D-Xylose Sensing in Saccharomyces cerevisiae: Insights from D-Glucose Signaling and Native D-Xylose Utilizers
Abstract
:1. Introduction
2. What Is Sugar Sensing and Signaling?
2.1. Signaling Networks Control Cellular Functions in Response to Environmental Changes
2.2. Main Pathways Involved in S. cerevisiae Sugar Sensing and Signaling
3. What Happens on d-Glucose, the Model Case for Sugar Signaling?
3.1. d-Glucose Sensing by the Snf3p/Rgt2p Pathway Regulates Hexose Transporter Gene Expression
3.2. The SNF1/Mig1p Pathway Represses Transcription of Genes Related to Alternative Carbon Sources upon Sensing of d-Glucose
3.3. The cAMP/PKA Pathway Triggers a Phosphorylation Cascade after Sensing d-Glucose
3.4. The Effect of d-Glucose on Other Signaling Pathways
3.4.1. MAPK Pathways: The HOG Pathway and the Filamentous Growth Pathway
3.4.2. The TOR Pathway

3.4.3. The Galactose Regulon
3.5. Cross-Talk between the Different Sugar Signaling Pathways
3.6. Connections between Sugar Signaling and Glycolysis
4. What Happens on d-Xylose, and Why?
4.1. d-Xylose Signaling in Natural and Engineered S. cerevisiae
4.1.1. The Snf3p/Rgt2p Pathway Weakly Senses d-Xylose
4.1.2. d-Xylose Affects the SNF1/Mig1p Pathway Both Directly and Indirectly
4.1.3. Assimilation of d-Xylose Is Weakly Sensed by the Intracellular Branch of the cAMP/PKA Pathway
4.1.4. Effect of d-Xylose on Other d-Glucose-Responsive Signaling Pathways
4.1.5. Proposed Mechanisms for d-Xylose Sensing
4.2. d-Xylose Signaling in Other Xylose-Utilizing Species
4.2.1. d-Xylose Regulation in Other Yeast Species
4.2.2. XylR as an Inducer of the d-Xylose Operon (XylR-I)
4.2.3. XylR as a Repressor of the Xylose Operon (XylR-R)
5. Current Status of Engineering of S. cerevisiae d-Xylose Signaling
5.1. Modifications to the Existing Signaling Network
5.1.1. Engineering the Snf3p/Rgt2p Pathway
5.1.2. Engineering the SNF1/Mig1p Pathway
5.1.3. Engineering the cAMP/PKA Pathway
5.2. Synthetic d-Xylose Signaling Networks
5.2.1. XylR-Based Signaling Circuits

5.2.2. GAL-Based Signaling Circuits
6. Outlook
6.1. Towards Increased Understanding of d-Xylose Sensing
6.2. Future Directions for Synthetic d-Xylose Signaling Networks
6.3. Computational Modeling of Sugar Signaling?
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Abril, M.A.; Michan, C.; Timmis, K.N.; Ramos, J.L. Regulator and Enzyme Specificities of the Tol Plasmid-Encoded Upper Pathway for Degradation of Aromatic-Hydrocarbons and Expansion of the Substrate Range of the Pathway. J. Bacteriol. 1989, 171, 6782–6790. [Google Scholar] [CrossRef] [Green Version]
- Klemba, M.; Jakobs, B.; Wittich, R.M.; Pieper, D. Chromosomal integration of tcb chlorocatechol degradation pathway genes as a means of expanding the growth substrate range of bacteria to include haloaromatics. Appl. Environ. Microbiol. 2000, 66, 3255–3261. [Google Scholar] [CrossRef] [Green Version]
- Müller, J.E.N.; Meyer, F.; Litsanov, B.; Kiefer, P.; Potthoff, E.; Heux, S.; Quax, W.J.; Wendisch, V.F.; Brautaset, T.; Portais, J.C.; et al. Engineering Escherichia coli for methanol conversion. Metab. Eng. 2015, 28, 190–201. [Google Scholar] [CrossRef]
- Löwe, H.; Schmauder, L.; Hobmeier, K.; Kremling, A.; Pfluger-Grau, K. Metabolic engineering to expand the substrate spectrum of Pseudomonas putida toward sucrose. MicrobioliologyOpen 2017, 6, e00473. [Google Scholar] [CrossRef]
- Hong, K.K.; Nielsen, J. Metabolic engineering of Saccharomyces cerevisiae: A key cell factory platform for future biorefineries. Cell. Mol. Life Sci. 2012, 69, 2671–2690. [Google Scholar] [CrossRef] [PubMed]
- Ledesma-Amaro, R.; Nicaud, J.M. Metabolic Engineering for Expanding the Substrate Range of Yarrowia lipolytica. Trends Biotechnol. 2016, 34, 798–809. [Google Scholar] [CrossRef]
- Dangi, A.K.; Sharma, B.; Hill, R.T.; Shukla, P. Bioremediation through microbes: Systems biology and metabolic engineering approach. Crit. Rev. Biotechnol. 2019, 39, 79–98. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.J.; Nielsen, J. Advancing metabolic engineering through systems biology of industrial microorganisms. Curr. Opin. Biotechnol. 2015, 36, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Granados, R.; Lerma-Escalera, J.A.; Morones-Ramirez, J.R. Metabolic Engineering and Synthetic Biology: Synergies, Future, and Challenges. Front. Bioeng. Biotechnol. 2019, 7, 36. [Google Scholar] [CrossRef] [PubMed]
- Straathof, A.J.J.; Wahl, S.A.; Benjamin, K.R.; Takors, R.; Wierckx, N.; Noorman, H.J. Grand Research Challenges for Sustainable Industrial Biotechnology. Trends Biotechnol. 2019, 37, 1042–1050. [Google Scholar] [CrossRef] [PubMed]
- Rangel, A.E.T.; Ramirez, J.M.G.; Barrios, A.F.G. From industrial by-products to value-added compounds: The design of efficient microbial cell factories by coupling systems metabolic engineering and bioprocesses. Biofuels Bioprod. Biorefining 2020, 14, 1228–1238. [Google Scholar] [CrossRef]
- Nogue, V.S.; Karhumaa, K. Xylose fermentation as a challenge for commercialization of lignocellulosic fuels and chemicals. Biotechnol. Lett. 2015, 37, 761–772. [Google Scholar] [CrossRef]
- Hahn-Hägerdal, B.; Karhumaa, K.; Jeppsson, M.; Gorwa-Grauslund, M.F. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Biofuels 2007, 108, 147–177. [Google Scholar]
- Kötter, P.; Amore, R.; Hollenberg, C.P.; Ciriacy, M. Isolation and Characterization of the Pichia stipitis Xylitol Dehydrogenase Gene, XYL2, and Construction of a Xylose-Utilizing Saccharomyces cerevisiae Transformant. Curr. Genet. 1990, 18, 493–500. [Google Scholar] [CrossRef]
- Kuyper, M.; Harhangi, H.R.; Stave, A.K.; Winkler, A.A.; Jetten, M.S.M.; de Laat, W.T.A.M.; den Ridder, J.J.J.; Op den Camp, H.J.M.; van Dijken, J.P.; Pronk, J.T. High-level functional expression of a fungal xylose isomerase: The key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Res. 2003, 4, 69–78. [Google Scholar] [CrossRef] [Green Version]
- Salusjärvi, L.; Toivari, M.; Vehkomäki, M.-L.; Koivistoinen, O.; Mojzita, D.; Niemelä, K.; Penttilä, M.; Ruohonen, L. Production of ethylene glycol or glycolic acid from D-xylose in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2017, 101, 8151–8163. [Google Scholar] [CrossRef]
- Borgström, C.; Wasserstrom, L.; Almqvist, H.; Broberg, K.; Klein, B.; Noack, S.; Lidén, G.; Gorwa-Grauslund, M.F. Identification of modifications procuring growth on xylose in recombinant Saccharomyces cerevisiae strains carrying the Weimberg pathway. Metab. Eng. 2019, 55, 1–11. [Google Scholar] [CrossRef]
- Matsushika, A.; Inoue, H.; Kodaki, T.; Sawayama, S. Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: Current state and perspectives. Appl. Microbiol. Biotechnol. 2009, 84, 37–53. [Google Scholar] [CrossRef]
- Moyses, D.N.; Reis, V.C.B.; de Almeida, J.R.M.; de Moraes, L.M.P.; Torres, F.A.G. Xylose Fermentation by Saccharomyces cerevisiae: Challenges and Prospects. Int J. Mol. Sci. 2016, 17, 207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verhoeven, M.D.; Lee, M.; Kamoen, L.; van den Broek, M.; Janssen, D.B.; Daran, J.M.G.; van Maris, A.J.A.; Pronk, J.T. Mutations in PMR1 stimulate xylose isomerase activity and anaerobic growth on xylose of engineered Saccharomyces cerevisiae by influencing manganese homeostasis. Sci. Rep. UK 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banares, A.B.; Nisola, G.M.; Valdehuesa, K.N.G.; Lee, W.K.; Chung, W.J. Understanding D-xylonic acid accumulation: A cornerstone for better metabolic engineering approaches. Appl. Microbiol. Biotechnol. 2021, 105, 5309–5324. [Google Scholar] [CrossRef]
- Kötter, P.; Ciriacy, M. Xylose Fermentation by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 1993, 38, 776–783. [Google Scholar] [CrossRef]
- Leandro, M.J.; Fonseca, C.; Goncalves, P. Hexose and pentose transport in ascomycetous yeasts: An overview. FEMS Yeast Res. 2009, 9, 511–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Z.; Zhang, B.; Li, Y. Engineering Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: Reflections and perspectives. Biotechnol. J. 2012, 7, 34–46. [Google Scholar] [CrossRef]
- Eliasson, A.; Christensson, C.; Wahlbom, C.F.; Hahn-Hägerdal, B. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl. Environ. Microbiol. 2000, 66, 3381–3386. [Google Scholar] [CrossRef] [Green Version]
- Wahlbom, C.F.; Eliasson, A.; Hahn-Hägerdal, B. Intracellular fluxes in a recombinant xylose-utilizing Saccharomyces cerevisiae cultivated anaerobically at different dilution rates and feed concentrations. Biotechnol. Bioeng. 2001, 72, 289–296. [Google Scholar] [CrossRef]
- Hector, R.E.; Qureshi, N.; Hughes, S.R.; Cotta, M.A. Expression of a heterologous xylose transporter in a Saccharomyces cerevisiae strain engineered to utilize xylose improves aerobic xylose consumption. Appl. Microbiol. Biotechnol. 2008, 80, 675–684. [Google Scholar] [CrossRef]
- Nijland, J.G.; Driessen, A.J.M. Engineering of Pentose Transport in Saccharomyces cerevisiae for Biotechnological Applications. Front. Bioeng. Biotechnol. 2020, 7, 464. [Google Scholar] [CrossRef]
- Bueno, J.G.R.; Borelli, G.; Corrêa, T.L.R.; Fiamenghi, M.B.; José, J.; de Carvalho, M.; de Oliveira, L.C.; Pereira, G.A.; Dos Santos, L.V. Novel xylose transporter Cs4130 expands the sugar uptake repertoire in recombinant Saccharomyces cerevisiae strains at high xylose concentrations. Biotechnol. Biofuels 2020, 13, 1–20. [Google Scholar] [CrossRef]
- Chattopadhyay, A.; Singh, R.; Das, A.K.; Maiti, M.K. Characterization of two sugar transporters responsible for efficient xylose uptake in an oleaginous yeast Candida tropicalis SY005. Arch. Biochem. Biophys. 2020, 695, 108645. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Shen, Y.; Gu, L.C.; Wang, Z.Z.; Su, N.; Niu, K.L.; Guo, W.; Hou, S.L.; Bao, X.M.; Tian, C.G.; et al. Identification and Characterization of an Efficient D-Xylose Transporter in Saccharomyces cerevisiae. J. Agric. Food Chem. 2020, 68, 2702–2710. [Google Scholar] [CrossRef]
- Podolsky, I.A.; Seppala, S.; Xu, H.Q.; Jin, Y.S.; O’Malley, M.A. A SWEET surprise: Anaerobic fungal sugar transporters and chimeras enhance sugar uptake in yeast. Metab. Eng. 2021, 66, 137–147. [Google Scholar] [CrossRef]
- Qiao, Y.M.; Li, C.L.; Lu, X.Y.; Zong, H.; Zhuge, B. Transporter engineering promotes the co-utilization of glucose and xylose by Candida glycerinogenes for D-xylonate production. Biochem. Eng. J. 2021, 175, 108150. [Google Scholar] [CrossRef]
