Fine Breakpoint Mapping by Genome Sequencing Reveals the First Large X Inversion Disrupting the NHS Gene in a Patient with Syndromic Cataracts
Abstract
1. Introduction
2. Results
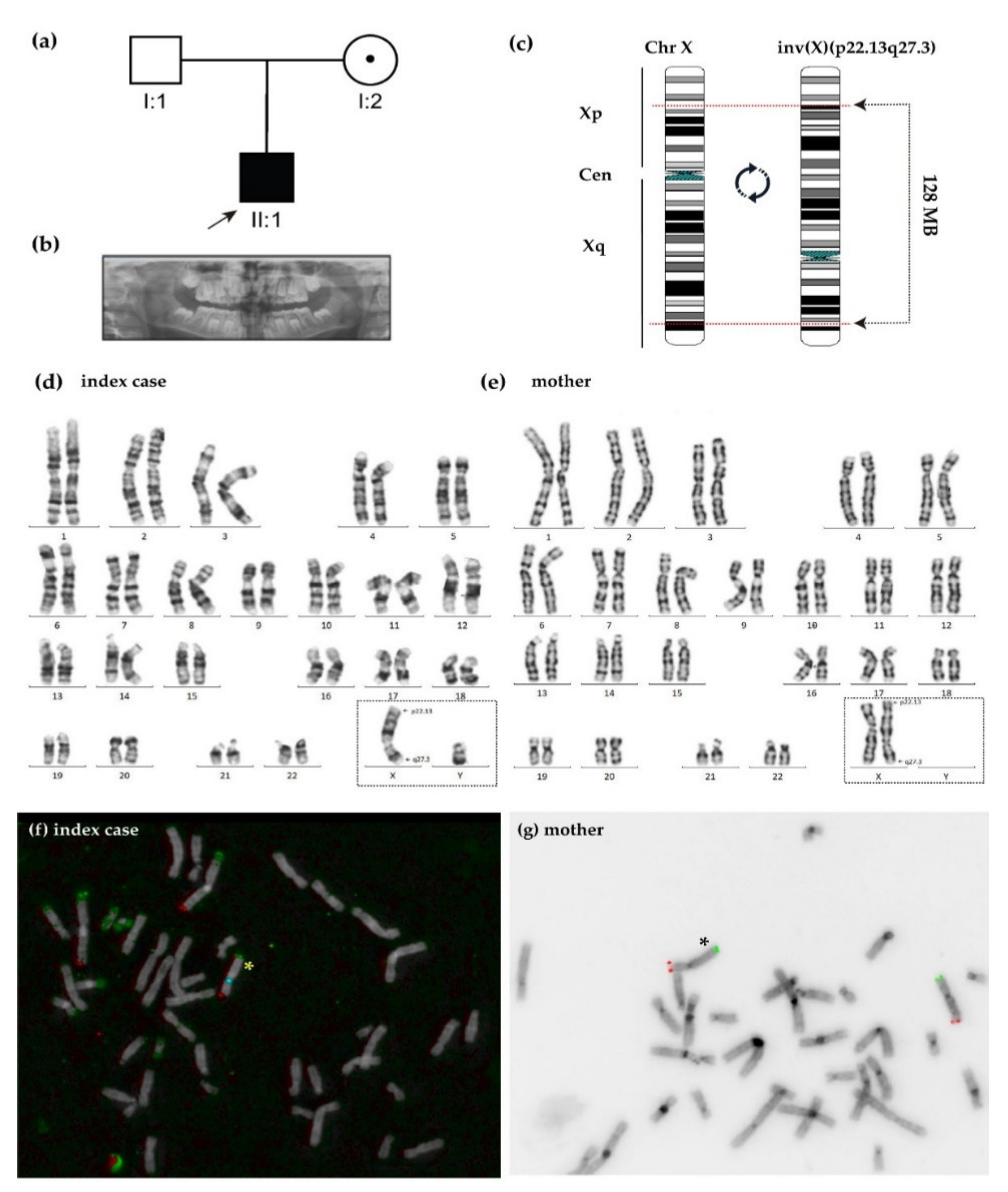
2.1. Clinical Features
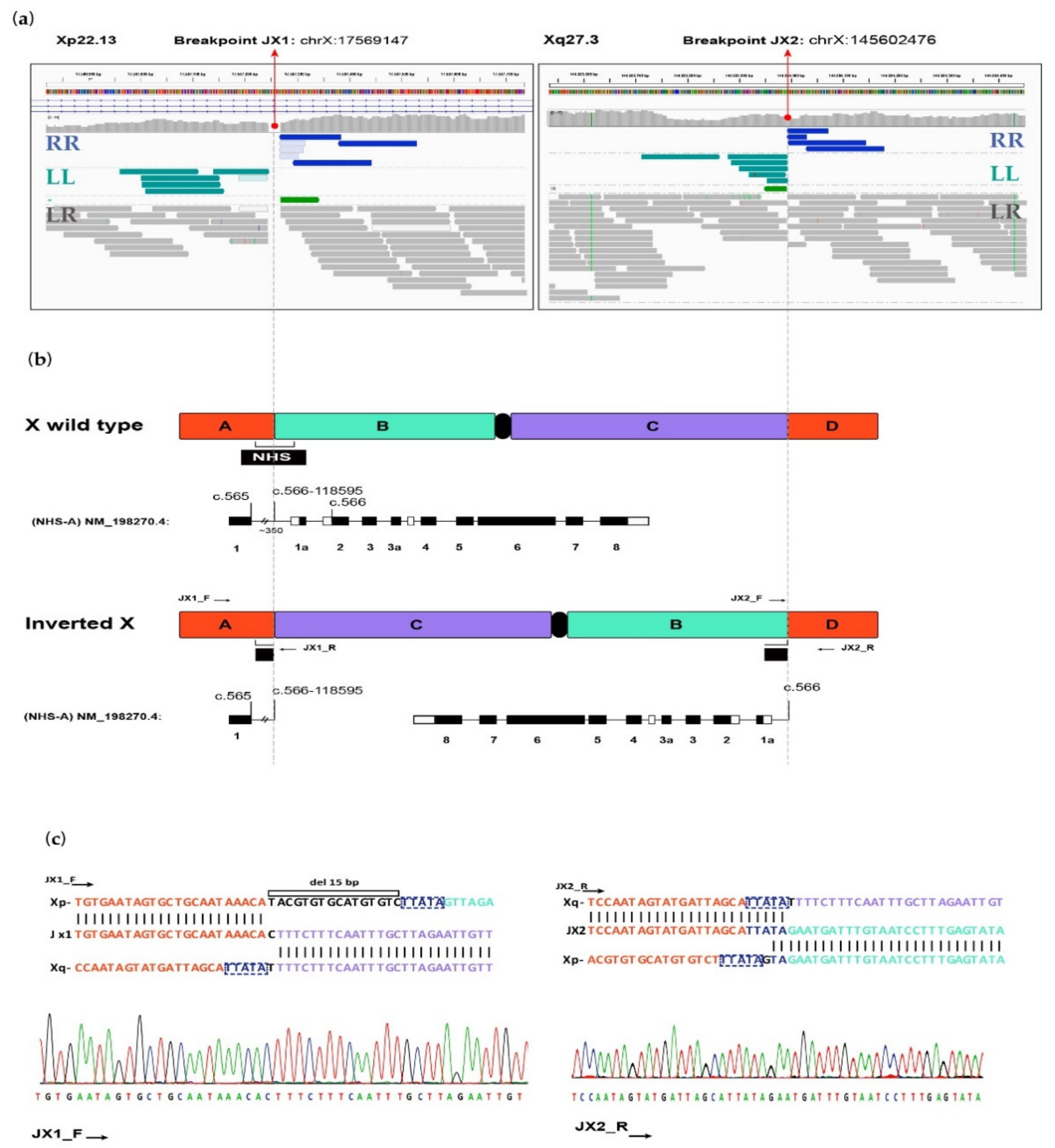
2.2. Genetic Analysis
2.3. Correlation Phenotype—Genotype
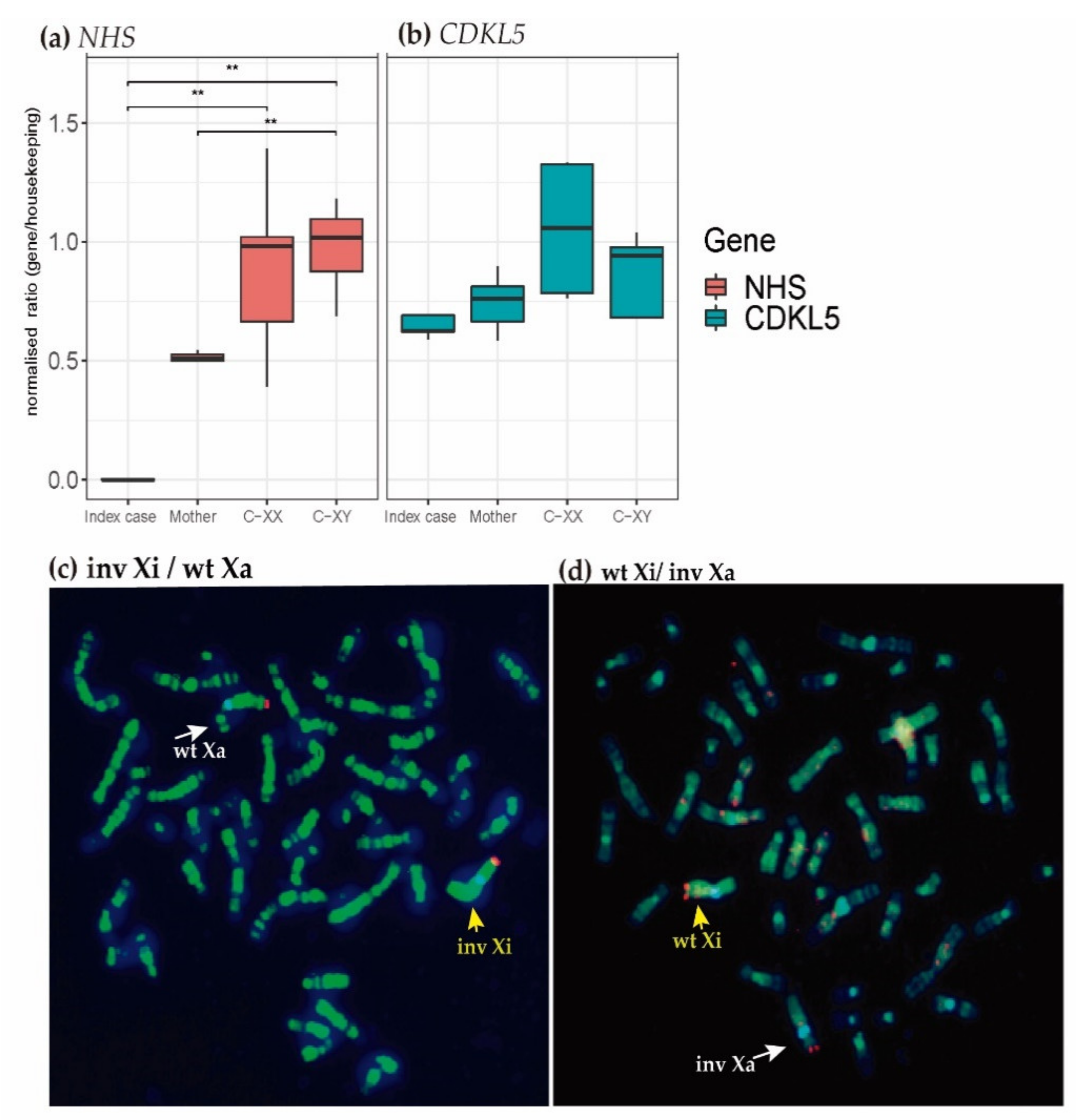
2.4. Transcriptional Analysis
2.5. X-Chromosome Inactivation (XCI) Analysis
3. Discussion
4. Materials and Methods
4.1. Cytogenetic Analysis
4.2. Molecular Analysis
4.3. RNA Expression Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

| Clinical Symptoms | Index Case |
|---|---|
| Mayor signs in 80–99% of NHS patients | |
| Cataract | Yes |
| Long face | No |
| Mandibular prognathia | Yes |
| Microcornea | Yes |
| Nystagmus | Yes |
| Prominent nasal bridge | Yes |
| Prominent nose | Yes |
| Visual loss | Yes |
| Signs in 30–79% of NHS patients | |
| Increased number of teeth | Yes |
| Intellectual disability | Yes |
| Protruding ear | Yes |
| Short metacarpal | No |
| Strabismus | Yes |
| Behavioral abnormality | No |
| Glaucoma | No |
| Microphthalmia | No |
| Retinal detachment | No |
| Minor signs in < 4% of NHS patients | |
| Intellectual disability, moderate | Yes |
| Mulberry molar | Unknown |
| Screwdriver-shaped incisors | No |
| Additional minor symptoms | |
| Autism | No |
| Broad finger | No |
| Developmental cataract | Yes |
| Diastema | No |
| Macrotia | Yes |
| Narrow face | No |
| Posterior Y-sutural cataract | Unknown |
| Short phalanx of finger | No |
| Supernumerary maxillary incisor | Yes |
| X-linked dominant inheritance | Yes |
| Abnormal heart morphology | Yes |
| Abnormality of the urogenital system | Yes |
| Seizures | Yes |
| Primers | Gene | Exons | Size (BP) | Sequence (5′–3′) |
|---|---|---|---|---|
| JX1_DNA_F | JX1 fragment | 454 | GAGAAAATGCGGTGTTTGGT | |
| JX1_DNA_R | GGGAGATTTCAACACCCCACT | |||
| JX2_DNA_F | JX2 fragment | 542 | AGGTGCTGGAGAGGATGTGG | |
| JX2_DNA_R | TTGAGGTTGCTTTGCTCTTG | |||
| NHS_RNA_F | NHS | E6-E7 | 99 | GAGACCCAAGGAAATGTGGA |
| NHS_RNA_R | TGCTTCAGCTTGGGAGTCTT | |||
| CDKL5_RNA_F | CDKL5 | E4-E5 | 101 | ACATGAAATTGTGGCGATCA |
