To Discover the Efficient and Novel Drug Targets in Human Cancers Using CRISPR/Cas Screening and Databases
Abstract
:1. The Evolution and Usefulness of Random Mutagenesis in Cancers
2. Application of CRISPR Libraries, Optimization of gRNA, and Efficient Next-Generation Libraries
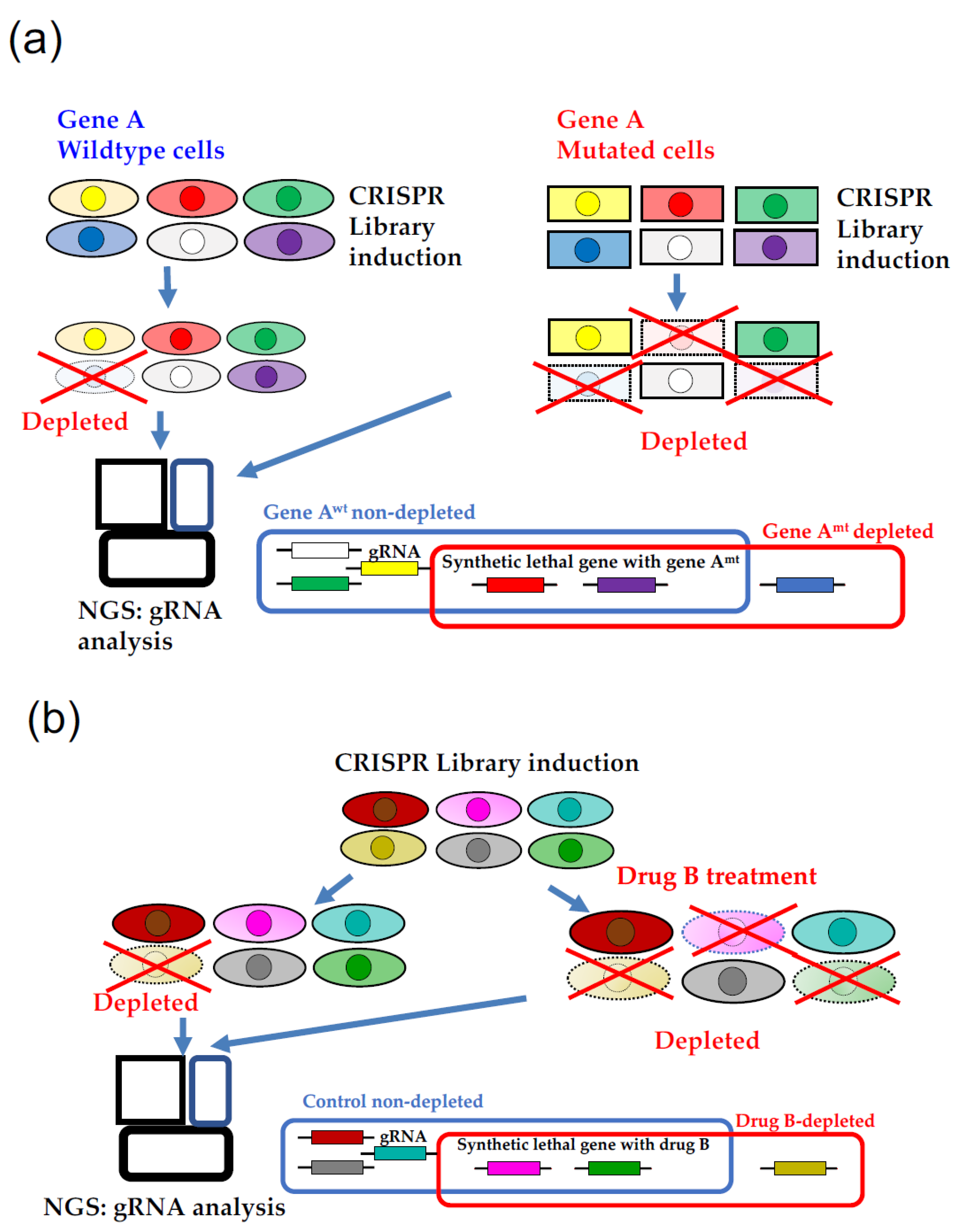
3. CRISPR Screening Focusing on Synthetic Lethality
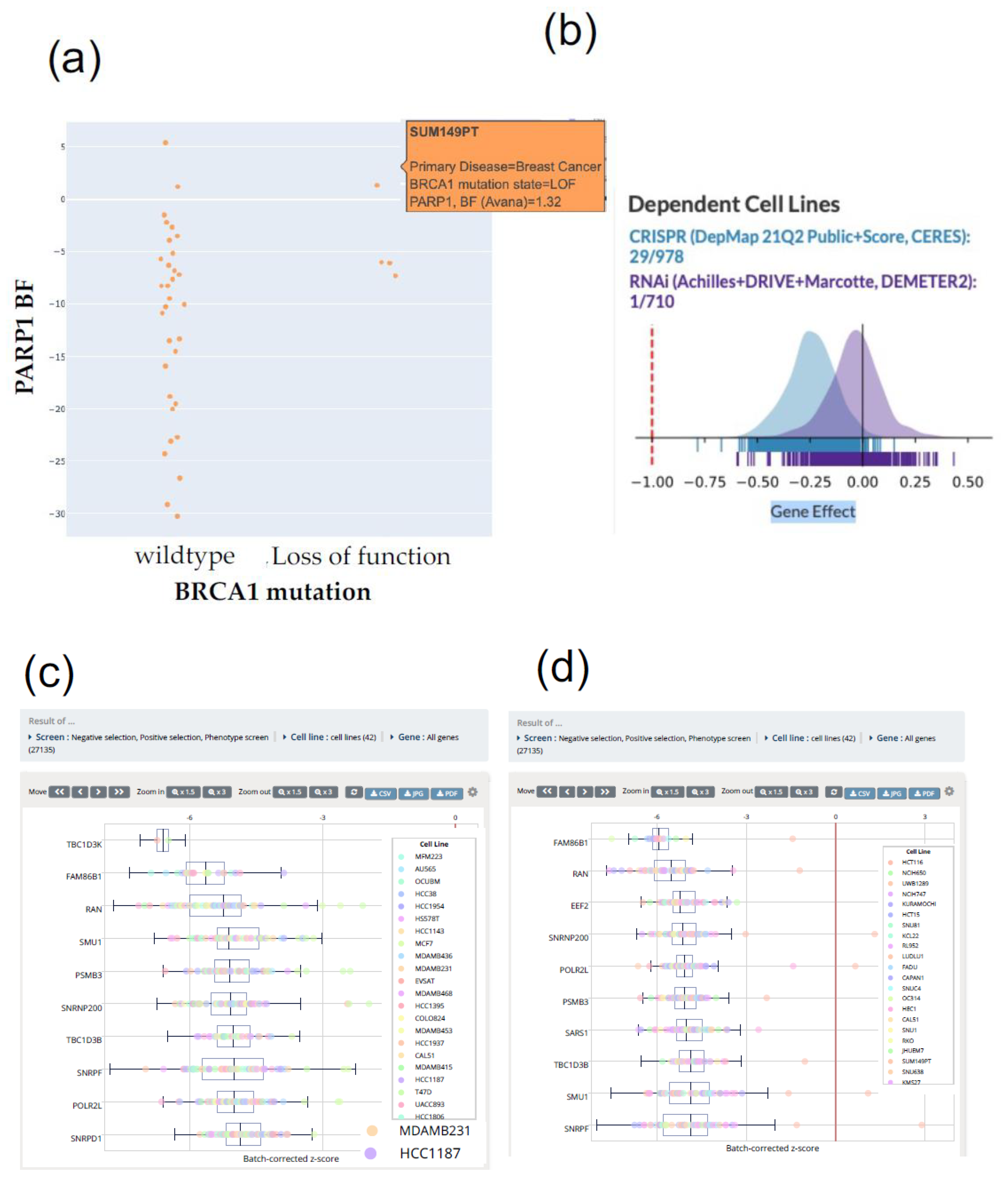
4. Efficient CRISPR Screening Using the Database to Avoid Pitfalls
5. Use of Database Focusing on Synthetic Lethality
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xia, Y.; Ji, X.; Jang, I.S.; Surka, C.; Hsu, C.; Wang, K.; Rolfe, M.; Bence, N.; Lu, G. Genetic and pharmacological interrogation of cancer vulnerability using a multiplexed cell line screening platform. Commun. Biol. 2021, 4, 834. [Google Scholar] [CrossRef]
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018, 173, 371–385.e18. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Jiménez, F.; Muiños, F.; Sentís, I.; Deu-Pons, J.; Reyes-Salazar, I.; Arnedo-Pac, C.; Mularoni, L.; Pich, O.; Bonet, J.; Kranas, H.; et al. A compendium of mutational cancer driver genes. Nat. Rev. Cancer 2020, 20, 555–572. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T. Retroviral insertional mutagenesis identifies oncogene cooperation. Cancer Sci. 2005, 96, 7–12. [Google Scholar] [CrossRef] [PubMed]
- McDonald, E.R., III; De Weck, A.; Schlabach, M.R.; Billy, E.; Mavrakis, K.J.; Hoffman, G.R.; Belur, D.; Castelletti, D.; Frias, E.; Gampa, K.; et al. Project DRIVE: A compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell 2017, 170, 577–592.e10. [Google Scholar] [CrossRef] [Green Version]
- Kolfschoten, I.G.; van Leeuwen, B.; Berns, K.; Mullenders, J.; Beijersbergen, R.L.; Bernards, R.; Voorhoeve, P.M.; Agami, R. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell 2005, 121, 849–858. [Google Scholar] [CrossRef] [Green Version]
- Brummelkamp, T.R.; Nijman, S.M.; Dirac, A.M.; Bernards, R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 2003, 424, 797–801. [Google Scholar] [CrossRef]
- Morgens, D.W.; Deans, R.M.; Li, A.; Bassik, M.C. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol. 2016, 34, 634–636. [Google Scholar] [CrossRef] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable dual-RNA—Guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [Green Version]
- Cheng, A.; Wang, H.; Yang, H.; Shi, L.; Katz, Y.; Theunissen, T.; Rangarajan, S.; Shivalila, C.S.; Dadon, D.B.; Jaenisch, R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013, 23, 1163–1171. [Google Scholar] [CrossRef]
- Gilbert, L.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-Mediated modular RNA-Guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.; Ewen-Campen, B.; Ni, X.; Housden, B.; Perrimon, N. In vivo transcriptional activation using crispr/cas9 in drosophila. Genetics 2015, 201, 433–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konermann, S.; Brigham, M.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.; Habib, N.; Gootenberg, J.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef] [Green Version]