- Salusjärvi, L.; Pitkanen, J.P.; Aristidou, A.; Ruohonen, L.; Penttilä, M. Transcription analysis of recombinant Saccharomyces cerevisiae reveals novel responses to xylose. Appl. Biochem. Biotechnol. 2006, 128, 237–261. [Google Scholar] [CrossRef]
- Salusjärvi, L.; Kankainen, M.; Soliymani, R.; Pitkanen, J.P.; Penttilä, M.; Ruohonen, L. Regulation of xylose metabolism in recombinant Saccharomyces cerevisiae. Microb. Cell Fact. 2008, 7, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Bergdahl, B.; Sandström, A.G.; Borgström, C.; Boonyawan, T.; van Niel, E.W.J.; Gorwa-Grauslund, M.F. Engineering Yeast Hexokinase 2 for Improved Tolerance Toward Xylose-Induced Inactivation. PLoS ONE 2013, 8, e75055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsushika, A.; Goshima, T.; Hoshino, T. Transcription analysis of recombinant industrial and laboratory Saccharomyces cerevisiae strains reveals the molecular basis for fermentation of glucose and xylose. Microb. Cell Fact. 2014, 13, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Y.S.; Laplaza, J.M.; Jeffries, T.W. Saccharomyces cerevisiae engineered for xylose metabolism exhibits a respiratory response. Appl. Environ. Microbiol. 2004, 70, 6816–6825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gopinarayanan, V.E.; Nair, N.U. Pentose Metabolism in Saccharomyces cerevisiae: The Need to Engineer Global Regulatory Systems. Biotechnol. J. 2019, 14, e1800364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostergaard, S.; Roca, C.; Ronnow, B.; Nielsen, J.; Olsson, L. Physiological studies in aerobic batch cultivations of Saccharomyces cerevisiae strains harboring the MEL1 gene. Biotechnol Bioeng. 2000, 68, 252–259. [Google Scholar] [CrossRef]
- Bracher, J. Engineering of metabolism and membrane transport in Saccharomyces cerevisiae for improved industrial performance. Ph.D. Thesis, Delft University of Technology, Delft, The Netherlands, 2019. [Google Scholar]
- Kuyper, M.; Hartog, M.M.P.; Toirkens, M.J.; Almering, M.J.H.; Winkler, A.A.; van Dijken, J.P.; Pronk, J.T. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 2005, 5, 399–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Cheng, J.S.; Wang, B.L.; Fink, G.R.; Stephanopoulos, G. Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab. Eng. 2012, 14, 611–622. [Google Scholar] [CrossRef]
- Cadete, R.M.; Alejandro, M.; Sandström, A.G.; Ferreira, C.; Gírio, F.; Gorwa-Grauslund, M.-F.; Rosa, C.A.; Fonseca, C. Exploring xylose metabolism in Spathaspora species: XYL1. 2 from Spathaspora passalidarum as the key for efficient anaerobic xylose fermentation in metabolic engineered Saccharomyces cerevisiae. Biotechnology for Biofuels 2016, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.R.; Skerker, J.M.; Kang, W.; Lesmana, A.; Wei, N.; Arkin, A.P.; Jin, Y.S. Rational and Evolutionary Engineering Approaches Uncover a Small Set of Genetic Changes Efficient for Rapid Xylose Fermentation in Saccharomyces cerevisiae. PLoS ONE 2013, 8, e57048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goudreau, P.N.; Stock, A.M. Signal transduction in bacteria: Molecular mechanisms of stimulus-response coupling. Curr. Opin. Microbiol. 1998, 1, 160–169. [Google Scholar] [CrossRef]
- Milanesi, R.; Coccetti, P.; Tripodi, F. The Regulatory Role of Key Metabolites in the Control of Cell Signaling. Biomolecules 2020, 10, 862. [Google Scholar] [CrossRef]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; Van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruis, H.; Schüller, C. Stress signaling in yeast. Bioessays 1995, 17, 959–965. [Google Scholar] [CrossRef]
- Hohmann, S. Osmotic stress signaling and osmoadaptation in Yeasts. Microbiol. Mol. Biol. R 2002, 66, 300. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.E.; Thorner, J. Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae. BBA-Mol. Cell Res. 2007, 1773, 1311–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knorre, D.A.; Sokolov, S.S.; Zyrina, A.N.; Severin, F.F. How do yeast sense mitochondrial dysfunction? Microbiol. Cell 2016, 3, 401–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardo, B.; Crabbe, L.; Pasero, P. Signaling pathways of replication stress in yeast. FEMS Yeast Res. 2017, 17, fow101. [Google Scholar] [CrossRef]
- Gonzalez, A.; Hall, M.N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017, 36, 397–408. [Google Scholar] [CrossRef] [Green Version]
- Elion, E.A. Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 2000, 3, 573–581. [Google Scholar] [CrossRef]
- Hyduke, D.R.; Palsson, B.O. Towards genome-scale signalling-network reconstructions. Nat. Rev. Genet. 2010, 11, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, D.; Braberg, H.; Mehta, M.; Chechik, G.; Cagney, G.; Mukherjee, P.; Silva, A.C.; Shales, M.; Collins, S.R.; van Wageningen, S.; et al. Functional Organization of the S. cerevisiae Phosphorylation Network. Cell 2009, 136, 952–963. [Google Scholar] [CrossRef] [Green Version]
- Woelk, T.; Sigismund, S.; Penengo, L.; Polo, S. The ubiquitination code: A signalling problem. Cell Div. 2007, 2, 11. [Google Scholar] [CrossRef] [Green Version]
- Pawson, T.; Nash, P. Protein-protein interactions define specificity in signal transduction. Gene Dev. 2000, 14, 1027–1047. [Google Scholar] [CrossRef] [PubMed]
- Teruel, M.N.; Meyer, T. Translocation and reversible localization of signaling proteins: A dynamic future for signal transduction. Cell 2000, 103, 181–184. [Google Scholar] [CrossRef] [Green Version]
- Newton, A.C.; Bootman, M.D.; Scott, J.D. Second messengers. Cold Spring Harb. Perspect. Biol. 2016, 8, a005926. [Google Scholar] [CrossRef]
- Hofer, A.M.; Lefkimmiatis, K. Extracellular calcium and cAMP: Second messengers as “third messengers”? Physiology 2007, 22, 320–327. [Google Scholar] [CrossRef]
- Welkenhuysen, N.; Borgqvist, J.; Backman, M.; Bendrioua, L.; Goksor, M.; Adiels, C.B.; Cvijovic, M.; Hohmann, S. Single-cell study links metabolism with nutrient signaling and reveals sources of variability. BMC Syst. Biol. 2017, 11, 1–10. [Google Scholar]
- Kinnunen, P.C.; Luker, K.E.; Luker, G.D.; Linderman, J.J. Computational methods for characterizing and learning from heterogeneous cell-signaling data. Curr. Opin. Syst. Biol. 2021, 26, 98–108. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Mukerjea, R.; Robyt, J.F. Specificity of yeast (Saccharomyces cerevisiae) in removing carbohydrates by fermentation. Carbohydr. Res. 2003, 338, 1127–1132. [Google Scholar] [CrossRef]
- Smets, B.; Ghillebert, R.; De Snijder, P.; Binda, M.; Swinnen, E.; De Virgilio, C.; Winderickx, J. Life in the midst of scarcity: Adaptations to nutrient availability in Saccharomyces cerevisiae. Curr. Genet. 2010, 56, 1–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santangelo, G.M. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol Rev. 2006, 70, 253–282. [Google Scholar] [CrossRef] [Green Version]
- Gancedo, J.M. The early steps of glucose signalling in yeast. FEMS Microbiol. Rev. 2008, 32, 673–704. [Google Scholar] [CrossRef] [Green Version]
- Gancedo, J.M. Yeast carbon catabolite repression. Microbiol. Mol. Biol Rev. 1998, 62, 334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dynesen, J.; Smits, H.P.; Olsson, L.; Nielsen, J. Carbon catabolite repression of invertase during batch cultivations of Saccharomyces cerevisiae: The role of glucose, fructose, and mannose. Appl. Microbiol. Biotechnol. 1998, 50, 579–582. [Google Scholar] [CrossRef]
- Meinander, N.Q.; Boels, I.; Hahn-Hägerdal, B. Fermentation of xylose/glucose mixtures by metabolically engineered Saccharomyces cerevisiae strains expressing XYL1 and XYL2 from Pichia stipitis with and without overexpression of TAL1. Bioresour. Technol. 1999, 68, 79–87. [Google Scholar] [CrossRef]
- Sonderegger, M.; Jeppsson, M.; Larsson, C.; Gorwa-Grauslund, M.F.; Boles, E.; Olsson, L.; Spencer-Martins, I.; Hahn-Hagerdal, B.; Sauer, U. Fermentation performance of engineered and evolved xylose-fermenting Saccharomyces cerevisiae strains. Biotechnol. Bioeng. 2004, 87, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Karhumaa, K.; Fromanger, R.; Hahn-Hägerdal, B.; Gorwa-Grauslund, M.F. High activity of xylose reductase and xylitol dehydrogenase improves xylose fermentation by recombinant Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2007, 73, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Krahulec, S.; Petschacher, B.; Wallner, M.; Longus, K.; Klimacek, M.; Nidetzky, B. Fermentation of mixed glucose-xylose substrates by engineered strains of Saccharomyces cerevisiae: Role of the coenzyme specificity of xylose reductase, and effect of glucose on xylose utilization. Microb. Cell Fact. 2010, 9, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.R.; Kwee, N.R.; Kim, H.; Jin, Y.S. Feasibility of xylose fermentation by engineered Saccharomyces cerevisiae overexpressing endogenous aldose reductase (GRE3), xylitol dehydrogenase (XYL2), and xylulokinase (XYL3) from Scheffersomyces stipitis. FEMS Yeast Res. 2013, 13, 312–321. [Google Scholar] [CrossRef] [Green Version]
- Carlson, M. Glucose repression in yeast. Curr. Opin. Microbiol. 1999, 2, 202–207. [Google Scholar] [CrossRef]
- Osiro, K.O.; Brink, D.P.; Borgström, C.; Wasserstrom, L.; Carlquist, M.; Gorwa-Grauslund, M.F. Assessing the effect of D-xylose on the sugar signaling pathways of Saccharomyces cerevisiae in strains engineered for xylose transport and assimilation. FEMS Yeast Res. 2018, 18, fox096. [Google Scholar] [CrossRef] [Green Version]
- Osiro, K.O. Used but not Sensed-The Paradox of D-xylose Metabolism in Saccharomyces cerevisiae. Ph.D. Thesis, Lund University, Lund, Sweden, 2019. [Google Scholar]
- Borgström, C. The role of sugar sensing and pathway selection on D-xylose utilization by Saccharomyces cerevisiae. Ph.D. Thesis, Lund University, Lund, Sweden, 2020. [Google Scholar]
- van Dam, K. Role of glucose signaling in yeast metabolism. Biotechnol. Bioeng. 1996, 52, 161–165. [Google Scholar] [CrossRef]
- Johnston, M. Feasting, fasting and fermenting—Glucose sensing in yeast and other cells. Trends Genet. 1999, 15, 29–33. [Google Scholar] [CrossRef]
- Peeters, K.; Thevelein, J.M. Glucose sensing and signal transduction in Saccharomyces cerevisiae. In Molecular Mechanisms in Yeast Carbon Metabolism; Springer: Berlin/Heidelberg, Germany, 2014; pp. 21–56. [Google Scholar]
- Kayikci, O.; Nielsen, J. Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 2015, 15, fov068. [Google Scholar] [CrossRef] [Green Version]
