Weakening the IF2-fMet-tRNA Interaction Suppresses the Lethal Phenotype Caused by GTPase Inactivation
Abstract
1. Introduction
2. Results
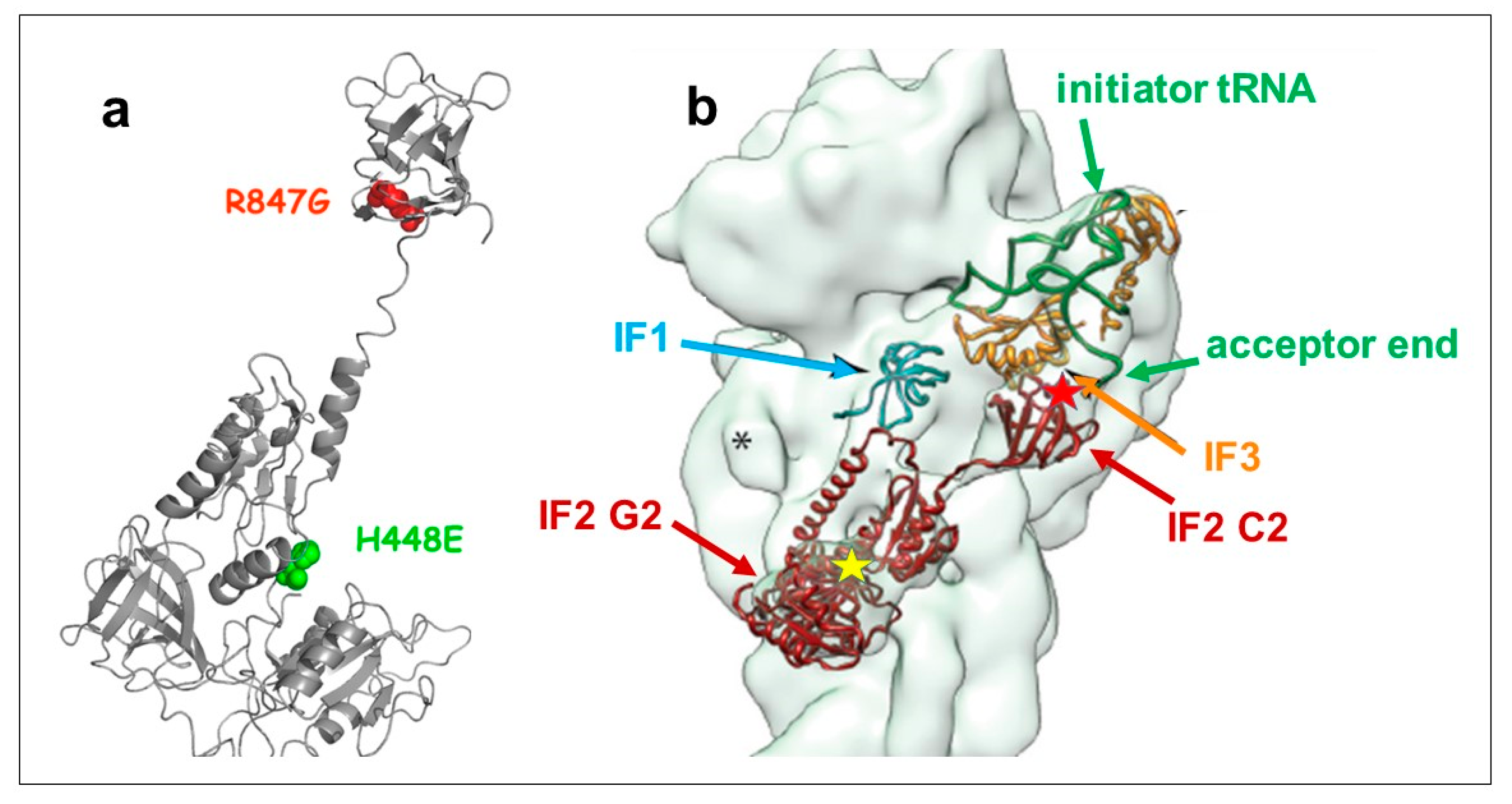
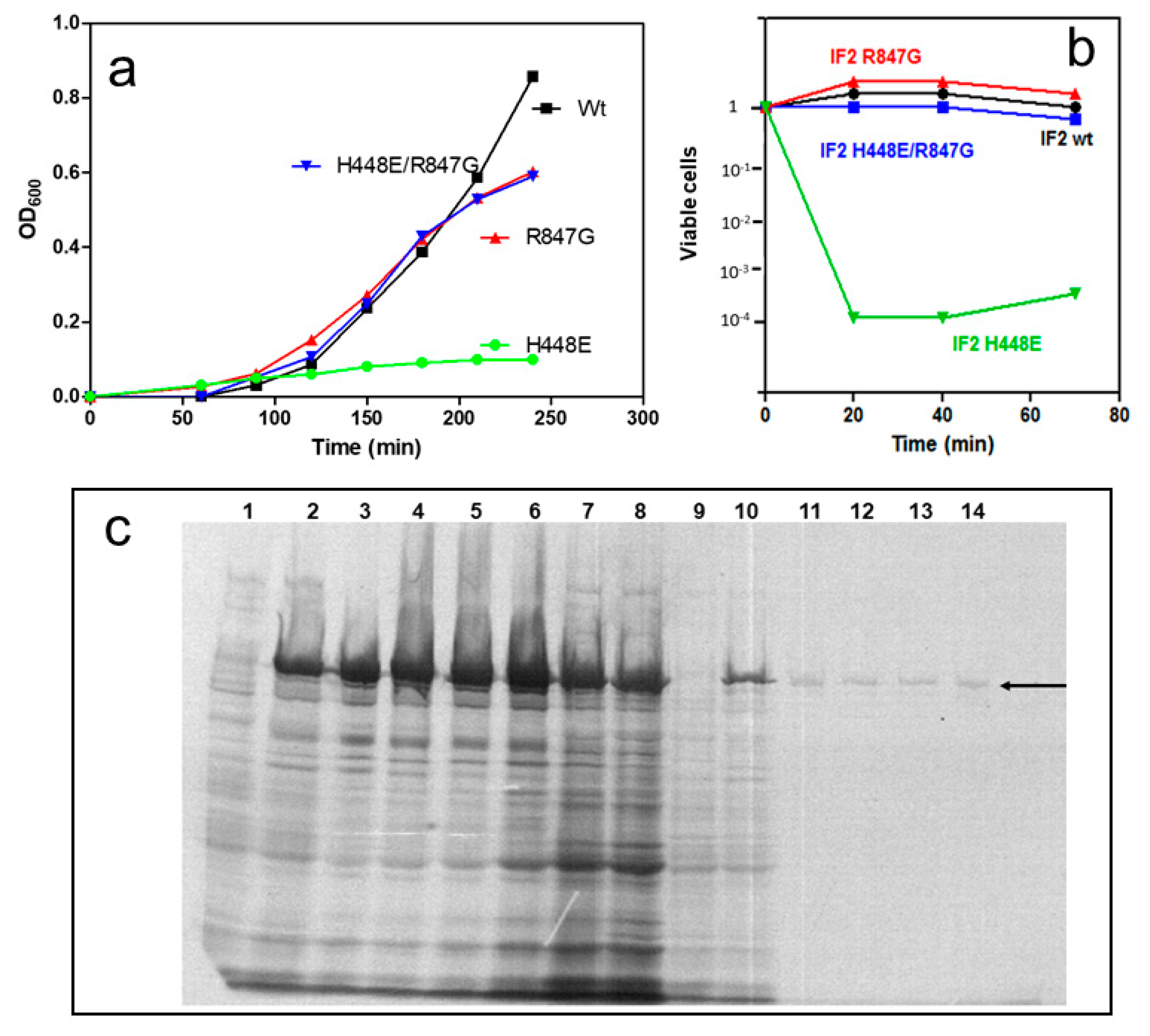
