1. Introduction
The immunotherapy targeting cytotoxic T lymphocyte-associated protein 4 (CTLA4) and programmed cell death 1 (PD-1) has recently been demonstrated to be an effective approach to activate immune response and treat a wide range of cancers [
1]. However, the response to immunotherapy is not observed in all types of cancers. Moreover, progressive disease eventually develops in most patients who initially respond to immunotherapy. The mechanisms of the primary and secondary resistance are complex and largely determined by the interaction between cancer and immune cells in the microenvironment [
2]. The most dominant and import immune infiltrates in the tumor microenvironment are macrophages, which are often termed as tumor-associated macrophages [
3]. In majority of cancers, tumor-associated macrophages are pro-tumoral macrophages. These macrophages exhibit pro-tumorigenic effects by suppressing anti-tumor immune response of effector cells, including T cells and NK cells, and promoting tumor proliferation, survival, and metastasis. Furthermore, the production of angiogenic factors and stimulation of angiogenesis by macrophages are demonstrated in a wide range of models [
4]. Numerous studies have shown the relationship between tumor-associated macrophages and poor clinical outcome [
5]. Therefore, targeting tumor-associated macrophages, especially the tumor-promoting M2-like macrophages, has become a promising therapeutic strategy to improve the efficacy of current immunotherapy in recent years.
The key pathway regulating the macrophages is colony-stimulating factor 1 receptor (CSF-1R) signaling. CSF-1R, consisting of five extracellular immunoglobulin-like domains, a transmembrane helix, and an intracellular autoinhibitory juxtamembrane domain linked to split tyrosine kinase domains [
6], is a class III receptor tyrosine kinase encoded by the
CSF1R gene [
7]. This receptor contains 972 amino acids with a molecular weight of approximately 108 kDa. Our prior study identified an important germline
CSF1R genetic variant, c.1085A>G, causing the change of amino acid from histidine to arginine in the domain of receptor dimerization [
8]. Macrophages with different
CSF1R c.1085 genotypes displayed different response to CSF-1 stimulation. CSF-1 induced less CSF-1R phosphorylation and endocytosis in macrophages with
CSF1R c.1085 genotype A_G. The weak CSF-1R phosphorylation was accompanied by less macrophage differentiation and M2 polarization. In addition, cancer patients with