- Stasyk, O.G.; Stasyk, V. Glucose sensing and regulation in yeasts. In Non-Conventional Yeasts: From Basic Research to Application; Springer: Berlin/Heidelberg, Germany, 2019; pp. 477–519. [Google Scholar]
- Brewster, J.L.; Gustin, M.C. Hog1: 20 years of discovery and impact. Sci. Signal. 2014, 7, re7. [Google Scholar] [CrossRef]
- Karunanithi, S.; Cullen, P.J. The Filamentous Growth MAPK Pathway Responds to Glucose Starvation Through the Mig1/2 Transcriptional Repressors in Saccharomyces cerevisiae. Genetics 2012, 192, 869–887. [Google Scholar] [CrossRef] [Green Version]
- Kunkel, J.; Luo, X.X.; Capaldi, A.P. Integrated TORC1 and PKA signaling control the temporal activation of glucose-induced gene expression in yeast. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Lohr, D.; Venkov, P.; Zlatanova, J. Transcriptional Regulation in the Yeast Gal Gene Family—A Complex Genetic Network. FASEB J. 1995, 9, 777–787. [Google Scholar] [CrossRef]
- Bhat, P.J.; Murthy, T.V.S. Transcriptional control of the GAL/MEL regulon of yeast Saccharomyces cerevisiae: Mechanism of galactose-mediated signal transduction. Mol. Microbiol. 2001, 40, 1059–1066. [Google Scholar] [CrossRef]
- Latchman, D.S. Transcription factors: An overview. Int. J. Biochem. Cell Biol. 1997, 29, 1305–1312. [Google Scholar] [CrossRef] [Green Version]
- Treitel, M.A.; Kuchin, S.; Carlson, M. Snf1 protein kinase regulates phosphorylation of the mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998, 18, 6273–6280. [Google Scholar] [CrossRef] [Green Version]
- Thevelein, J.M.; de Winde, J.H. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 1999, 33, 904–918. [Google Scholar] [CrossRef] [PubMed]
- Moriya, H.; Johnston, M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. Sci. USA 2004, 101, 1572–1577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Özcan, S.; Leong, T.; Johnston, M. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol. Cell. Biol 1996, 16, 6419–6426. [Google Scholar] [CrossRef] [Green Version]
- Lemaire, K.; de Velde, S.V.; Van Dijck, P.; Thevelein, J.M. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 2004, 16, 293–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, M.C.; McCartney, R.R.; Zhang, X.D.; Tillman, T.S.; Solimeo, H.; Wolfl, S.; Almonte, C.; Watkins, S.C. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999, 19, 4561–4571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Özcan, S.; Johnston, M. Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 1999, 63, 554–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Roy, A.; Jouandot, D.; Cho, K.H. The glucose signaling network in yeast. BBA-Gen. Subj. 2013, 1830, 5204–5210. [Google Scholar] [CrossRef] [Green Version]
- Özcan, S.; Dover, J.; Johnston, M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998, 17, 2566–2573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karhumaa, K.; Wu, B.Q.; Kielland-Brandt, M.C. Conditions With High Intracellular Glucose Inhibit Sensing Through Glucose Sensor Snf3 in Saccharomyces cerevisiae. J. Cell. Biochem. 2010, 110, 920–925. [Google Scholar] [CrossRef]
- Lafuente, M.J.; Gancedo, C.; Jauniaux, J.C.; Gancedo, J.M. Mth1 receives the Signal given by the glucose sensors Snf3 and Rgt2 in Saccharomyces cerevisiae. Mol. Microbiol. 2000, 35, 161–172. [Google Scholar] [CrossRef]
- Lakshmanan, J.; Mosley, A.L.; Özcan, S. Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1. Curr. Genet. 2003, 44, 19–25. [Google Scholar] [CrossRef]
- Willems, A.R.; Schwab, M.; Tyers, M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. BBA-Mol. Cell Res. 2004, 1695, 133–170. [Google Scholar] [CrossRef] [Green Version]
- Flick, K.M.; Spielewoy, N.; Kalashnikova, T.I.; Guaderrama, M.; Zhu, Q.Z.; Chang, H.C.; Wittenberg, C. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol. Biol. Cell 2003, 14, 3230–3241. [Google Scholar] [CrossRef] [Green Version]
- Mosley, A.L.; Lakshmanan, J.; Aryal, B.K.; Özcan, S. Glucose-mediated phosphorylation converts the transcription factor Rgt1 from a repressor to an activator. J. Biol. Chem. 2003, 278, 10322–10327. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Johnston, M. Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J. Biol. Chem. 2006, 281, 26144–26149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Özcan, S.; Johnston, M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol. Cell. Biol. 1995, 15, 1564–1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, H.; Gaber, R.F. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol. Biol. Cell. 1996, 7, 1953–1966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Özcan, S.; Dover, J.; Rosenwald, A.G.; Wolfl, S.; Johnston, M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. USA 1996, 93, 12428–12432. [Google Scholar] [CrossRef] [Green Version]
- Reifenberger, E.; Boles, E.; Ciriacy, M. Kinetic characterization of individual hexose transporters of Saccharomyces cerevisiae and their relation to the triggering mechanisms of glucose repression. Eur. J. Biochem. 1997, 245, 324–333. [Google Scholar] [CrossRef]
- Hamacher, T.; Becker, J.; Gardonyi, M.; H. Hahn-Hägerdal, B.; Boles, E. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiol. SGM 2002, 148, 2783–2788. [Google Scholar] [CrossRef]
- Verwaal, R.; Paalman, J.W.G.; Hogenkamp, A.; Verkleij, A.J.; Verrips, C.T.; Boonstra, J. HXT5 expression is determined by growth rates in Saccharomyces cerevisiae. Yeast 2002, 19, 1029–1038. [Google Scholar] [CrossRef] [Green Version]
- Lutfiyya, L.L.; Iyer, V.R.; DeRisi, J.; DeVit, M.J.; Brown, P.O.; Johnston, M. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics 1998, 150, 1377–1391. [Google Scholar] [CrossRef]
- Kaniak, A.; Xue, Z.; Macool, D.; Kim, J.H.; Johnston, M. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot. Cell 2004, 3, 221–231. [Google Scholar] [CrossRef] [Green Version]
- Diderich, J.A.; Raamsdonk, L.M.; Kruckeberg, A.L.; Berden, J.A.; Van Dam, K. Physiological properties of Saccharomyces cerevisiae from which hexokinase II has been deleted. Appl. Environ. Microbiol. 2001, 67, 1587–1593. [Google Scholar] [CrossRef] [Green Version]
- Rose, M.; Albig, W.; Entian, K.D. Glucose repression in Saccharomyces cerevisiae is directly associated with hexose phosphorylation by hexokinases PI and PII. Eur J. Biochem. 1991, 199, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Bloom, L.M.; Walsh, C.T.; Botstein, D. The Residual Enzymatic Phosphorylation Activity of Hexokinase-Ii Mutants Is Correlated with Glucose Repression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1989, 9, 5643–5649. [Google Scholar]
- Fernandez-Garcia, P.; Pelaez, R.; Herrero, P.; Moreno, F. Phosphorylation of Yeast Hexokinase 2 Regulates Its Nucleocytoplasmic Shuttling. J. Biol. Chem. 2012, 287, 42151–42164. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Jiang, R.; Carlson, M. A family of proteins containing a conserved domain that mediates interaction with the yeast SNF1 protein kinase complex. EMBO J. 1994, 13, 5878–5886. [Google Scholar] [CrossRef]
- Erickson, J.R.; Johnston, M. Genetic and molecular characterization of GAL83: Its interaction and similarities with other genes involved in glucose repression in Saccharomyces cerevisiae. Genetics 1993, 135, 655–664. [Google Scholar] [CrossRef]
- Celenza, J.L.; Carlson, M. Mutational analysis of the Saccharomyces cerevisiae Snf1 protein kinase and evidence for functional interaction with the Snf4 protein. Mol. Cell. Biol. 1989, 9, 5034–5044. [Google Scholar] [PubMed] [Green Version]
- Jiang, R.; Carlson, M. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 1996, 10, 3105–3115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincent, O.; Townley, R.; Kuchin, S.; Carlson, M. Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev. 2001, 15, 1104–1114. [Google Scholar] [CrossRef] [Green Version]
- Sanz, P.; Alms, G.R.; Haystead, T.A.J.; Carlson, M. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 2000, 20, 1321–1328. [Google Scholar] [CrossRef] [Green Version]
- Bu, Y.; Schmidt, M.C. Identification of cis-acting elements in the SUC2 promoter of Saccharomyces cerevisiae required for activation of transcription. Nucleic Acids Res. 1998, 26, 1002–1009. [Google Scholar] [CrossRef] [Green Version]
- Braun, K.A.; Parua, P.K.; Dombek, K.M.; Miner, G.E.; Young, E.T. 14-3-3 (Bmh) proteins regulate combinatorial transcription following RNA polymerase II recruitment by binding at Adr1-dependent promoters in Saccharomyces cerevisiae. Mol. Cell. Biol. 2013, 33, 712–724. [Google Scholar] [CrossRef] [Green Version]
- Bloecher, A.; Tatchell, K. Defects in Saccharomyces cerevisiae protein phosphatase type I activate the spindle/kinetochore checkpoint. Genes Dev. 1999, 13, 517–522. [Google Scholar] [CrossRef] [Green Version]
- Chandrashekarappa, D.G.; McCartney, R.R.; Schmidt, M.C. Subunit and Domain Requirements for Adenylate-mediated Protection of Snf1 Kinase Activation Loop from Dephosphorylation. J. Biol. Chem. 2011, 286, 44532–44541. [Google Scholar] [CrossRef] [Green Version]
- Rubio-Texeira, M.; Van Zeebroeck, G.; Voordeckers, K.; Thevelein, J.M. Saccharomyces cerevisiae plasma membrane nutrient sensors and their role in PKA signaling. FEMS Yeast Res. 2010, 10, 134–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraakman, L.; Lemaire, K.; Ma, P.; Teunissen, A.W.; Donaton, M.C.; Van Dijck, P.; Winderickx, J.; de Winde, J.H.; Thevelein, J.M. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 1999, 32, 1002–1012. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.; Struhl, K. Protein-Kinase-a Mediates Growth-Regulated Expression of Yeast Ribosomal-Protein Genes by Modulating Rap1 Transcriptional Activity. Mol. Cell. Biol. 1994, 14, 1920–1928. [Google Scholar]
- Yin, Z.K.; Wilson, S.; Hauser, N.C.; Tournu, H.; Hoheisel, J.D.; Brown, A.J.P. Glucose triggers different global responses in yeast, depending on the strength of the signal, and transiently stabilizes ribosomal protein mRNAs. Mol. Microbiol. 2003, 48, 713–724. [Google Scholar] [CrossRef] [Green Version]
- Robertson, L.S.; Causton, H.C.; Young, R.A.; Fink, G.R. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc. Natl. Acad. Sci. USA 2000, 97, 5984–5988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zurita-Martinez, S.A.; Cardenas, M.E. Tor and cyclic AMP-protein kinase A: Two parallel pathways regulating expression of genes required for cell growth. Eukaryot. Cell 2005, 4, 63–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belinchon, M.M.; Gancedo, J.M. Glucose controls multiple processes in Saccharomyces cerevisiae through diverse combinations of signaling pathways. FEMS Yeast Res. 2007, 7, 808–818. [Google Scholar] [CrossRef] [Green Version]
