Microsatellite Instability Analysis (MSA) for Bladder Cancer: Past History and Future Directions
Abstract
1. Introduction
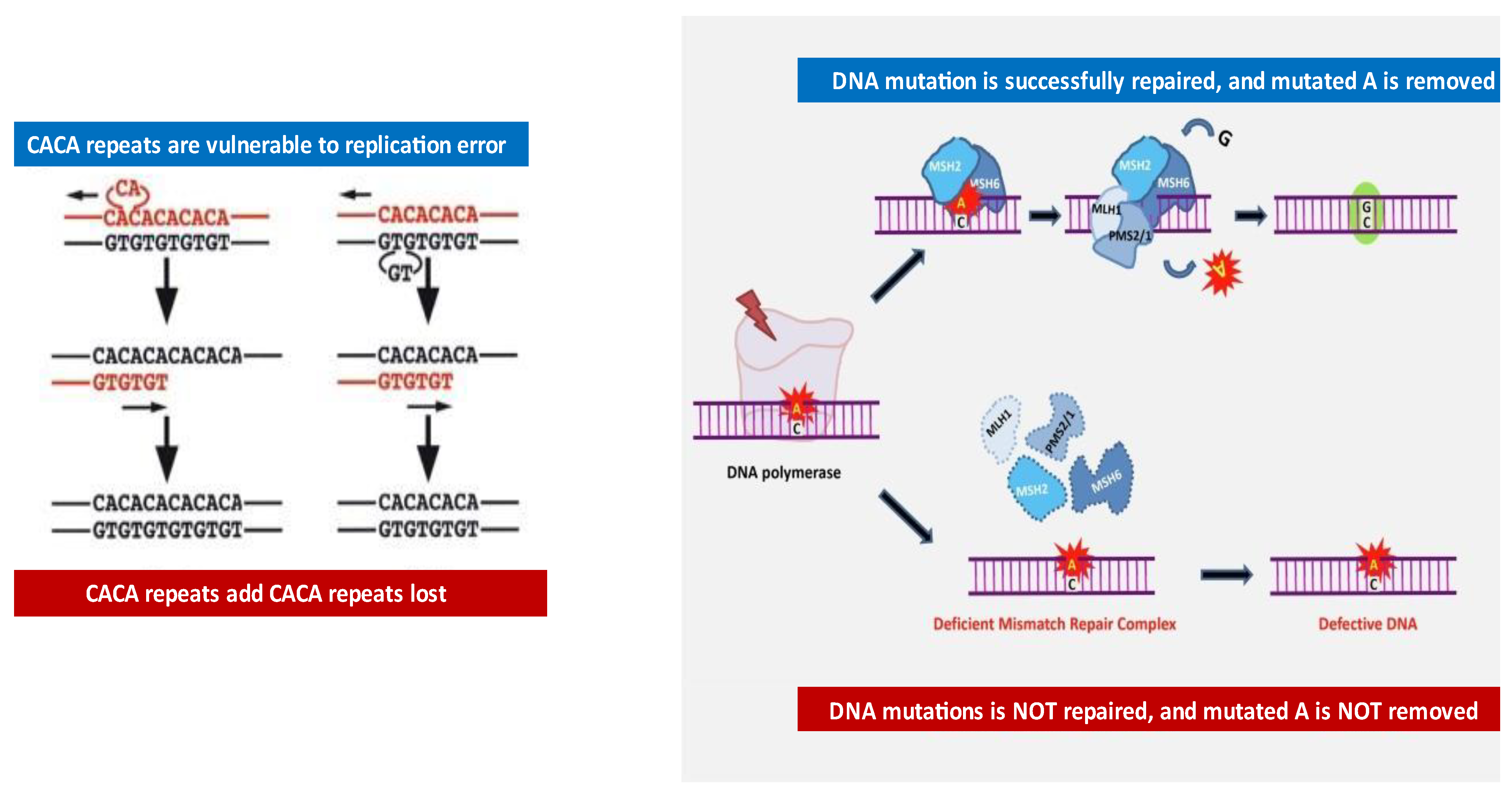
2. An Overview of MSI
2.1. Initial Discoveries and Clinical Applications
2.2. Evolution of MSA Methods
2.3. Loss of Heterozygosity (LOH) in Bladder Cancer Patients
3. Use of Microsatellite Assay for Bladder Cancer Detection
3.1. Bladder Cancer Overview and Diagnostic Biomarkers
3.2. Overview: MSI and LOH in Bladder Cancer Detection
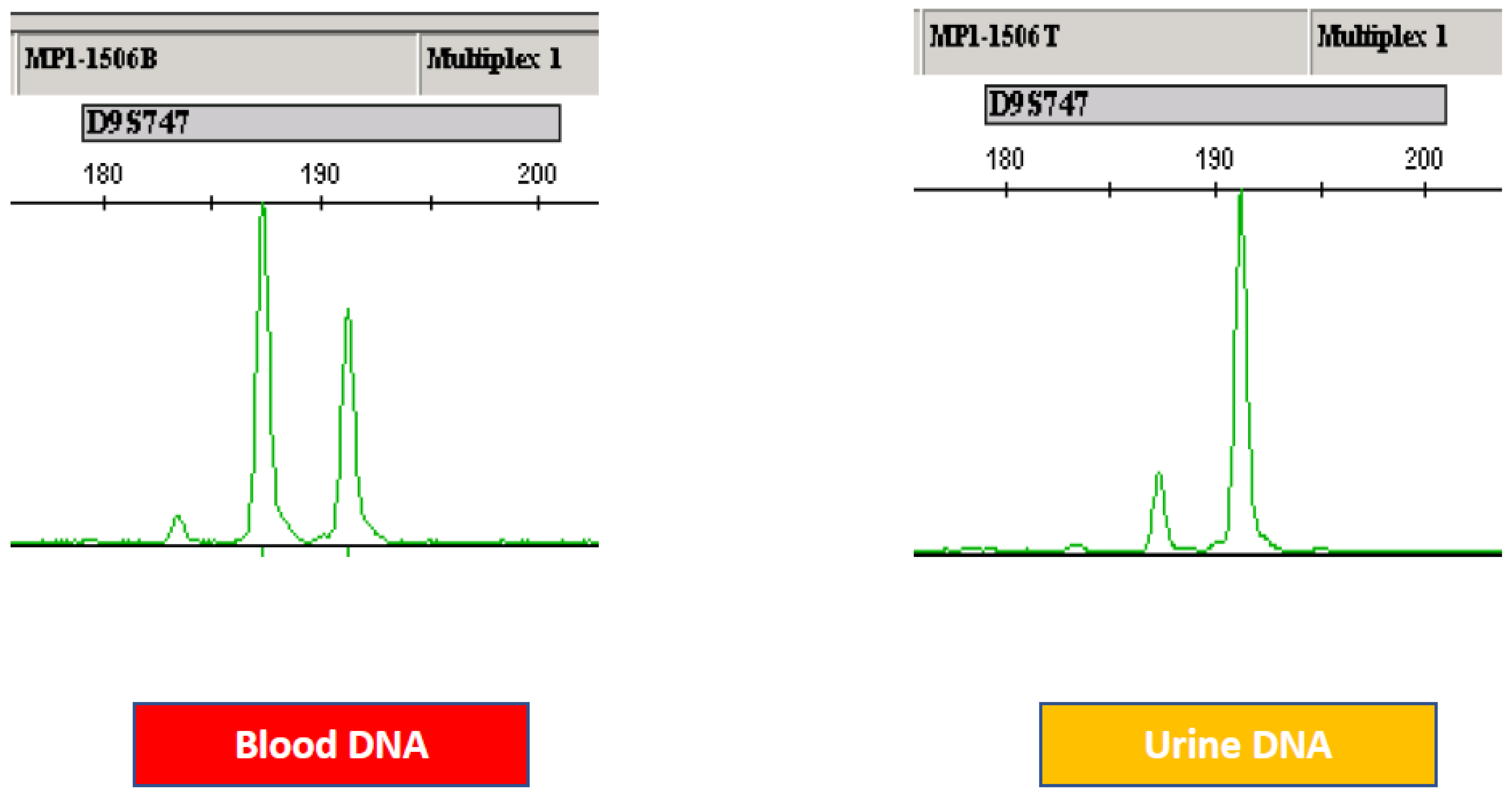
3.3. Microsatellite Analysis: Initial Studies
3.4. Microsatellite Analysis: Follow Up Studies
3.5. MSA Assay for Surveillance for Recurrent Bladder Cancer
3.6. MSA Assay as a Tool Predicting Recurrent Bladder Cancer
3.7. MSA Assay for Different Ethnic Group
3.8. Automated MSA Assay for Detection of Bladder Cancer
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aaltonen, L.A.; Peltomäki, P.; Leach, F.S.; Sistonen, P.; Pylkkänen, L.; Mecklin, J.P.; Järvinen, H.; Powell, S.M.; Jen, J.; Hamilton, S.R.; et al. Clues to the pathogenesis of familial colorectal cancer. Science 1993, 260, 812–816. [Google Scholar] [CrossRef]
- Ionov, Y.; Peinado, M.A.; Malkhosyan, S.; Shibata, D.; Perucho, M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 1993, 363, 558–561. [Google Scholar] [CrossRef]
- Thibodeau, S.; Bren, G.; Schaid, D. Microsatellite instability in cancer of the proximal colon. Science 1993, 260, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Imai, K.; Yamamoto, H. Carcinogenesis and microsatellite instability: The interrelationship between genetics and epigenetics. Carcinogenesis 2008, 29, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Adachi, Y.; Taniguchi, H.; Kunimoto, H.; Nosho, K.; Suzuki, H.; Shinomura, Y. Interrelationship between microsatellite instability and microRNA in gastrointestinal cancer. World J. Gastroenterol. 2012, 18, 2745–2755. [Google Scholar] [CrossRef]
- Yamamoto, H.; Watanabe, Y.; Maehata, T.; Morita, R.; Yoshida, Y.; Oikawa, R.; Ishigooka, S.; Ozawa, S.-I.; Matsuo, Y.; Hosoya, K.; et al. An updated review of gastric cancer in the next-generation sequencing era: Insights from bench to bedside and vice versa. World J. Gastroenterol. 2014, 20, 3927–3937. [Google Scholar] [CrossRef]
- Yamamoto, H.; Imai, K. Microsatellite instability: An update. Arch. Toxicol. 2015, 89, 899–921. [Google Scholar] [CrossRef] [PubMed]
- Gelsomino, F.; Barbolini, M.; Spallanzani, A.; Pugliese, G.; Cascinu, S. The evolving role of microsatellite instability in colorectal cancer: A review. Cancer Treat. Rev. 2016, 51, 19–26. [Google Scholar] [CrossRef]
- de la Chapelle, A.; Hampel, H. Clinical relevance of microsatellite instability in colorectal cancer. J. Clin. Oncol. 2010, 28, 3380–3387. [Google Scholar] [CrossRef]
- Bacher, J.W.; Flanagan, L.A.; Smalley, R.L.; Nassif, N.A.; Burgart, L.J.; Halberg, R.B.; Megid, W.M.A.; Thibodeau, S.N. Development of a fluorescent multiplex assay for detection of MSI-High tumors. Dis. Markers 2004, 20, 237–250. [Google Scholar] [CrossRef]
- Kim, T.-M.; Laird, P.W.; Park, P.J. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell 2013, 155, 858–868. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Soong, T.D.; Elemento, O. A novel approach for characterizing microsatellite instability in cancer cells. PLoS ONE 2013, 8, e63056. [Google Scholar] [CrossRef]
- McIver, L.J.; Fonville, N.C.; Karunasena, E.; Garner, H.R. Microsatellite genotyping reveals a signature in breast cancer exomes. Breast Cancer Res. Treat. 2014, 145, 791–798. [Google Scholar] [CrossRef]
- Niu, B.; Ye, K.; Zhang, Q.; Lu, C.; Xie, M.; McLellan, M.D.; Wendl, M.C.; Ding, L. MSIsensor: Microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2014, 30, 1015–1016. [Google Scholar] [CrossRef] [PubMed]
- Salipante, S.J.; Scroggins, S.M.; Hampel, H.L.; Turner, E.H.; Pritchard, C.C. Microsatellite instability detection by next generation sequencing. Clin. Chem. 2014, 60, 1192–1199. [Google Scholar] [CrossRef]
- Huang, M.N.; McPherson, J.R.; Cutcutache, I.; Teh, B.T.; Tan, P.; Rozen, S.G. MSIseq: Software for assessing microsatellite instability from catalogs of somatic mutations. Sci. Rep. 2015, 5, 13321. [Google Scholar] [CrossRef]
- Yamamoto, H.; Imai, K.; Perucho, M. Gastrointestinal cancer of the microsatellite mutator phenotype pathway. J. Gastroenterol. 2002, 37, 153–163. [Google Scholar] [CrossRef]