- Tanenbaum, M.E.; Gilbert, L.A.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014, 159, 635–646. [Google Scholar] [CrossRef] [Green Version]
- Hilton, I.B.; D’ippolito, A.M.; Vockley, C.M.; Thakore, P.I.; Crawford, G.E.; Reddy, T.E.; Gersbach, C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015, 33, 510–517. [Google Scholar] [CrossRef] [Green Version]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knock-out screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Wei, J.J.; Sabatini, D.M.; Lander, E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 2014, 343, 80–84. [Google Scholar] [CrossRef] [Green Version]
- Joung, J.; Engreitz, J.M.; Konermann, S.; Abudayyeh, O.O.; Verdine, V.K.; Aguet, F.; Gootenberg, J.S.; Sanjana, N.E.; Wright, J.B.; Fulco, C.P.; et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 2017, 548, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Esposito, R.; Bosch, N.; Lanzós, A.; Polidori, T.; Pulido-Quetglas, C.; Johnson, R. Hacking the cancer genome: Profiling therapeutically actionable long non-coding rnas using crispr-cas9 screening. Cancer Cell 2019, 35, 545–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonçalves, E.; Thomas, M.; Behan, F.M.; Picco, G.; Pacini, C.; Allen, F.; Vinceti, A.; Sharma, M.; Jackson, D.A.; Price, S.; et al. Minimal genome-wide human CRISPR-Cas9 library. Genome Biol. 2021, 22, 40. [Google Scholar] [CrossRef]
- DeWeirdt, P.C.; Sangree, A.K.; Hanna, R.E.; Sanson, K.R.; Hegde, M.; Strand, C.; Persky, N.S.; Doench, J.G. Genetic screens in isogenic mammalian cell lines without single cell cloning. Nat. Commun. 2020, 11, 752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 2014, 11, 783–784. [Google Scholar] [CrossRef] [Green Version]
- Sanson, K.R.; Hanna, R.E.; Hegde, M.; Donovan, K.F.; Strand, C.; Sullender, M.E.; Vaimberg, E.W.; Goodale, A.; Root, D.E.; Piccioni, F.; et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun. 2018, 9, 5416. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Dang, Y.; Wu, Y.; Jia, G.; Anaya, E.; Zhang, J.; Abraham, S.; Choi, J.-G.; Shi, G.; Qi, L.; et al. A CRISPR-based screen identifies genes essential for west-nile-virus-induced cell death. Cell Rep. 2015, 12, 673–683. [Google Scholar] [CrossRef] [Green Version]
- Tzelepis, K.; Koike-Yusa, H.; De Braekeleer, E.; Li, Y.; Metzakopian, E.; Dovey, O.M.; Mupo, A.; Grinkevich, V.; Li, M.; Mazan, M.; et al. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 2016, 17, 1193–1205. [Google Scholar] [CrossRef] [Green Version]
- Peets, E.M.; Crepaldi, L.; Zhou, Y.; Allen, F.; Elmentaite, R.; Noell, G.; Turner, G.; Iyer, V.; Parts, L. Minimized double guide RNA libraries enable scale-limited CRISPR/Cas9 screens. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Cao, Z.; Liu, Z.; He, Y.; Wang, Y.; Yuan, P.; Li, W.; Tian, F.; Bao, Y.; Wei, W. Guide RNAs with embedded barcodes boost CRISPR-pooled screens. Genome Biol. 2019, 20, 20. [Google Scholar] [CrossRef]