2.1. Dominant Lethal Phenotype of IF2 H448E Mutant and Suppression of Lethality
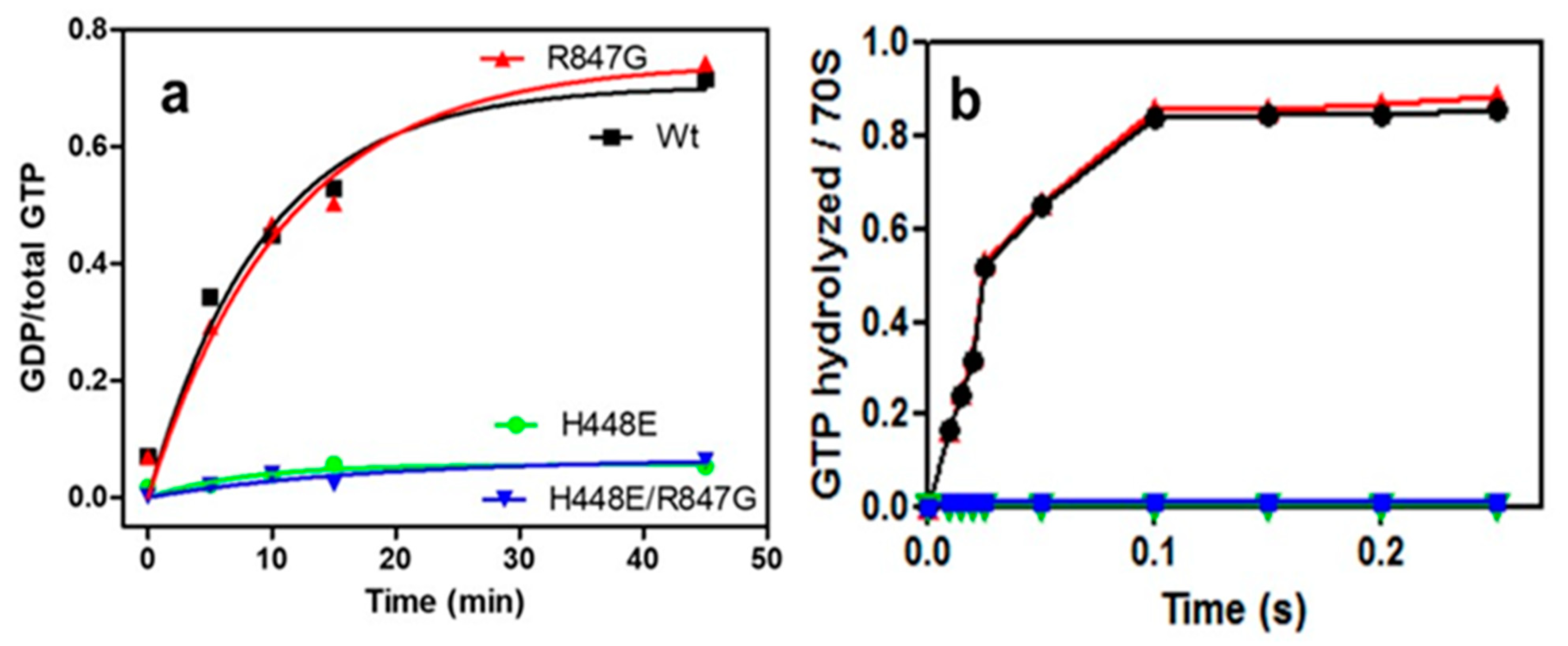
2.2. Both H448E and H448E/R847G IF2 Mutants Are Inactive in GTP Hydrolysis
