Modulation of Neurolipid Signaling and Specific Lipid Species in the Triple Transgenic Mouse Model of Alzheimer’s Disease
Abstract
:1. Introduction
2. Results
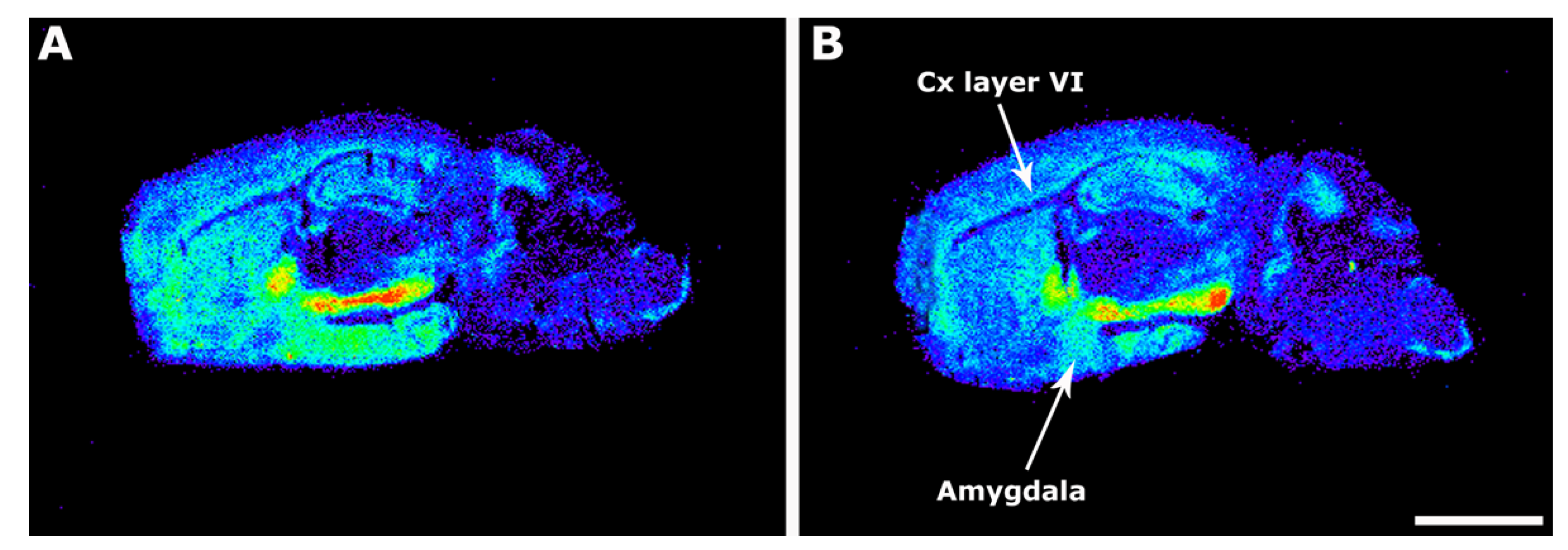
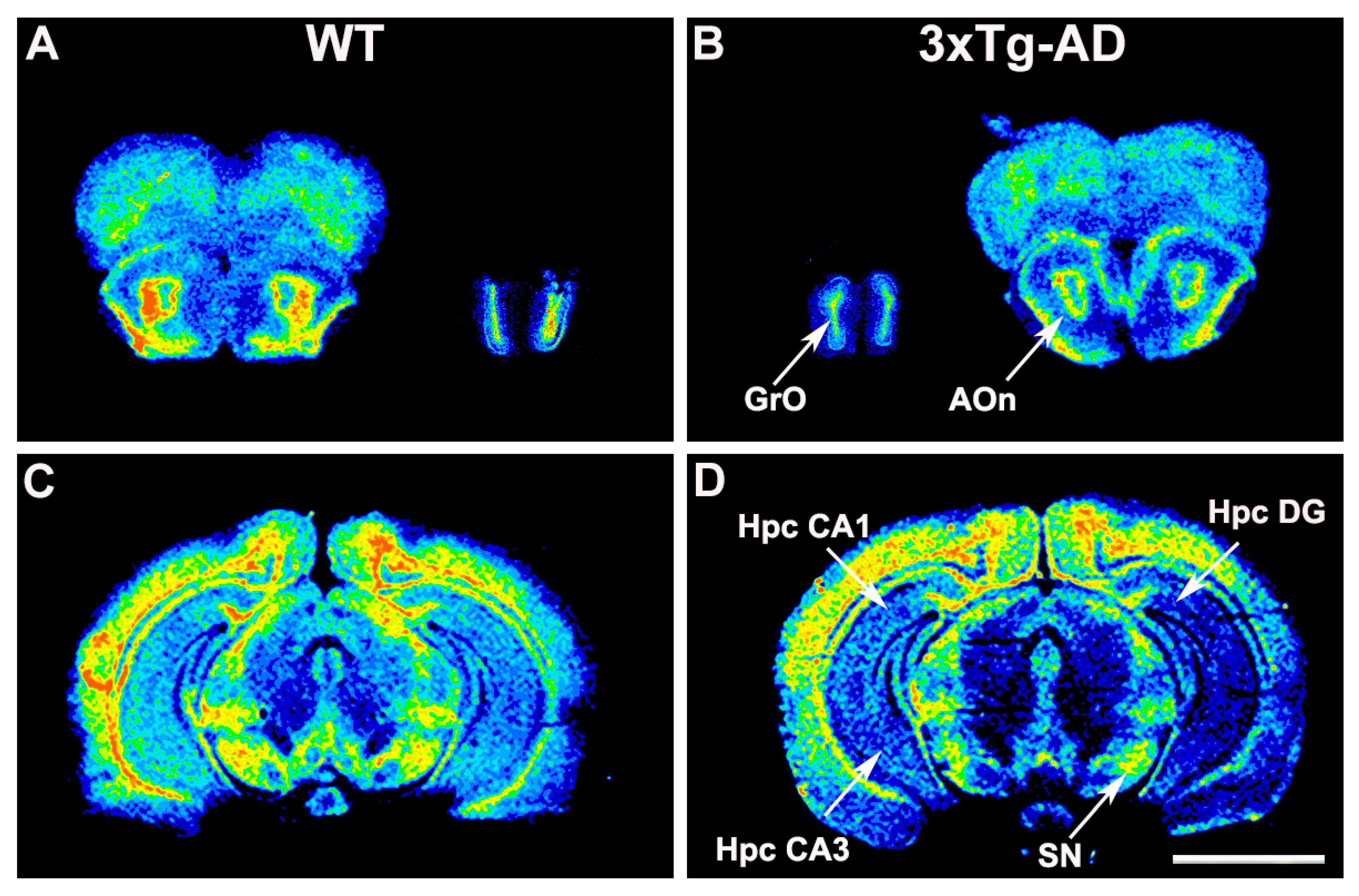
2.1. [35S]GTPγS Binding Assay in 3xTg-AD Mice Brain Sections
2.2. Cannabinoid Receptor Density
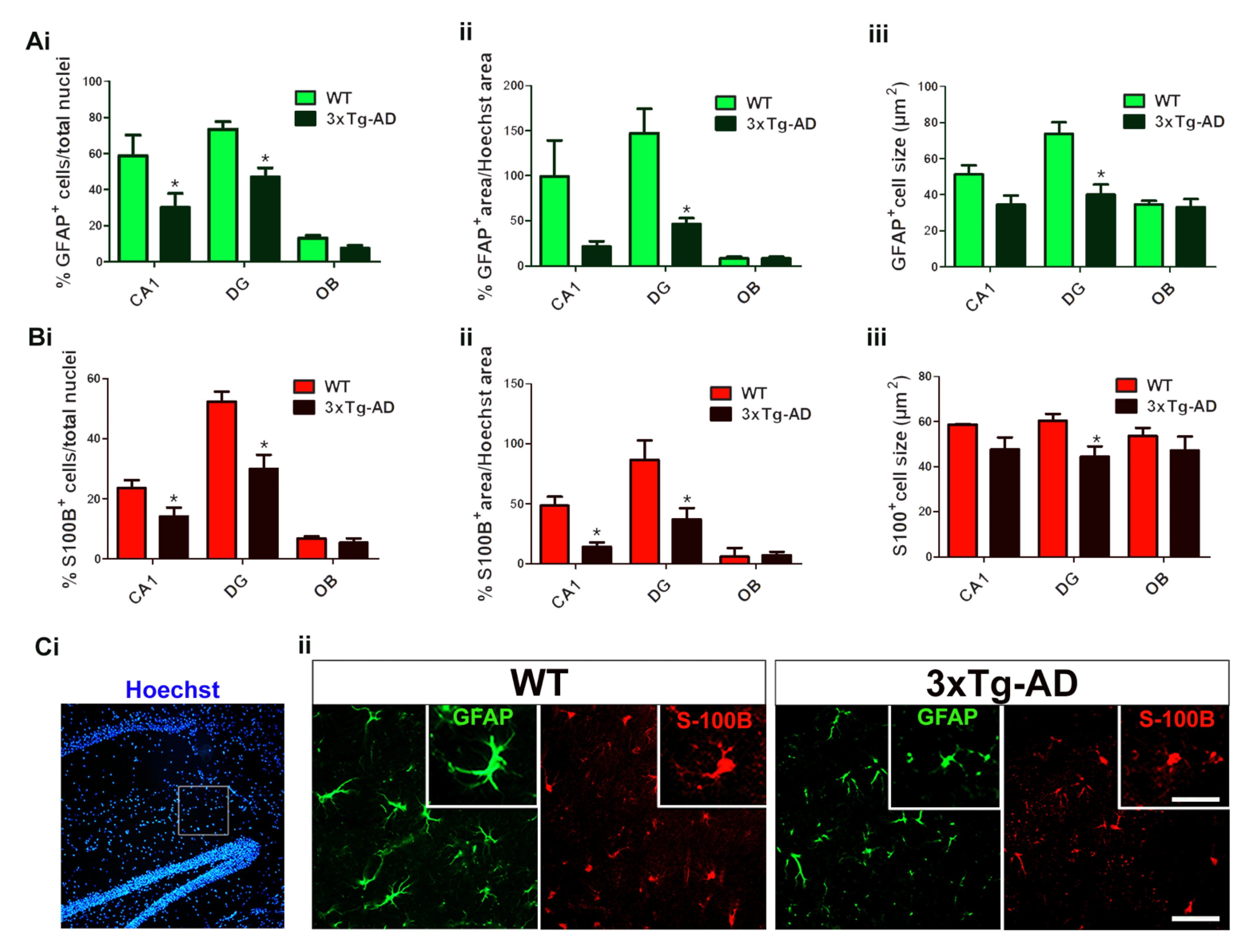
2.3. GPCR-Immunoreactivity and Astrocyte Density
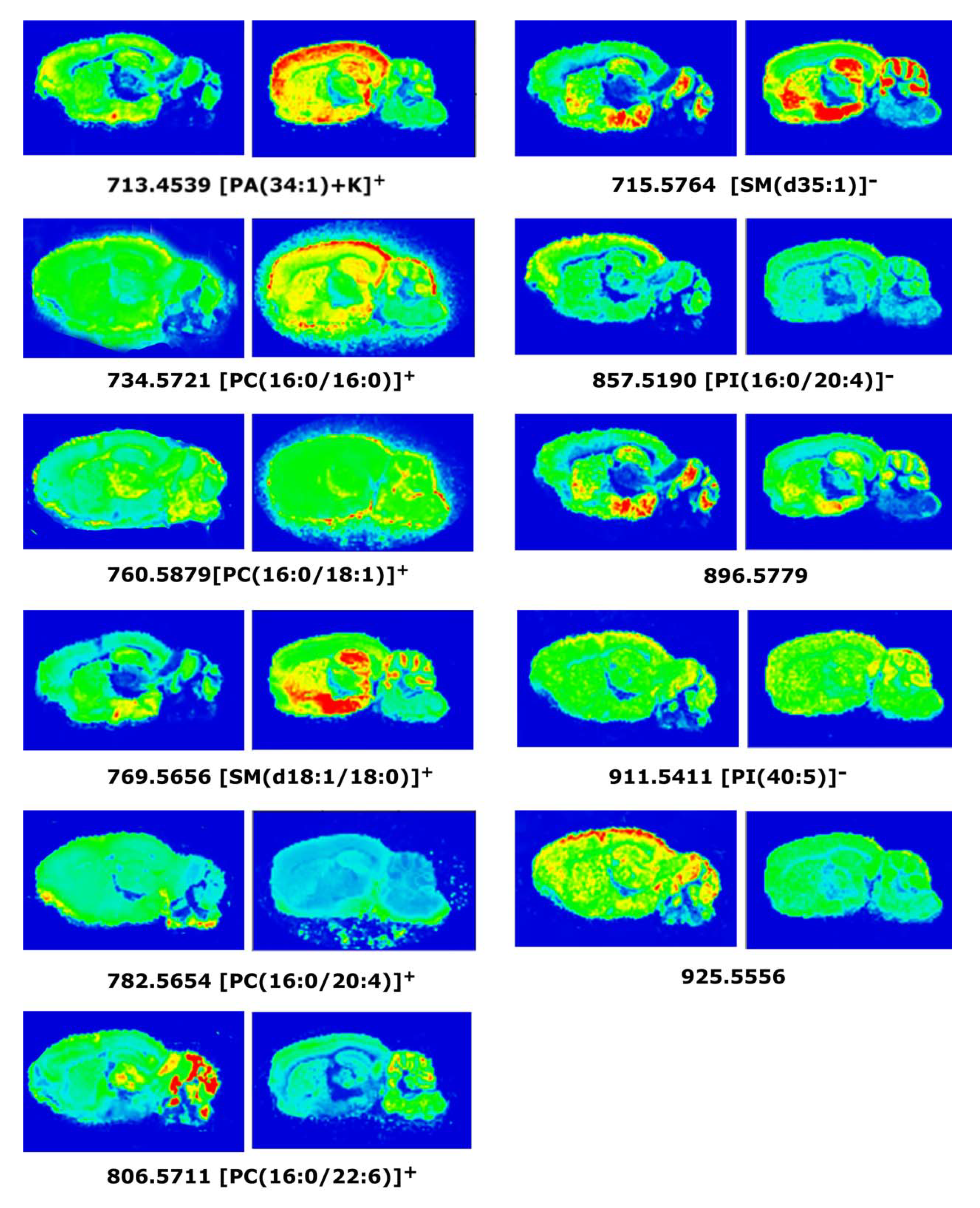
2.4. MALDI-MSI Assay in 3xTg-AD Mice Brain Sections
3. Discussion
3.1. Modulation of CB1 Receptor Activity
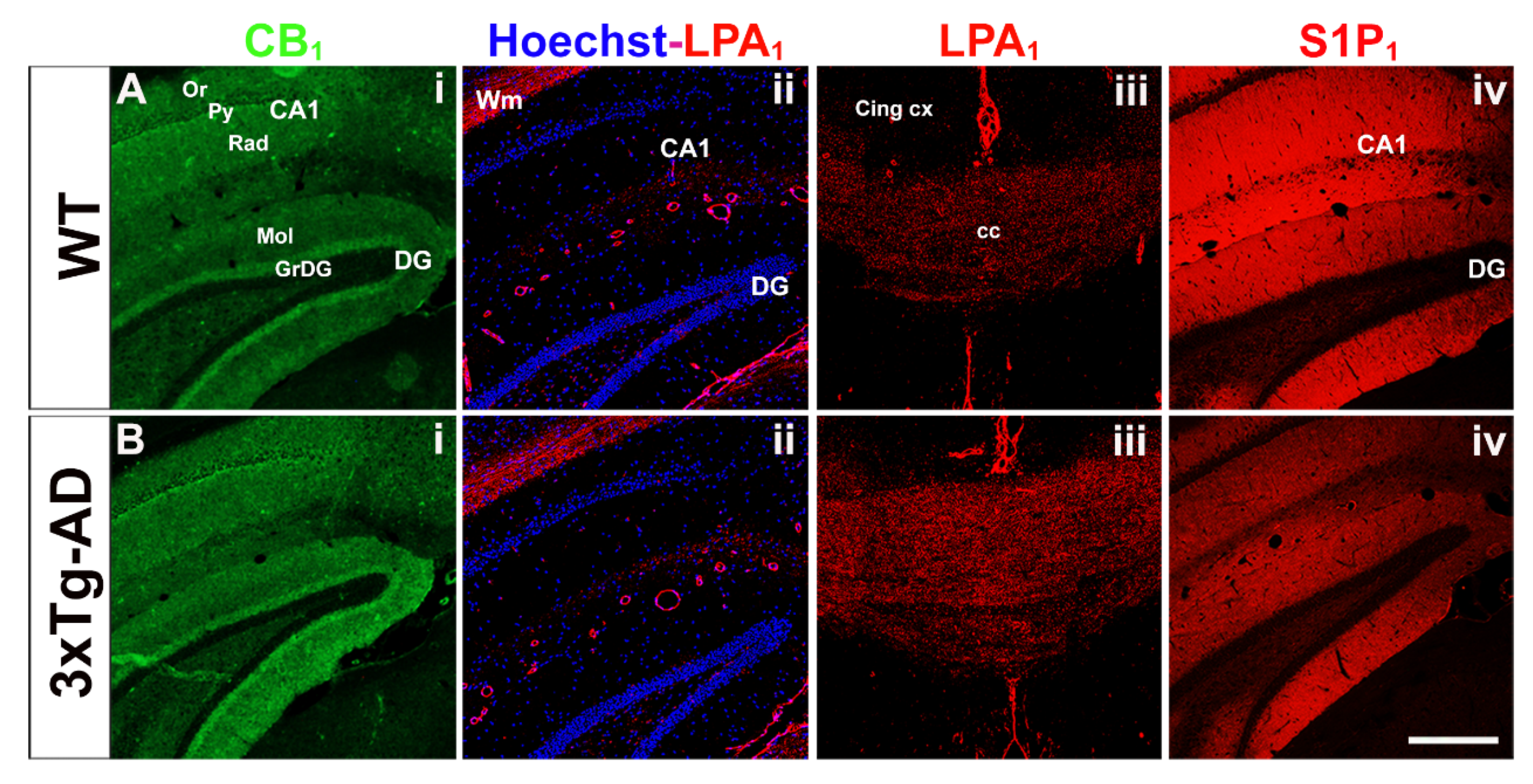
3.2. Modulation of LPA1 Receptor Activity
3.3. Modulation of S1P1 Receptor Activity
3.4. Anatomical Localization of Lipid Species in 3xTg-AD Mice Brain by MALDI-MSI Assay
4. Materials and Methods
4.1. Chemicals
4.2. Animals and Tissue Preparation
4.3. Tissue Preparation
4.4. [35S]GTPγS Binding Assay
4.5. Quantitative Image Analysis of Film Autoradiograms
4.6. Immunofluorescence Studies
4.7. Quantitative Analyses of Astrocytes
4.8. Sample Preparation for MALDI-MSI