| CDKL5_RNA_R | GCTTGAGAGTCCGAAGCATT | |||
| GAPDH_RNA_F | GADPH | E8-E9 | 203 | CGACCACTTTGTCAAGCTCA |
| GAPDH_RNA_R | AGGGGAGATTCAGTGTGGTG | |||
| RPS18_RNA_F | RPS18 | E2–E3 | 130 | GTTCCAGCATATTTTGCGAGT |
| RPS18_RNA_R | GTCAATGTCTGCTTTCCTCAAC | |||
| ACTB_RNA_F | ACTB | E1–E2 | 110 | ACAGAGCCTCGCCTTTG |
| ACTB_RNA_R | CCTTGCACATGCCGGAG | |||
| UBE2D2_RNA_F | UBE2D2 | E7–E8 | 120 | GTACTCTTGTCCATCTGTTCTCTG |
| UBE2D2_RNA_R | CCATTCCCGAGCTATTCTGTT | |||
References
- Sudmant, P.H.; Rausch, T.; Gardner, E.J.; Handsaker, R.E.; Abyzov, A.; Huddleston, J.; Zhang, Y.; Ye, K.; Jun, G.; The 1000 Genomes Project Consortium; et al. An integrated map of structural variation in 2,504 human genomes. Nature 2015, 526, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Feuk, L.; Carson, A.R.; Scherer, S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006, 7, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Spielmann, M.; Lupiáñez, D.G.; Mundlos, S. Structural variation in the 3D genome. Nat. Rev. Genet. 2018, 19, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, M.; Grochowski, C.M.; Wincent, J.; Eisfeldt, J.; Breman, A.M.; Cheung, S.W.; Krepischi, A.C.V.; Rosenberg, C.; Lupski, J.R.; Ottosson, J.; et al. Cytogenetically visible inversions are formed by multiple molecular mechanisms. Hum. Mutat. 2020, 41, 1979–1998. [Google Scholar] [CrossRef]
- Giner-Delgado, C.; Villatoro, S.; Lerga-Jaso, J.; Gayà-Vidal, M.; Oliva, M.; Castellano, D.; Pantano, L.; Bitarello, B.D.; Izquierdo, D.; Noguera, I.; et al. Evolutionary and functional impact of common polymorphic inversions in the human genome. Nat. Commun. 2019, 10, 4222. [Google Scholar] [CrossRef]
- Puig, M.; Casillas, S.; Villatoro, S.; Cáceres, M. Human inversions and their functional consequences. Brief. Funct. Genom. 2015, 14, 369–379. [Google Scholar] [CrossRef]
- Koumbaris, G.; Hatzisevastou-Loukidou, H.; Alexandrou, A.; Ioannides, M.; Christodoulou, C.; Fitzgerald, T.; Rajan, D.; Clayton, S.; Kitsiou-Tzeli, S.; Vermeesch, J.R.; et al. FoSTeS, MMBIR and NAHR at the human proximal Xp region and the mechanisms of human Xq isochromosome formation. Hum. Mol. Genet. 2011, 20, 1925–1936. [Google Scholar] [CrossRef]