2.3. Formation of 30S and 70S Initiation Complexes in the Presence of IF2 Mutants
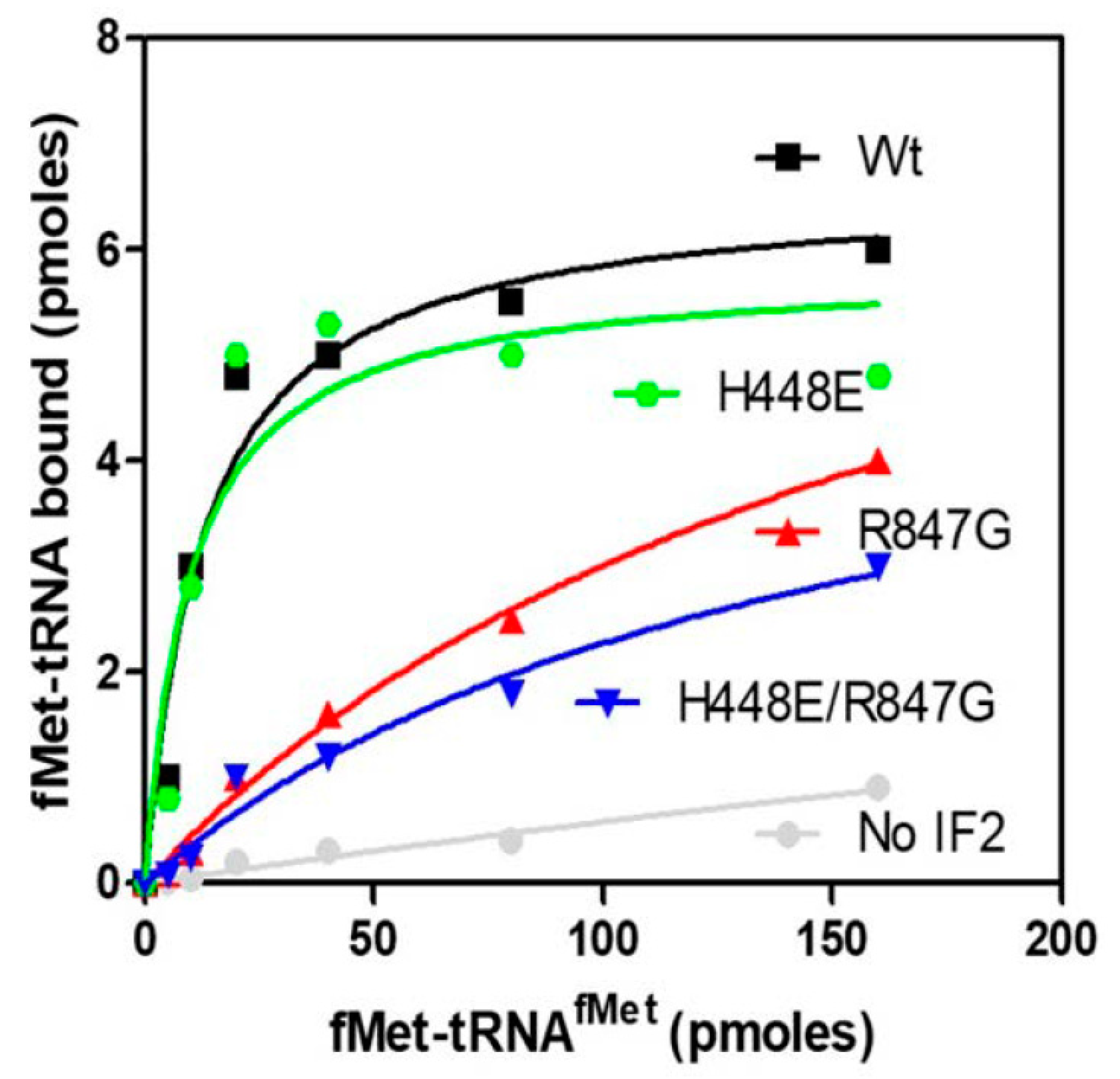
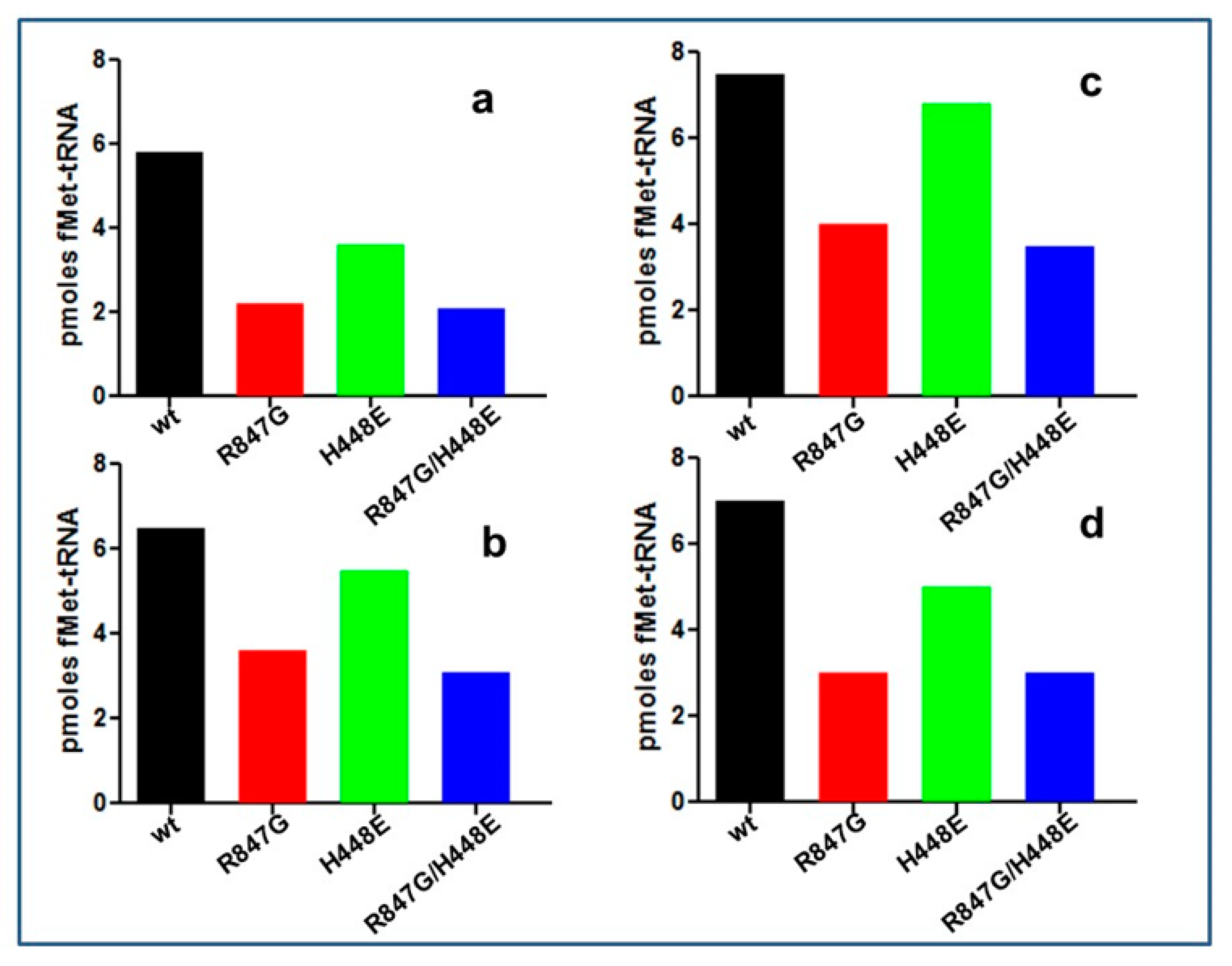
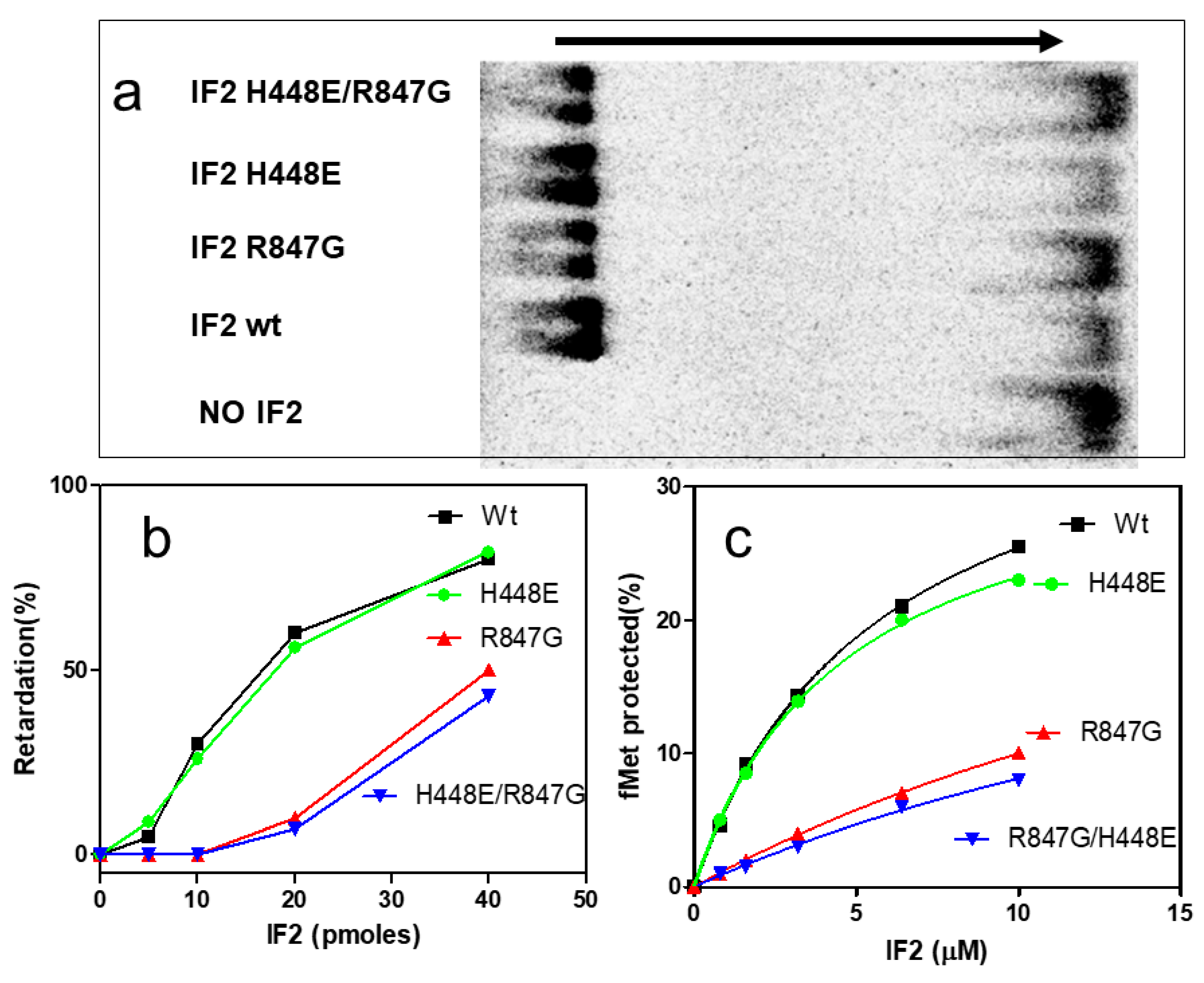
