Clinical Translation of Combined MAPK and Autophagy Inhibition in RAS Mutant Cancer
Abstract
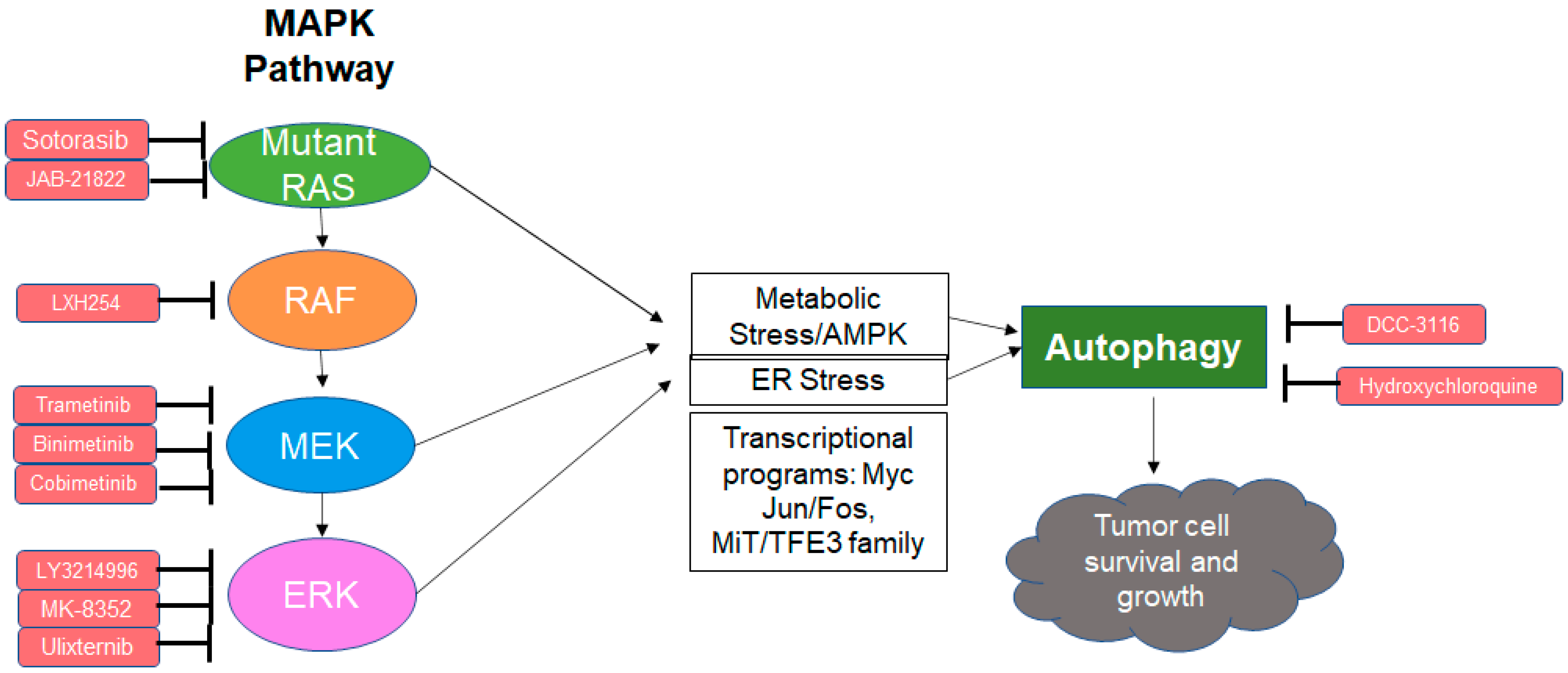
1. Introduction
2. Allosteric RAS Inhibitors
3. Inhibition of Post Translational Modification of RAS
4. RAF Inhibitors
5. MEK Inhibitors
6. ERK Inhibitors
7. Resistance to MEK Inhibitors in RAS Mutant Cancers
8. Autophagy as a Resistance Mechanism to MAPK Inhibition
9. Combined MAPK and Autophagy Inhibition Is Synergistic in RAS Mutant Cancer
10. Clinical Trials with Autophagy and MAPK Inhibitors in RAS Mutant Cancers:
11. Open Questions
12. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bos, J.L. Ras oncogenes in human cancer: A review. Cancer Res. 1989, 49, 4682–4689. [Google Scholar] [PubMed]
- Thein, K.Z.; Biter, A.B.; Hong, D.S. Therapeutics Targeting Mutant KRAS. Annu. Rev. Med. 2021, 72, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Prior, I.A.; Lewis, P.D.; Mattos, C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012, 72, 2457–2467. [Google Scholar] [CrossRef]
- Li, S.; Balmain, A.; Counter, C.M. A model for RAS mutation patterns in cancers: Finding the sweet spot. Nat. Rev. Cancer 2018, 18, 767–777. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Vander Heiden, M.G.; McCormick, F. The Metabolic Landscape of RAS-Driven Cancers from biology to therapy. Nat. Cancer 2021, 2, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, G.A.; Der, C.J.; Rossman, K.L. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 2016, 129, 1287–1292. [Google Scholar] [CrossRef]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e310. [Google Scholar] [CrossRef]
- Prior, I.A.; Hood, F.E.; Hartley, J.L. The Frequency of Ras Mutations in Cancer. Cancer Res. 2020, 80, 2969–2974. [Google Scholar] [CrossRef]
- Munoz-Maldonado, C.; Zimmer, Y.; Medova, M. A Comparative Analysis of Individual RAS Mutations in Cancer Biology. Front. Oncol. 2019, 9, 1088. [Google Scholar] [CrossRef]
- Moore, A.R.; Rosenberg, S.C.; McCormick, F.; Malek, S. RAS-targeted therapies: Is the undruggable drugged? Nat. Rev. Drug Discov. 2020, 19, 533–552. [Google Scholar] [CrossRef]
- Zenonos, K.; Kyprianou, K. RAS signaling pathways, mutations and their role in colorectal cancer. World J. Gastrointest. Oncol. 2013, 5, 97–101. [Google Scholar] [CrossRef]
- Nakamura, K.; Ichise, H.; Nakao, K.; Hatta, T.; Otani, H.; Sakagami, H.; Kondo, H.; Katsuki, M. Partial functional overlap of the three ras genes in mouse embryonic development. Oncogene 2008, 27, 2961–2968. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Johnson, L.; Greenbaum, D.; Cichowski, K.; Mercer, K.; Murphy, E.; Schmitt, E.; Bronson, R.T.; Umanoff, H.; Edelmann, W.; Kucherlapati, R.; et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997, 11, 2468–2481. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.S.; Fakih, M.G.; Strickler, J.H.; Desai, J.; Durm, G.A.; Shapiro, G.I.; Falchook, G.S.; Price, T.J.; Sacher, A.; Denlinger, C.S.; et al. KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 2020, 383, 1207–1217. [Google Scholar] [CrossRef]