- Liu, J.; Srinivasan, S.; Li, C.-Y.; Ho, I.-L.; Rose, J.; Shaheen, M.; Wang, G.; Yao, W.; Deem, A.; Bristow, C.; et al. Pooled library screening with multiplexed Cpf1 library. Nat. Commun. 2019, 10, 3144. [Google Scholar] [CrossRef] [Green Version]
- Hart, T.; Chandrashekhar, M.; Aregger, M.; Steinhart, Z.; Brown, K.; MacLeod, G.; Mis, M.; Zimmermann, M.; Fradet-Turcotte, A.; Sun, S.; et al. High-Resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 2015, 163, 1515–1526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horlbeck, M.A.; Gilbert, L.; Villalta, J.; Adamson, B.; Pak, R.; Chen, Y.; Fields, A.P.; Park, C.Y.; Corn, J.; Kampmann, M.; et al. Compact and highly active next- generation libraries for CRISPR-mediated gene repression and activation. Elife 2016, 5, e19760. [Google Scholar] [CrossRef]
- Joung, J.; Konermann, S.; Gootenberg, J.S.; Abudayyeh, O.O.; Platt, R.J.; Brigham, M.D.; Sanjana, N.E.; Zhang, F. Genome-scale CRISPR-Cas9 knock-out and transcriptional activation screening. Nat. Protoc. 2017, 12, 828–863. [Google Scholar] [CrossRef] [Green Version]
- Kurata, M.; Yamamoto, K.; Moriarity, B.S.; Kitagawa, M.; Largaespada, D.A. CRISPR/Cas9 library screening for drug target discovery. J. Hum. Genet. 2018, 63, 179–186. [Google Scholar] [CrossRef]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R.; et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef] [Green Version]
- Bester, A.C.; Lee, J.; Chavez, A.; Lee, Y.-R.; Nachmani, D.; Vora, S.; Victor, J.; Sauvageau, M.; Monteleone, E.; Rinn, J.L.; et al. An integrated genome-wide CRISPRa approach to functionalize lncrnas in drug resistance. Cell 2018, 173, 649–664.e20. [Google Scholar] [CrossRef]
- He, C.; Han, S.; Chang, Y.; Wu, M.; Zhao, Y.; Chen, C.; Chu, X. CRISPR screen in cancer: Status quo and future perspectives. Am. J. Cancer Res. 2021, 11, 1031. [Google Scholar]
- Dhanjal, J.K.; Radhakrishnan, N.; Sundar, D. Identifying synthetic lethal targets using CRISPR/Cas9 system. Methods 2017, 131, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, M.; Cho, T.; Álvarez-Quilón, A.; Li, K.; Schellenberg, M.J.; Zimmermann, M.; Hustedt, N.; Rossi, S.E.; Adam, S.; Melo, H.; et al. A genetic map of the response to DNA damage in human cells. Cell 2020, 182, 481–496.e21. [Google Scholar] [CrossRef] [PubMed]
- Setton, J.; Zinda, M.; Riaz, N.; Durocher, D.; Zimmermann, M.; Koehler, M.; Reis-Filho, J.S.; Powell, S.N. Synthetic lethality in cancer therapeutics: The next generation. Cancer Discov. 2021, 11, 1626–1635. [Google Scholar] [CrossRef] [PubMed]
- Yau, E.H.; Kummetha, I.R.; Lichinchi, G.; Tang, R.; Zhang, Y.; Rana, T.M. Genome-wide CRISPR screen for essential cell growth mediators in mutant KRAS Colorectal cancers. Cancer Res. 2017, 77, 6330–6339. [Google Scholar] [CrossRef] [Green Version]
- Steinhart, Z.; Pavlovic, Z.; Chandrashekhar, M.; Hart, T.; Wang, X.; Zhang, X.; Robitaille, M.; Brown, K.; Jaksani, S.; Overmeer, R.; et al. Genome-wide CRISPR screens reveal a Wnt-FZD5 signaling circuit as a druggable vulnerability of RNF43-mutant pancreatic tumors. Nat. Med. 2016, 23, 60–68. [Google Scholar] [CrossRef]