CSF1R c.1085A>G variant had less tumor-associated macrophages, M2-like macrophages, and VEGF expression in tumor tissues. In parallel with the less M2-like macrophages, the presence of this germline variant was significantly associated with better disease-free survival in patients with colorectal, endometrial, and ovarian cancer. Despite the known relationship between tumor-associated macrophages and poor clinical outcome, the detailed mechanisms mediating different outcome between patients with
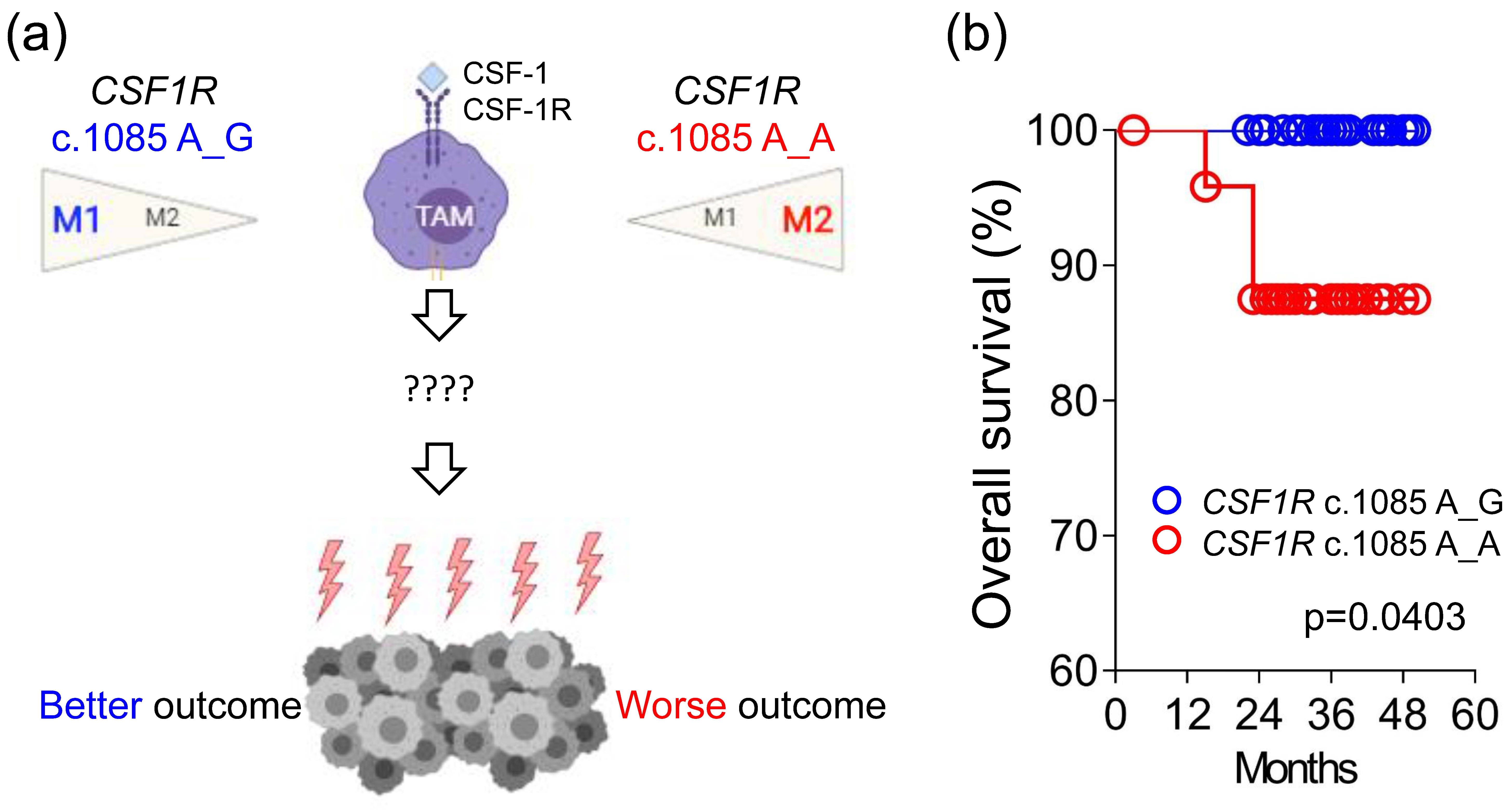
CSF1R c.1085 genotypes A_A and A_G remain unclear (
Figure 1a).
Here, we extensively analyzed the expression of genes involved in immune response from 66 primary tumor samples of colorectal cancer (CRC) patients and compared the difference between patients with different CSF1R c.1085 genotypes. The results showed CRCs harboring CSF1R c.1085 A>G variant had higher CD40LG and IL-2 expression in tumor tissues than those with CSF1R c.1085 genotype A_A. More CD3+CD40L+ T cells and higher IL-2 expression observed in tumors with CSF1R c.1085 genotype A_G confirmed the finding of mRNA expression. These results, together with prior reports, suggest higher IL-2 expression generated by the interaction of CD40 ligand and CD40 between helper T cells and macrophages with CSF1R c.1085 genotype A_G might be the mechanism explaining why CRC patients with this germline variant had a better clinical outcome.
3. Discussion
Immunotherapy has shown a great advance in cancer treatment. However, primary and secondary resistances are still important clinical issues to be resolved. Better understanding of the tumor immune microenvironment is essential to overcome the resistance and establish efficient cancer immunotherapies. By analyzing the clinical outcome and tumor immune microenvironment from 109 stage III or high-risk stage II CRC patients, this study identified several important findings: (i) CRC patients with
CSF1R c.1085 genotype A_A had a worse overall survival than patients with
CSF1R genotype A_G. (ii) Higher
CD40LG and
IL-2 expression was observed in tumor tissues with genotype A_G than those with genotype A_A. (iii) Tumors with
CSF1R c.1085 genotype A_G contained more CD3
+CD40L
+ T cells. Moreover, higher IL-2 expression was also observed in tumors with
CSF1R c.1085 genotype A_G compared to genotype A_A. These confirmed the finding of mRNA expression. The CD40 pathway has been demonstrated to be a key regulator of cytokine production, including IL-2, and anti-tumor immune response [
11]. Taking the results of prior studies and our studies together, we found that higher IL-2 expression generated by the interaction of CD40 ligand and CD40 between helper T cells and macrophages with
CSF1R c.1085 genotype A_G is the potential mechanism explaining why CRC patients with this germline variant had a better clinical outcome.