- Ottaviani, D.; LeCain, M.; Sheer, D. The role of microhomology in genomic structural variation. Trends Genet. 2014, 30, 85–94. [Google Scholar] [CrossRef]
- Lee, J.A.; Carvalho, C.M.B.; Lupski, J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 2007, 131, 1235–1247. [Google Scholar] [CrossRef]
- Weckselblatt, B.; Rudd, M.K. Human structural variation: Mechanisms of chromosome rearrangements. Trends Genet. 2015, 31, 587–599. [Google Scholar] [CrossRef]
- Feuk, L. Inversion variants in the human genome: Role in disease and genome architecture. Genome. Med. 2010, 2, 1–8. [Google Scholar] [CrossRef]
- Schmidt, S.; Claussen, U.; Liehr, T.; Weise, A. Evolution versus constitution: Differences in chromosomal inversion. Hum. Genet. 2005, 117, 213–219. [Google Scholar] [CrossRef]
- Liehr, T.; Weise, A.; Mrasek, K.; Ziegler, M.; Padutsch, N.; Wilhelm, K.; Al-Rikabi, A. Recombinant chromosomes resulting from parental pericentric inversions—Two new cases and a review of the literature. Front. Genet. 2019, 10, 1165. [Google Scholar] [CrossRef]
- Sismani, C.; Rapti, S.-M.; Iliopoulou, P.; Spring, A.; Neroutsou, R.; Lagou, M.; Robola, M.; Tsitsopoulos, E.; Kousoulidou, L.; Alexandrou, A.; et al. Novel pericentric inversion Inv(9)(P23q22.3) in unrelated individuals with fertility problems in the Southeast European population. J. Hum. Genet. 2020, 65, 783–795. [Google Scholar] [CrossRef]
- Schluth-Bolard, C.; Diguet, F.; Chatron, N.; Rollat-Farnier, P.-A.; Bardel, C.; Afenjar, A.; Amblard, F.; Amiel, J.; Blesson, S.; Callier, P.; et al. Whole genome paired-end sequencing elucidates functional and phenotypic consequences of balanced chromosomal rearrangement in patients with developmental disorders. J. Med. Genet. 2019, 56, 526–535. [Google Scholar] [CrossRef]
- McArthur, E.; Capra, J.A. Topologically associating domain boundaries that are stable across diverse cell types are evolutionarily constrained and enriched for heritability. Am. J. Hum. Genet. 2021, 108, 269–283. [Google Scholar] [CrossRef]
- Shanta, O.; Noor, A.; Sebat, J.; Human genome structural variation consortium (HGSVC). The effects of common structural variants on 3D chromatin structure. BMC Genom. 2020, 21, 95. [Google Scholar] [CrossRef]