- Canon, J.; Rex, K.; Saiki, A.Y.; Mohr, C.; Cooke, K.; Bagal, D.; Gaida, K.; Holt, T.; Knutson, C.G.; Koppada, N.; et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019, 575, 217–223. [Google Scholar] [CrossRef]
- Lito, P.; Solomon, M.; Li, L.S.; Hansen, R.; Rosen, N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 2016, 351, 604–608. [Google Scholar] [CrossRef] [PubMed]
- Patricelli, M.P.; Janes, M.R.; Li, L.S.; Hansen, R.; Peters, U.; Kessler, L.V.; Chen, Y.; Kucharski, J.M.; Feng, J.; Ely, T.; et al. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discov. 2016, 6, 316–329. [Google Scholar] [CrossRef]
- Campbell, S.L.; Philips, M.R. Post-translational modification of RAS proteins. Curr. Opin. Struct. Biol. 2021, 71, 180–192. [Google Scholar] [CrossRef]
- Baines, A.T.; Xu, D.; Der, C.J. Inhibition of Ras for cancer treatment: The search continues. Future Med. Chem. 2011, 3, 1787–1808. [Google Scholar] [CrossRef]
- Busquets-Hernandez, C.; Triola, G. Palmitoylation as a Key Regulator of Ras Localization and Function. Front. Mol. Biosci. 2021, 8, 659861. [Google Scholar] [CrossRef]
- Simanshu, D.K.; Nissley, D.V.; McCormick, F. RAS Proteins and Their Regulators in Human Disease. Cell 2017, 170, 17–33. [Google Scholar] [CrossRef]
- James, G.; Goldstein, J.L.; Brown, M.S. Resistance of K-RasBV12 proteins to farnesyltransferase inhibitors in Rat1 cells. Proc. Natl. Acad. Sci. USA 1996, 93, 4454–4458. [Google Scholar] [CrossRef]
- Whyte, D.B.; Kirschmeier, P.; Hockenberry, T.N.; Nunez-Oliva, I.; James, L.; Catino, J.J.; Bishop, W.R.; Pai, J.K. K-and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem. 1997, 272, 14459–14464. [Google Scholar] [CrossRef]
- Cox, A.D.; Der, C.J.; Philips, M.R. Targeting RAS Membrane Association: Back to the Future for Anti-RAS Drug Discovery? Clin. Cancer Res. 2015, 21, 1819–1827. [Google Scholar] [CrossRef] [PubMed]
- Remsberg, J.R.; Suciu, R.M.; Zambetti, N.A.; Hanigan, T.W.; Firestone, A.J.; Inguva, A.; Long, A.; Ngo, N.; Lum, K.M.; Henry, C.L.; et al. ABHD17 regulation of plasma membrane palmitoylation and N-Ras-dependent cancer growth. Nat. Chem. Biol. 2021, 17, 856–864. [Google Scholar] [CrossRef]
- Lin, D.T.S.; Davis, N.G.; Conibear, E. Targeting the Ras palmitoylation/depalmitoylation cycle in cancer. Biochem. Soc. Trans. 2017, 45, 913–921. [Google Scholar] [CrossRef]
- Karoulia, Z.; Gavathiotis, E.; Poulikakos, P.I. New perspectives for targeting RAF kinase in human cancer. Nat. Rev. Cancer 2017, 17, 676–691. [Google Scholar] [CrossRef]
- Grey, A.; Cooper, A.; McNeil, C.; O′Toole, S.; Thompson, J.; Grimison, P. Progression of KRAS mutant pancreatic adenocarcinoma during vemurafenib treatment in a patient with metastatic melanoma. Intern. Med. J. 2014, 44, 597–600. [Google Scholar] [CrossRef] [PubMed]