- Perucho, M. Tumors with microsatellite instability: Many mutations, targets and paradoxes. Oncogene 2003, 22, 2223–2225. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oda, S.; Zhao, Y.; Maehara, Y. Microsatellite instability in gastrointestinal tract cancers: A brief update. Surg. Today 2005, 35, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Söreide, K.; Janssen, E.A.M.; Söiland, H.; Körner, H.; Baak, J.P.A. Microsatellite instability in colorectal cancer. Br. J. Surg. 2006, 93, 395–406. [Google Scholar] [CrossRef]
- Suzuki, K.; Suzuki, I.; Leodolter, A.; Alonso, S.; Horiuchi, S.; Yamashita, K.; Perucho, M. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell 2006, 9, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.; Carey, F.A.; Beattie, J.; Wilkie, M.J.V.; Lightfoot, T.J.; Coxhead, J.; Garner, R.C.; Steele, R.J.C.; Wolf, C.R. Mutations in APC, Kirsten-ras, and p53-alternative genetic pathways to colorectal cancer. Proc. Natl. Acad. Sci. USA 2002, 99, 9433–9438. [Google Scholar] [CrossRef]
- Vilar, E.; Gruber, S.B. Microsatellite instability in colorectal cancer-the stable evidence. Nat. Rev. Clin. Oncol. 2010, 7, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Puliga, E.; Corso, S.; Pietrantonio, F.; Giordano, S. Microsatellite instability in Gastric Cancer: Between lights and shadows. Cancer Treat Rev. 2021, 95, 102175. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef]
- Timmermann, B.; Kerick, M.; Roehr, C.; Fischer, A.; Isau, M.; Boerno, S.T.; Wunderlich, A.; Barmeyer, C.; Seemann, P.; Koenig, J.; et al. Somatic mutation profiles of MSI and MSS colorectal cancer identified by whole exome next generation sequencing and bioinformatics analysis. PLoS ONE 2010, 5, e15661. [Google Scholar] [CrossRef]
- Woerner, S.M.; Yuan, Y.P.; Benner, A.; Korff, S.; von Knebel Doeberitz, M.; Bork, P. SelTarbase, a database of human mononucleotide-microsatellite mutations and their potential impact to tumorigenesis and immunology. Nucleic Acids Res. 2010, 38, D682–D689. [Google Scholar] [CrossRef]
- Boland, C.R.; Thibodeau, S.N.; Hamilton, S.R.; Sidransky, D.; Eshleman, J.R.; Burt, R.W.; Meltzer, S.J.; Rodriguez-Bigas, M.A.; Fodde, R.; Ranzani, G.N.; et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: Development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998, 58, 5248–5257. [Google Scholar] [PubMed]
- Onda, M.; Nakamura, I.; Suzuki, S.; Takenoshita, S.; Brogren, C.H.; Stampanoni, S.; Li, D.; Rampino, N. Microsatellite instability in thyroid cancer: Hot spots, clinicopathological implications, and prognostic significance. Clin. Cancer Res. 2001, 7, 3444–3449. [Google Scholar]
- Forgacs, E.; Wren, J.D.; Kamibayashi, C.; Kondo, M.; Xu, X.L.; Markowitz, S.; Tomlinson, G.E.; Muller, C.Y.; Gazdar, A.F.; Garner, H.R.; et al. Searching for microsatellite mutations in coding regions in lung, breast, ovarian and colorectal cancers. Oncogene 2001, 20, 1005–1009. [Google Scholar] [CrossRef][Green Version]
- Duval, A.; Reperant, M.; Compoint, A.; Seruca, R.; Ranzani, G.N.; Iacopetta, B.; Hamelin, R. Target gene mutation profile differs between gastrointestinal and endometrial tumors with mismatch repair deficiency. Cancer Res. 2002, 62, 1609–1612. [Google Scholar] [PubMed]
- Mori, Y.; Sato, F.; Selaru, F.M.; Olaru, A.; Perry, K.; Kimos, M.C.; Tamura, G.; Matsubara, N.; Wang, S.; Xu, Y.; et al. Instabilotyping reveals unique mutational spectra in microsatellite-unstable gastric cancers. Cancer Res. 2002, 62, 3641–3645. [Google Scholar] [PubMed]
- Sonay, T.B.; Koletou, M.; Wagner, A. A survey of tandem repeat instabilities and associated gene expression changes in 35 colorectal cancers. BMC Genomics 2015, 16, 702. [Google Scholar]
- Yoon, K.; Lee, S.; Han, T.-S.; Moon, S.Y.; Yun, S.M.; Kong, S.-H.; Jho, S.; Choe, J.; Yu, J.; Lee, H.-J.; et al. Comprehensive genome- and transcriptome-wide analyses of mutations associated with microsatellite instability in Korean gastric cancers. Genome Res. 2013, 23, 1109–1117. [Google Scholar] [CrossRef]
- Umar, A.; Boland, C.R.; Terdiman, J.P.; Syngal, S.; de la Chapelle, A.; Rüschoff, J.; Fishel, R.; Lindor, N.M.; Burgart, L.J.; Hamelin, R.; et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J. Natl. Cancer Inst. 2004, 96, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Pasin, E.; Josephson, D.Y.; Mitra, A.P.; Cote, R.J.; Stein, J.P. Superficial bladder cancer: An update on etiology, molecular development, classification, and natural history. Rev. Urol. 2008, 10, 31–43. [Google Scholar]