- Suzuki, T.; Tsurusaki, Y.; Nakashima, M.; Miyake, N.; Saitsu, H.; Takeda, S.; Matsumoto, N. Precise detection of chromosomal translocation or inversion breakpoints by whole-genome sequencing. J. Hum. Genet. 2014, 59, 649–654. [Google Scholar] [CrossRef]
- Sanchis-Juan, A.; Stephens, J.; French, C.E.; Gleadall, N.; Mégy, K.; Penkett, C.; Shamardina, O.; Stirrups, K.; Delon, I.; Dewhurst, E.; et al. Complex structural variants in mendelian disorders: Identification and breakpoint resolution using short- and long-read genome sequencing. Genome Med. 2018, 10, 95. [Google Scholar] [CrossRef]
- Silva, M.; de Leeuw, N.; Mann, K.; Schuring-Blom, H.; Morgan, S.; Giardino, D.; Rack, K.; Hastings, R. European Guidelines for Constitutional Cytogenomic Analysis. Eur. J. Hum. Genet. 2019, 27, 1–16. [Google Scholar] [CrossRef]
- Pös, O.; Radvanszky, J.; Buglyó, G.; Pös, Z.; Rusnakova, D.; Nagy, B.; Szemes, T. Copy number variation: Characteristics, evolutionary and pathological aspects. Biomed. J. in press. 2021. [Google Scholar] [CrossRef]
- Lindstrand, A.; Eisfeldt, J.; Pettersson, M.; Carvalho, C.M.B.; Kvarnung, M.; Grigelioniene, G.; Anderlid, B.-M.; Bjerin, O.; Gustavsson, P.; Hammarsjö, A.; et al. From cytogenetics to cytogenomics: Whole-genome sequencing as a first-line test comprehensively captures the diverse spectrum of disease-causing genetic variation underlying intellectual disability. Genome Med. 2019, 11, 68. [Google Scholar] [CrossRef]
- Coccia, M.; Brooks, S.P.; Webb, T.R.; Christodoulou, K.; Wozniak, I.O.; Murday, V.; Balicki, M.; Yee, H.A.; Wangensteen, T.; Riise, R.; et al. X-linked cataract and Nance-Horan syndrome are allelic disorders. Hum. Mol. Genet. 2009, 18, 2643–2655. [Google Scholar] [CrossRef]
- Brooks, S.P. Identification of the gene for nance-horan syndrome (NHS). J. Med. Genet. 2004, 41, 768–771. [Google Scholar] [CrossRef]
- Burdon, K.P.; McKay, J.D.; Sale, M.M.; Russell-Eggitt, I.M.; Mackey, D.A.; Wirth, M.G.; Elder, J.E.; Nicoll, A.; Clarke, M.P.; FitzGerald, L.M.; et al. Mutations in a novel gene, NHS, cause the pleiotropic effects of Nance-Horan syndrome, including severe congenital cataract, dental anomalies, and mental retardation. Am. J. Hum. Genet. 2003, 73, 1120–1130. [Google Scholar] [CrossRef]