- Callahan, M.K.; Rampal, R.; Harding, J.J.; Klimek, V.M.; Chung, Y.R.; Merghoub, T.; Wolchok, J.D.; Solit, D.B.; Rosen, N.; Abdel-Wahab, O.; et al. Progression of RAS-mutant leukemia during RAF inhibitor treatment. N. Engl. J. Med. 2012, 367, 2316–2321. [Google Scholar] [CrossRef] [PubMed]
- Cook, F.A.; Cook, S.J. Inhibition of RAF dimers: It takes two to tango. Biochem. Soc. Trans. 2021, 49, 237–251. [Google Scholar] [CrossRef]
- Monaco, K.A.; Delach, S.; Yuan, J.; Mishina, Y.; Fordjour, P.; Labrot, E.; McKay, D.; Guo, R.; Higgins, S.; Wang, H.Q.; et al. LXH254, a Potent and Selective ARAF-Sparing Inhibitor of BRAF and CRAF for the Treatment of MAPK-Driven Tumors. Clin. Cancer Res. 2021, 27, 2061–2073. [Google Scholar] [CrossRef]
- Cheng, Y.; Tian, H. Current Development Status of MEK Inhibitors. Molecules 2017, 22, 1551. [Google Scholar] [CrossRef] [PubMed]
- Blumenschein, G.R., Jr.; Smit, E.F.; Planchard, D.; Kim, D.W.; Cadranel, J.; De Pas, T.; Dunphy, F.; Udud, K.; Ahn, M.J.; Hanna, N.H.; et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC)dagger. Ann. Oncol. 2015, 26, 894–901. [Google Scholar] [CrossRef] [PubMed]
- Thumar, J.; Shahbazian, D.; Aziz, S.A.; Jilaveanu, L.B.; Kluger, H.M. MEK targeting in N-RAS mutated metastatic melanoma. Mol. Cancer 2014, 13, 45. [Google Scholar] [CrossRef]
- Dummer, R.; Schadendorf, D.; Ascierto, P.A.; Arance, A.; Dutriaux, C.; Di Giacomo, A.M.; Rutkowski, P.; Del Vecchio, M.; Gutzmer, R.; Mandala, M.; et al. Binimetinib versus dacarbazine in patients with advanced NRAS-mutant melanoma (NEMO): A multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2017, 18, 435–445. [Google Scholar] [CrossRef]
- Mincu, R.I.; Mahabadi, A.A.; Michel, L.; Mrotzek, S.M.; Schadendorf, D.; Rassaf, T.; Totzeck, M. Cardiovascular Adverse Events Associated With BRAF and MEK Inhibitors: A Systematic Review and Meta-analysis. JAMA Netw. Open 2019, 2, e198890. [Google Scholar] [CrossRef]
- Mendez-Martinez, S.; Calvo, P.; Ruiz-Moreno, O.; Pardinas Baron, N.; Lecinena Bueno, J.; Gil Ruiz, M.D.R.; Pablo, L. Ocular Adverse Events Associated with Mek Inhibitors. Retina 2019, 39, 1435–1450. [Google Scholar] [CrossRef]
- Russo, I.; Zorzetto, L.; Chiarion Sileni, V.; Alaibac, M. Cutaneous Side Effects of Targeted Therapy and Immunotherapy for Advanced Melanoma. Scientifica 2018, 2018, 5036213. [Google Scholar] [CrossRef]
- Kohler, J.; Zhao, Y.; Li, J.; Gokhale, P.C.; Tiv, H.L.; Knott, A.R.; Wilkens, M.K.; Soroko, K.M.; Lin, M.; Ambrogio, C.; et al. ERK Inhibitor LY3214996-Based Treatment Strategies for RAS-Driven Lung Cancer. Mol. Cancer Ther. 2021, 20, 641–654. [Google Scholar] [CrossRef]