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Thun, M.J. Cancer statistics, 2009. Cancer J. Clin. 2009, 59, 225–249. [Google Scholar] [CrossRef]
- Botteman, M.F.; Pashos, C.L.; Redaelli, A.; Laskin, B.; Hauser, R. The health economics of bladder cancer. PharmacoEconomics 2003, 22, 1315–1330. [Google Scholar] [CrossRef]
- Lotan, Y.; Kamat, A.M.; Porter, M.P.; Robinson, V.L.; Shore, N.; Jewett, M.; Schelhammer, P.F.; White, R.D.; Quale, D.; Lee, C.T. Key concerns about the current state of bladder cancer. Cancer 2009, 115, 4096–4103. [Google Scholar] [CrossRef]
- Lotan, Y.; Svatek, R.S.; Malats, N. Screening for bladder cancer: A perspective. World J. Urol. 2008, 26, 13–18. [Google Scholar] [CrossRef]
- Shariat, S.F.; Lotan, Y.; Vickers, A.; Karakiewicz, P.I.; Schmitz-Dräger, B.J.; Goebell, P.J.; Malats, N. Statistical consideration for clinical biomarker research in bladder cancer. Urol. Oncol. 2010, 28, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Lotan, Y.; Roehrborn, C.G. Sensitivity and specificity of commonly available bladder tumor markers versus cytology: Results of a comprehensive literature review and meta-analyses. Urology 2003, 61, 109–118. [Google Scholar] [CrossRef]
- Bensalah, K.; Montorsi, F.; Shariat, S.F. Challenges of cancer biomarker profiling. Eur. Urol. 2007, 52, 1601–1609. [Google Scholar] [CrossRef] [PubMed]
- Babjuk, M.; Oosterlinck, W.; Sylvester, R.; Kaasinen, E.; Böhle, A.; Palou-Redorta, J.; Rouprêt, M. EAU guidelines on non–muscle-invasive urothelial carcinoma of the bladder, the 2011 update. Eur. Urol. 2011, 59, 997–1008. [Google Scholar] [CrossRef]
- Hall, M.C.; Chang, S.S.; Dalbagni, G.; Pruthi, R.S.; Seigne, J.D.; Skinner, E.C.; Wolf, J.S.; Schellhammer, P.F. Guideline for the management of nonmuscle invasive bladder cancer (Stages Ta, T1, and Tis): 2007 Update. J. Urol. 2007, 178, 2314–2330. [Google Scholar] [CrossRef]
- Mitra, A.P.; Datar, R.H.; Cote, R.J. Molecular pathways in invasive bladder cancer: New insights into mechanisms, progression, and target identification. J. Clin. Oncol. 2006, 24, 5552–5564. [Google Scholar] [CrossRef]
- Mitra, A.P.; Cote, R.J. Molecular pathogenesis and diagnostics of bladder cancer. Annu. Rev. Pathol. 2009, 4, 251–285. [Google Scholar] [CrossRef]
- Bryan, R.T.; Wallace, D.M.A. ‘Superficial’ bladder cancer—time to uncouple pT1 tumours from pTa tumours. BJU Int. 2002, 90, 846–852. [Google Scholar] [CrossRef]
- Stein, J.P.; Lieskovsky, G.; Cote, R.; Groshen, S.; Feng, A.-C.; Boyd, S.; Skinner, E.; Bochner, B.; Thangathurai, D.; Mikhail, M.; et al. Radical cystectomy in the treatment of invasive bladder cancer: Long-term results in 1054 patients. J. Clin. Oncol. 2001, 19, 666–675. [Google Scholar] [CrossRef]
- Foresman, W.H.; Messing, E.M. Bladder cancer: Natural history, tumor markers, and early detection strategies. Semin. Surg. Oncol. 1997, 13, 299–306. [Google Scholar] [CrossRef]
- Parekh, D.J.; Bochner, B.H.; Dalbagni, G. Superficial and muscle-invasive bladder cancer: Principles of management for outcomes assessments. J. Clin. Oncol. 2006, 24, 5519–5527. [Google Scholar] [CrossRef]
- Wakui, M.; Shiigai, T. Urinary tract cancer screening through analysis of urinary red blood cell volume distribution. Int. J. Urol. 2000, 7, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Grossfeld, G.D.; Wolf, J.S., Jr.; Litwan, M.S.; Hricak, H.; Shuler, C.L.; Agerter, D.C.; Carroll, P.R. Asymptomatic microscopic hematuria in adults: Summary of the AUA best practice policy recommendations. Am. Fam. Physician 2001, 63, 1145–1154. [Google Scholar] [PubMed]
- Messing, E.M.; Young, T.B.; Hunt, V.B.; Wehbie, J.M.; Rust, P. Urinary tract cancers found by homescreening with hematuria dipsticks in healthy men over 50 years of age. Cancer 1989, 64, 2361–2367. [Google Scholar] [CrossRef]
- Messing, E.M.; Young, T.B.; Hunt, V.B.; Newton, M.A.; Bram, L.L.; Vaillancourt, A.; Hisgen, W.J.; Greenberg, E.B.; Kuglitsch, M.E.; Wegenke, J.D. Hematuria home screening: Repeat testing results. J. Urol. 1995, 154, 57–61. [Google Scholar] [CrossRef]
- Messing, E.M.; Young, T.B.; Hunt, V.B.; Roecker, E.B.; Vaillancourt, A.M.; Hisgen, W.J.; Greenberg, E.B.; Kuglitsch, M.E.; Wegenke, J.D. Home screening for hematuria: Results of a multi-clinic study. J. Urol. 1992, 148, 289–292. [Google Scholar] [CrossRef]