The clinical impacts of
CSF1R c.1085 genotype had been investigated in several studies [
12,
13]. However, the association between
CSF1R c.1085 genotype and clinical outcome of cancer patients was rarely reported. Our prior study demonstrated germline
CSF1R c.1085A>G variant was associated with disease-free survival in stage III or high-risk stage II CRC patients [
8]. In addition, a better overall survival and a trend showing better outcome in the variant group were observed in patients with endometrial cancer and ovarian cancer, respectively. With a longer follow-up period and more enrollment for the CRC cohort, we demonstrated patients with
CSF1R c.1085 A_G had a better overall survival compared to patients with
CSF1R genotype A_A in this study, consistent with our prior findings. Only one genome-wide association study using a Korean non-small cell lung cancer cohort reported the impact of
CSF1R c.1085A>G variant (rs10079250) on clinical outcome. In agreement with our findings, Yoo et al. showed
CSF1R rs10079250 A>G was associated with a better disease-free survival in never-smoking females [
13]. These data suggested
CSF1R c.1085A>G variant had an impact on clinical outcome of cancer patients.
A more understanding of the mechanism mediating the different outcome of patients with and without
CSF1R c.1085A>G variant may help to develop new therapeutic strategies for patients with different genotypes. In the present study, higher
CD40LG expression was observed in tumor tissues with
CSF1R c.1085 genotype A_G compared to those with genotype A_A. The protein encoded by
CD40LG is CD40 ligand (CD40L) expressed on the surface of T cell. In parallel with the gene expression, higher CD3
+CD40L
+ T cells infiltrates were also noted in tumors with
CSF1R c.1085 genotype A_G. CD40 is the receptor found on macrophages. Our gene expression data showed
CD40 expression was slightly higher in CRCs with
CSF1R genotype A_G compared to genotype A_A (log
2 fold of change: 0.04849 and
p = 0.772221), and our prior study demonstrated the number of macrophages was significantly higher in tumor tissues with the genetic background of
CSF1R c.1085 genotype A_A compared to genotype A_G [
8]. The higher CD40 expression and lower macrophages in CRCs with
CSF1R c.1085 genotype A_G suggest the level of CD40 expression might be higher in macrophages with
CSF1R c.1085 genotype A_G than genotype A_A. Further studies are still needed to confirm this hypothesis. The interaction between CD40 on macrophages and CD40L on lymphocytes had been known to induce intracellular signaling pathways and subsequent IL-2 expression, which is important for cytotoxic T cell priming. Our study also demonstrated CRC tumors with
CSF1R c.1085 genotype A_G had higher IL-2 gene and protein expression. Therefore, higher IL-2 expression generated by the interaction of CD40 ligand and CD40 between helper T cells and macrophages with
CSF1R c.1085 genotype A_G was the potential mechanism explaining why CRC patients with this germline variant had a better clinical outcome. These findings provide a rationale to test the utility of IL-2 in CRC patients with
CSF1R c.1085 genotype A_A.
CRCs with CSF1R c.1085 genotype A_G had higher IL-2 gene expression than those with genotype A_A; however, the difference in IL-2 protein expression was not statistically significant but trending. This may be explained by the case number and different stage of the lesions distributed in different groups of CSF1R genotyping. Moreover, the IL-2 gene and protein expression measured in this study were the total IL-2 expressed by both tumor and non-tumor cells in the tumor immune microenvironment. It is not easy to specifically determine IL-2 level generated by immune cells in tumor tissues. Further studies, such as co-culture of CD4+ T cells with macrophages with and without CSF1R c.1085A>G variant, will be needed to confirm this hypothesis.