- Moschos, S.J.; Sullivan, R.J.; Hwu, W.J.; Ramanathan, R.K.; Adjei, A.A.; Fong, P.C.; Shapira-Frommer, R.; Tawbi, H.A.; Rubino, J.; Rush, T.S., 3rd; et al. Development of MK-8353, an orally administered ERK1/2 inhibitor, in patients with advanced solid tumors. JCI Insight 2018, 3, e92352. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, R.J.; Infante, J.R.; Janku, F.; Wong, D.J.L.; Sosman, J.A.; Keedy, V.; Patel, M.R.; Shapiro, G.I.; Mier, J.W.; Tolcher, A.W.; et al. First-in-Class ERK1/2 Inhibitor Ulixertinib (BVD-523) in Patients with MAPK Mutant Advanced Solid Tumors: Results of a Phase I Dose-Escalation and Expansion Study. Cancer Discov. 2018, 8, 184–195. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, B.; Liu, Y.; Yu, X.; Cheng, G. Dual effects of active ERK in cancer: A potential target for enhancing radiosensitivity. Oncol. Lett. 2020, 20, 993–1000. [Google Scholar] [CrossRef]
- Zhan, T.; Ambrosi, G.; Wandmacher, A.M.; Rauscher, B.; Betge, J.; Rindtorff, N.; Haussler, R.S.; Hinsenkamp, I.; Bamberg, L.; Hessling, B.; et al. MEK inhibitors activate Wnt signalling and induce stem cell plasticity in colorectal cancer. Nat. Commun. 2019, 10, 2197. [Google Scholar] [CrossRef]
- Tsubaki, M.; Takeda, T.; Noguchi, M.; Jinushi, M.; Seki, S.; Morii, Y.; Shimomura, K.; Imano, M.; Satou, T.; Nishida, S. Overactivation of Akt Contributes to MEK Inhibitor Primary and Acquired Resistance in Colorectal Cancer Cells. Cancers 2019, 11, 1866. [Google Scholar] [CrossRef] [PubMed]
- Echevarria-Vargas, I.M.; Reyes-Uribe, P.I.; Guterres, A.N.; Yin, X.; Kossenkov, A.V.; Liu, Q.; Zhang, G.; Krepler, C.; Cheng, C.; Wei, Z.; et al. Co-targeting BET and MEK as salvage therapy for MAPK and checkpoint inhibitor-resistant melanoma. EMBO Mol. Med. 2018, 10, e8446. [Google Scholar] [CrossRef] [PubMed]
- Brown, W.S.; McDonald, P.C.; Nemirovsky, O.; Awrey, S.; Chafe, S.C.; Schaeffer, D.F.; Li, J.; Renouf, D.J.; Stanger, B.Z.; Dedhar, S. Overcoming Adaptive Resistance to KRAS and MEK Inhibitors by Co-targeting mTORC1/2 Complexes in Pancreatic Cancer. Cell Rep. Med. 2020, 1, 100131. [Google Scholar] [CrossRef]
- Jones, G.G.; Del Rio, I.B.; Sari, S.; Sekerim, A.; Young, L.C.; Hartig, N.; Areso Zubiaur, I.; El-Bahrawy, M.A.; Hynds, R.E.; Lei, W.; et al. SHOC2 phosphatase-dependent RAF dimerization mediates resistance to MEK inhibition in RAS-mutant cancers. Nat. Commun. 2019, 10, 2532. [Google Scholar] [CrossRef]
- Sun, C.; Hobor, S.; Bertotti, A.; Zecchin, D.; Huang, S.; Galimi, F.; Cottino, F.; Prahallad, A.; Grernrum, W.; Tzani, A.; et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 2014, 7, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Kun, E.; Tsang, Y.T.M.; Ng, C.W.; Gershenson, D.M.; Wong, K.K. MEK inhibitor resistance mechanisms and recent developments in combination trials. Cancer Treat. Rev. 2021, 92, 102137. [Google Scholar] [CrossRef]
- Dai, B.; Meng, J.; Peyton, M.; Girard, L.; Bornmann, W.G.; Ji, L.; Minna, J.D.; Fang, B.; Roth, J.A. STAT3 mediates resistance to MEK inhibitor through microRNA miR-17. Cancer Res. 2011, 71, 3658–3668. [Google Scholar] [CrossRef]
- Yao, W.; Yue, P.; Zhang, G.; Owonikoko, T.K.; Khuri, F.R.; Sun, S.Y. Enhancing therapeutic efficacy of the MEK inhibitor, MEK162, by blocking autophagy or inhibiting PI3K/Akt signaling in human lung cancer cells. Cancer Lett. 2015, 364, 70–78. [Google Scholar] [CrossRef]