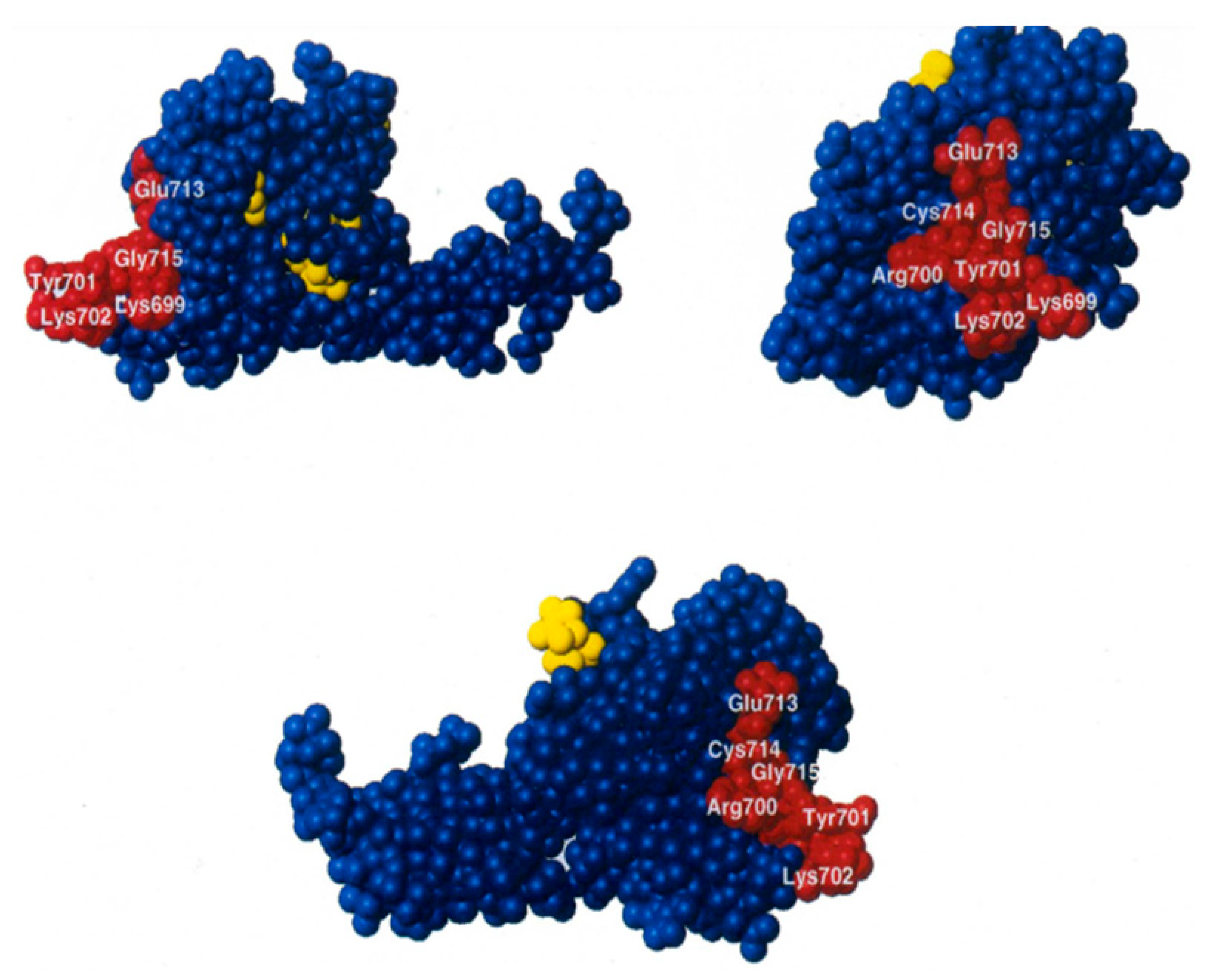
2.4. The IF2 Mutants Bearing the R847G Substitution Have a Reduced Affinity for fMet-tRNA
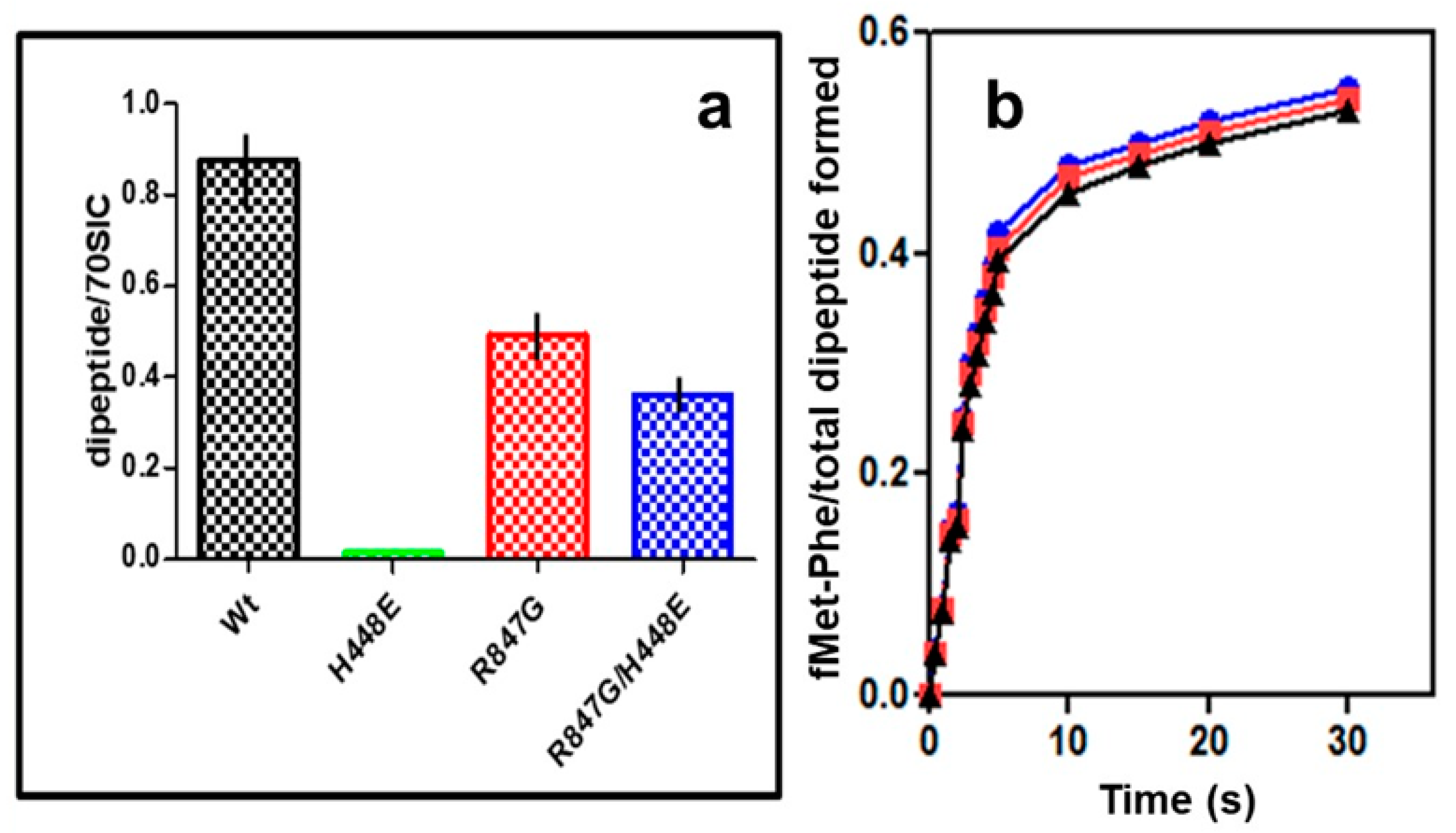
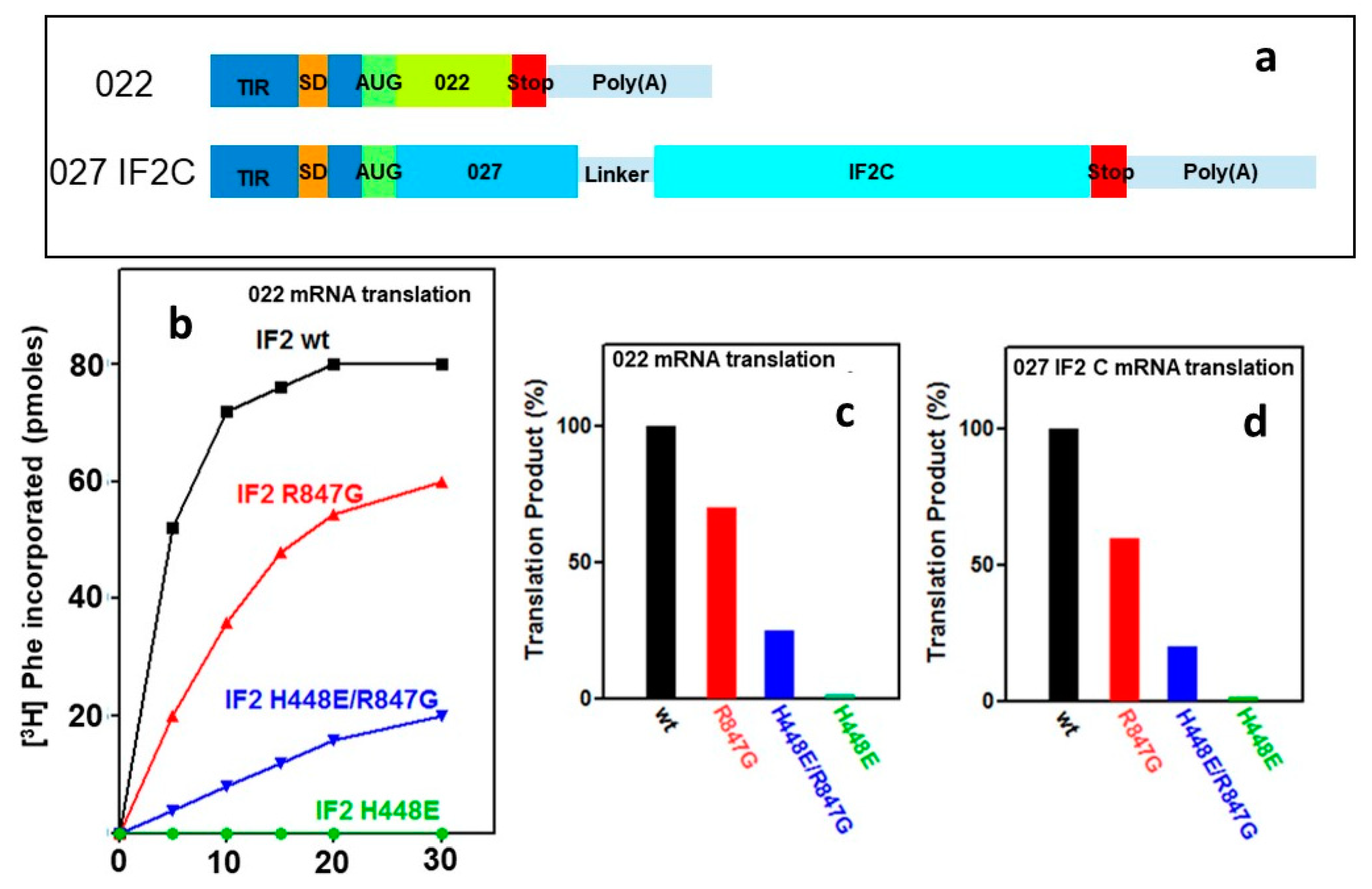
2.5. “Initiation Dipeptide” Formation and mRNA Translation with IF2 Mutants
3. Discussion
4. Materials and Methods
4.1. Buffers
- Buffer A: 10 mM Tris-HCl, pH 7.7, 60 mM NH4Cl, 5 mM Mg Acetate2, 5 mM β-mercaptoethanol, 0.2 mM PMSF.