4.9. Mass Spectrometer
4.10. Image and Spectra Analysis for MALDI-MSI
4.11. Peak Assignment
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Tierney, M.C.; Fisher, R.H.; Lewis, A.J.; Zorzitto, M.L.; Snow, W.G.; Reid, D.W.; Nieuwstraten, P. The NINCDS-ADRDA Work Group Criteria for the Clinical Diagnosis of Probable Alzheimer’s Disease: A Clinicopathologic Study of 57 Cases. Neurology 1988, 38, 359–364. [Google Scholar] [CrossRef]
- Dickson, D.W. The Pathogenesis of Senile Plaques. J. Neuropathol. Exp. Neurol. 1997, 56, 321–339. [Google Scholar] [CrossRef] [Green Version]
- Selkoe, D.J. Alzheimer’s Disease Results from the Cerebral Accumulation and Cytotoxicity of Amyloid Beta-Protein. J. Alzheimer’s Dis. 2001, 3, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Surguchov, A. Caveolin: A New Link Between Diabetes and AD. Cell. Mol. Neurobiol. 2020, 40, 1059–1066. [Google Scholar] [CrossRef]
- Goate, A.; Chartier-Harlin, M.C.; Mullan, M.; Brown, J.; Crawford, F.; Fidani, L.; Giuffra, L.; Haynes, A.; Irving, N.; James, L. Segregation of a Missense Mutation in the Amyloid Precursor Protein Gene with Familial Alzheimer’s Disease. Nature 1991, 349, 704–706. [Google Scholar] [CrossRef] [PubMed]