- Moon, J.J.; Lu, A.; Moon, C. Role of genomic instability in human carcinogenesis. Exp. Biol. Med. 2019, 244, 227–240. [Google Scholar] [CrossRef]
- Saran, K.K.; Gould, D.; Godec, C.J.; Verma, R.S. Genetics of bladder cancer. J. Mol. Med. 1996, 74, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Skacel, M.; Pettay, J.D.; Tsiftsakis, E.K.; Procop, G.W.; Biscotti, C.V.; Tubbs, R.R. Validation of a multicolor interphase fluorescence in situ hybridization assay for detection of transitional cell carcinoma on fresh and archival thin-layer, liquid-based cytology slides. Anal. Quant. Cytol. Histol. 2001, 23, 381–387. [Google Scholar]
- Lopez-Beltran, A.; Amin, M.B.; Oliveira, P.S.; Montironi, R.; Algaba, F.; McKenney, J.K.; de Torres, I.; Mazerolles, C.; Wang, M.; Cheng, L. Urothelial carcinoma of the bladder, lipid cell variant: Clinicopathologic findings and LOH analysis. Am. J. Surg. Pathol. 2010, 34, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Ploussard, G.; Dubosq, F.; Soliman, H.; Verine, J.; Desgrandchamps, F.; De Thé, H.; Mongiat-Artus, P. Prognostic value of loss of heterozygosity at chromosome 9p in non–muscle-invasive bladder cancer. Urology 2010, 76, 513.e513–513.e518. [Google Scholar] [CrossRef]
- Cai, T.; Nesi, G.; Canto, M.D.; Mondaini, N.; Mauro, P.; Bartoletti, R. Prognostic role of loss of heterozygosity on chromosome 18 in patients with low-risk nonmuscle-invasive bladder cancer: Results from a prospective study. J. Surg. Res. 2010, 161, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Sibley, K.; Cuthbert-Heavens, D.; Knowles, M.A. Loss of heterozygosity at 4p16.3 and mutation of FGFR3 in transitional cell carcinoma. Oncogene 2001, 20, 686–691. [Google Scholar] [CrossRef]
- Yoon, D.-S.; Li, L.; Zhang, R.-D.; Kram, A.; Ro, J.Y.; Johnston, D.; Grossman, H.B.; Scherer, S.; Czerniak, B. Genetic mapping and DNA sequence-based analysis of deleted regions on chromosome 16 involved in progression of bladder cancer from occult preneoplastic conditions to invasive disease. Oncogene 2001, 20, 5005–5014. [Google Scholar] [CrossRef]
- Docimo, S.G.; Chow, N.-H.; Steiner, G.; Silver, R.I.; Rodriguez, R.; Kinsman, S.; Sidransky, D.; Schoenberg, M. Detection of adenocarcinoma by urinary microsatellite analysis after augmentation cystoplasty. Urology 1999, 54, 561. [Google Scholar] [CrossRef]
- Szarvas, T. The diagnostic value of microsatellite LOH analysis and the prognostic relevance of angiogenic gene expression in urinary bladder cancer. Magy. Onkol. 2009, 53, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Bartoletti, R.; Cai, T.; Nesi, G.; Roberta Girardi, L.; Baroni, G.; Dal Canto, M. Loss of P16 expression and chromosome 9p21 LOH in predicting outcome of patients affected by superficial bladder cancer. J. Surg. Res. 2007, 143, 422–427. [Google Scholar] [CrossRef]
- Ørntoft, T.F.; Wolf, H. Molecular alterations in bladder cancer. Urol. Res. 1998, 26, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Schoenberg, M.P.; Scicchitano, M.; Erozan, Y.S.; Merlo, A.; Schwab, D.; Sidransky, D. Molecular detection of primary bladder cancer by microsatellite analysis. Science 1996, 271, 659–662. [Google Scholar] [CrossRef]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef]
- Seripa, D.; Parrella, P.; Gallucci, M.; Gravina, C.; Papa, S.; Fortunato, P.; Alcini, A.; Flammia, G.; Lazzari, M.; Fazio, V.M. Sensitive detection of transitional cell carcinoma of the bladder by microsatellite analysis of cells exfoliated in urine. Int. J. Cancer Res. 2001, 95, 364–369. [Google Scholar]
- Frigerio, S.; Padberg, B.C.; Strebel, R.T.; Lenggenhager, D.M.; Messthaler, A.; Abdou, M.-T.; Moch, H.; Zimmermann, D.R. Improved detection of bladder carcinoma cells in voided urine by standardized microsatellite analysis. Int. J. Cancer Res. 2007, 121, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.O.; Lee, J.; Begum, S.; Yamashita, K.; Engles, J.M.; Schoenberg, M.; Westra, W.H.; Sidransky, D. High-throughput molecular analysis of urine sediment for the detection of bladder cancer by high-density single-nucleotide polymorphism array. Cancer Res. 2003, 63, 5723–5726. [Google Scholar] [PubMed]
- de Bekker-Grob, E.W.; van der Aa, M.N.M.; Zwarthoff, E.C.; Eijkemans, M.J.C.; van Rhijn, B.W.; van der Kwast, T.H.; Steyerberg, E.W. Non-muscle-invasive bladder cancer surveillance for which cystoscopy is partly replaced by microsatellite analysis of urine: A cost-effective alternative? BJU Int. 2009, 104, 41–47. [Google Scholar] [CrossRef] [PubMed]