- Norton, H.K.; Phillips-Cremins, J.E. Crossed wires: 3D genome misfolding in human disease. J. Cell Biol. 2017, 216, 3441–3452. [Google Scholar] [CrossRef]
- Plesser Duvdevani, M.; Pettersson, M.; Eisfeldt, J.; Avraham, O.; Dagan, J.; Frumkin, A.; Lupski, J.R.; Lindstrand, A.; Harel, T. Whole-genome sequencing reveals complex chromosome rearrangement disrupting NIPBL in infant with Cornelia de Lange syndrome. Am. J. Med. Genet. 2020, 182, 1143–1151. [Google Scholar] [CrossRef]
- Wu, Z.; Wu, Y.; Gao, J. InvBFM: Finding genomic inversions from high-throughput sequence data based on feature mining. BMC Genom. 2020, 21, 173. [Google Scholar] [CrossRef]
- Vuillaume, M.-L.; Cogné, B.; Jeanne, M.; Boland, A.; Ung, D.-C.; Quinquis, D.; Besnard, T.; Deleuze, J.-F.; Redon, R.; Bézieau, S.; et al. Whole genome sequencing identifies a de NOVO 2.1 Mb balanced paracentric inversion disrupting FOXP1 and leading to severe intellectual disability. Clinica Chimica Acta 2018, 485, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Pons, L.; Bouvagnet, P.; Bakloul, M.; Di Filippo, S.; Buisson, A.; Chatron, N.; Labalme, A.; Metton, O.; Mitchell, J.; Diguet, F.; et al. Supravalvular aortic stenosis caused by a familial chromosome 7 inversion disrupting the ELN gene uncovered by whole-genome sequencing. Mol. Syndromol. 2019, 10, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Spiegler, S.; Rath, M.; Hoffjan, S.; Dammann, P.; Sure, U.; Pagenstecher, A.; Strom, T.; Felbor, U. First large genomic inversion in familial cerebral cavernous malformation identified by whole genome sequencing. Neurogenetics 2018, 19, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Shoshany, N.; Avni, I.; Morad, Y.; Weiner, C.; Einan-Lifshitz, A.; Pras, E. NHS gene mutations in Ashkenazi Jewish families with Nance–Horan syndrome. null 2017, 42, 1240–1244. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.-M.; Niu, D.-M.; Chen, Y.-J.; Fang, J.-S.; Chen, S.-J.; Chen, C.-H. Identification of a microdeletion at Xp22.13 in a Taiwanese family presenting with Nance-Horan syndrome. J. Hum. Genet. 2011, 56, 8–11. [Google Scholar] [CrossRef]