- Sherrington, R.; Rogaev, E.I.; Liang, Y.; Rogaeva, E.A.; Levesque, G.; Ikeda, M.; Chi, H.; Lin, C.; Li, G.; Holman, K.; et al. Cloning of a Gene Bearing Missense Mutations in Early-Onset Familial Alzheimer’s Disease. Nature 1995, 375, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Levy-Lahad, E.; Wasco, W.; Poorkaj, P.; Romano, D.M.; Oshima, J.; Pettingell, W.H.; Yu, C.E.; Jondro, P.D.; Schmidt, S.D.; Wang, K. Candidate Gene for the Chromosome 1 Familial Alzheimer’s Disease Locus. Science 1995, 269, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J. Amyloid, the Presenilins and Alzheimer’s Disease. Trends Neurosci. 1997, 20, 154–159. [Google Scholar] [CrossRef]
- Price, D.L.; Tanzi, R.E.; Borchelt, D.R.; Sisodia, S.S. Alzheimer’s Disease: Genetic Studies and Transgenic Models. Annu. Rev. Genet. 1998, 32, 461–493. [Google Scholar] [CrossRef]
- McGeer, P.L.; McGeer, E.G.; Yasojima, K. Alzheimer Disease and Neuroinflammation. J. Neural Transm. Suppl. 2000, 59, 53–57. [Google Scholar] [CrossRef]
- Wyss-Coray, T.; Mucke, L. Inflammation in Neurodegenerative Disease--a Double-Edged Sword. Neuron 2002, 35, 419–432. [Google Scholar] [CrossRef] [Green Version]
- Braak, H.; Braak, E. Neuropathological Stageing of Alzheimer-Related Changes. Acta Neuropathol. 1991, 82, 239–259. [Google Scholar] [CrossRef]
- Tanaka, M.; Toldi, J.; Vécsei, L. Exploring the Etiological Links behind Neurodegenerative Diseases: Inflammatory Cytokines and Bioactive Kynurenines. Int. J. Mol. Sci. 2020, 21, 2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vignon, A.; Salvador-Prince, L.; Lehmann, S.; Perrier, V.; Torrent, J. Deconstructing Alzheimer’s Disease: How to Bridge the Gap between Experimental Models and the Human Pathology? Int. J. Mol. Sci. 2021, 22, 8769. [Google Scholar] [CrossRef] [PubMed]
- Marsicano, G.; Goodenough, S.; Monory, K.; Hermann, H.; Eder, M.; Cannich, A.; Azad, S.C.; Cascio, M.G.; Gutiérrez, S.O.; van der Stelt, M.; et al. CB1 Cannabinoid Receptors and On-Demand Defense against Excitotoxicity. Science 2003, 302, 84–88. [Google Scholar] [CrossRef] [Green Version]