- Kochetkova, E.Y.; Blinova, G.I.; Bystrova, O.A.; Martynova, M.G.; Pospelov, V.A.; Pospelova, T.V. Targeted elimination of senescent Ras-transformed cells by suppression of MEK/ERK pathway. Aging 2017, 9, 2352–2375. [Google Scholar] [CrossRef]
- Verykiou, S.; Alexander, M.; Edwards, N.; Plummer, R.; Chaudhry, B.; Lovat, P.E.; Hill, D.S. Harnessing autophagy to overcome mitogen-activated protein kinase kinase inhibitor-induced resistance in metastatic melanoma. Br. J. Dermatol. 2019, 180, 346–356. [Google Scholar] [CrossRef]
- Yun, C.W.; Lee, S.H. The Roles of Autophagy in Cancer. Int. J. Mol. Sci. 2018, 19, 3466. [Google Scholar] [CrossRef] [PubMed]
- Ichimiya, T.; Yamakawa, T.; Hirano, T.; Yokoyama, Y.; Hayashi, Y.; Hirayama, D.; Wagatsuma, K.; Itoi, T.; Nakase, H. Autophagy and Autophagy-Related Diseases: A Review. Int. J. Mol. Sci. 2020, 21, 8974. [Google Scholar] [CrossRef]
- Lamark, T.; Johansen, T. Mechanisms of Selective Autophagy. Annu. Rev. Cell Dev. Biol. 2021, 37, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.Y.; Xia, B.; White, E. Autophagy-mediated tumor promotion. Cell 2013, 155, 1216–1219. [Google Scholar] [CrossRef] [PubMed]
- Karsli-Uzunbas, G.; Guo, J.Y.; Price, S.; Teng, X.; Laddha, S.V.; Khor, S.; Kalaany, N.Y.; Jacks, T.; Chan, C.S.; Rabinowitz, J.D.; et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014, 4, 914–927. [Google Scholar] [CrossRef]
- Yang, A.; Rajeshkumar, N.V.; Wang, X.; Yabuuchi, S.; Alexander, B.M.; Chu, G.C.; Von Hoff, D.D.; Maitra, A.; Kimmelman, A.C. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014, 4, 905–913. [Google Scholar] [CrossRef]
- Rangel, M.; Kong, J.; Bhatt, V.; Khayati, K.; Guo, J.Y. Autophagy and tumorigenesis. FEBS J. 2021. [Google Scholar] [CrossRef]
- Guo, J.Y.; Chen, H.Y.; Mathew, R.; Fan, J.; Strohecker, A.M.; Karsli-Uzunbas, G.; Kamphorst, J.J.; Chen, G.; Lemons, J.M.; Karantza, V.; et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011, 25, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, V.; Khayati, K.; Hu, Z.S.; Lee, A.; Kamran, W.; Su, X.; Guo, J.Y. Autophagy modulates lipid metabolism to maintain metabolic flexibility for Lkb1-deficient Kras-driven lung tumorigenesis. Genes Dev. 2019, 33, 150–165. [Google Scholar] [CrossRef] [PubMed]
- Poillet-Perez, L.; Xie, X.; Zhan, L.; Yang, Y.; Sharp, D.W.; Hu, Z.S.; Su, X.; Maganti, A.; Jiang, C.; Lu, W.; et al. Autophagy maintains tumour growth through circulating arginine. Nature 2018, 563, 569–573. [Google Scholar] [CrossRef]
- Sousa, C.M.; Biancur, D.E.; Wang, X.; Halbrook, C.J.; Sherman, M.H.; Zhang, L.; Kremer, D.; Hwang, R.F.; Witkiewicz, A.K.; Ying, H.; et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 2016, 536, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Katheder, N.S.; Khezri, R.; O’Farrell, F.; Schultz, S.W.; Jain, A.; Rahman, M.M.; Schink, K.O.; Theodossiou, T.A.; Johansen, T.; Juhasz, G.; et al. Microenvironmental autophagy promotes tumour growth. Nature 2017, 541, 417–420. [Google Scholar] [CrossRef]