- Melo, U.S.; Schöpflin, R.; Acuna-Hidalgo, R.; Mensah, M.A.; Fischer-Zirnsak, B.; Holtgrewe, M.; Klever, M.-K.; Türkmen, S.; Heinrich, V.; Pluym, I.D.; et al. Hi-C identifies complex genomic rearrangements and TAD-shuffling in developmental diseases. Am. J. Hum. Genet. 2020, 106, 872–884. [Google Scholar] [CrossRef]
- Van Bemmel, J.G.; Galupa, R.; Gard, C.; Servant, N.; Picard, C.; Davies, J.; Szempruch, A.J.; Zhan, Y.; Żylicz, J.J.; Nora, E.P.; et al. The bipartite TAD organization of the X-inactivation center ensures opposing developmental regulation of Tsix and Xist. Nat. Genet. 2019, 51, 1024–1034. [Google Scholar] [CrossRef]
- Tokoro, M.; Tamura, S.; Suzuki, N.; Kakihara, M.; Hattori, Y.; Odaira, K.; Suzuki, S.; Takagi, A.; Katsumi, A.; Hayakawa, F.; et al. Aberrant X chromosomal rearrangement through multi-step template switching during sister chromatid formation in a patient with severe hemophilia A. Mol. Genet. Genomic. Med. 2020, 8, e1390. [Google Scholar] [CrossRef]
- Brooks, S.P.; Coccia, M.; Tang, H.R.; Kanuga, N.; Machesky, L.M.; Bailly, M.; Cheetham, M.E.; Hardcastle, A.J. The nance–horan syndrome protein encodes a functional WAVE homology domain (WHD) and is important for Co-ordinating actin remodelling and maintaining cell morphology. Hum. Mol. Genet. 2010, 19, 2421–2432. [Google Scholar] [CrossRef]
- Gómez-Laguna, L.; Martínez-Herrera, A.; Reyes-de la Rosa, A.D.P.; García-Delgado, C.; Nieto-Martínez, K.; Fernández-Ramírez, F.; Valderrama-Atayupanqui, T.Y.; Morales-Jiménez, A.B.; Villa-Morales, J.; Kofman, S.; et al. Nance–Horan syndrome in females due to a balanced X;1 translocation that disrupts the NHS gene: Familial case report and review of the literature. Ophthalmic Genet. 2018, 39, 56–62. [Google Scholar] [CrossRef]
- Accogli, A.; Traverso, M.; Madia, F.; Bellini, T.; Vari, M.S.; Pinto, F.; Capra, V. A novel Xp22.13 microdeletion in Nance-Horan syndrome: Xp22.13 microdeletion in NHS. Birth Defects Res. 2017, 109, 866–868. [Google Scholar] [CrossRef]
- Milunsky, A.; Milunsky, J.M.; Dong, W.; Hovhannisyan, H.; Oates, R.D. A contiguous microdeletion syndrome at Xp23.13 with non-obstructive azoospermia and congenital cataracts. J. Assist. Reprod. Genet. 2020, 37, 471–475. [Google Scholar] [CrossRef]