- Francois, J.; Vanschaftingen, E.; Hers, H.G. The Mechanism by Which Glucose Increases Fructose 2,6-Bisphosphate Concentration in Saccharomyces cerevisiae—A Cyclic-AMP-Dependent Activation of Phosphofructokinase 2. Eur. J. Biochem. 1984, 145, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Oritz, C.H.; Maia, J.C.C.; Tenan, M.N.; Brazpadrao, G.R.; Mattoon, J.R.; Panek, A.D. Regulation of Yeast Trehalase by a Monocyclic, Cyclic AMP-Dependent Phosphorylation-Dephosphorylation Cascade System. J. Bacteriol. 1983, 153, 644–651. [Google Scholar] [CrossRef] [Green Version]
- Castermans, D.; Somers, I.; Kriel, J.; Louwet, W.; Wera, S.; Versele, M.; Janssens, V.; Thevelein, J.M. Glucose-induced posttranslational activation of protein phosphatases PP2A and PP1 in yeast. Cell Res. 2012, 22, 1058–1077. [Google Scholar] [CrossRef] [Green Version]
- Gancedo, J.M.; Mazón, M.J.; Gancedo, C. Inactivation and phosphorylation of yeast fructose 1, 6-bisphosphatase. Biochem. Soc. Trans. 1982, 10, 326–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ordiz, I.; Herrero, P.; Rodicio, R.; Moreno, F. Glucose-induced inactivation of isocitrate lyase in Saccharomyces cerevisiae is mediated by the cAMP-dependent protein kinase catalytic subunits Tpk1 and Tpk2. FEBS Lett. 1996, 385, 43–46. [Google Scholar] [CrossRef] [Green Version]
- Pohlig, G.; Holzer, H. Phosphorylation and Inactivation of Yeast Fructose-1,6-Bisphosphatase by Cyclic AMP-Dependent Protein-Kinase from Yeast. J. Biol. Chem. 1985, 260, 3818–3823. [Google Scholar] [CrossRef]
- Smith, A.; Ward, M.P.; Garrett, S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998, 17, 3556–3564. [Google Scholar] [CrossRef] [Green Version]
- Reinders, A.; Burckert, N.; Boller, T.; Wiemken, A.; De Virgilio, C. Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Gene Dev. 1998, 12, 2943–2955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferguson, S.B.; Anderson, E.S.; Harshaw, R.B.; Thate, T.; Craig, N.L.; Nelson, H.C.M. Protein kinase A regulates constitutive expression of small heat-shock genes in an Msn2/4p-independent and Hsf1p-dependent manner in Saccharomyces cerevisiae. Genetics 2005, 169, 1203–1214. [Google Scholar] [CrossRef] [Green Version]
- Apweiler, E.; Sameith, K.; Margaritis, T.; Brabers, N.; van de Pasch, L.; Bakker, L.V.; van Leenen, D.; Holstege, F.C.P.; Kemmeren, P. Yeast glucose pathways converge on the transcriptional regulation of trehalose biosynthesis. BMC Genom. 2012, 13, 239. [Google Scholar] [CrossRef] [Green Version]
- Trevisol, E.T.V.; Panek, A.D.; De Mesquita, J.F.; Eleutherio, E.C.A. Regulation of the yeast trehalose-synthase complex by cyclic AMP-dependent phosphorylation. BBA-Gen. Subj. 2014, 1840, 1646–1650. [Google Scholar] [CrossRef]
- Gancedo, J.M.; Flores, C.L.; Gancedo, C. The repressor Rgt1 and the cAMP-dependent protein kinases control the expression of the SUC2 gene in Saccharomyces cerevisiae. BBA Gen. Subj. 2015, 1850, 1362–1367. [Google Scholar] [CrossRef] [Green Version]
- Nicastro, R.; Tripodi, F.; Gaggini, M.; Castoldi, A.; Reghellin, V.; Nonnis, S.; Tedeschi, G.; Coccetti, P. Snf1 Phosphorylates Adenylate Cyclase and Negatively Regulates Protein Kinase A-dependent Transcription in Saccharomyces cerevisiae. J. Biol. Chem. 2015, 290, 24715–24726. [Google Scholar] [CrossRef] [Green Version]
- Bos, J.L.; Rehmann, H.; Wittinghofer, A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell 2007, 130, 385. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Batlle, M.; Hirsch, J.P. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p G(alpha) subunit and functions in a Ras-independent pathway. EMBO J. 1998, 17, 1996–2007. [Google Scholar] [CrossRef]
- Peeters, T.; Louwet, W.; Geladé, R.; Nauwelaers, D.; Thevelein, J.M.; Versele, M. Kelch-repeat proteins interacting with the Gα protein Gpa2 bypass adenylate cyclase for direct regulation of protein kinase A in yeast. Proc. Natl. Acad. Sci. USA 2006, 103, 13034–13039. [Google Scholar] [CrossRef] [Green Version]
- Shima, F.; Okada, T.; Kido, M.; Sen, H.; Tanaka, Y.; Tamada, M.; Hu, C.D.; Yamawaki-Kataoka, Y.; Kariya, K.; Kataoka, T. Association of yeast adenylyl cyclase with cyclase-associated protein CAP forms a second Ras-binding site which mediates its Ras-dependent activation. Mol. Cell. Biol. 2000, 20, 26–33. [Google Scholar] [CrossRef] [Green Version]
- Colombo, S.; Ma, P.; Cauwenberg, L.; Winderickx, J.; Crauwels, M.; Teunissen, A.; Nauwelaers, D.; de Winde, J.H.; Gorwa, M.F.; Colavizza, D.; et al. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 1998, 17, 3326–3341. [Google Scholar] [CrossRef] [Green Version]
- Broggi, S.; Martegani, E.; Colombo, S. Live-cell imaging of endogenous Ras-GTP shows predominant Ras activation at the plasma membrane and in the nucleus in Saccharomyces cerevisiae. Int. J. Biochem. Cell Biol. 2013, 45, 384–394. [Google Scholar] [CrossRef] [PubMed]
- Colombo, S.; Ronchetti, D.; Thevelein, J.M.; Winderickx, J.; Martegani, E. Activation state of the Ras2 protein and glucose-induced signaling in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 46715–46722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos, S.; Pardo, L.A.; Sanchez, L.M.; Lazo, P.S. Upstream regulation of Saccharomyces cerevisiae adenylate cyclase. Biochem. Soc. Trans. 1989, 17, 976–978. [Google Scholar] [CrossRef] [Green Version]
- Toda, T.; Cameron, S.; Sass, P.; Zoller, M.; Scott, J.D.; McMullen, B.; Hurwitz, M.; Krebs, E.G.; Wigler, M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1987, 7, 1371–1377. [Google Scholar]
- Hixson, C.S.; Krebs, E.G. Characterization of a cyclic AMP-binding protein from bakers’ yeast. Identification as a regulatory subunit of cyclic AMP-dependent protein kinase. J. Biol. Chem. 1980, 255, 2137–2145. [Google Scholar] [CrossRef]
- Solari, C.A.; Tudisca, V.; Pugliessi, M.; Nadra, A.D.; Moreno, S.; Portela, P. Regulation of PKA activity by an autophosphorylation mechanism in Saccharomyces cerevisiae. Biochem. J. 2014, 462, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Vanhalewyn, M.; Dumortier, F.; Debast, G.; Colombo, S.; Ma, P.; Winderickx, J.; van Dijck, P.; Thevelein, J.M. A mutation in Saccharomyces cerevisiae adenylate cyclase, Cyr1K1876M, specifically affects glucose- and acidification-induced cAMP signalling and not the basal cAMP level. Mol. Microbiol. 1999, 33, 363–376. [Google Scholar] [CrossRef] [Green Version]
- Nijkamp, J.F.; van den Broek, M.; Datema, E.; de Kok, S.; Bosman, L.; Luttik, M.A.; Daran-Lapujade, P.; Vongsangnak, W.; Nielsen, J.; Heijne, W.H.M.; et al. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb. Cell Fact. 2012, 11, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dihazi, H.; Kessler, R.; Eschrich, K. High osmolarity glycerol (HOG) pathway-induced phosphorylation and activation of 6-phosphofructo-2-kinase are essential for glycerol accumulation and yeast cell proliferation under hyperosmotic stress. J. Biol. Chem. 2004, 279, 23961–23968. [Google Scholar] [CrossRef] [Green Version]
- Inoue, Y.; Tsujimoto, Y.; Kimura, A. Expression of the glyoxalase I gene of Saccharomyces cerevisiae is regulated by high osmolarity glycerol mitogen-activated protein kinase pathway in osmotic stress response. J. Biol. Chem. 1998, 273, 2977–2983. [Google Scholar] [CrossRef] [Green Version]
- Mösch, H.-U.; Roberts, R.L.; Fink, G.R. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1996, 93, 5352–5356. [Google Scholar] [CrossRef] [Green Version]
- Posas, F.; WurglerMurphy, S.M.; Maeda, T.; Witten, E.A.; Thai, T.C.; Saito, H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 1996, 86, 865–875. [Google Scholar] [CrossRef] [Green Version]
- Maeda, T.; Takekawa, M.; Saito, H. Activation of Yeast Pbs2 Mapkk by Mapkkks or by Binding of an Sh3-Containing Osmosensor. Science 1995, 269, 554–558. [Google Scholar] [CrossRef] [PubMed]
- Tatebayashi, K.; Tanaka, K.; Yang, H.Y.; Yamamoto, K.; Matsushita, Y.; Tomida, T.; Imai, M.; Saito, H. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J. 2007, 26, 3521–3533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirayarna, T.; Maeda, T.; Saito, H.; Shinozaki, K. Cloning and characterization of seven cDNAs for hyperosmolarity-responsive (HOR) genes of Saccharomyces cerevisiae. Mol. Gen. Genet. 1995, 249, 127–138. [Google Scholar] [CrossRef]
- Greatrix, B.W.; van Vuuren, H.J.J. Expression of the HXT13, HXT15 and HXT17 genes in Saccharomyces cerevisiae and stabilization of the HXT1 gene transcript by sugar-induced osmotic stress. Curr. Genet. 2006, 49, 205–217. [Google Scholar] [CrossRef]
- Tomas-Cobos, L.; Casadome, L.; Mas, G.; Sanz, P.; Posas, F. Expression of the HXT1 low affinity glucose transporter requires the coordinated activities of the HOG and glucose signalling pathways. J. Biol. Chem. 2004, 279, 22010–22019. [Google Scholar] [CrossRef] [Green Version]
- Cullen, P.J.; Sprague, G.F. The Regulation of Filamentous Growth in Yeast. Genetics 2012, 190, 23–49. [Google Scholar] [CrossRef]
- Klipp, E.; Schaber, J. Modelling of signal transduction in yeast–sensitivity and model analysis. In Understanding and Exploiting Systems Biology in Bioprocesses and Biomedicine; Fundación Cajamurcia: Murcia, Spain, 2006; pp. 15–30. [Google Scholar]
- Kumar, A. The Complex Genetic Basis and Multilayered Regulatory Control of Yeast Pseudohyphal Growth. Annu. Rev. Genet. 2021, 55, 1. [Google Scholar] [CrossRef]
- Pitoniak, A.; Birkaya, B.; Dionne, H.M.; Vadaie, N.; Cullen, P.J. The Signaling Mucins Msb2 and Hkr1 Differentially Regulate the Filamentation Mitogen-activated Protein Kinase Pathway and Contribute to a Multimodal Response. Mol. Biol. Cell. 2009, 20, 3101–3114. [Google Scholar] [CrossRef] [Green Version]
- Saito, H. Regulation of cross-talk in yeast MAPK signaling pathways. Curr. Opin. Microbiol. 2010, 13, 677–683. [Google Scholar] [CrossRef]
- Pan, X.W.; Heitman, J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999, 19, 4874–4887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuchin, S.; Vyas, V.K.; Carlson, M. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 2002, 22, 3994–4000. [Google Scholar] [CrossRef] [Green Version]