- van der Aa, M.N.M.; Zwarthoff, E.C.; Steyerberg, E.W.; Boogaard, M.W.; Nijsen, Y.; van der Keur, K.A.; van Exsel, A.J.A.; Kirkels, W.J.; Bangma, C.; van der Kwast, T.H. Microsatellite analysis of voided-urine samples for surveillance of low-grade non-muscle-invasive urothelial carcinoma: Feasibility and clinical utility in a prospective multicenter study (cost-effectiveness of follow-up of urinary bladder cancer trial [CEFUB]). Eur. Urol. 2009, 55, 659–668. [Google Scholar]
- Wild, P.J.; Fuchs, T.; Stoehr, R.; Zimmermann, D.; Frigerio, S.; Padberg, B.; Steiner, I.; Zwarthoff, E.C.; Burger, M.; Denzinger, S.; et al. Detection of urothelial bladder cancer cells in voided urine can be improved by a combination of cytology and standardized microsatellite analysis. Cancer Epidemiol. Biomark. Prev. 2009, 18, 1798–1806. [Google Scholar] [CrossRef] [PubMed]
- Steiner, G.; Schoenberg, M.P.; Linn, J.F.; Mao, L.; Sidransky, D. Detection of bladder cancer recurrence by microsatellite analysis of urine. Nat. Med. 1997, 3, 621–624. [Google Scholar] [CrossRef]
- van Rhijn, B.W.G.; Lurkin, I.; Kirkels, W.J.; van der Kwast, T.H.; Zwarthoff, E.C. Microsatellite analysis? DNA test in urine competes with cystoscopy in follow-up of superficial bladder carcinoma. Cancer 2001, 92, 768–775. [Google Scholar] [CrossRef]
- Amira, N.; Mourah, S.; Rozet, F.; Teillac, P.; Fiet, J.; Aubin, P.; Cortesse, A.; Desgrandchamps, F.; Le Duc, A.; Cussenot, O.; et al. Non-invasive molecular detection of bladder cancer recurrence. Int. J. Cancer Res. 2002, 101, 293–297. [Google Scholar] [CrossRef]
- Schmitz-Dräger, B.J.; Beiche, B.; Tirsar, L.-A.; Schmitz-Dräger, C.; Bismarck, E.; Ebert, T. Immunocytology in the assessment of patients with asymptomatic microhaematuria. Eur. Urol. 2008, 51, 1582–1588. [Google Scholar] [CrossRef]
- Linn, J.F.; Lango, M.; Halachmi, S.; Schoenberg, M.P.; Sidransky, D. Microsatellite analysis and telomerase activity in archived tissue and urine samples of bladder cancer patients. Int. J. Cancer Res. 1997, 74, 625–629. [Google Scholar] [CrossRef]
- Mourah, S.; Cussenot, O.; Vimont, V.; Desgrandchamps, F.; Teillac, P.; Cochant-Priollet, B.; Le Duc, A.; Fiet, J.; Soliman, H. Assessment of microsatellite instability in urine in the detection of transitional-cell carcinoma of the bladder. Int. J. Cancer Res. 1998, 79, 629–633. [Google Scholar] [CrossRef]
- Steiner, G.; Reinschmidt, G.; Müller, S.C. Molecular genetic diagnosis of de novo and recurrent bladder cancer. Electrophoresis 1999, 20, 280–282. [Google Scholar] [CrossRef]
- Baron, A.; Mastroeni, F.; Moore, P.S.; Bonetti, F.; Orlandini, S.; Manfrin, E.; Schiavone, D.; Migliorini, F.; Lusuardi, L.; Mobilio, G.; et al. Detection of bladder cancer by semi-automated microsatellite analysis of urine sediment. Adv. Clin. Path. 2000, 4, 19–24. [Google Scholar]
- Christensen, M.; Wolf, H.; Orntoft, T.F. Microsatellite alterations in urinary sediments from patients with cystitis and bladder cancer. Int. J. Cancer Res. 2000, 85, 614–617. [Google Scholar] [CrossRef]
- Schneider, A.; Borgnat, S.; Lang, H.; Régine, O.; Lindner, V.; Kassem, M.; Saussine, C.; Oudet, P.; Jacqmin, D.; Gaub, M.P. Evaluation of microsatellite analysis in urine sediment for diagnosis of bladder cancer. Cancer Res. 2000, 60, 4617–4622. [Google Scholar]
- Zhang, J.; Fan, Z.; Gao, Y.; Xiao, Z.; Li, C.; An, Q.; Cheng, S. Detecting bladder cancer in the Chinese by microsatellite analysis: Ethnic and etiologic considerations. J. Natl. Cancer Inst. 2001, 93, 45–50. [Google Scholar] [CrossRef]
- Mao, L.; Lee, D.J.; Tockman, M.S.; Erozan, Y.S.; Askin, F.; Sidransky, D. Microsatellite alterations as clonal markers for the detection of human cancer. Proc. Natl. Acad. Sci. USA 1994, 91, 9871–9875. [Google Scholar] [CrossRef]
- Zeger, S.; Liang, K.-Y. An overview of methods for the analysis of longitudinal data. Stat. Med. 1992, 11, 1825–1839. [Google Scholar] [CrossRef]
- Bartoletti, R.; Dal Canto, M.; Cai, T.; Piazzini, M.; Travaglini, F.; Gavazzi, A.; Rizzo, M. Early diagnosis and monitoring of superficial transitional cell carcinoma by microsatellite analysis on urine sediment. Oncol. Rep. 2005, 13, 531–537. [Google Scholar] [CrossRef]
