, K.J.; Pandha, H.S. Targeting HOX/PBX dimers in cancer. Oncotarget 2015, 8, 32322–32331. [Google Scholar] [CrossRef] [PubMed]


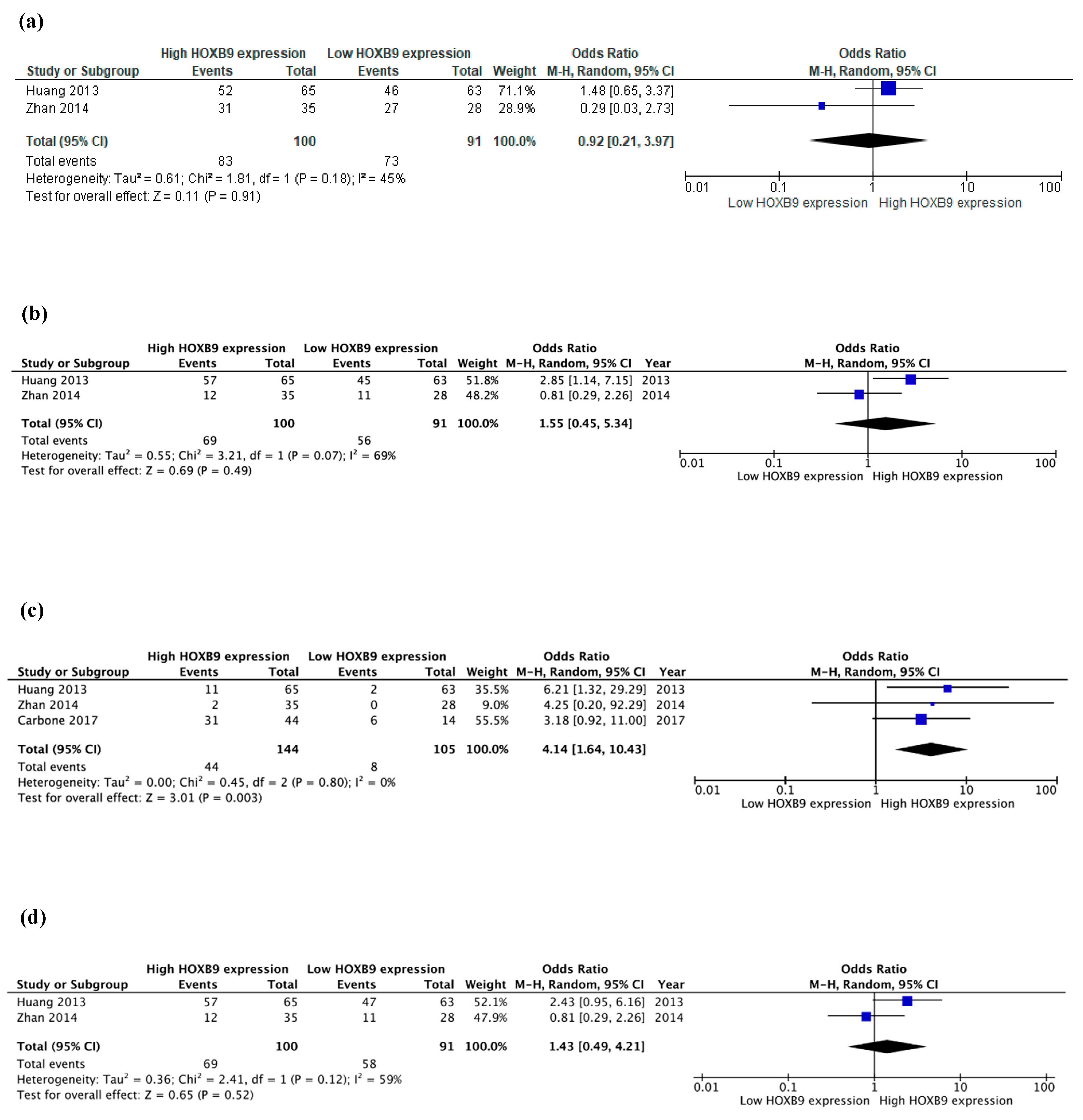
| Author (Year) | Gene | Patients (%M) | Age (ys) | Stage | FUP (m) (max) | Sample | Methods | DE (C vs. N) | HOX Overexpression Association with Clinicopathological Characteristics (Positive or Negative) | DFS (High vs. Low Expression) | OS (High vs. Low Expression) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | Sex | T | N | M | S | G | CEA | |||||||||||
| Liu et al. [24] (2020) | HOXD9 | 100 (59%) | NR | I–IV | NR | FFPE | IHC | ↑ *** | NS | NS | NS | ↑ * | NR | ↑ * | ↑ *** | NR | NR | 5y: Worse (p = 0.000) |
| Cui et al. [25] (2019) | HOXC11 | 265 (NR) | NR | NR | NR | NR | Data mining | ↑ * | NR | NR | NR | NR | NR | NR | NR | NR | NR | 10y: Worse (p = 0.021) |
| Ying et al. [28] (2019) | HOXB8 | 80 (59%) | NR | I–IV | 120 | NR | qRT-PCR | ↑ * | NS | NS | NR | ↑ | NR | ↑ | NS | NR | NR | 10y: Worse (p = 0.048) |
| 510 (NR) | NR | NR | 120 | NR | Data mining | ↑ *** | NS | NS | ↑ | NS | ↑ | NS | NS | NR | 10y: Worse (p = 0.047) | 10ys: Worse (p = 0.013) | ||
| Wu et al. [34] (2018) | HOXA6 | 16 (63%) | 49–80 | NR | NR | NR | qRT-PCR | ↑ * | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| Yuan et al. [35] (2018) | HOXA10 | 85 (58%) | 26–80 | II–IV | 60 | FFPE | IHC | ↑ *** | NS | NS | NS | NS | NR | NS | NS | NS | 5y: Worse (HR = 4.485, 95%CI:1.163–17.829, p = 0.015) | NR |
| Tatangelo et al. [37] (2018) | HOXA13 | 82 (54%) | 50–91 | I–IV | NR | FFPE | IHC | ↑ (NR) | NS | NS | NS | NS | NR | NR | NS | NS | NR | NR |
| HOXB13 | ↑ (NR) | ↑ | ↑ | NS | ↑ ** | NR | ||||||||||||
| HOXC13 | ↑ (NR) | NS | NS | NS | ↑ | NR | ||||||||||||
| HOXD13 | ↑ (NR) | NS | NS | NS | NS | NR | ||||||||||||
| Song et al. [38] (2018) | AcK27-HOXB9 | 90 (51%) | 24–90 | I–IV | 73 | FFPE | IHC | ↓ *** | ↑ * | NS | NS | NS | NR | ↓ * | NR | NR | NR | 5y: Better (p = 0.0007) |
| Bhatlekar et al. [39] (2018) | HOXA4, HOXD10 | 3 (NR) | NR | NR | NR | FT | qRT-PCR/IHC | ↑ (NR) | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| Watanabe et al. [40] (2017) | HOXA9 | 231 (58.9%) | NR | I–IV | 100 | FT FFPE | qRT-PCR/IHC | ↑ *** | NS | NS | NS | ↑ * | NS | ↑ * | NR | NR | NR | 5y: NS (p = 0.80) |