In addition to the differential
CD40LG and
IL-2 expression shown in this study, our prior work demonstrated the impact of germline
CSF1R c.1085A>G variant on CSF-1R signaling and macrophage functions [
8]. Macrophages with
CSF1R c.1085A>G variant displayed a poor response to CSF-1 stimulation. In addition, patients harboring this variant had fewer total macrophages and M2-like macrophages in tumor tissues. Interestingly, the gene expression data also showed higher
CSF1R mRNA expression in CRCs with
CSF1R c.1085 genotype A_A compared to genotype A_G. It is very likely that CRCs with
CSF1R c.1085 genotype A_A had higher CSF-1R protein expression than those with genotype A_G. CSF-1R is primarily expressed on macrophages, and it has been known that CSF-1R signaling is important to main macrophages in an M2-like state. Higher proportion of M2 macrophages associated with advanced stage and poor clinical outcome has been reported in many different types of cancer [
14]. The underlying mechanism mediating this correlation has also been extensively studied in vitro and in vivo [
15]. Lower CSF-1R expression, less tumor-associated macrophages, and less M2-like macrophages in tumors with
CSF1R genotype A_G also provide an explanation why patients with this germline variant had better clinical outcome. The CSF-1R expression on macrophages and the underlying mechanism mediating higher M2-like macrophage and less CD3
+CD40L
+ T cell infiltrates in tumors with
CSF1R c.1085 genotype A_G is worth further exploration.
Taking this and our prior study together, we find that germline
CSF1R c.1085A>G variant had a great impact on macrophage functions, tumor immune microenvironment, and clinical outcome (summarized in
Figure 5). Macrophages with
CSF1R c.1085A>G variant displayed a poor response to CSF-1 stimulation in terms of CSF-1R phosphorylation, macrophage differentiation, and M2 polarization. CRCs with
CSF1R c.1085 genotype A_G contained less M2-like macrophage infiltrates in tumor tissues. In addition to less M2 distribution, higher
CD40LG expression and CD3
+CD40L
+ T cells infiltrates were also observed in CRCs with
CSF1R c.1085A>G variant and accompanied by higher IL-2 expression, which is important in activation of nature killer (NK) cells and cytotoxic lymphocytes. These findings explained how
CSF1R germline c.1085 genetic variant impacted the tumor microenvironment and why patients with
CSF1R c.1085 genotype A_G had a better clinical outcome than those with genotype A_A. Since IL-2 had been applied in cancer immunotherapy clinically, it is worth testing the utility of IL-2 in CRC patients with
CSF1R c.1085 genotype A_A.
4. Materials and Methods
4.1. Database Used for Analysis
The database generated from a retrospective pilot study and an ongoing prospective study evaluating the association of germline genetic variants and chemotherapy-induced peripheral neuropathy (CIPN) [
16] was used for analysis in this study. These 2 studies were approved by the institutional review board (A-ER-103-395 and A-ER-1040153) of National Cheng Kung University Hospital (NCKUH), Tainan, Taiwan, and conducted in accordance with the Declaration of Helsinki. Written informed consent was provided by all patients. The information of these 2 CIPN studies was described in detail in the prior study [
8]. In brief, stage III or high-risk stage II CRC patients receiving standard surgical resection followed by adjuvant chemotherapy with mFOLFOX6 were enrolled. Whole genome sequencing was performed on DNA generated from peripheral blood to determine germline genetic variants. Until July 2017, 109 CRC patients, including 37 patients in the pilot study and 72 patients in the prospective study, were enrolled. Primary tumor samples of these patients were used to investigate the expression of immune response gene expression, and 72 patients enrolled in the prospective study were used to study the impact of germline
CSF1R c.1085 genetic variant on clinical outcome.
4.2. RNA Sequencing
The RNA was isolated from paraformaldehyde-fixed paraffin-embedded (FFPE) primary CRC tumor tissue using RecoverAll Total Nucleic Acid Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA). RNA concentration was determined by Invitrogen™ Qubit™ Fluorometer with the Qubit™ RNA High Sensitivity Assay (Thermo Fisher Scientific). SuperScript™ IV VILO™ Master Mix Kit was used to perform reverse transcription with 20ng RNA. Immune Response libraries were prepared using the Ion AmpliSeq™ Kit for Chef DL8 with the Ion Chef™ System. The conditions of library preparation on the Ion Chef™ System was performed according to the Oncomine™ Immune Response Research Assay user guide (pub. no. MAN0015867). Template preparation, chip loading, and sequencing were carried out on the Ion Chef™ System and Ion S5 XL sequencing system with Ion 510 and Ion 520 and Ion 530 Kit-Chef Kit (Thermo Fisher Scientific, Waltham, MA, USA). The Ion Chef™ and Ion S5 XL sequencing system (Thermo Fisher Scientific, Waltham, MA, USA) were used as described in the User Guide: Ion 510 and Ion 520 and Ion 530 Chef Kit—Instructions for automated template preparation, chip loading, and sequencing (pub. no. MAN0016854). The raw gene expression data were preprocessed using Torrent Suite (Thermo Fisher Scientific, Waltham, MA, USA) and analyzed using DESeq2 package in R. Data normalization was performed via function DESeq. Candidate genes that were differentially expressed between the groups of CSF1R c.1085 genotype A_A and A_G were selected for further study.