- Yang, A.; Herter-Sprie, G.; Zhang, H.; Lin, E.Y.; Biancur, D.; Wang, X.; Deng, J.; Hai, J.; Yang, S.; Wong, K.K.; et al. Autophagy Sustains Pancreatic Cancer Growth through Both Cell-Autonomous and Nonautonomous Mechanisms. Cancer Discov. 2018, 8, 276–287. [Google Scholar] [CrossRef]
- Khezri, R.; Holland, P.; Schoborg, T.A.; Abramovich, I.; Takats, S.; Dillard, C.; Jain, A.; O’Farrell, F.; Schultz, S.W.; Hagopian, W.M.; et al. Host autophagy mediates organ wasting and nutrient mobilization for tumor growth. EMBO J. 2021, 40, e107336. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Koh, J.Y.; Price, S.; White, E.; Mehnert, J.M. Atg7 Overcomes Senescence and Promotes Growth of BrafV600E-Driven Melanoma. Cancer Discov. 2015, 5, 410–423. [Google Scholar] [CrossRef]
- Ma, X.H.; Piao, S.F.; Dey, S.; McAfee, Q.; Karakousis, G.; Villanueva, J.; Hart, L.S.; Levi, S.; Hu, J.; Zhang, G.; et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J. Clin. Investig. 2014, 124, 1406–1417. [Google Scholar] [CrossRef]
- Levy, J.M.; Thompson, J.C.; Griesinger, A.M.; Amani, V.; Donson, A.M.; Birks, D.K.; Morgan, M.J.; Mirsky, D.M.; Handler, M.H.; Foreman, N.K.; et al. Autophagy inhibition improves chemosensitivity in BRAF(V600E) brain tumors. Cancer Discov. 2014, 4, 773–780. [Google Scholar] [CrossRef]
- Viale, A.; Pettazzoni, P.; Lyssiotis, C.A.; Ying, H.; Sanchez, N.; Marchesini, M.; Carugo, A.; Green, T.; Seth, S.; Giuliani, V.; et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 2014, 514, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Ojha, R.; Leli, N.M.; Onorati, A.; Piao, S.; Verginadis, I.I.; Tameire, F.; Rebecca, V.W.; Chude, C.I.; Murugan, S.; Fennelly, C.; et al. ER Translocation of the MAPK Pathway Drives Therapy Resistance in BRAF-Mutant Melanoma. Cancer Discov. 2019, 9, 396–415. [Google Scholar] [CrossRef]
- Lee, C.S.; Lee, L.C.; Yuan, T.L.; Chakka, S.; Fellmann, C.; Lowe, S.W.; Caplen, N.J.; McCormick, F.; Luo, J. MAP kinase and autophagy pathways cooperate to maintain RAS mutant cancer cell survival. Proc. Natl. Acad. Sci. USA 2019, 116, 4508–4517. [Google Scholar] [CrossRef] [PubMed]
- Bryant, K.L.; Stalnecker, C.A.; Zeitouni, D.; Klomp, J.E.; Peng, S.; Tikunov, A.P.; Gunda, V.; Pierobon, M.; Waters, A.M.; George, S.D.; et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med. 2019, 25, 628–640. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1. Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef] [PubMed]
- Kinsey, C.G.; Camolotto, S.A.; Boespflug, A.M.; Guillen, K.P.; Foth, M.; Truong, A.; Schuman, S.S.; Shea, J.E.; Seipp, M.T.; Yap, J.T.; et al. Protective autophagy elicited by RAF→MEK→ERKinhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med. 2019, 25, 620–627. [Google Scholar] [CrossRef] [PubMed]