| Mansour et al. [41] (2017) | HOXD8 | 26 (NR) | 30–60 | II–IV | NR | FT | qRT-PCR and data mining | ↓ * | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| Zhang et al. [36] (2017) | HOXA3 | 232 (61%) | NR | I–IV | 140 | FFT | qRT-PCR | ↑ ** | NR | NR | NR | NR | NR | ↑ ** | NR | NR | 10y: Worse (p = 0.022) | 10y: Worse (p = 0.024) |
| Carbone et al. [42] (2017) | HOXB9 | 58 (53%) | 25–84 | I–IV | NR | FFPE | IHC | NR | NS | NR | ↑ * | NR | ↑ | NR | NR | NR | 5y: Worse, (HR = 2.552, 95%CI:1.180–5.518, p = 0.017) | NR |
| Ji et al. [43] (2016) | HOXC6 | 462 (61%) | NR | I–IV | 84 | FFPE | IHC | ↑ *** | NS | NS | ↑ *** | ↑ *** | NS | NR | NS | NS | NS | 5y: Worse, (HR = 2.14, 95%CI: 1.487–3.088, p < 0.001) |
| Wang et al. [44] (2016) | HOXD10 | 126 (59%) | NR | I–III | NR | FFT | qRT-PCR/IHC | ↓ ** | NR | NR | NR | ↓ ** | NR | NR | NR | NR | NR | NR |
| Shen et al. [45] (2016) | HOXB8 | 30 (63%) | 20–90 | I–IV | NR | FFT | qRT-PCR/WB | EQ | NS | NS | NS | NS | NS | NS | NS | NR | NR | NR |
| Hoshino et al. [28] (2014) | HOXB9 | 93 (NR) | NR | II–III | NR | FFT FFPE | qRT-PCR/IHC | ↑ (NR) | NR | NR | NR | NR | NR | NR | ↑ *** | NR | NS | 5y: Worse (p = 0.038) |
| Zhan et al. [47] (2014) | HOXB9 | 63 (54%) | 24–90 | I–IV | 73 | FFPE | IHC | NR | NR | NS | NS | NS | NS | NS | ↓ * | NR | NR | 5y: Better (p = 0.040) |
| Huang et al. [48] (2014) | HOXB9 | 128 (47%) | NR | I–IV | 60 | FFT FFPE | IHC/WB | ↑ * | NS | NS | NS | ↑ * | ↑ ** | ↑ | NS | NS | NR | 5y: Worse (p = 0.013) |
| Liao et al. [26] (2011) | HOXB7 | 224 (57%) | 23–86 | I–IV | 87 | FFT FFPE | qRT-PCR/IHC | ↑ (NR) | NS | NS | ↑ * | ↑ | ↑ * | ↑ *** | NR | NR | NR | 5y: Worse, (HR = 2.279, 95%CI: 1.062–2.687, p = 0.027) |
| Kanai et al. [27] (2010) | All HOX | 40 (68%) | 48–89 | I–IV | NR | FFT | qRT-PCR | ↑*: A9,B3, B8,B9 ↓**: B2,B13, D1,D3, D4,D8, D12 | NR | NR | NR | ↑* A3D1 | NR | NR | NR | NR | NR | NR |
| Cantile et al. [29] (2009) | HOXD13 | 48 (NR) | NR | NR | NR | FFT | qRT-PCR | ↑ *** | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| Groene et al. [30] (2006) | HOXA9 | 36 (50%) | NR | II–III | NR | FFT | qRT-PCR | NR | NR | NR | NR | NR | NR | ↑ * | NR | NR | NR | NR |