- Bartoletti, R.; Cai, T.; Dal Canto, M.; Boddi, V.; Nesi, G.; Piazzini, M. Multiplex polymerase chain reaction for microsatellite analysis of urine sediment cells: A rapid and inexpensive method for diagnosing and monitoring superficial transitional bladder cell carcinoma. J. Urol. 2006, 175, 2032–2037. [Google Scholar] [CrossRef]
- van Rhijn, B.W.; Lurkin, I.; Chopin, D.K.; Kirkels, W.J.; Thiery, J.P.; van der Kwast, T.H.; Radvanyi, F.; Zwarthoff, E.C. Combined microsatellite and FGFR3 mutation analysis enables a highly sensitive detection of urothelial cell carcinoma in voided urine. Clin. Cancer Res. 2003, 9, 257–263. [Google Scholar]
- Vanrhijn, B.; Smit, M.; Vangeenen, D.; Wijnmaalen, A.; Kirkels, W.; Vanderkwast, T.; Kuenenboumeester, V.; Zwarthoff, E. Surveillance with microsatellite analysis of urine in bladder cancer patients treated by radiotherapy. Eur. Urol. 2003, 43, 369–373. [Google Scholar] [CrossRef]
- van Rhijn, B.W.G.; van der Poel, H.G.; van der Kwast, T.H. Urine markers for bladder cancer surveillance: A systematic review. Eur. Urol. 2005, 47, 736–748. [Google Scholar] [CrossRef]
- van Oers, J.M.M.; Lurkin, I.; van Exsel, A.J.A.; Nijsen, Y.; van Rhijn, B.W.G.; van der Aa, M.N.M.; Zwarthoff, E.C. A simple and fast method for the simultaneous detection of nine fibroblast growth factor receptor 3 mutations in bladder cancer and voided urine. Clin. Cancer Res. 2005, 11, 7743–7748. [Google Scholar] [CrossRef]
- van der Poel, H.G.; van Rhijn, B.W.G.; Peelen, P.; Debruyne, F.M.J.; Boon, M.E.; Schalken, J.A. Consecutive quantitative cytology in bladder cancer. Urology 2000, 56, 584–588. [Google Scholar] [CrossRef]
- Wallerand, H. Mutations in TP53, but not FGFR3, in urothelial cell carcinoma of the bladder are influenced by smoking: Contribution of exogenous versus endogenous carcinogens. Carcinogenesis 2005, 26, 177–184. [Google Scholar] [CrossRef][Green Version]
- Rieger-Christ, K.M.; Medina, A.; Lee, P.J.; Rieger-Christ, K.M.; Mourtzinos, A.; Lee, P.J.; Libertino, J.A.; Summerhayes, I.C. Identification of fibroblast growth factor receptor 3 mutations in urine sediment DNA samples complements cytology in bladder tumor detection. Cancer 2003, 98, 737–744. [Google Scholar] [CrossRef]
- van Rhijn, B.W.G.; Vis, A.N.; van der Kwast, T.H.; Kirkels, W.J.; Radvanyi, F.; Ooms, E.C.M.; Chopin, D.K.; Boevé, E.R.; Jöbsis, A.C.; Zwarthoff, E.C. Molecular grading of urothelial cell carcinoma with fibroblast growth factor receptor 3 and MIB-1 is superior to pathologic grade for the prediction of clinical outcome. J. Clin. Oncol. 2003, 21, 1912–1921. [Google Scholar] [CrossRef]
- Hernández, S.; López-Knowles, E.; Lloreta, J.; Kogevinas, M.; Amorós, A.; Tardón, A.; Carrato, A.; Serra, C.; Malats, N.; Real, F.X. Prospective study of FGFR3 mutations as a prognostic factor in nonmuscle invasive urothelial bladder carcinomas. J. Clin. Oncol. 2006, 24, 3664–3671. [Google Scholar] [CrossRef]
- Montironi, R. Editorial comment on: FGFR3 mutations and a normal CK20 staining pattern define low-grade noninvasive urothelial bladder tumours. Eur. Urol. 2007, 52, 768. [Google Scholar] [CrossRef]
- Burger, M.; van der Aa, M.N.M.; van Oers, J.M.M.; Brinkmann, A.; van der Kwast, T.H.; Steyerberg, E.C.; Stoehr, R.; Kirkels, W.J.; Denzinger, S.; Wild, P.J.; et al. Prediction of progression of non–muscle-invasive bladder cancer by WHO 1973 and 2004 grading and by FGFR3 mutation status: A prospective study. Eur. Urol. 2008, 54, 835–844. [Google Scholar] [CrossRef]
- Knowles, M.A. Role of FGFR3 in urothelial cell carcinoma: Biomarker and potential therapeutic target. World J. Urol. 2007, 25, 581–593. [Google Scholar] [CrossRef][Green Version]
- Song, X.-B.; Zhou, Y.; Ying, B.-W.; Wang, L.-L.; Li, Y.-S.; Liu, J.-F.; Bai, X.-G.; Zhang, L.; Lu, X.-J.; Wang, J.; et al. Short-tandem repeat analysis in seven Chinese regional populations. Genet. Mol. Biol. 2010, 33, 605–609. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cai, G.Q.; Chen, L.X.; Tong, D.Y.; Ou, J.H.; Wu, X.Y. Mutations of 15 short tandem repeat loci in Chinese population. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2005, 22, 507–509. [Google Scholar] [PubMed]
- Chakraborty, R.; Smouse, P.E.; Neel, J.V. Population amalgamation and genetic variation: Observations on artificially agglomerated tribal populations of Central and South America. Am. J. Hum. Genet. 1988, 43, 709–725. [Google Scholar] [PubMed]