- Orlova, M.; Ozcetin, H.; Barrett, L.; Kuchin, S. Roles of the Snf1-Activating Kinases during Nitrogen Limitation and Pseudohyphal Differentiation in Saccharomyces cerevisiae. Eukaryot. Cell 2010, 9, 208–214. [Google Scholar] [CrossRef] [Green Version]
- O’Rourke, S.M.; Herskowitz, I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Gene Dev. 1998, 12, 2874–2886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, Y.; Nomura, W. TOR Signaling in Budding Yeast. In The Yeast Role in Medical Applications; Abdulkhair, W.M.H., Ed.; IntechOpen: London, UK, 2018; Volume 55. [Google Scholar]
- Stracka, D.; Jozefczuk, S.; Rudroff, F.; Sauer, U.; Hall, M.N. Nitrogen Source Activates TOR (Target of Rapamycin) Complex 1 via Glutamine and Independently of Gtr/Rag Proteins. J. Biol. Chem. 2014, 289, 25010–25020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.P.; Du, G.C.; Zhou, J.W.; Chen, J. Regulation of Sensing, Transportation, and Catabolism of Nitrogen Sources in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2018, 82, e00040-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duvel, K.; Santhanam, A.; Garrett, S.; Schneper, L.; Broach, J.R. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell 2003, 11, 1467–1478. [Google Scholar] [CrossRef]
- Hallett Hallet, J.E.; Luo, X.X.; Capaldi, A.P. State Transitions in the TORC1 Signaling Pathway and Information Processing in Saccharomyces cerevisiae. Genetics 2014, 198, 773–786. [Google Scholar] [CrossRef] [Green Version]
- Tate, J.J.; Cooper, T.G. Five conditions commonly used to down-regulate tor complex 1 generate different physiological situations exhibiting distinct requirements and outcomes. J. Biol. Chem. 2013, 288, 27243–27262. [Google Scholar] [CrossRef] [Green Version]
- Conway, M.K.; Grunwald, D.; Heideman, W. Glucose, Nitrogen, and Phosphate Repletion in Saccharomyces cerevisiae: Common Transcriptional Responses to Different Nutrient Signals. G3 Genes Genom. Genet. 2012, 2, 1003–1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lippman, S.I.; Broach, J.R. Protein kinase A and TORC1 activate genes for ribosomal biogenesis by inactivating repressors encoded by Dot6 and its homolog Tod6. Proc. Natl. Acad. Sci. USA 2009, 106, 19928–19933. [Google Scholar] [CrossRef] [Green Version]
- Alfatah, M.; Wong, J.H.; Krishnan, V.G.; Lee, Y.C.; Sin, Q.F.; Goh, C.J.H.; Kong, K.W.; Lee, W.T.; Lewis, J.; Hoon, S.; et al. TORC1 regulates the transcriptional response to glucose and developmental cycle via the Tap42-Sit4-Rrd1/2 pathway in Saccharomyces cerevisiae. BMC Biol. 2021, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, K.; Kayikci, O.; Schaeffer, D.G.; Magwene, P.M. Modeling mutant phenotypes and oScillatory dynamics in the Saccharomyces cerevisiae cAMP-PKA pathway. BMC Syst. Biol. 2013, 7, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallett Hallet, J.E.; Luo, X.X.; Capaldi, A.P. Snf1/AMPK promotes the formation of Kog1/Raptor-bodies to increase the activation threshold of TORC1 in budding yeast. Elife 2015, 4, e09181. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.L.; Shen, Y.B.; Gao, W.X.; Dong, J. Role of Sch9 in regulating Ras-cAMP signal pathway in Saccharomyces cerevisiae. FEBS Lett. 2011, 585, 3026–3032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilms, T.; Swinnen, E.; Eskes, E.; Dolz-Edo, L.; Uwineza, A.; Van Essche, R.; Rosseels, J.; Zabrocki, P.; Cameroni, E.; Franssens, V.; et al. The yeast protein kinase Sch9 adjusts V-ATPase assembly/disassembly to control pH homeostasis and longevity in response to glucose availability. PLoS Genet. 2017, 13, e1006835. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.Y.; Lin, Y.Y.; Sheu, J.C.; Wu, J.T.; Lee, F.J.; Chen, Y.; Lin, M.I.; Chiang, F.T.; Tai, T.Y.; Berger, S.L.; et al. Acetylation of Yeast AMPK Controls Intrinsic Aging Independently of Caloric Restriction. Cell 2011, 146, 968–978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostergaard, S.; Olsson, L.; Johnston, M.; Nielsen, J. Increasing galactose consumption by Saccharomyces cerevisiae through metabolic engineering of the GAL gene regulatory network. Nat. Biotechnol. 2000, 18, 1283–1286. [Google Scholar] [CrossRef]
- Bhat, P.J.; Oh, D.; Hopper, J.E. Analysis of the Gal3 Signal Transduction Pathway Activating Gal4 Protein-Dependent Transcription in Saccharomyces cerevisiae. Genetics 1990, 125, 281–291. [Google Scholar] [CrossRef]
- Winge, Ø.; Roberts, C. Inheritance of Enzymatic Characters in Yeasts, and the Phenomenon of Long-Term Adaptation; Hagerup in Komm.: Copenhagen, Denmark, 1948. [Google Scholar]
- Platt, A.; Reece, R.J. The yeast galactose genetic switch is mediated by the formation of a Gal4p-Gal80p-Gal3p complex. EMBO J. 1998, 17, 4086–4091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.Q.; Moffat, J.; Wilson, W.A.; Moore, L.; Cheng, C.; Roach, P.J.; Andrews, B. Cyclin partners determine Pho85 protein kinase substrate specificity in vitro and in vivo: Control of glycogen biosynthesis by Pcl8 and Pcl10. Mol. Cell. Biol. 1998, 18, 3289–3299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jund, R.; Weber, E.; Chevallier, M.R. Primary Structure of the Uracil Transport Protein of Saccharomyces cerevisiae. Eur. J. Biochem. 1988, 171, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Robert, F.; Wyrick, J.J.; Aparicio, O.; Jennings, E.G.; Simon, I.; Zeitlinger, J.; Schreiber, J.; Hannett, N.; Kanin, E.; et al. Genome-wide location and function of DNA binding proteins. Science 2000, 290, 2306–2309. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.M.; Johnston, S.A. Are all DNA binding and transcription regulation by an activator physiologically relevant? Mol. Cell. Biol. 2001, 21, 2467–2474. [Google Scholar] [CrossRef] [Green Version]
- Schneider, J.C.; Guarente, L. Vectors for Expression of Cloned Genes in Yeast—Regulation, Overproduction, and Underproduction. Methods Enzymol. 1991, 194, 373–388. [Google Scholar]
- Mumberg, D.; Muller, R.; Funk, M. Regulatable Promoters of Saccharomyces cerevisiae—Comparison of Transcriptional Activity and Their Use for Heterologous Expression. Nucleic Acids Res. 1994, 22, 5767–5768. [Google Scholar] [CrossRef]
- Polish, J.A.; Kim, J.-H.; Johnston, M. How the Rgt1 transcription factor of Saccharomyces cerevisiae is regulated by glucose. Genetics 2005, 169, 583–594. [Google Scholar] [CrossRef] [Green Version]
- Shashkova, S.; Welkenhuysen, N.; Hohmann, S. Molecular communication: Crosstalk between the Snf1 and other signaling pathways. FEMS Yeast Res. 2015, 15, fov026. [Google Scholar] [CrossRef] [Green Version]
- Görner, W.; Durchschlag, E.; Wolf, J.; Brown, E.L.; Ammerer, G.; Ruis, H.; Schuller, C. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 2002, 21, 135–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Wever, V.; Reiter, W.; Ballarini, A.; Ammerer, G.; Brocard, C. A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation. EMBO J. 2005, 24, 4115–4123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coccetti, P.; Nicastro, R.; Tripodi, F. Conventional and emerging roles of the energy sensor Snf1/AMPK in Saccharomyces cerevisiae. Microbiol. Cell 2018, 5, 482–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avonce, N.; Leyman, B.; Mascorro-Gallardo, J.O.; Van Dijck, P.; Thevelein, J.M.; Iturriaga, G. The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol. 2004, 136, 3649–3659. [Google Scholar] [CrossRef] [Green Version]
- Fichtner, F.; Lunn, J.E. The Role of Trehalose 6-Phosphate (Tre6P) in Plant Metabolism and Development. Annu. Rev. Plant Biol. 2021, 72, 737–760. [Google Scholar] [CrossRef]
- Deroover, S.; Ghillebert, R.; Broeckx, T.; Winderickx, J.; Rolland, F. Trehalose-6-phosphate synthesis controls yeast gluconeogenesis downstream and independent of SNF1. FEMS Yeast Res. 2016, 16, fow036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peeters, K.; Van Leemputte, F.; Fischer, B.; Bonini, B.M.; Quezada, H.; Tsytlonok, M.; Haesen, D.; Vanthienen, W.; Bernardes, N.; Gonzalez-Blas, C.B.; et al. Fructose-1,6-bisphosphate couples glycolytic flux to activation of Ras. Nat. Commun. 2017, 8, 1–5. [Google Scholar] [CrossRef]
- van Heerden, J.H.; Wortel, M.T.; Bruggeman, F.J.; Heijnen, J.J.; Bollen, Y.J.M.; Planque, R.; Hulshof, J.; O’Toole, T.G.; Wahl, S.A.; Teusink, B. Lost in Transition: Start-Up of Glycolysis Yields Subpopulations of Nongrowing Cells. Science 2014, 343, 1245114. [Google Scholar] [CrossRef]
- Runquist, D.; Hahn-Hägerdal, B.; Bettiga, M. Increased expression of the oxidative pentose phosphate pathway and gluconeogenesis in anaerobically growing xylose-utilizing Saccharomyces cerevisiae. Microb. Cell Fact. 2009, 8, 49. [Google Scholar] [CrossRef] [Green Version]
- Bergdahl, B.; Heer, D.; Sauer, U.; Hahn-Hägerdal, B.; van Niel, E.W.J. Dynamic metabolomics differentiates between carbon and energy starvation in recombinant Saccharomyces cerevisiae fermenting xylose. Biotechnol. Biofuels 2012, 5, 34. [Google Scholar] [CrossRef] [Green Version]
- Alff-Tuomala, S.; Salusjärvi, L.; Barth, D.; Oja, M.; Penttilä, M.; Pitkanen, J.P.; Ruohonen, L.; Jouhten, P. Xylose-induced dynamic effects on metabolism and gene expression in engineered Saccharomyces cerevisiae in anaerobic glucose-xylose cultures. Appl. Microbiol. Biotechnol. 2016, 100, 969–985. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.Y.; Tang, Y.Q.; Gou, M.; Xia, Z.Y.; Kida, K. Transcriptomes of a xylose-utilizing industrial flocculating Saccharomyces cerevisiae strain cultured in media containing different sugar sources. AMB Express 2016, 6, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atkinson, D.E. Energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry-US 1968, 7, 4030–4034. [Google Scholar] [CrossRef]
- Belinchon, M.M.; Gancedo, J.M. Xylose and some non-sugar carbon sources cause catabolite repression in Saccharomyces cerevisiae. Arch. Microbiol. 2003, 180, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Wasylenko, T.M.; Stephanopoulos, G. Metabolomic and 13C-metabolic flux analysis of a xylose-consuming Saccharomyces cerevisiae strain expressing xylose isomerase. Biotechnol. Bioeng. 2015, 112, 470–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietvorst, J.; Karhumaa, K.; Kielland-Brandt, M.C.; Brandt, A. Amino acid residues involved in ligand preference of the Snf3 transporter-like sensor in Saccharomyces cerevisiae. Yeast 2010, 27, 131–138. [Google Scholar] [CrossRef]