4.3. Chemicals and Antibodies
The antibodies used for immunofluorescent staining were mouse anti-CD3 (Thermo Fisher Scientific Cat# 14-0037-82, RRID: AB_467057), rabbit anti-CD40L (Abcam Cat# ab2391, RRID: AB_303034), and Hoechst 33258 (Thermo Fisher Scientific Cat# H3569, RRID: AB_2651133). Alexa Fluor 594- (Thermo Fisher Scientific, Waltham, MA, USA, Cat# A-21203, RRID: AB_2535789) and Alexa 488-conjugated secondary antibodies (Thermo Fisher Scientific, Waltham, MA, USA, Cat# A-11008, RRID: AB_143165) were purchased from Thermo Fisher Scientific Inc (Waltham, MA, USA). Rabbit anti-IL-2 polyclonal antibody (Bioss, Woburn, MA, USA, Cat# bs-0605R, RRID: AB_10856306) was the primary antibody used for immunohistochemical staining.
4.4. Immunofluorescent Staining, Confocal Images, and Tissue Scanning
Primary tumor specimens of CRC patients enrolled in CIPN studies were used for immunofluorescent staining. We used anti-CD3 and anti-CD40L to stain T cells and CD40 ligand, respectively. Alexa Fluor 594-conjugated secondary antibody was used for CD3, and Alexa 488-conjugated secondary antibody was used for CD40L. Tissue was co-stained with Hoechst to detect the nucleus. Whole tissue was scanned automatically using a TissueFAXS PLUS microscope (TissueGnostics, Vienna, Austria). In each sample, the region of interest (ROI) was randomly selected from 5 fields of the tumor microenvironment. Each field was 1.2 × 1.2 mm2. The number of CD3+CD40L+ cells was quantitated automatically by TissueQuest analysis software (RRID: SCR_014822).
4.5. Immunohistochemical Staining
Immunohistochemical (IHC) staining was performed on 4 µm thick formalin-fixed paraffin-embedded sections. We used rabbit anti-human IL-2 polyclonal antibody (1:100 dilution) as the primary antibody. The procedures were conducted with the Bond-Max Automated IHC stainer (Leica Biosystems, Lincolnshire, IL, USA) according to the following protocol. Tissues were deparaffinized with xylene and pre-treated with the Epitope Retrieval Solution 2 (EDTA buffer, pH 9·0) at 100 °C for 20 min. After deparaffinization, the tissues were incubated with primary antibody at room temperature for 30 min. Subsequently, tissues were incubated with polymer at room temperature for 8 min using the Bond Polymer Refine Detection Kit (Leica Biosystems, Lincolnshire, IL, USA) and then developed with 3,3′-diaminobenzidine chromogen for 10 min. Counterstaining was carried out with hematoxylin. In each sample, the region of interest (ROI) was randomly selected from 5 fields of the tumor microenvironment to determine the IL-2 expression level. Each field was 1.2 × 1.2 mm2. The staining intensity of IL-2 was categorized into 5 categories as follows: no staining (0+), faint (1+), weak (2+), moderate (3+), and intense (4+). The percentage of positive staining area for each intensity was analyzed by HistoQuest analysis software (RRID: SCR_014822). The IL-2 expression level was scored by multiplying the percentage of positive areas by the intensity.
4.6. Statistical Analysis
Kaplan–Meier survival analysis was used to estimate the survival, and the log-rank test was used to compare the difference between groups. All values were presented as mean +/− SEM. Unpaired t-test was used to compare the difference between groups. A p-value < 0.05 was considered statistically significant.