- Chu, J.Y.; Huang, W.; Kuang, S.Q.; Wang, J.M.; Xu, J.J.; Chu, Z.T.; Yang, Z.Q.; Lin, K.Q.; Li, P.; Wu, M.; et al. Genetic relationship of populations in China. Proc. Natl. Acad. Sci. USA 1998, 95, 11763–11768. [Google Scholar] [CrossRef]
- Deng, Y.-J.; Yan, J.-W.; Yu, X.-G.; Li, Y.-Z.; Mu, H.-F.; Huang, Y.-Q.; Shi, X.-T.; Sun, W.-M. Genetic analysis of 15 STR loci in Chinese Han population from West China. Genomics Proteomics Bioinform. 2007, 5, 66–69. [Google Scholar] [CrossRef]
- Edwards, A.; Hammond, H.A.; Jin, L.; Caskey, C.T.; Chakraborty, R. Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genomics 1992, 12, 241–253. [Google Scholar] [CrossRef]
- Grunbaum, B.W.; Selvin, S.; Pace, N.; Black, D.M. Frequency distribution and discrimination probability of twelve protein genetic variants in human blood as functions of race, sex, and age. J. Forensic Sci. 1978, 23, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Y.; Guo, J.; Fu, X.; Wang, Z.; Liu, Y.; Cai, J.; Zha, L. Genetic polymorphism of 29 STR loci in the Hunan Han population from China. Forensic Sci. Res. 2017, 4, 351–353. [Google Scholar] [CrossRef]
- Hyun, J.S.; Jo, B.-K.; Park, C.J.; Yi, J.Y.; Lee, J.Y.; Rhyu, M.-G. Loss of heterozygosity and PCR artifacts in a microsatellite analysis of psoriasis and colorectal cancer. J. Korean Med. Sci. 2002, 17, 641–647. [Google Scholar] [CrossRef]
- Sieben, N.L.G.; ter Haar, N.T.; Cornelisse, C.J.; Fleuren, G.J.; Cleton-Jansen, A.-M. PCR artifacts in LOH and MSI analysis of microdissected tumor cells. Hum. Pathol. 2000, 31, 1414–1419. [Google Scholar] [CrossRef]
- Bocker, T.; Diermann, J.; Friedl, W.; Gebert, J.; Holinski-Feder, E.; Karner-Hanusch, J.; von Knebel-Doeberitz, M.; Koelble, K.; Moeslein, G.; Schackert, H.K.; et al. Microsatellite instability analysis: A multicenter study for reliability and quality control. Cancer Res. 1997, 57, 4739–4743. [Google Scholar]
- Cawkwell, L.; Li, D.; Lewis, F.A.; Martin, I.; Dixon, M.F.; Quirke, P. Microsatellite instability in colorectal cancer: Improved assessment using fluorescent polymerase chain reaction. Gastroenterology 1995, 109, 465–471. [Google Scholar] [CrossRef]
- Canzian, F.; Salovaara, R.; Hemminki, A.; Kristo, P.; Chadwick, R.B.; Aaltonen, L.A.; de la Chapelle, A. Semiautomated assessment of loss of heterozygosity and replication error in tumors. Cancer Res. 1996, 56, 3331–3337. [Google Scholar]
- Suraweera, N.; Duval, A.; Reperant, M.; Vaury, C.; Furlan, D.; Leroy, K.; Seruca, R.; Iacopetta, B.; Hamelin, R. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology 2002, 123, 1804–1811. [Google Scholar] [CrossRef]
- Goel, A.; Nagasaka, T.; Hamelin, R.; Boland, C.R. An optimized pentaplex PCR for detecting DNA mismatch repair-deficient colorectal cancers. PLoS ONE 2010, 5, e9393. [Google Scholar] [CrossRef]
- Takehara, Y.; Nagasaka, T.; Nyuya, A.; Haruma, T.; Haraga, J.; Mori, Y.; Nakamura, K.; Fujiwara, T.; Boland, C.R.; Goel, A. Accuracy of four mononucleotide-repeat markers for the identification of DNA mismatch-repair deficiency in solid tumors. J. Transl. Med. 2018, 16, 5. [Google Scholar] [CrossRef] [PubMed]
- Laghi, L.; Bianchi, P.; Malesci, A. Differences and evolution of the methods for the assessment of microsatellite instability. Oncogene 2008, 27, 6313–6321. [Google Scholar] [CrossRef]
- Dal Canto, M.; Bartoletti, R.; Travaglini, F.; Piazzini, M.; Lodovichi, G.; Rizzo, M.; Selli, C. Molecular urinary sediment analysis in patients with transitional cell bladder carcinoma. Anticancer Res. 2003, 23, 5095–5100. [Google Scholar] [PubMed]
- Choi, C.; Kim, M.H.; Juhng, S.-W.; Oh, B.-R. Loss of heterozygosity at chromosome segments 8p22 and 8p11.2-21.1 in transitional-cell carcinoma of the urinary bladder. Int. J. Cancer Res. 2000, 86, 501–505. [Google Scholar] [CrossRef]
- van Tilborg, A.A.G.; de Vries, A.; de Bont, M.; Groenfeld, L.E.; Zwarthoff, E.C. The random development of LOH on chromosome 9q in superficial bladder cancers. J. Pathol. 2002, 198, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Knowles, M.A. Identification of novel bladder tumour suppressor genes. Electrophoresis 1999, 20, 269–279. [Google Scholar] [CrossRef]
| A | ||||
|---|---|---|---|---|
| Study | No. of Cancers Detected | Sensitivity | Healthy Controls with Neg MSA Result | Specificity |
| by MSA | (%) | (%) | ||
| Mao et al. (1996) ♦ Science 271:659–662 | 19/20 | 95 | 5 out of 5 | 100 |
| Wild et al. (2009) ♦ Cancer Epidemiol. Biomark. Prev, 18, 1798–1806 | 71/81 | 88 | 37 out of 38 | 97 |