- Brink, D.P.; Borgström, C.; Tueros, F.G.; Gorwa-Grauslund, M.F. Real-time monitoring of the sugar sensing in Saccharomyces cerevisiae indicates endogenous mechanisms for xylose signaling. Microbiol. Cell Fact. 2016, 15, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Li, H.; Wei, S.; Wu, H.; Wu, X.; Bao, X.; Hou, J.; Liu, W.; Shen, Y. Simulating extracellular glucose signals enhances xylose metabolism in recombinant Saccharomyces cerevisiae. Microorganisms 2020, 8, 100. [Google Scholar] [CrossRef] [Green Version]
- Özcan, S.; Vallier, L.G.; Flick, J.S.; Carlson, M.; Johnston, M. Expression of the SUC2 gene of Saccharomyces cerevisiae is induced by low levels of glucose. Yeast 1997, 13, 127–137. [Google Scholar] [CrossRef]
- Winderickx, J.; deWinde, J.H.; Crauwels, M.; Hino, A.; Hohmann, S.; VanDijck, P.; Thevelein, J.M. Regulation of genes encoding subunits of the trehalose synthase complex in Saccharomyces cerevisiae: Novel variations of STRE-mediated transcription control? Mol. Gen. Genet. 1996, 252, 470–482. [Google Scholar]
- Parrou, J.L.; Enjalbert, B.; Plourde, L.; Bauche, A.; Gonzalez, B.; Francois, J. Dynamic responses of reserve carbohydrate metabolism under carbon and nitrogen limitations in Saccharomyces cerevisiae. Yeast 1999, 15, 191–203. [Google Scholar] [CrossRef]
- Fernandez, R.; Herrero, P.; Fernandez, M.T.; Moreno, F. Mechanism of Inactivation of Hexokinase Pii of Saccharomyces Cerevisiae by D-Xylose. Microbiology 1986, 132, 3467–3472. [Google Scholar] [CrossRef] [Green Version]
- Heidrich, K.; Otto, A.; Behlke, J.; Rush, J.; Wenzel, K.W.; Kriegel, T. Autophosphorylation-inactivation site of hexokinase 2 in Saccharomyces cerevisiae. Biochemistry-US 1997, 36, 1960–1964. [Google Scholar] [CrossRef]
- Kuser, P.R.; Krauchenco, S.; Antunes, O.A.; Polikarpov, I. The high resolution crystal structure of yeast hexokinase PII with the correct primary sequence provides new insights into its mechanism of action. J. Biol. Chem. 2000, 275, 20814–20821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neigeborn, L.; Carlson, M. Genes Affecting the Regulation of Suc2 Gene-Expression by Glucose Repression in Saccharomyces cerevisiae. Genetics 1984, 108, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Trumbly, R.J. Glucose Repression in the Yeast Saccharomyces cerevisiae. Mol. Microbiol. 1992, 6, 15–21. [Google Scholar] [CrossRef]
- Lutfiyya, L.L.; Johnston, M. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol. Cell. Biol. 1996, 16, 4790–4797. [Google Scholar] [CrossRef] [Green Version]
- Meijer, M.M.C.; Boonstra, J.; Verkleij, A.J.; Verrips, C.T. Glucose repression in Saccharomyces cerevisiae is related to the glucose concentration rather than the glucose flux. J. Biol. Chem. 1998, 273, 24102–24107. [Google Scholar] [CrossRef] [Green Version]
- Carlson, M.; Taussig, R.; Kustu, S.; Botstein, D. The secreted form of invertase in Saccharomyces cerevisiae is synthesized from mRNA encoding a signal sequence. Mol. Cell. Biol. 1983, 3, 439–447. [Google Scholar]
- Myers, K.S.; Riley, N.M.; MacGilvray, M.E.; Sato, T.K.; McGee, M.; Heilberger, J.; Coon, J.J.; Gasch, A.P. Rewired cellular signaling coordinates sugar and hypoxic responses for anaerobic xylose fermentation in yeast. PLoS Genet. 2019, 15, e1008037. [Google Scholar] [CrossRef]
- Roca, C.; Haack, M.B.; Olsson, L. Engineering of carbon catabolite repression in recombinant xylose fermenting Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2004, 63, 578–583. [Google Scholar] [CrossRef]
- Rolland, F.; de Winde, J.H.; Lemaire, K.; Boles, E.; Thevelein, J.M.; Winderickx, J. Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol. Microbiol. 2000, 38, 348–358. [Google Scholar] [CrossRef]
- Matsushika, A.; Nagashima, A.; Goshima, T.; Hoshino, T. Fermentation of Xylose Causes Inefficient Metabolic State Due to Carbon/Energy Starvation and Reduced Glycolytic Flux in Recombinant Industrial Saccharomyces cerevisiae. PLoS ONE 2013, 8, e69005. [Google Scholar]
- Pitkanen, J.P.; Aristidou, A.; Salusjärvi, L.; Ruohonen, L.; Penttilä, M. Metabolic flux analysis of xylose metabolism in recombinant Saccharomyces cerevisiae using continuous culture. Metab. Eng. 2003, 5, 16–31. [Google Scholar] [CrossRef] [Green Version]
- Harashima, T.; Anderson, S.; Yates III, J.R.; Heitman, J. The kelch proteins Gpb1 and Gpb2 inhibit Ras activity via association with the yeast RasGAP neurofibromin homologs Ira1 and Ira2. Mol. Cell 2006, 22, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Klimacek, M.; Krahulec, S.; Sauer, U.; Nidetzky, B. Limitations in Xylose-Fermenting Saccharomyces cerevisiae, Made Evident through Comprehensive Metabolite Profiling and Thermodynamic Analysis. Appl. Environ. Microbiol. 2010, 76, 7566–7574. [Google Scholar] [CrossRef] [Green Version]
- Babazadeh, R.; Lahtvee, P.J.; Adiels, C.B.; Goksor, M.; Nielsen, J.B.; Hohmann, S. The yeast osmostress response is carbon source dependent. Sci. Rep. UK 2017, 7, 990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ando, A.; Tanaka, F.; Murata, Y.; Takagi, H.; Shima, J. Identification and classification of genes required for tolerance to high-sucrose stress revealed by genome-wide screening of Saccharomyces cerevisiae. FEMS Yeast Res. 2006, 6, 249–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karhumaa, K.; Hahn-Hägerdal, B.; Gorwa-Grauslund, M.F. Investigation of limiting metabolic steps in the utilization of xylose by recombinant Saccharomyces cerevisiae using metabolic engineering. Yeast 2005, 22, 359–368. [Google Scholar] [CrossRef]
- Matsushika, A.; Inoue, H.; Murakami, K.; Takimura, O.; Sawayama, S. Bioethanol production performance of five recombinant strains of laboratory and industrial xylose-fermenting Saccharomyces cerevisiae. Bioresour. Technol. 2009, 100, 2392–2398. [Google Scholar] [CrossRef]
- Träff, K.L.; Cordero, R.R.O.; van Zyl, W.H.; Hahn-Hägerdal, B. Deletion of the GRE3 aldose reductase gene and its influence on xylose metabolism in recombinant strains of Saccharomyces cerevisiae expressing the xylA and XKS1 genes. Appl. Environ. Microbiol. 2001, 67, 5668–5674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguilera, J.; Prieto, J.A. The Saccharomyces cerevisiae aldose, reductase is implied in the metabolism of methylglyoxal in response to stress conditions. Curr. Genet. 2001, 39, 273–283. [Google Scholar] [CrossRef]
- dos Santos, L.V.; Carazzolle, M.F.; Nagamatsu, S.T.; Sampaio, N.M.V.; Almeida, L.D.; Pirolla, R.A.S.; Borelli, G.; Correa, T.L.R.; Argueso, J.L.; Pereira, G.A.G. Unraveling the genetic basis of xylose consumption in engineered Saccharomyces cerevisiae strains. Sci. Rep. UK 2016, 6, 38676. [Google Scholar] [CrossRef]
- Sato, T.K.; Tremaine, M.; Parreiras, L.S.; Hebert, A.S.; Myers, K.S.; Higbee, A.J.; Sardi, M.; McIlwain, S.J.; Ong, I.M.; Breuer, R.J.; et al. Directed Evolution Reveals Unexpected Epistatic Interactions That Alter Metabolic Regulation and Enable Anaerobic Xylose Use by Saccharomyces cerevisiae. PLoS Genet. 2016, 12, e1006447. [Google Scholar]
- Wagner, E.R.; Myers, K.S.; Riley, N.M.; Coon, J.J.; Gasch, A.P. PKA and HOG signaling contribute separable roles to anaerobic xylose fermentation in yeast engineered for biofuel production. PLoS ONE 2019, 14, e0212389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, L.Y.; Zhao, X.Q.; Ge, X.M.; Bai, F.W. Identification and functional study of a new FLO10-derivative gene from the industrial flocculating yeast SPSC01. J. Ind Microbiol. Biotechnol. 2012, 39, 1135–1140. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Jiao, C.L.; Peng, B.; Shen, Y.; Bao, X.M. Mutation of a regulator Ask10p improves xylose isomerase activity through up-regulation of molecular chaperones in Saccharomyces cerevisiae. Metab. Eng. 2016, 38, 241–250. [Google Scholar] [CrossRef]
- Sedlak, M.; Ho, N.W.Y. Characterization of the effectiveness of hexose transporters for transporting xylose during glucose and xylose co-fermentation by a recombinant Saccharomyces yeast. Yeast 2004, 21, 671–684. [Google Scholar] [CrossRef] [Green Version]
- Tani, T.; Taguchi, H.; Fujimori, K.E.; Sahara, T.; Ohgiya, S.; Kamagata, Y.; Akamatsu, T. Isolation and characterization of xylitol-assimilating mutants of recombinant Saccharomyces cerevisiae. J. BioSci. Bioeng. 2016, 122, 446–455. [Google Scholar] [CrossRef]
- Farwick, A.; Bruder, S.; Schadeweg, V.; Oreb, M.; Boles, E. Engineering of yeast hexose transporters to transport D-xylose without inhibition by D-glucose. Proc. Natl. Acad. Sci. USA 2014, 111, 5159–5164. [Google Scholar] [CrossRef] [Green Version]
- Reznicek, O.; Facey, S.J.; de Waal, P.P.; Teunissen, A.W.R.H.; de Bont, J.A.M.; Nijland, J.G.; Driessen, A.J.M.; Hauer, B. Improved xylose uptake in Saccharomyces cerevisiae due to directed evolution of galactose permease Gal2 for sugar co-consumption. J. Appl. Microbiol. 2015, 119, 99–111. [Google Scholar] [CrossRef]
- Sanchez, R.G.; Hahn-Hägerdal, B.; Gorwa-Grauslund, M.F. PGM2 overexpression improves anaerobic galactose fermentation in Saccharomyces cerevisiae. Microbiol. Cell Fact. 2010, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, R.G.; Hahn-Hägerdal, B.; Gorwa-Grauslund, M.F. Cross-reactions between engineered xylose and galactose pathways in recombinant Saccharomyces cerevisiae. Biotechnol. Biofuels 2010, 3, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gopinarayanan, V.E.; Nair, N.U. A semi-synthetic regulon enables rapid growth of yeast on xylose. Nat. Commun. 2018, 9, 1–12. [Google Scholar]
- Selim, K.A.; Easa, S.M.; El-Diwany, A.I. The Xylose Metabolizing Yeast Spathaspora passalidarum is a Promising Genetic Treasure for Improving Bioethanol Production. Fermentation 2020, 6, 33. [Google Scholar] [CrossRef] [Green Version]
- Passoth, V.; Zimmermann, M.; Klinner, U. Peculiarities of the regulation of fermentation and respiration in the crabtree-negative, xylose-fermenting yeast Pichia stipitis. Appl. Biochem. Biotech. 1996, 57–58, 201–212. [Google Scholar] [CrossRef]
- Shi, N.Q.; Jeffries, T.W. Anaerobic growth and improved fermentation of Pichia stipitis bearing a URA1 gene from Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 1998, 50, 339–345. [Google Scholar] [CrossRef] [PubMed]
- du Preez, J.C.; Bosch, M.; Prior, B.A. The Fermentation of Hexose and Pentose Sugars by Candida shehatae and Pichia stipitis. Appl. Microbiol. Biotechnol. 1986, 23, 228–233. [Google Scholar] [CrossRef]