| Jung et al. [31] (2005) | HOXB13 | 53 (NR) | NR | NR | NR | FFT | qRT-PCR | ↓ (NR) | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| Toiyama et al. [32] (2005) | HOXA4 | 4 | 40–68 | NR | NR | FT | qRT-PCR | ↓ ** | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| Vider et al. [33] (1997) | HOXB5, B6, B7, B8, B9, C9 | 11 (NR) | NR | NR | NR | FFT | qRT-PCR | ↑ (NR) | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| Author (Year) | Gene | Intervention | Outcomes (Intervention vs. Control Cell Line Group) | ||||
|---|---|---|---|---|---|---|---|
| PR | CLF | AP | INV | MIGR | |||
| Studies Performed in vitro Experiments, only | |||||||
| Cui et al. [25] (2019) | HOXC11 | KD | ↓ * | NR | ↑ * | NR | NR |
| Li et al. [49] (2019) | HOXB2 | OE | ↑ *** | NR | NR | ↑ ** | ↑ ** |
| Wu et al. [34] (2018) | HOXA6 | OE | ↑ *** | ↑ ** | ↓ ** | ↑ *** | ↑ *** |
| Li et al. [50] (2018) | HOXA1 | KD | ↓ ** | NR | NR | ↓ ** | NR |
| Watanabe et al. [40] (2018) | HOXA9 | KD | NS | NR | NR | NR | NR |
| Bhatlekar et al. [51] (2018) | HOXA4 HOXA9 | KD | ↓ ** | ↓ ** | NR | NR | NR |
| Mansour et al. [41] (2017) | HOXD8 | OE | ↓ * | ↓ * | ↑ * | ↓ * | NR |
| Han et al. [52] (2017) | HOXA5 | OE | ↓ ** | ↓ ** | NR | ↓ ** | ↓ ** |
| Chen et al. [53] (2016) | HOXD3 | KD | ↓ ** | ↓ ** | ↑ ** | NR | NR |
| Kasiri et al. [54] (2013) | HOXC13 | KD | ↓ * | NR | ↑ (NR) | NR | NR |
| Jung et al. [31] (2005) | HOXB13 | OE | ↓ (NR) | NR | NR | NR | NR |
| Author (Year) | Gene | Intervention | Outcomes (Intervention vs. Control Cell Line Group) | Nude Mice (Type, n) | Outcomes (Intervention vs. Control Mice Group) | ||||
|---|---|---|---|---|---|---|---|---|---|
| PR | CLF | AP | INV | MIGR | |||||
| Studies Performed in vitro and in vivo Experiments | |||||||||
| Liu et al. [24] (2020) | HOXD9 | OE | ↑ *** | ↑ ** | NR | ↑ *** | ↑ *** | BALB/c (n = NR) | Lung mets: ↑ *** Liver mets: ↑ *** |
| Ying et al. [28] (2019) | HOXB8 | KD | ↓ ** | ↓ ** | NR | ↓ * | ↓ ** | BALB/c (n = 24) | TV (mm3): ↓ ** TW (gr): ↓ ** Liver mets: NS |