- Abraham, C.G.; Ludwig, M.P.; Andrysik, Z.; Pandey, A.; Joshi, M.; Galbraith, M.D.; Sullivan, K.D.; Espinosa, J.M. ΔNp63α suppresses TGFB2 expression and RHOA activity to drive cell proliferation in squamous cell carcinomas. Cell Rep. 2018, 24, 3224–3236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oser, M.G.; Fonseca, R.; Chakraborty, A.A.; Brough, R.; Spektor, A.; Jennings, R.; Flaifel, A.; Novak, J.S.; Gulati, A.; Buss, E.; et al. Cells lacking the rb1 tumor suppressor gene are hyperdependent on aurora b kinase for survival. Cancer Discov. 2021, 9, 230–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, J.; Zhao, H.; Diplas, B.H.; Liu, S.; Liu, J.; Wang, D.; Lu, Y.; Zhu, Q.; Wu, J.; Wang, W. Genome-Wide CRISPR-Cas9 screen reveals selective vulnerability of atrx-mutant cancers to wee1 inhibition. Cancer Res. 2019, 80, 510–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, K.; Jeng, E.; Hess, G.; Morgens, D.W.; Li, A.; Bassik, M.C. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol. 2017, 35, 463–474. [Google Scholar] [CrossRef]
- Hinze, L.; Pfirrmann, M.; Karim, S.; Degar, J.; McGuckin, C.; Vinjamur, D.; Sacher, J.; Stevenson, K.E.; Neuberg, D.S.; Orellana, E.; et al. Synthetic lethality of wnt pathway activation and asparaginase in drug-resistant acute leukemias. Cancer Cell 2019, 35, 664–676.e7. [Google Scholar] [CrossRef] [Green Version]
- Sulahian, R.; Kwon, J.J.; Walsh, K.H.; Pailler, E.; Bosse, T.L.; Thaker, M.; Almanza, D.; Dempster, J.M.; Pan, J.; Piccioni, F.; et al. Synthetic lethal interaction of SHOC2 depletion with MEK inhibition in ras-driven Cancers. Cell Rep. 2019, 29, 118–134.e8. [Google Scholar]
- Wang, C.; Wang, G.; Feng, X.; Shepherd, P.; Zhang, J.; Tang, M.; Chen, Z.; Srivastava, M.; McLaughlin, M.E.; Navone, N.M.; et al. Genome-wide CRISPR screens reveal synthetic lethality of RNASEH2 deficiency and ATR inhibition. Oncogene 2019, 38, 2451–2463. [Google Scholar] [PubMed]
- Shu, S.; Wu, H.J.; Jennifer, Y.G.; Zeid, R.; Harris, I.S.; Jovanović, B.; Murphy, K.; Wang, B.; Qiu, X.; Endress, J.E.; et al. Synthetic lethal and resistance interactions with bet bromodomain inhibitors in triple-negative breast cancer. Mol. Cell 2020, 78, 1096–1113.e8. [Google Scholar] [CrossRef]
- Gier, R.A.; Budinich, K.A.; Evitt, N.H.; Cao, Z.; Freilich, E.S.; Chen, Q.; Qi, J.; Lan, Y.; Kohli, R.M.; Shi, J. High-performance CRISPR-Cas12a genome editing for combinatorial genetic screening. Nat. Commun. 2020, 11, 3455. [Google Scholar] [CrossRef]
- Bersuker, K.; Hendricks, J.M.; Li, Z.; Magtanong, L.; Ford, B.; Tang, P.H.; Roberts, M.A.; Tong, B.; Maimone, T.J.; Zoncu, R.; et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 2019, 575, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Einstein, J.M.; Perelis, M.; Chaim, I.A.; Meena, J.K.; Nussbacher, J.K.; Tankka, A.T.; Yee, B.A.; Li, H.; Madrigal, A.A.; Neill, N.J.; et al. Inhibition of YTHDF2 triggers proteotoxic cell death in MYC-driven breast cancer. Mol. Cell 2021, 81, 3048–3064.e9. [Google Scholar] [CrossRef] [PubMed]