- Van Brunt, J. Fermentation economics. Bio/Technology 1986, 4, 395–401. [Google Scholar] [CrossRef]
- Delgenès, J.P.; Moletta, R.; Navarro, J.M. The Ethanol Tolerance of Pichia stipitis Y-7124 Grown on a D-Xylose, D-Glucose and L-Arabinose Mixture. J. Ferment. Bioeng. 1988, 66, 417–422. [Google Scholar] [CrossRef]
- Delgenès, J.-P.; Moletta, R.; Navarro, J. Effects of lignocellulose degradation products on ethanol fermentations of glucose and xylose by Saccharomyces cerevisiae, Zymomonas mobilis, Pichia stipitis, and Candida shehatae. Enzym. Microbiol. Technol. 1996, 19, 220–225. [Google Scholar] [CrossRef]
- du Preez, J.C.; Bosch, M.; Prior, B.A. Temperature Profiles of Growth and Ethanol Tolerance of the Xylose-Fermenting Yeasts Candida shehatae and Pichia stipitis. Appl. Microbiol. Biotechnol. 1987, 25, 521–525. [Google Scholar] [CrossRef]
- Jeffries, T.W.; Van Vleet, J.R.H. Pichia stipitis genomics, transcriptomics, and gene clusters. FEMS Yeast Res. 2009, 9, 793–807. [Google Scholar] [CrossRef] [Green Version]
- Shi, N.Q.; Davis, B.; Sherman, F.; Cruz, J.; Jeffries, T.W. Disruption of the cytochrome c gene in xylose-utilizing yeast Pichia stipitis leads to higher ethanol production. Yeast 1999, 15, 1021–1030. [Google Scholar] [CrossRef]
- Yang, Z.M.; Bisson, L.F. The SKS1 protein kinase is a multicopy suppressor of the snf3 mutation of Saccharomyces cerevisiae. Yeast 1996, 12, 1407–1419. [Google Scholar] [CrossRef]
- Hou, X. Anaerobic xylose fermentation by Spathaspora passalidarum. Appl. Microbiol. Biotechnol. 2012, 94, 205–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, L.E.; Albuini, F.M.; Castro, A.G.; Campos, V.J.; de Souza, G.B.; Mendonça, J.G.; Rosa, C.A.; Mendes, T.A.; Santana, M.F.; da Silveira, W.B. Influence of glucose on xylose metabolization by Spathaspora passalidarum. Fungal Genet. Biol. 2021, 103624. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, J.E.; Long, C.P.; Antoniewicz, M.R. Comprehensive analysis of glucose and xylose metabolism in Escherichia coli under aerobic and anaerobic conditions by C-13 metabolic flux analysis. Metab. Eng. 2017, 39, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Fox, K.J.; Prather, K.L.J. Carbon catabolite repression relaxation in Escherichia coli: Global and sugar-specific methods for glucose and secondary sugar co-utilization. Curr. Opin. Chem. Eng. 2020, 30, 9–16. [Google Scholar] [CrossRef]
- Ni, L.; Tonthat, N.K.; Chinnam, N.; Schumacher, M.A. Structures of the Escherichia coli transcription activator and regulator of diauxie, XylR: An AraC DNA-binding family member with a LacI/GalR ligand-binding domain. Nucleic Acids Res. 2013, 41, 1998–2008. [Google Scholar] [CrossRef] [Green Version]
- Song, S.G.; Park, C. Organization and regulation of the D-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. J. Bacteriol. 1997, 179, 7025–7032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievert, C.; Nieves, L.M.; Panyon, L.A.; Loeffler, T.; Morris, C.; Cartwright, R.A.; Wang, X. Experimental evolution reveals an effective avenue to release catabolite repression via mutations in XylR. Proc. Natl. Acad. Sci. USA 2017, 114, 7349–7354. [Google Scholar] [CrossRef] [Green Version]
- Stephens, C.; Christen, B.; Watanabe, K.; Fuchs, T.; Jenal, U. Regulation of D-xylose metabolism in Caulobacter crescentus by a LacI-type repressor. J. Bacteriol. 2007, 189, 8828–8834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meisenzahl, A.C.; Shapiro, L.; Jenal, U. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J. Bacteriol. 1997, 179, 592–600. [Google Scholar] [CrossRef] [Green Version]
- Hottes, A.K.; Meewan, M.; Yang, D.; Arana, N.; Romero, P.; McAdams, H.H.; Stephens, C. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J. Bacteriol. 2004, 186, 1448–1461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreuzer, P.; Gartner, D.; Allmansberger, R.; Hillen, W. Identification and Sequence-Analysis of the Bacillus subtilis W23 Xylr Gene and Xyl Operator. J. Bacteriol. 1989, 171, 3840–3845. [Google Scholar] [CrossRef] [Green Version]
- Lokman, B.C.; Heerikhuisen, M.; Leer, R.J.; vanderBroek, A.; Borsboom, Y.; Chaillou, S.; Postma, P.W.; Pouwels, P.H. Regulation of expression of the Lactobacillus pentosus xylAB operon. J. Bacteriol. 1997, 179, 5391–5397. [Google Scholar] [CrossRef] [Green Version]
- Sizemore, C.; Wieland, B.; Gotz, F.; Hillen, W. Regulation of Staphylococcus xylosus Xylose Utilization Genes at the Molecular-Level. J. Bacteriol. 1992, 174, 3042–3048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apel, A.R.; Ouellet, M.; Szmidt-Middleton, H.; Keasling, J.D.; Mukhopadhyay, A. Evolved hexose transporter enhances xylose uptake and glucose/xylose co-utilization in Saccharomyces cerevisiae. Sci. Rep. UK 2016, 6, 19512. [Google Scholar] [CrossRef] [PubMed]
- Nijland, J.G.; Shin, H.Y.; de Jong, R.M.; De Waal, P.P.; Klaassen, P.; Driessen, A.J.M. Engineering of an endogenous hexose transporter into a specific D-xylose transporter facilitates glucose-xylose co-consumption in Saccharomyces cerevisiae. Biotechnol. Biofuels 2014, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Shin, H.Y.; Nijland, J.G.; de Waal, P.P.; de Jong, R.M.; Klaassen, P.; Driessen, A.J.M. An engineered cryptic Hxt11 sugar transporter facilitates glucose-xylose co-consumption in Saccharomyces cerevisiae. Biotechnol. Biofuels 2015, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Nijland, J.G.; Vos, E.; Shin, H.Y.; de Waal, P.P.; Klaassen, P.; Driessen, A.J.M. Improving pentose fermentation by preventing ubiquitination of hexose transporters in Saccharomyces cerevisiae. Biotechnol. Biofuels 2016, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Wei, S.; Wu, M.; Zhu, X.; Bao, X.; Hou, J.; Liu, W.; Shen, Y. Improving Xylose Fermentation in Saccharomyces cerevisiae by Expressing Nuclear-Localized Hexokinase 2. Microorganisms 2020, 8, 856. [Google Scholar] [CrossRef] [PubMed]
- Hacisalihoglu, B.; Holyavkin, C.; Topaloglu, A.; Kisakesen, H.I.; Cakar, Z.P. Genomic and transcriptomic analysis of a coniferyl aldehyde-resistant Saccharomyces cerevisiae strain obtained by evolutionary engineering. FEMS Yeast Res. 2019, 19, foz021. [Google Scholar] [CrossRef] [PubMed]
- Satomura, A.; Miura, N.; Kuroda, K.; Ueda, M. Reconstruction of thermotolerant yeast by one-point mutation identified through whole-genome analyses of adaptively-evolved strains. Sci. Rep. UK 2016, 6, 23157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osiro, K.O.; Borgström, C.; Brink, D.P.; Fjölnisdóttir, B.L.; Gorwa-Grauslund, M.F. Exploring the xylose paradox in Saccharomyces cerevisiae through in vivo sugar signalomics of targeted deletants. Microb. Cell Fact. 2019, 18, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Robinson, G.; Butcher, R.W.; Sutherland, E.W. Cyclic AMP. Annu. Rev. Biochem. 1968, 37, 149–174. [Google Scholar] [CrossRef]
- Tanaka, K.; Nakafuku, M.; Tamanoi, F.; Kaziro, Y.; Matsumoto, K.; Toh-e, A. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol. Cell. Biol. 1990, 10, 4303–4313. [Google Scholar]
- Auesukaree, C.; Damnernsawad, A.; Kruatrachue, M.; Pokethitiyook, P.; Boonchird, C.; Kaneko, Y.; Harashima, S. Genome-wide identification of genes involved in tolerance to various environmental stresses in Saccharomyces cerevisiae. J. Appl. Genet. 2009, 50, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-I.; Grant, C.M.; Dawes, I.W. The high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae is the major determinant of cAMP levels in stationary phase: Involvement of different branches of the Ras–cyclic AMP pathway in stress responses. Biochem. Biophys. Res. Commun. 2005, 327, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Giardina, D.M.; Siegal, M.L. Control of nongenetic heterogeneity in growth rate and stress tolerance of Saccharomyces cerevisiae by cyclic AMP-regulated transcription factors. PLoS Genet. 2018, 14, e1007744. [Google Scholar] [CrossRef]
- Lim, W.A. Designing customized cell signalling circuits. Nat. Rev. Mol. Cell Bio. 2010, 11, 393–403. [Google Scholar] [CrossRef] [Green Version]
- Qian, S.; Cirino, P.C. Using metabolite-responsive gene regulators to improve microbial biosynthesis. Curr. Opin. Chem. Eng. 2016, 14, 93–102. [Google Scholar] [CrossRef] [Green Version]
- Taylor, N.D.; Garruss, A.S.; Moretti, R.; Chan, S.; Arbing, M.A.; CaScio, D.; Rogers, J.K.; Isaacs, F.J.; Kosuri, S.; Baker, D.; et al. Engineering an allosteric transcription factor to respond to new ligands. Nat. Methods 2016, 13, 177–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castano-Cerezo, S.; Fournie, M.; Urban, P.; Faulon, J.L.; Truan, G. Development of a Biosensor for Detection of Benzoic Acid Derivatives in Saccharomyces cerevisiae. Front. Bioeng. Biotechol. 2020, 7, 372. [Google Scholar] [CrossRef]
- Teo, W.S.; Chang, M.W. Bacterial XylRs and synthetic promoters function as genetically encoded xylose biosensors in Saccharomyces cerevisiae. Biotechnol. J. 2015, 10, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, S.J.; Zhao, H.M. Design and engineering of intracellular-metabolite-sensing/regulation gene circuits in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2016, 113, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.P.; Shang, Y.Z.; Zhang, P.; Liu, Y.; You, D.; Yin, B.C.; Ye, B.C. Engineering Prokaryotic Transcriptional Activator XylR as a Xylose-Inducible Biosensor for Transcription Activation in Yeast. Acs Synth. Biol. 2020, 9, 1022–1029. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Shin, J.; Cho, B.K. Applications of CRISPR/Cas System to Bacterial Metabolic Engineering. Int. J. Mol. Sci. 2018, 19, 1089. [Google Scholar] [CrossRef] [Green Version]
- Hector, R.E.; Mertens, J.A. A Synthetic Hybrid Promoter for Xylose-Regulated Control of Gene Expression in Saccharomyces Yeasts. Mol. Biotechnol. 2017, 59, 24–33. [Google Scholar] [CrossRef]
- Qiu, C.X.; Chen, X.X.; Rexida, R.; Shen, Y.; Qi, Q.S.; Bao, X.M.; Hou, J. Engineering transcription factor-based biosensors for repressive regulation through transcriptional deactivation design in Saccharomyces cerevisiae. Microb. Cell Fact. 2020, 19, 1–10. [Google Scholar] [CrossRef]