- Buffer B: 50 mM Tris-HCl, (pH 7.5), 80 mM NH4Cl, 30 mM KCl, 7 mM MgCl2,1 mM DTT.
- Buffer C: 50 mM HEPES/KOH (pH 7.5), 10 mM MgCl2, 50 mM NH4Cl.
- Buffer D: 20 mM Tris-HCl (pH 7.7), 7 mM Mg Acetate2, 60 mM NH4Cl, 1 mM DTT.
- Buffer E: 150 mM Tris-HCl (pH 7.5), 75 mM NH4Cl, 10 mM MgCl2, 5 mM β-mercaptoethanol
4.2. General Preparations
4.3. Strains, Growth Media, and Plasmid Constructs
4.4. Heterologous Expression of IF2 wt and Mutants in Yeast
4.5. Transformation of S. cerevisiae
4.6. Heterologous Expression of IF2 wt and Mutants
4.7. Pulse–Chase Experiment
4.8. GTPase Activity of IF2wt and Mutants
4.9. fMet-tRNA Protection from Ribonuclease Hydrolysis by IF2
4.10. Gel-Retardation Experiments
4.11. IF2-Dependent Formation of 30S and 70S Initiation Complexes
4.12. Preparation of EF-Tu-GTP-Phe-tRNA Ternary Complex
4.13. Initiation Dipeptide Formation
4.14. HPLC Analysis of fMet-Phe Dipeptide
4.15. mRNA Translation In Vitro
5. Bullets
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bourne, H.R.; Sanders, D.A.; McCormick, F. The GTPase superfamily: Conserved structure and molecular mechanism. Nature 1991, 349, 117–127. [Google Scholar] [CrossRef]
- Cenatiempo, Y.; Deville, F.; Dondon, J.; Grunberg-Manago, M.; Sacerdot, C.; Hershey, J.W.; Hansen, H.F.; Petersen, H.U.; Clark, B.F.C.; Kjeldgaard, M.; et al. The protein synthesis initiation factor 2 G-domain. Study of a functionally active C-terminal 65-kilodalton fragment of IF2 from Escherichia coli. Biochemistry 1987, 26, 5070–5076. [Google Scholar] [CrossRef] [PubMed]
- Simonetti, A.; Marzi, S.; Fabbretti, A.; Hazemann, I.; Jenner, L.; Urzhumtsev, A.; Gualerzi, C.O.; Klaholz, B.P. Structure of the protein core of translation initiation factor 2 in apo, GTP-bound and GDP-bound forms. Acta Crystall. D Biol. Crystallogr. 2013, 69, 925–933. [Google Scholar] [CrossRef]
- Wienk, H.; Tishchenko, E.; Belardinelli, R.; Tomaselli, S.; Dongre, R.; Spurio, R.; Folkers, G.E.; Gualerzi, C.O.; Boelens, R. Structural dynamics of bacterial translation initiation factor IF2. J. Biol. Chem. 2012, 287, 10922–10932. [Google Scholar] [CrossRef]
- Milon, P.; Carotti, M.; Konevega, A.L.; Wintermeyer, W.; Rodnina, M.V.; Gualerzi, C.O. The ribosome- bound initiation factor 2 recruits initiator tRNA to the 30S initiation complex. EMBO Rep. 2010, 11, 312–316. [Google Scholar] [CrossRef]
- Goyal, A.; Belardinelli, R.; Maracci, C.; Milon, P.; Rodnina, M.V. Directional transition from initiation to elongation in bacterial translation. Nucleic Acids Res. 2015, 43, 10700–10712. [Google Scholar] [CrossRef]
- Gualerzi, C.O.; Pon, C.L. Initiation of mRNA translation in bacteria—Structural and dynamic aspects. Cell. Mol. Life Sci. 2015, 72, 4341–4367. [Google Scholar] [CrossRef]
- Myasnikov, A.G.; Marzi, S.; Simonetti, A.; Giuliodori, A.M.; Gualerzi, C.O.; Yusupova, G.; Yusupov, M.; Klaholz, B.P. Conformational transition of initiation factor 2 from the GTP- to GDP-state visualized on the ribosome. Nat. Struct. Mol. Biol. 2005, 12, 1145–1149. [Google Scholar] [CrossRef]
- Allen, G.S.; Zavialov, A.; Gursky, R.; Ehrenberg, M.; Frank, J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 2005, 121, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Simonetti, A.; Marzi, S.; Myasnikov, A.G.; Yusupov, M.; Gualerzi, C.O.; Klaholz, B.P. Structure of a 30S ribosomal initiation complex. Nature 2008, 455, 416–420. [Google Scholar] [CrossRef]
- Julian, P.; Milon, P.; Agirrezabala, X.; Lasso, G.; Gil, D.; Rodnina, M.V.; Valle, M. The Cryo-EM Structure of a Complete 30S Translation Initiation Complex from Escherichia coli. PLoS Biol. 2011, 9, e1001095. [Google Scholar] [CrossRef]
- Simonetti, A.; Marzi, S.; Billas, I.M.; Tsai, A.; Fabbretti, A.; Myasnikov, A.G.; Roblin, P.; Vaiana, A.C.; Hazemann, I.; Eiler, D.; et al. Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor. Proc. Natl. Acad. Sci. USA 2013, 110, 15656–15661. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Llacer, J.L.; Wimberly, B.T.; Kieft, J.S.; Ramakrishnan, V. Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation. Cell 2016, 167, 133–144. [Google Scholar] [CrossRef]
- Sprink, T.; Ramrath, D.J.; Yamamoto, H.; Yamamoto, K.; Loerke, J.; Ismer, J.; Hildebrand, P.W.; Scheerer, P.; Bürger, J.; Mielke, T.; et al. Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association. Sci. Adv. 2016, 2, e1501502. [Google Scholar] [CrossRef]
- Kaledhonkar, S.; Fu, Z.; Caban, K.; Li, W.; Chen, B.; Sun, M.; Gonzalez, R.L., Jr.; Frank, J. Late steps in bacterial translation initiation visualized using time-resolved cryo-EM. Nature 2019, 570, 400–404. [Google Scholar] [CrossRef]
- Tomsic, J.; Vitali, L.A.; Daviter, T.; Savelsbergh, A.; Spurio, R.; Striebeck, P.; Wintermeyer, W.; Rodnina, M.V.; Gualerzi, C.O. Late events of translation initiation in bacteria: A kinetic analysis. EMBO J. 2000, 19, 2127–2136. [Google Scholar] [CrossRef] [PubMed]
- Antoun, A.; Pavlov, M.Y.; Andersson, K.; Tenson, T.; Ehrenberg, M. The roles of initiation factor 2 and guanosine triphosphate in initiation of protein synthesis. EMBO J. 2003, 22, 5593–5601. [Google Scholar] [CrossRef]