- Mengwasser, K.E.; Adeyemi, R.O.; Leng, Y.; Choi, M.Y.; Clairmont, C.; D’Andrea, A.D.; Elledge, S.J. Genetic Screens reveal FEN1 and APEX2 as BRCA2 synthetic lethal targets. Mol. Cell 2019, 73, 885–899.e6. [Google Scholar] [CrossRef] [Green Version]
- Bakke, J.; Wright, W.C.; Zamora, A.E.; Oladimeji, P.; Crawford, J.C.; Brewer, C.T.; Autry, R.J.; Evans, W.E.; Thomas, P.G.; Chen, T. Genome-wide CRISPR screen reveals PSMA6 to be an essential gene in pancreatic cancer cells. BMC Cancer 2019, 19, 253. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Krall, E.B.; Aguirre, A.J.; Kim, M.; Widlund, H.; Doshi, M.B.; Sicinska, E.; Sulahian, R.; Goodale, A.; Cowley, G.S.; et al. ATXN1L, CIC, and ETS transcription factors modulate sensitivity to mapk pathway inhibition. Cell Rep. 2017, 18, 1543–1557. [Google Scholar] [CrossRef] [Green Version]
- Szlachta, K.; Kuscu, C.; Tufan, T.; Adair, S.J.; Shang, S.; Michaels, A.D.; Mullen, M.G.; Fischer, N.L.; Yang, J.; Liu, L. CRISPR knock-out screening identifies combinatorial drug targets in pancreatic cancer and models cellular drug response. Nat. Commun. 2018, 9, 4275. [Google Scholar] [CrossRef] [Green Version]
- Sarr, A.; Bré, J.; Um, I.H.; Chan, T.H.; Mullen, P.; Harrison, D.J.; Reynolds, P.A. Genome-scale CRISPR/Cas9 screen determines factors modulating sensitivity to ProTide NUC-1031. Sci. Rep. 2019, 9, 7643. [Google Scholar] [CrossRef]
- Toledo, C.M.; Ding, Y.; Hoellerbauer, P.; Davis, R.J.; Basom, R.; Girard, E.; Lee, E.; Corrin, P.; Hart, T.; Bolouri, H.; et al. Genome-wide CRISPR-Cas9 screens reveal loss of redundancy between PKMYT1 and wee1 in glioblastoma stem-like cells. Cell Rep. 2015, 13, 2425–2439. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Bailey, P.; Pilarsky, C. CRISPR Cas9 in pancreatic cancer research. Front. Cell Dev. Biol. 2019, 7, 239. [Google Scholar] [CrossRef] [PubMed]
- Kinowaki, Y.; Kurata, M.; Ishibashi, S.; Ikeda, M.; Tatsuzawa, A.; Yamamoto, M.; Miura, O.; Kitagawa, M.; Yamamoto, K. Glutathione peroxidase 4 overexpression inhibits ROS-induced cell death in diffuse large B-cell lymphoma. Lab. Investig. 2018, 98, 609–619. [Google Scholar] [CrossRef]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.; et al. Defining a cancer dependency map. Cell 2017, 170, 564–576.e16. [Google Scholar] [CrossRef] [Green Version]
- Dwane, L.; Behan, F.M.; Gonçalves, E.; Lightfoot, H.; Yang, W.; van der Meer, D.; Shepherd, R.; Pignatelli, M.; Iorio, F.; Garnett, M.J.; et al. Project score database: A resource for investigating cancer cell dependencies and prioritizing therapeutic targets. Nucleic Acids Res. 2021, 49, D1365–D1372. [Google Scholar] [CrossRef] [PubMed]
- Lenoir, W.F.; Lim, T.L.; Hart, T. PICKLES: The database of pooled in-vitro CRISPR knock-out library essentiality screens. Nucleic Acids Res. 2018, 46, D776–D780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, A.; Jang, I.; Han, H.; Kim, M.-S.; Choi, J.; Lee, J.; Cho, S.-Y.; Jun, Y.; Lee, C.; Kim, J.; et al. iCSDB: An integrated database of CRISPR screens. Nucleic Acids Res. 2021, 49, D956–D961. [Google Scholar] [CrossRef]