- Van Esch, H.; Jansen, A.; Bauters, M.; Froyen, G.; Fryns, J.-P. Encephalopathy and bilateral cataract in a boy with an interstitial deletion of Xp22 comprising the CDKL5 and NHS genes. Am. J. Med. Genet. 2007, 143, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Westbrook, V.A.; Schoppee, P.D.; Vanage, G.R.; Klotz, K.L.; Diekman, A.B.; Flickinger, C.J.; Coppola, M.A.; Herr, J.C. hominoid-specific SPANXA/D genes demonstrate differential expression in individuals and protein localization to a distinct nuclear envelope domain during spermatid morphogenesis. Mol. Hum. Reprod. 2006, 12, 703–716. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aruga, J.; Mikoshiba, K. Identification and characterization of Slitrk, a novel neuronal transmembrane protein family controlling neurite outgrowth. Mol. Cell. Neurosci. 2003, 24, 117–129. [Google Scholar] [CrossRef]
- Khan, A.O.; Aldahmesh, M.A.; Mohamed, J.Y.; Alkuraya, F.S. Phenotype-genotype correlation in potential female carriers of x-linked developmental cataract (Nance-Horan syndrome). Ophthalmic Genet. 2012, 33, 89–95. [Google Scholar] [CrossRef]
- Migeon, B.R. X-linked diseases: Susceptible females. Genet. Med. 2020, 22, 1156–1174. [Google Scholar] [CrossRef]
- Allen, R.C.; Zoghbi, H.Y.; Moseley, I.A.B.; Rosenblatt, H.M.; Belmont, J.W. Methylation of Hpall and Hhal sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am. J. Hum. Genet. 1992, 51, 1229–1239. [Google Scholar]
- Sisdelli, L.; Vidi, A.C.; Moysés-Oliveira, M.; Di Battista, A.; Bortolai, A.; Moretti-Ferreira, D.; Dias da Silva, M.R.; Melaragno, M.I.; Carvalheira, G. Incorporation of 5-ethynyl-2′-deoxyuridine (EdU) as a novel strategy for identification of the skewed X inactivation pattern in balanced and unbalanced X-rearrangements. Hum. Genet. 2016, 135, 185–192. [Google Scholar] [CrossRef]
- Ceroni, F.; Aguilera-Garcia, D.; Chassaing, N.; Bax, D.A.; Blanco-Kelly, F.; Ramos, P.; Tarilonte, M.; Villaverde, C.; da Silva, L.R.J.; Ballesta-Martínez, M.J.; et al. New GJA8 variants and phenotypes highlight its critical role in a broad spectrum of eye anomalies. Hum. Genet. 2019, 138, 1027–1042. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef]