- Antoun, A.; Pavlov, M.Y.; Tenson, T.; Ehrenberg, M. Ribosome formation from subunits studied by stopped-flow Rayleigh light scattering. Biol. Proced. Online 2004, 6, 35–54. [Google Scholar] [CrossRef]
- Fabbretti, A.; Pon, C.L.; Hennelly, S.P.; Hill, W.E.; Lodmell, J.S.; Gualerzi, C.O. The real-time path of translation factor IF3 onto and off the ribosome. Mol. Cell 2007, 25, 285–296. [Google Scholar] [CrossRef]
- Grigoriadou, C.; Marzi, S.; Kirillov, S.; Gualerzi, C.O.; Cooperman, B.S. A qualitative kinetic scheme for 70S initiation complex formation. J. Mol. Biol. 2007, 373, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Milón, P.; Konevega, A.L.; Gualerzi, C.O.; Rodnina, M.V. Kinetic checkpoint at a late step in translation initiation. Mol. Cell 2008, 30, 712–720. [Google Scholar] [CrossRef]
- Milon, P.; Maracci, C.; Filonava, L.; Gualerzi, C.O.; Rodnina, M.V. Kinetic landscape of the 30S translation initiation complex formation. Nat. Struct. Mol. Biol. 2012, 19, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Caban, K.; Gonzalez, R.L., Jr. Ribosomal initiation complex-driven changes in the stability and dynamics of initiation factor 2 regulate the fidelity of translation initiation. J. Mol. Biol. 2015, 427, 1819–1834. [Google Scholar] [CrossRef]
- Eiler, D.; Lin, J.; Simonetti, A.; Klaholz, B.P.; Steitz, T.A. Initiation factor 2 crystal structure reveals a different domain organization from eukaryotic initiation factor 5B and mechanism among translational GTPases. Proc. Natl. Acad. Sci. USA 2013, 110, 15662–15667. [Google Scholar] [CrossRef] [PubMed]
- Pon, C.L.; Paci, M.; Pawlik, R.T.; Gualerzi, C.O. Structure-function relationship in Escherichia coli initiation factors. Biochemical and biophysical characterization of the interaction between IF-2 and guanosine nucleotides. J. Biol. Chem. 1985, 260, 8918–8924. [Google Scholar] [CrossRef]
- Marshall, R.A.; Aitken, C.E.; Puglisi, J.D. GTP Hydrolysis by IF2 Guides Progression of the Ribosome into Elongation. Mol. Cell 2009, 35, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Frank, J.; Gao, H.; Sengupta, J.; Gao, N.; Taylor, D.J. The process of mRNA–tRNA translocation. Proc. Natl. Acad. Sci. USA 2007, 104, 19671–19678. [Google Scholar] [CrossRef]
- Ermolenko, D.N.; Cornish, P.V.; Ha, T.; Noller, H.F. Antibiotics that bind to the A site of the large ribosomal subunit can induce mRNA translocation. RNA 2013, 19, 158–166. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, J.; Lancaster, L.; Donohuea, J.P.; Noller, H.F. Spontaneous ribosomal translocation of mRNA and tRNAs into a chimeric hybrid state. Proc. Natl. Acad. Sci. USA 2019, 116, 7813–7818. [Google Scholar] [CrossRef]
- Frank, J.; Gonzalez, R.L., Jr. Structure and Dynamics of a Processive Brownian Motor: The Translating Ribosome. Annu. Rev. Biochem. 2010, 79, 381–412. [Google Scholar] [CrossRef] [PubMed]
- Cornish, P.V.; Ermolenko, D.N.; Noller, H.F.; Ha, T. Spontaneous Intersubunit Rotation in Single Ribosomes. Mol. Cell 2008, 30, 578–588. [Google Scholar]
- Guo, Z.; Noller, H.F. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc. Natl. Acad. Sci. USA 2012, 109, 20391–20394. [Google Scholar] [CrossRef]
- Dever, T.E.; Glynias, M.J.; Merrick, W.C. GTP-binding domain: Three consensus sequence elements with distinct spacing. Proc. Natl. Acad. Sci. USA 1987, 84, 1814–1818. [Google Scholar] [CrossRef]
- Roll-Mecak, A.; Cao, C.; Dever, T.E.; Burley, S.K. X-Ray Structures of the Universal Translation Initiation Factor IF2/eIF5B. Cell 2000, 103, 781–792. [Google Scholar] [CrossRef]
- Cool, R.H.; Parmeggiani, A. Substitution of histidine-84 and the GTPase mechanism of elongation factor Tu. Biochemistry 1991, 30, 362–366. [Google Scholar] [CrossRef]
- Scarano, G.; Krab, I.M.; Bocchini, V.; Parmeggiani, A. Relevance of histidine-84 in the elongation factor Tu GTPase activity and in poly(Phe) synthesis: Its substitution by glutamine and alanine. FEBS Lett. 1995, 365, 214–218. [Google Scholar] [CrossRef]
- Zeidler, W.; Egle, C.; Ribeiro, S.; Wagner, A.; Katunin, V.; Kreutzer, R.; Rodnina, M.; Wintermeyer, W.; Sprinzl, M. Site-directed mutagenesis of Thermus thermophilus elongation factor Tu. Replacement of His85, Asp81 and Arg300. Eur. J. Biochem. 1995, 229, 596–604. [Google Scholar] [CrossRef]
- Laalami, S.; Timofeev, A.V.; Putzer, H.; Leautey, J.; Grunberg-Manago, M. In vivo study of engineered G-domain mutants of Escherichia coli translation initiation factor IF2. Mol. Microbiol. 1994, 11, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Luchin, S.; Putzer, H.; Cenatiempo, Y.; Grunberg-Manago, M.; Laalami, S. In Vitro study of two dominant inhibitory GTPase mutants of Escherichia coli translation initiation factor IF2. Direct evidence that GTP hydrolysis is necessary for factor recycling. J. Biol. Chem. 1999, 274, 6074–6079. [Google Scholar] [CrossRef]
- Caserta, E.; Ferrara, C.; Milon, P.; Fabbretti, A.; Rocchetti, A.; Tomsic, J.; Pon, C.L.; Gualerzi, C.O.; La Teana, A. Ribosomal interaction of Bacillus stearothermophilus translation initiation factor IF2: Characterization of the active sites. J. Mol. Biol. 2010, 396, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Fabbretti, A.; Brandi, L.; Milon, P.; Spurio, R.; Pon, C.L.; Gualerzi, C.O. Translation initiation without IF2-dependent GTP hydrolysis. Nucleic Acids Res. 2012, 40, 7946–7955. [Google Scholar] [CrossRef]