- Cui, Y.; Cheng, X.; Chen, Q.; Song, B.; Chiu, A.; Gao, Y.; Dawson, T.; Chao, L.; Zhang, W.; Li, D.; et al. CRISP-view: A database of functional genetic screens spanning multiple phenotypes. Nucleic Acids Res. 2021, 49, D848–D854. [Google Scholar] [CrossRef]
- Pacini, C.; Dempster, J.M.; Boyle, I.; Gonçalves, E.; Najgebauer, H.; Karakoc, E.; van der Meer, D.; Barthorpe, A.; Lightfoot, H.; Jaaks, P.; et al. Integrated cross-study datasets of genetic dependencies in cancer. Nat. Commun. 2021, 12, 1661. [Google Scholar] [CrossRef] [PubMed]
- Corsello, S.M.; Nagari, R.T.; Spangler, R.D.; Rossen, J.; Kocak, M.; Bryan, J.G.; Humeidi, R.; Peck, D.; Wu, X.; Tang, A.A.; et al. Discovering the anticancer potential of non-oncology drugs by systematic viability profiling. Nat. Cancer 2020, 1, 235–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Name | Advantage | sgRNAS/Gene | Total gRNAs | Ref. |
|---|---|---|---|---|
| Garnett Lab MinLibCas9 Library | Minimal genome-wide human CRISPR-Cas9 library | 2 | 37,722 | [20] |
| Human CRISPR Knockout Pooled Library (Gattinara) | Minimal genome-wide human CRISPR-Cas9 library compatible with the Brunello library | 2 | 40,964 | [21] |
| Human GeCKO v2 | Targets early consecutive exons Contains 1000 control (non-targeting) sgRNAs | 3 or 6 | 123,411 | [22] |
| Broad GPP genome-wide Brunello | Improved on-target activity predictions and off-target scores compared to the GeCKOv2 library | 4 | 76,441 | [23] |
| Human genome-wide library v1 | Targets sites in a region close to the translation initiation site for complete gene disruption | 4 | 77,406 | [24] |
| Human improved genome-wide library v1 | gRNAs redesigned using pipeline with a new design Improved on-target sensitivity and reduced off-target effect scaffold | 5 | 90,709 | [25] |
| Human genome-wide reduced double-gRNA library | Optimization of guide RNA designs and delivery of two gRNAs with each construct | 3 | 59,576 | [26] |
| Human whole genome sgRNA iBAR library | Incorporates four 6-basepair internal barcodes (iBARs) in each sgRNA Efficient and accurate screening at high MOI | 3 | 58,630 | [27] |
| Mini-human AsCpf1-based human genome-wide knockout library | Each gene targeted by an AsCpf1(AsCas12a)-based array containing 3–4 guides concatenated in one vector | 3–4 | 17,032 | [28] |
| Toronto KnockOut (TKO) version3 | Improved accuracy, efficiency, and scalability for CRISPR screens compared to TKO version 1 | 4 | 70,948 | [29] |
| Name | Advantages | sgRNAs/Gene | Total gRNAs | Ref. |
|---|---|---|---|---|
| Activation | ||||
| CRISPRa-v2 | SunTag-VP64 activation system | 5 or 10 | 104,540 or 209,080 | [30] |
| SAM (Synergistic Activation Mediator) v1–3 plasmid system | Comprises three plasmids (Cas9-VP64 fusion, gRNA incorporating two MS2 RNA aptamers at the tetraloop and stem-loop 2, and MS2-P65-HST) Efficient gene upregulation | 3 | 70,290 | [13] |
| SAM v2–2 plasmid system | Comprises two plasmids (gRNA library–lenti SAM v2 backbone and MS2-P65-HST) Efficient gene upregulation | 3 | 70,290 | [31] |