| Linn et al. (1997) ♦ Int J Cancer 74:625–629 | 13/15 | 87 | N/A | N/A |
| Schneider et al. (2000) ♦ Cancer Res 60:4617–4622 | 87/103 | 84 | N/A | N/A |
| Sourvinos et al. (2001) ♦ J Urol 165:249–252 | 26/28 | 93 | 10 out of 10 | 100 |
| Zhang et al. (2001) ♦ Cancer Lett. 172:55–58 | 73/81 | 90 | 19/19 | 100 |
| Seripa et al. (2001) ♦ Int J Cancer 95:364–369 | 33/34 | 97 | 11 out of 11 | 100 |
| Zhang et al. (2001) ♦ JNCI 93:45–50 | 22/23 | 96 | 17/17 | 100 |
| van Rhijn et al. (2003) ♦ Clin. Cancer Res. 9, 257–263 | 29/32 | 91 | 14 out of 15 | 93 |
| OVERALL | 373/417 | 90% | 113/115 | 98% |
| B | ||||
| Study | No. of Cancers Detected | Sensitivity | Healthy Controls with Neg MSA Result | Specificity |
| by MSA | (%) | (%) | ||
| van Rhijn et al. (2001) ♦ Cancer 92:768–775.e 271:659–662 | 23/29 | 79 | 66 out of 70 | 94 |
| Steiner et al. (1997) ♦ Nat Med. 3:621–624 | 10/11 | 91 | 10 out of 10 | 100 |
| Baron et al. (2000) ♦ Adv Clin Path.4 (1):19–24 | 21/25 | 84 | N/A | N/A |
| Bartoletti et al. (2005) ♦ Oncol Rep;13:531–537 | 25/30 | 84 | 30 out of 30 | 100 |
| Bartoletti et al. (2006) ♦ J Urol175:2032–2037 | 59/73 | 81 | 36 out of 43 | 84 |
| Bas et al. (2003) ♦ European Urology 43, 369–373 | 83 | 93 | ||
| Frigerio et al. (2007) ♦ Int. J. Cancer Res, 121, 329–338 | 59/63 | 93 | 28 out of 28 | 100 |
| Mourah et al. (1998) ♦ Int. J. Cancer Res. 79, 629–633. | 10/12 | 96 | 15 out of 15 | 100 |
| Amira et al. (2002) ♦ Int J Cancer 101:293–297 | 44/47 | 94 | N/A | N/A |
| OVERALL | 251/290 | 87% | 185/196 | 94% |
| Marker | Median Sensitivity | Range (Min–Max) | 5% Difference | Median Specificity | Range (Min–Max) | 5% Difference |
|---|---|---|---|---|---|---|
| BTAstat | 70 | 24–89 | Yes | 75 | 52–93 | – |
| BTAtrak | 69 | 57–79 | – | 65 | 48–95 | – |
| NMP22 | 73 | 47–100 | – | 80 | 56–95 | Yes |
| FDP | 61 | 52–81 | Yes | 79 | 75–96 | Yes |
| ImmunoCyt | 83 | 50–100 | Yes | 80 | 69–90 | Yes |
| Cytometry | 60 | 45–83 | – | 80 | 36–87 | – |
| Quanticyt | 59 | 45–69 | – | 79 | 70–93 | – |
| Hb-dipstick | 52 | 41–95 | Yes | 82 | 68–93 | – |
| LewisX | 83 | 80–89 | Yes | 85 | 80–86 | – |
| FISH | 84 | 73–92 | Yes | 95 | 92–100 | Yes |
| Telomerase | 75 | 7–100 | Yes | 86 | 24–93 | na |
| Microsatellite | 91 | 83–95 | Yes | 94 | 89–100 | Yes |
| CYFRA21-1 | 94 | 74–99 | Yes | 86 | 67–100 | – |
| UBC | 78 | 66–87 | Yes | 91 | 80–97 | – |
| Cytokeratin20 | 91 | 82–96 | Yes | 84 | 67–97 | Yes |
| BTA | 50 | 28–80 | – | 86 | 66–95 | – |
| TPS | 72 | 64–88 | Yes | 78 | 55–95 | – |
| Cytology | 48 | 31–100 | Yes | 94 | 62–100 | – |
| Marker (Reference Number) | No. pts./Median Sensitivity | No. pts./Median Specificity | ||
|---|---|---|---|---|
| G1 | G2 | G3 | ||
| BTAstat | 228/45 | 206/60 | 208/75 | 972/79 |
| BTAtrak | 60/55 | 61/59 | 101/74 | 195/66 |
| NMP22 | 56/41 | 77/53 | 81/80 | 235/59 |
| FDP | 13/62 | 36/64 | 22/86 | 113/80 |
| ImmunoCyt | 23/78 | 10/90 | 18/100 | 83/62 |
| Cytometry | 18/11 | 54/41 | 38/66 | 52/87 |
| Quanticyt | - | 11/64 | 5/80 | 56/68 |
| Hb-dipstick | 13/15 | 36/39 | 22/73 | 113/87 |
| FISH | 25/56 | 9/78 | 20/95 | 130/70 |
| Microsatellite | 27/67 | 21/86 | 30/93 | 138/88 |
| UBC | 29/38 | 29/41 | 16/69 | 79/72 |
| Cytokeratin20 | 14/71 | 35/80 | 35/100 | na |
| BTA | 31/16 | 43/47 | 50/52 | 91/91 |
| TPS | 29/32 | 35/54 | 15/74 | 72/63 |
| Cytology | 239/17 | 274/34 | 201/58 | 861/95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moon, C.; Gordon, M.; Moon, D.; Reynolds, T. Microsatellite Instability Analysis (MSA) for Bladder Cancer: Past History and Future Directions. Int. J. Mol. Sci. 2021, 22, 12864. https://doi.org/10.3390/ijms222312864
Moon C, Gordon M, Moon D, Reynolds T. Microsatellite Instability Analysis (MSA) for Bladder Cancer: Past History and Future Directions. International Journal of Molecular Sciences. 2021; 22(23):12864. https://doi.org/10.3390/ijms222312864
Chicago/Turabian StyleMoon, Chulso, Maxie Gordon, David Moon, and Thomas Reynolds. 2021. "Microsatellite Instability Analysis (MSA) for Bladder Cancer: Past History and Future Directions" International Journal of Molecular Sciences 22, no. 23: 12864. https://doi.org/10.3390/ijms222312864
APA StyleMoon, C., Gordon, M., Moon, D., & Reynolds, T. (2021). Microsatellite Instability Analysis (MSA) for Bladder Cancer: Past History and Future Directions. International Journal of Molecular Sciences, 22(23), 12864. https://doi.org/10.3390/ijms222312864