| Zhang et al. [36] (2018) | HOXA3 | KD | ↓ ** | ↓ ** | ↑ *** | NR | NR | Nod N = 10 | TW (gr): ↓ *** |
| Yuan et al. [35] (2018) | HOXA10 | KD | NR | ↓ (NR) | ↑ (NR) | NR | NR | BALB/c (n = 10) | TV (mm3): ↓ ** |
| Ji et al. [43] (2016) | HOXC6 | KD | ↓ *** | ↓ *** | NS | NR | NR | Nu/Nu (n = 8) | TS (cm): ↓ * |
| Sun et al. [55] (2016) | HOXA10 | OE | NR | NR | NR | ↓ * | NR | BALB/c (n = 10) | Lung mets: ↓ ** |
| Hoshino et al. [46] (2014) | HOXB9 | OE | NR | NR | NR | NR | NR | BALB/c (n = 8) | TV (mm3): ↑ *** TW (gr): ↑ *** |
| Zhan et al. [47] (2014) | HOXB9 | OE | ↓ ** | NR | NR | ↓ ** | ↓ ** | BALB/c (n = 19) | TW (gr): ↓ ** Lung mets: ↓ (NR) (37.5% vs. 50%) Liver mets: ↓ (NR) (37.5% vs. 70%) |
| Huang et al. [48] (2013) | HOXB9 | KD | NR | NR | NR | ↓ * | ↓ * | BALB/c (n = 24) | Lung mets: ↓ (NR) (0% vs. 56%) Liver mets: ↓ (NR) (12% vs. 81%) |
| Liao et al. [26] (2011) | HOXB7 | OE | ↑ * | ↑ ** | NR | NR | NR | BALB/c (n = 10) | TV (mm3): ↑ * |
| Ghoshal et al. [56] (2010) | HOXB13 | OE | ↓ ** | ↓ ** | NR | NR | NR | NR | TW (gr): ↓ *** TV (mm3): ↓ *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinou, E.; Falgari, G.; Bagwan, I.; Angelidi, A.M. A Systematic Review on HOX Genes as Potential Biomarkers in Colorectal Cancer: An Emerging Role of HOXB9. Int. J. Mol. Sci. 2021, 22, 13429. https://doi.org/10.3390/ijms222413429
Martinou E, Falgari G, Bagwan I, Angelidi AM. A Systematic Review on HOX Genes as Potential Biomarkers in Colorectal Cancer: An Emerging Role of HOXB9. International Journal of Molecular Sciences. 2021; 22(24):13429. https://doi.org/10.3390/ijms222413429
Chicago/Turabian StyleMartinou, Eirini, Giulia Falgari, Izhar Bagwan, and Angeliki M. Angelidi. 2021. "A Systematic Review on HOX Genes as Potential Biomarkers in Colorectal Cancer: An Emerging Role of HOXB9" International Journal of Molecular Sciences 22, no. 24: 13429. https://doi.org/10.3390/ijms222413429
APA StyleMartinou, E., Falgari, G., Bagwan, I., & Angelidi, A. M. (2021). A Systematic Review on HOX Genes as Potential Biomarkers in Colorectal Cancer: An Emerging Role of HOXB9. International Journal of Molecular Sciences, 22(24), 13429. https://doi.org/10.3390/ijms222413429