| Human CRISPR lncRNA activation pooled library | SAM library for transcriptional activation of lncRNAs | 10 | 96,458 | [32] |
| Broad GPP activation Calabrese p65-HSF | Modified tracrRNA with two MS2 loops and two PP7 loops Better concordance of sgRNAs compared to the SAM v2 library | 3 or 6 | 56,762 (Set A) 56,476 (Set B) | [23] |
| Inhibition | ||||
| CRISPRi-v2 | dCas9-KRAB represses TSS downstream of TSS sites | 5 or 10 | 104,535 209,070 | [30] |
| Broad GPP inhibition Dolcetto | gRNAs redesigned based on FANTOM5 CAGE data Gene regulation equal to the CRISPR KO library | 3 or 6 | 57,050 (Set A) 57,011 (Set B) | [23] |
| Cancer Type (Cell Line) | Altered Gene/Drug | CRISPR Type | Library | Synthetic Lethal Hits | Ref. |
|---|---|---|---|---|---|
| Colorectal cancer (HCT116) | KRAS (G13D) | Knockout | GeCKOv2 | NADK, KHK, INO80C | [39] |
| Pancreatic cancer (HPAF-II) | RNF43 | Knockout | TKO | FZD5, Wnt pathway genes | [40] |
| Lung squamous cell carcinoma (H226 shp63) | ΔNp63α | Knockout | GeCKOv2 | RHOA, TGFBR2 | [41] |
| Small-cell lung cancer (NCI-H82) | RB1−/− | Knockout | Custom | Aurora kinase B | [42] |
| Hepatocellular carcinoma (PLC/PRF/5) | ATRX loss | Knockout | GeCKOv2 | WEE1 | [43] |
| Chronic myelogenous leukemia (K562) | – | Double knockout | Paired sgRNA | BCL2L1–MCL1 combination | [44] |
| T-acute lymphocytic leukemia (CCRF-CEM) | Asparaginase-resistant | Knockout | GeCKO | NKD2, LGR6, ASNS | [45] |
| Pancreatic cancer, non-small-cell lung cancer (CFPAC-1, A549, NCIH23) | MEK1/2 inhibition | Knockout | Avana-4 barcoded sgRNA | SHOC2 | [46] |
| Colorectal cancer, breast cancer (HCT116, MCF10A) | ATR inhibition | Knockout | TKOv3 | RNASEH2 | [47] |
| Triple-negative breast cancer (SUM159, SUM149) | BET bromodomain inhibitor | Knockout | H1 and H2 | CDK4 and BRD2 | [48] |
| Murine acute myelogenous leukemia (RN2) | – | Cas12a (Cpf1) multigene knockout | Custom | BRD9 & JMJD6, KAT6A & JMJD6, BRPF1 & JMJD6 | [49] |
| Osteosarcoma (U2) | GPX4 (ferroptosis-resistant) | Knockout | Custom | FSP1 (AIFM2) | [50] |
| Myc-driven breast cancer model (MYC-ER HMECs) | MYC | Knockout | RNA-binding protein pooled CRISPR knockout | YTHDF2 | [51] |
| Colorectal cancer (BRCA2−/− DLD1) | BRCA2 mutation | Knockout | Custom | FEN1, APEX2 | [52] |
| Pancreatic cancer (PANC-1) | Gemcitabine | Knockout | Brunello | PSMA6 | [53] |
| Pancreatic cancer (PATU8902) | Trametinib | Knockout | GeCKOv2 Avana | CIC, ATXN1L | [54] |
| Pancreatic cancer (PDX366) | MEK and CENPE inhibitor | Knockout | Nuclear proteins gRNA sub-pool | CENPE, RRM1 | [55] |
| Pancreatic cancer (Mia PaCa-2, A2780) | Gemcitabine, NUC-1031 | Knockout | GeCKOv2 | DCK, DCTPP1 | [56] |
| Glioblastoma stem-like cells (2 patient-derived cells) | EGFR+PI3K signaling | Knockout | GeCKOv1 | PKMYT1, WEE1 | [57] |
| Primary human retinal pigment epithelial cells (RPE1-hTERT p53−/− Flag-Cas9 cells) | 27 DNA-damaging agents | Knockout | TKO v2 TKO v3 | ERCC6L2, TOP2, ELOF1, STK19 | [58] |