- Hector, R.E.; Mertens, J.A.; Nichols, N.N. Development and characterization of vectors for tunable expression of both xylose-regulated and constitutive gene expression in Saccharomyces yeasts. New Biotechnol. 2019, 53, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.W.; Wang, M.Y.; Yu, G.L.; Zhu, S.H.; Yu, Y.; Heng, B.C.; Wu, J.L.; Ye, H.F. Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation. Proc. Natl Acad Sci. USA 2018, 115, E6722–E6730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chavez, A.; Scheiman, J.; Vora, S.; Pruitt, B.W.; Tuttle, M.; Iyer, E.P.R.; Lin, S.L.; Kiani, S.; Guzman, C.D.; Wiegand, D.J.; et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 2015, 12, 326–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, R.Q.; Wagner, J.M.; Alper, H.S.; Zhao, X.Q.; Bai, F.W. Design, Evolution, and Characterization of a Xylose Biosensor in Escherichia coli Using the XylR/xylO System with an Expanded Operating Range. Acs Synth. Biol. 2020, 9, 2714–2722. [Google Scholar] [CrossRef] [PubMed]
- Hohenschuh, W.; Hector, R.E.; Chaplen, F.; Murthy, G.S. Using high-throughput data and dynamic flux balance modeling techniques to identify points of constraint in xylose utilization in Saccharomyces cerevisiae. Syst. Microbiol. Biomanuf. 2021, 1, 58–75. [Google Scholar] [CrossRef]
- Tarazona, S.; Balzano-Nogueira, L.; Gomez-Cabrero, D.; Schmidt, A.; Imhof, A.; Hankemeier, T.; Tegner, J.; Westerhuis, J.A.; Conesa, A. Harmonization of quality metrics and power calculation in multi-omic studies. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, K.; Hohmann, S. Synthetic biology: Lessons from engineering yeast MAPK signalling pathways. Mol. Microbiol. 2013, 88, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.B.; Kwon, D.H.; Park, J.B.; Ha, S.J. Alleviation of catabolite repression in Kluyveromyces marxianus: The thermotolerant SBK1 mutant simultaneously coferments glucose and xylose. Biotechnol. Biofuels 2019, 12, 90. [Google Scholar] [CrossRef]
- Lane, S.; Xu, H.Q.; Oh, E.J.; Kim, H.; Lesmana, A.; Jeong, D.; Zhang, G.C.; Tsai, C.S.; Jin, Y.S.; Kim, S.R. Glucose repression can be alleviated by reducing glucose phosphorylation rate in Saccharomyces cerevisiae. Sci. Rep. UK 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergmann, J.C.; Trichez, D.; de Morais Junior, W.G.; Ramos, T.G.S.; Pacheco, T.F.; Carneiro, C.V.G.; Honorato, V.M.; Serra, L.A.; Almeida, J.R.M. Biotechnological application of non-conventional yeasts for xylose valorization. In Non-Conventional Yeasts: From Basic Research to Application; Springer: Berlin/Heidelberg, Germany, 2019; pp. 23–74. [Google Scholar]
- Karanicolas, J. Designing orthogonal signaling pathways: How to fit in with the surroundings. Proc. Natl. Acad. Sci. USA 2012, 109, 5140–5141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClune, C.J.; Alvarez-Buylla, A.; Voigt, C.A.; Laub, M.T. Engineering orthogonal signalling pathways reveals the sparse occupancy of sequence space. Nature 2019, 574, 702–706. [Google Scholar] [CrossRef]
- Jeong, D.E.; Park, S.H.; Pan, J.G.; Kim, E.J.; Choi, S.K. Genome engineering using a synthetic gene circuit in Bacillus subtilis. Nucleic Acids Res. 2015, 43, e42. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, E.J.; Monk, J.M.; Palsson, B.O. Using Genome-scale Models to Predict Biological Capabilities. Cell 2015, 161, 971–987. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.S.; Jeffries, T.W. Stoichiometric network constraints on xylose metabolism by recombinant Saccharomyces cerevisiae. Metab. Eng. 2004, 6, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Bro, C.; Regenberg, B.; Forster, J.; Nielsen, J. In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab. Eng. 2006, 8, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Zhao, H.M.; Price, N.D. Genome-Scale Consequences of Cofactor Balancing in Engineered Pentose Utilization Pathways in Saccharomyces cerevisiae. PLoS ONE 2011, 6, e27316. [Google Scholar] [CrossRef]
- Lee, J.M.; Gianchandani, E.P.; Eddy, J.A.; Papin, J.A. Dynamic analysis of integrated signaling, metabolic, and regulatory networks. PLoS Comput. Biol. 2008, 4, e1000086. [Google Scholar] [CrossRef]
- Goncalves, E.; Bucher, J.; Ryll, A.; Niklas, J.; Mauch, K.; Klamt, S.; Rocha, M.; Saez-Rodriguez, J. Bridging the layers: Towards integration of signal transduction, regulation and metabolism into mathematical models. Mol. Biosyst. 2013, 9, 1576–1583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banos, D.T.; Trebulle, P.; Elati, M. Integrating transcriptional activity in genome-scale models of metabolism. BMC Syst. Biol. 2017, 11, 81–90. [Google Scholar] [CrossRef] [Green Version]
- Österberg, L.; Domenzain, I.; Munch, J.; Nielsen, J.; Hohmann, S.; Cvijovic, M. A novel yeast hybrid modeling framework integrating Boolean and enzyme-constrained networks enables exploration of the interplay between signaling and metabolism. PLoS Comput. Biol. 2021, 17, e1008891. [Google Scholar] [CrossRef]
- Lubitz, T.; Welkenhuysen, N.; Shashkova, S.; Bendrioua, L.; Hohmann, S.; Klipp, E.; Krantz, M. Network reconstruction and validation of the Snf1/AMPK pathway in baker’s yeast based on a comprehensive literature review. NPJ Syst. Biol. Appl. 2015, 1, 15007. [Google Scholar] [CrossRef] [Green Version]
- Christensen, T.S.; Oliveira, A.P.; Nielsen, J. Reconstruction and logical modeling of glucose repression signaling pathways in Saccharomyces cerevisiae. BMC Syst. Biol. 2009, 3, 7. [Google Scholar] [CrossRef] [Green Version]
- Klipp, E.; Nordlander, B.; Krüger, R.; Gennemark, P.; Hohmann, S. Integrative model of the response of yeast to osmotic shock. Nat. Biotechnol. 2005, 23, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Welkenhuysen, N.; Schnitzer, B.; Osterberg, L.; Cvijovic, M. Robustness of Nutrient Signaling Is Maintained by Interconnectivity Between Signal Transduction Pathways. Front. Physiol. 2019, 9, 1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
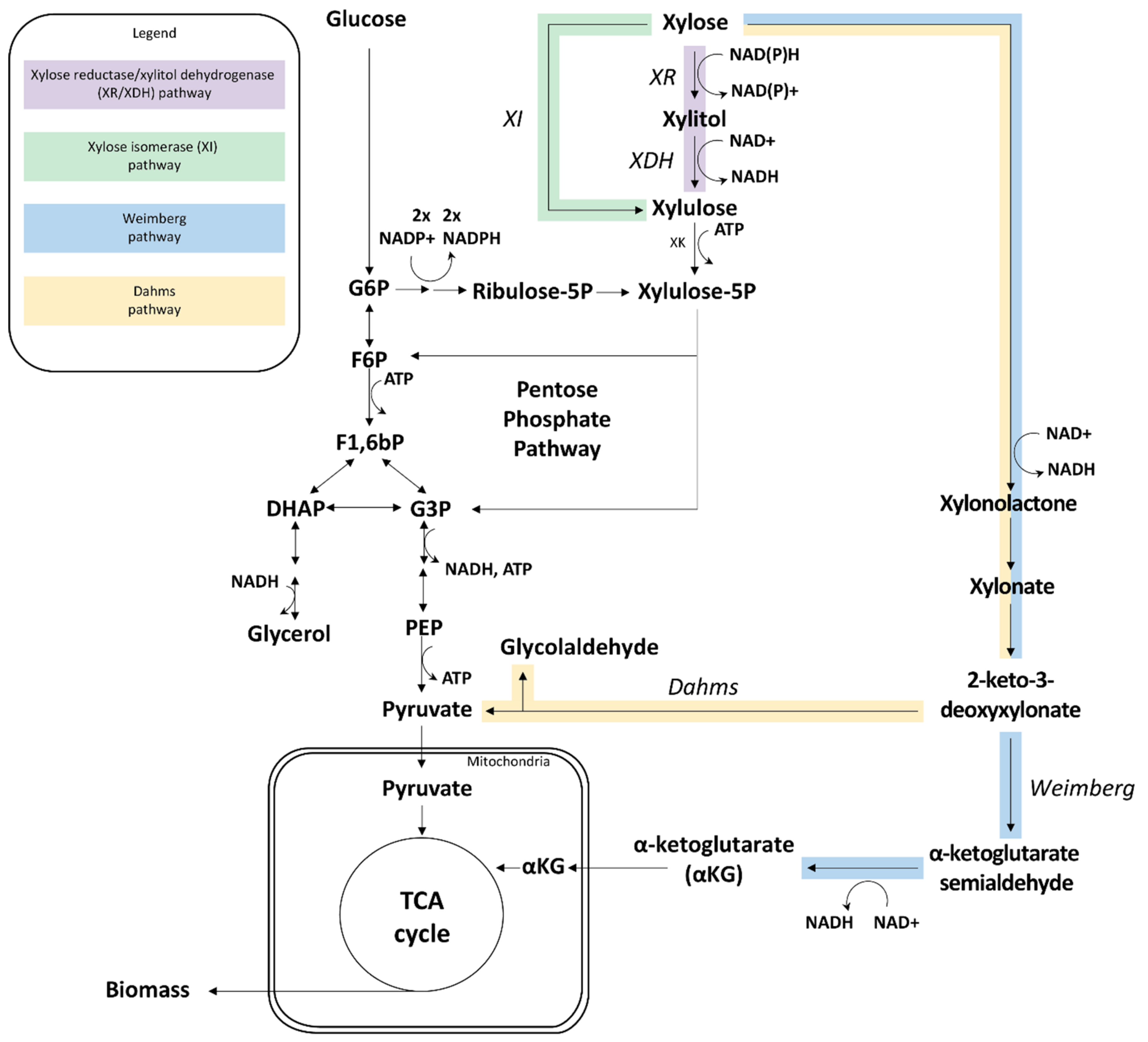




| Strain | d-Xylose Pathway (and Subsequent Evolution) | Oxygenation | Maximum Specific Growth Rate (µmax) on d-Xylose (h−1) | d-Xylose Consumption Rate (g d-Xylose g −1 CDW h −1) | Yield (g EtOH g−1 d-Xylose) | Specific Ethanol Production Rate (g EtOH g−1 CDW h−1) | Reference(s) |
|---|---|---|---|---|---|---|---|
| Anaerobic d-xylose assimilation via the pentose phosphate pathway | |||||||
| RWB 217 | XI (non-evolved) | Anaerobic | 0.09 | 1.06 | 0.43 | 0.46 | [41,42] |
| H131-A3-ALCS | XI (evolved) | Anaerobic | 0.20 | 1.87 | 0.41 | 0.77 | [43] |
| TMB 3504 | XR-XDH (non-evolved) | Anaerobic | 0.11 | 0.76 | 0.40 | 0.33 | [44] |
| SR8 | XR-XDH (evolved) | Anaerobic | 0.09 | 0.87 | 0.31 | 0.28 | [45] |
| Aerobic d-xylose oxidation | |||||||
| TMB4590 | Weimberg pathway | Aerobic | 0.08 | 0.16 | N/A | [17] | |
| H4099 | Dahms pathway | Aerobic | No growth | Specific rate not reported * | N/A | [16] | |
| Targets induced/activated by active PKA | Ribosome biogenesis and ribosomal protein genes | [131,132] |
| BAT1 gene (Tpk1p regulation): gene involved in exit from stationary phase, iron homeostasis and mitochondrial DNA stability | [133] | |
| Pseudohyphal growth (Tpk2p regulation) | [133] | |
| Genes involved in trehalose degradation and water homeostasis (Tpk2p regulation) | [133] | |
| Growth and increase of biomass | [131,134] | |
| Low-affinity hexose transporters via Rgt1p phosphorylation (e.g., HXT1) | [106] | |
| Glycolytic enzyme, e.g., by phosphorylation of Pfk26p and Nth1p, and transcriptional upregulation of Pdc1p | [67,135,136,137] | |
| Protein phosphatases (PP2A and PP1), specifically dephosphorylating serine/threonine amino acids | [138] | |
| Targets repressed/inactivated by active PKA | Enzymes involved in gluconeogenesis (fructose 1,6-bisphosphatase, isocitrate lyase) | [139,140,141] |
| Stress-responsive genes (e.g., MSN2/4) | [142] | |
| Glycogen accumulation | [142] | |
| Rim15p (a protein kinase involved in adaptation process to enter in the stationary phase) | [143] | |
| Genes involved in iron uptake (Tpk2p regulation) | [133] | |
| Heat-shock genes (e.g., HSP12, HSP26) by inactivating the transcriptional activator Hsf1 | [144] | |
| Transcription of genes involved in trehalose synthesis and accumulation (TPS1/2); Trehalose-6-phosphate synthase activity through phosphorylation of one of the regulatory subunits (Tps3p) | [145,146] | |
| SUC2 (encoding invertase) | [147] | |
| Sak1p and SNF1 proteins | [148] |
| Genes Related to: | References | |
|---|---|---|
| Upregulation | Gluconeogenesis | [35,37,214,217] |
| Genes related to the oxidative pentose phosphate pathway | [214,217] | |
| TCA and glyoxylate cycle | [35,37,38,217] | |
| Respiration | [35,37,38,217] | |
| Acetaldehyde and acetyl-CoA metabolism | [35,217] | |
| Genes typically expressed on non-fermentable carbon sources: SUC2, HXK1, HXT5, HXT13, maltose metabolism genes | [35,38,217] | |
| Sugar signaling: MTH1 *, ADR1, CAT8, RGT1 | [35,38,217] | |
| High-affinity d-glucose transporters (e.g., HXT2 *, HXT6 and HXT7) | [35,38] | |
| Downregulation | Glycolysis | [35] |
| Low-affinity d-glucose transporters (e.g., HXT1 and HXT3) | [38,217] | |
| Sulfur metabolism | [217] | |
| Heme biosynthesis from uroporphyrinogen | [217] | |
| Tryptophan degradation | [217] | |
| Sugar signaling: MTH1 *, STD1, MIG1, HXK2 | [35,217] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brink, D.P.; Borgström, C.; Persson, V.C.; Ofuji Osiro, K.; Gorwa-Grauslund, M.F. D-Xylose Sensing in Saccharomyces cerevisiae: Insights from D-Glucose Signaling and Native D-Xylose Utilizers. Int. J. Mol. Sci. 2021, 22, 12410. https://doi.org/10.3390/ijms222212410
Brink DP, Borgström C, Persson VC, Ofuji Osiro K, Gorwa-Grauslund MF. D-Xylose Sensing in Saccharomyces cerevisiae: Insights from D-Glucose Signaling and Native D-Xylose Utilizers. International Journal of Molecular Sciences. 2021; 22(22):12410. https://doi.org/10.3390/ijms222212410
Chicago/Turabian StyleBrink, Daniel P., Celina Borgström, Viktor C. Persson, Karen Ofuji Osiro, and Marie F. Gorwa-Grauslund. 2021. "D-Xylose Sensing in Saccharomyces cerevisiae: Insights from D-Glucose Signaling and Native D-Xylose Utilizers" International Journal of Molecular Sciences 22, no. 22: 12410. https://doi.org/10.3390/ijms222212410
APA StyleBrink, D. P., Borgström, C., Persson, V. C., Ofuji Osiro, K., & Gorwa-Grauslund, M. F. (2021). D-Xylose Sensing in Saccharomyces cerevisiae: Insights from D-Glucose Signaling and Native D-Xylose Utilizers. International Journal of Molecular Sciences, 22(22), 12410. https://doi.org/10.3390/ijms222212410