- Xavier, C.B.; Marchetti, K.R.; Castria, T.B.; Jardim, D.L.F.; Fernandes, G.S. Trametinib and Hydroxychloroquine (HCQ) Combination Treatment in KRAS-Mutated Advanced Pancreatic Adenocarcinoma: Detailed Description of Two Cases. J. Gastrointest. Cancer 2021, 52, 374–380. [Google Scholar] [CrossRef]
- Orlov, S.V.; Urtenova, M.A.; Sviridenko, M.A.; Nesterov, D.V.; Sokolova, T.N.; Imyanitov, E.N. Rapid Improvement of the Performance Status and Reduction of the Tumor Size in KRAS-Mutated Colorectal Cancer Patient Receiving Binimetinib, Hydroxychloroquine, and Bevacizumab. Case Rep. Oncol. 2020, 13, 985–989. [Google Scholar] [CrossRef]
- Buzun, K.; Gornowicz, A.; Lesyk, R.; Bielawski, K.; Bielawska, A. Autophagy Modulators in Cancer Therapy. Int. J. Mol. Sci. 2021, 22, 5804. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Yang, F.; Fu, R.; Li, X.; French, R.; Mose, E.; Pu, X.; Trinh, B.; Kumar, A.; Liu, J.; et al. Cancer cells escape autophagy inhibition via NRF2-induced macropinocytosis. Cancer Cell 2021, 39, 678–693.e611. [Google Scholar] [CrossRef]
| Treatment | Condition | Phase | ClinicalTrials.gov Registration |
|---|---|---|---|
| Sotorasib + Trametinib + Others | Advanced KRAS G12C Solid Tumors | I/II | NCT04185883 |
| Sotorasib + Docetaxel | Advanced KRAS G12C NSCLC | III | NCT04303780 |
| JAB-21822 + Cetuximab | Advanced KRAS G12C NSCLC + CRC | I/II | NCT05002270 |
| JAB-21822 | Advanced KRAS G12C NSCLC + CRC | I/II | NCT05009329 |
| Treatment | Condition | Phase | ClinicalTrials.gov Registration |
|---|---|---|---|
| Binimetinib and HCQ | Stage IV KRAS mutant NSCLC | II | NCT04735068 |
| Binimetinib and HCQ | Stage IV Pancreatic adenocarcinoma | I | NCT04132505 |
| Trametinib and HCQ | Stage IV Pancreatic adenocarcinoma | I | NCT03825289 |
| Trametinib and HCQ | Stage IV KRAS mutant biliary tract carcinoma | II | NCT04566133 |
| Cobimetinib + Atezolizumab + HCQ | Stage IV KRAS mutant gastrointestinal cancers | I/II | NCT04214418 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.J.; Jain, V.; Amaravadi, R.K. Clinical Translation of Combined MAPK and Autophagy Inhibition in RAS Mutant Cancer. Int. J. Mol. Sci. 2021, 22, 12402. https://doi.org/10.3390/ijms222212402
Lee JJ, Jain V, Amaravadi RK. Clinical Translation of Combined MAPK and Autophagy Inhibition in RAS Mutant Cancer. International Journal of Molecular Sciences. 2021; 22(22):12402. https://doi.org/10.3390/ijms222212402
Chicago/Turabian StyleLee, Jennifer J., Vaibhav Jain, and Ravi K. Amaravadi. 2021. "Clinical Translation of Combined MAPK and Autophagy Inhibition in RAS Mutant Cancer" International Journal of Molecular Sciences 22, no. 22: 12402. https://doi.org/10.3390/ijms222212402
APA StyleLee, J. J., Jain, V., & Amaravadi, R. K. (2021). Clinical Translation of Combined MAPK and Autophagy Inhibition in RAS Mutant Cancer. International Journal of Molecular Sciences, 22(22), 12402. https://doi.org/10.3390/ijms222212402