| Database | Characteristics | Number of Cell Lines | Usage | Ref. | URL |
|---|---|---|---|---|---|
| DepMap portal | Integrates CRISPR KO screening databases (DepMap, Sanger, and GeCKO) and unifies cellular model (CCLE) and drug sensitivity (PRISM) databases | 786 cell lines 42 cancer types | Discovering genetic and pharmacological dependenciesPrioritizing tumor contexts and predictive biomarkers Exploring over 2000 cancer models | [60] | https://depmap.org/portal/ (accessed on 24 Octorber 2021) |
| Project Score | Genetic screens for identifying cancer dependencies | 914 cell lines 25 tissues 7470 fitness genes | Investigating specific genes, cancer cell models, or tissue types Browsing all gene fitness scores | [61] | https://score.depmap.sanger.ac.uk/ (accessed on 24 Octorber 2021) |
| PICKLES (Pooled In-Vitro CRISPR Knockout Library Essentiality Screens) | Cell line essentiality profiles for CRISPR KO library and shRNA datasets | More than 50 cell lines for CRISPR screening and 100 cell lines for shRNA library | Easily exploring cell line essentiality and co-essentiality | [62] | http://pickles.hart-lab.org (accessed on 24 Octorber 2021) |
| iCSDB | Integrated DepMap portal and BioGRID ORCS Integrated database of CRISPR-CAS9 screening experiments for human cell lines and clinical information | 976 cell lines | Easily searching for cell line data associated with clinical and molecular data | [63] | https://www.kobic.re.kr/icsdb/ (accessed on 24 Octorber 2021) |
| CRISP-view | Data from 167 studies collected from PubMed, Gene Expression Omnibus (GEO), and Ensemble and Cancer Cell Line Encyclopedia (CCLE) | 321 human samples 825 mouse samples | Web interface visualizing datasets, allowing the exploration of interesting genes, cell lines, tissues, studies, or conditions | [64] | http://crispview.weililab.org (accessed on 24 Octorber 2021) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Onishi, I.; Yamamoto, K.; Kinowaki, Y.; Kitagawa, M.; Kurata, M. To Discover the Efficient and Novel Drug Targets in Human Cancers Using CRISPR/Cas Screening and Databases. Int. J. Mol. Sci. 2021, 22, 12322. https://doi.org/10.3390/ijms222212322
Onishi I, Yamamoto K, Kinowaki Y, Kitagawa M, Kurata M. To Discover the Efficient and Novel Drug Targets in Human Cancers Using CRISPR/Cas Screening and Databases. International Journal of Molecular Sciences. 2021; 22(22):12322. https://doi.org/10.3390/ijms222212322
Chicago/Turabian StyleOnishi, Iichiroh, Kouhei Yamamoto, Yuko Kinowaki, Masanobu Kitagawa, and Morito Kurata. 2021. "To Discover the Efficient and Novel Drug Targets in Human Cancers Using CRISPR/Cas Screening and Databases" International Journal of Molecular Sciences 22, no. 22: 12322. https://doi.org/10.3390/ijms222212322
APA StyleOnishi, I., Yamamoto, K., Kinowaki, Y., Kitagawa, M., & Kurata, M. (2021). To Discover the Efficient and Novel Drug Targets in Human Cancers Using CRISPR/Cas Screening and Databases. International Journal of Molecular Sciences, 22(22), 12322. https://doi.org/10.3390/ijms222212322