- Fishilevich, S.; Nudel, R.; Rappaport, N.; Hadar, R.; Plaschkes, I.; Iny Stein, T.; Rosen, N.; Kohn, A.; Twik, M.; Safran, M.; et al. GeneHancer: Genome-wide integration of enhancers and target genes in GeneCards. Database 2017, 2017, bax028. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Damián, A.; Ionescu, R.O.; Rodríguez de Alba, M.; Tamayo, A.; Trujillo-Tiebas, M.J.; Cotarelo-Pérez, M.C.; Pérez Rodríguez, O.; Villaverde, C.; de la Fuente, L.; Romero, R.; et al. Fine Breakpoint Mapping by Genome Sequencing Reveals the First Large X Inversion Disrupting the NHS Gene in a Patient with Syndromic Cataracts. Int. J. Mol. Sci. 2021, 22, 12713. https://doi.org/10.3390/ijms222312713
Damián A, Ionescu RO, Rodríguez de Alba M, Tamayo A, Trujillo-Tiebas MJ, Cotarelo-Pérez MC, Pérez Rodríguez O, Villaverde C, de la Fuente L, Romero R, et al. Fine Breakpoint Mapping by Genome Sequencing Reveals the First Large X Inversion Disrupting the NHS Gene in a Patient with Syndromic Cataracts. International Journal of Molecular Sciences. 2021; 22(23):12713. https://doi.org/10.3390/ijms222312713
Chicago/Turabian StyleDamián, Alejandra, Raluca Oancea Ionescu, Marta Rodríguez de Alba, Alejandra Tamayo, María José Trujillo-Tiebas, María Carmen Cotarelo-Pérez, Olga Pérez Rodríguez, Cristina Villaverde, Lorena de la Fuente, Raquel Romero, and et al. 2021. "Fine Breakpoint Mapping by Genome Sequencing Reveals the First Large X Inversion Disrupting the NHS Gene in a Patient with Syndromic Cataracts" International Journal of Molecular Sciences 22, no. 23: 12713. https://doi.org/10.3390/ijms222312713
APA StyleDamián, A., Ionescu, R. O., Rodríguez de Alba, M., Tamayo, A., Trujillo-Tiebas, M. J., Cotarelo-Pérez, M. C., Pérez Rodríguez, O., Villaverde, C., de la Fuente, L., Romero, R., Núñez-Moreno, G., Mínguez, P., Ayuso, C., & Cortón, M. (2021). Fine Breakpoint Mapping by Genome Sequencing Reveals the First Large X Inversion Disrupting the NHS Gene in a Patient with Syndromic Cataracts. International Journal of Molecular Sciences, 22(23), 12713. https://doi.org/10.3390/ijms222312713