- Guenneugues, M.; Meunier, S.; Boelens, R.; Caserta, E.; Brandi, L.; Spurio, R.; Pon, C.L.; Gualerzi, C.O. Mapping the fMet-tRNA binding site of initiation factor IF2. EMBO J. 2000, 19, 5233–5249. [Google Scholar] [CrossRef] [PubMed]
- Gualerzi, C.O.; Brandi, L.; Caserta, E.; La Teana, A.; Spurio, R.; Tomšic, J.; Pon, C.L. Translation initiation in bacteria. In The Ribosome; Garrett, R.A., Douthwaite, S.R., Liljas, A., Matheson, A.T., Moore, P.B., Noller, H.F., Eds.; ASM Press: Washington, DC, USA, 2000; pp. 475–494. [Google Scholar]
- Tomsic, J.; Smorlesi, A.; Caserta, E.; Giuliodori, A.M.; Pon, C.L.; Gualerzi, C.O. Disparate Phenotypes Resulting from Mutations of a Single Histidine in Switch II of Geobacillus stearothermophilus Translation Initiation Factor IF2. Int. J. Mol. Sci. 2020, 21, 735. [Google Scholar] [CrossRef] [PubMed]
- Brandi, L.; Fabbretti, A.; Milon, P.; Carotti, M.; Pon, C.L.; Gualerzi, C.O. Methods for identifying compounds that specifically target translation. Meth. Enzymol. 2007, 431, 229–267. [Google Scholar]
- Meunier, S.; Spurio, R.; Czisch, M.; Wechselberger, R.; Guenneugues, M.; Gualerzi, C.O.; Boelens, R. Solution structure of the fMet-tRNA binding domain of B. stearothermophilus initiation factor IF2. EMBO J. 2000, 19, 1918–1926. [Google Scholar] [CrossRef]
- Gualerzi, C.O.; Brandi, L.; Caserta, E.; Garofalo, C.; Lammi, M.; La Teana, A.; Petrelli, D.; Spurio, R.; Tomsic, J.; Pon, C.L. Role of the initiation factors in the early events of mRNA translation in bacteria. Cold Spring Harbor Symp. Quant. Biol. 2001, 66, 363–376. [Google Scholar] [CrossRef]
- Milon, P.; Konevega, A.L.; Peske, F.; Fabbretti, A.; Gualerzi, C.O.; Rodnina, M.V. Transient kinetics, fluorescence, and FRET in studies of bacterial initiation. Meth. Enzymol. 2007, 430, 1–30. [Google Scholar]
- Pawlik, R.T.; Littlechild, J.; Pon, C.L.; Gualerzi, C. Purification and properties of Escherichia coli translational initiation factors. Biochem. Int. 1981, 2, 421–428. [Google Scholar]
- Spurio, R.; Severini, M.; La Teana, A.; Canonaco, M.A.; Pawlik, R.T.; Gualerzi, C.; Pon, C. Novel structural and functional aspects of translation initiation factor IF2. In The Translational Apparatus; Nierhaus, K.H., Ed.; Plenum Press: New York, NY, USA, 1993; pp. 433–444. [Google Scholar]
- Spurio, R.; Brandi, L.; Caserta, E.; Pon, C.L.; Gualerzi, C.O.; Misselwitz, R.; Krafft, C.; Welfle, K.; Welfle, H. The C-terminal subdomain (IF2 C-2) contains the entire fMet-tRNA binding site of initiation factor IF2. J. Biol. Chem. 2000, 275, 2447–2454. [Google Scholar] [CrossRef]
- Elish, M.E.; Pierce, J.R.; Earhart, C.F. Biochemical analysis of spontaneous fepA mutants of Escherichia coli. J. Gen. Microbiol. 1988, 134, 1355–1364. [Google Scholar] [CrossRef]
- Baldari, C.; Murray, J.A.; Ghiara, P.; Cesareni, G.; Galeotti, C.L. A novel leader peptide which allows efficient secretion of a fragment of human interleukin 1 beta in Saccharomyces cerevisiae. EMBO J. 1987, 6, 229–234. [Google Scholar] [CrossRef]
- Ito, H.; Fukuda, Y.; Murata, K.; Kimura, A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983, 153, 163–168. [Google Scholar] [CrossRef]
- Sheele, J.S.; Rhee, J.M.; Boss, G.R. Cell Biology Determination of absolute amounts of GDP and GTP bound to Ras in mammalian cells: Comparison of parental and Ras-overproducing NIH 3T3 fibroblasts. Proc. Natl. Acad. Sci. USA 1995, 92, 1097–1100. [Google Scholar] [CrossRef]
- Rodnina, M.V.; Savelsbergh, A.; Matassova, N.B.; Katunin, V.I.; Semenkov, Y.P.; Wintermeyer, W. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc. Natl. Acad. Sci. USA 1999, 96, 9586–9590. [Google Scholar] [CrossRef] [PubMed]
- Förster, C.; Krafft, C.; Welfle, H.; Gualerzi, C.O.; Heinemann, H. Preliminary characterization by X-ray diffraction and Raman spectroscopy of a crystalline complex of Bacillus stearothermophilus initiation factor 2 C-domain and fMet-tRNAfMet. Acta Crystallogr. D Biol. Crystallogr. 1999, 55, 712–716. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomsic, J.; Caserta, E.; Pon, C.L.; Gualerzi, C.O. Weakening the IF2-fMet-tRNA Interaction Suppresses the Lethal Phenotype Caused by GTPase Inactivation. Int. J. Mol. Sci. 2021, 22, 13238. https://doi.org/10.3390/ijms222413238
Tomsic J, Caserta E, Pon CL, Gualerzi CO. Weakening the IF2-fMet-tRNA Interaction Suppresses the Lethal Phenotype Caused by GTPase Inactivation. International Journal of Molecular Sciences. 2021; 22(24):13238. https://doi.org/10.3390/ijms222413238
Chicago/Turabian StyleTomsic, Jerneja, Enrico Caserta, Cynthia L. Pon, and Claudio O. Gualerzi. 2021. "Weakening the IF2-fMet-tRNA Interaction Suppresses the Lethal Phenotype Caused by GTPase Inactivation" International Journal of Molecular Sciences 22, no. 24: 13238. https://doi.org/10.3390/ijms222413238
APA StyleTomsic, J., Caserta, E., Pon, C. L., & Gualerzi, C. O. (2021). Weakening the IF2-fMet-tRNA Interaction Suppresses the Lethal Phenotype Caused by GTPase Inactivation. International Journal of Molecular Sciences, 22(24), 13238. https://doi.org/10.3390/ijms222413238