- Roher, A.E.; Weiss, N.; Kokjohn, T.A.; Kuo, Y.-M.; Kalback, W.; Anthony, J.; Watson, D.; Luehrs, D.C.; Sue, L.; Walker, D.; et al. Increased A Beta Peptides and Reduced Cholesterol and Myelin Proteins Characterize White Matter Degeneration in Alzheimer’s Disease. Biochemistry 2002, 41, 11080–11090. [Google Scholar] [CrossRef] [PubMed]
- Stella, N.; Schweitzer, P.; Piomelli, D. A Second Endogenous Cannabinoid That Modulates Long-Term Potentiation. Nature 1997, 388, 773–778. [Google Scholar] [CrossRef] [Green Version]
- Blondeau, N.; Lauritzen, I.; Widmann, C.; Lazdunski, M.; Heurteaux, C. A Potent Protective Role of Lysophospholipids against Global Cerebral Ischemia and Glutamate Excitotoxicity in Neuronal Cultures. J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab. 2002, 22, 821–834. [Google Scholar] [CrossRef] [Green Version]
- Oddo, S.; Caccamo, A.; Shepherd, J.D.; Murphy, M.P.; Golde, T.E.; Kayed, R.; Metherate, R.; Mattson, M.P.; Akbari, Y.; LaFerla, F.M. Triple-Transgenic Model of Alzheimer’s Disease with Plaques and Tangles: Intracellular Abeta and Synaptic Dysfunction. Neuron 2003, 39, 409–421. [Google Scholar] [CrossRef] [Green Version]
- Oddo, S.; Caccamo, A.; Kitazawa, M.; Tseng, B.P.; LaFerla, F.M. Amyloid Deposition Precedes Tangle Formation in a Triple Transgenic Model of Alzheimer’s Disease. Neurobiol. Aging 2003, 24, 1063–1070. [Google Scholar] [CrossRef]
- Mastrangelo, M.A.; Bowers, W.J. Detailed Immunohistochemical Characterization of Temporal and Spatial Progression of Alzheimer’s Disease-Related Pathologies in Male Triple-Transgenic Mice. BMC Neurosci. 2008, 9, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giménez-Llort, L.; Marin-Pardo, D.; Marazuela, P.; Hernández-Guillamón, M. Survival Bias and Crosstalk between Chronological and Behavioral Age: Age- and Genotype-Sensitivity Tests Define Behavioral Signatures in Middle-Aged, Old, and Long-Lived Mice with Normal and AD-Associated Aging. Biomedicines 2021, 9, 636. [Google Scholar] [CrossRef]
- Muntsant, A.; Jiménez-Altayó, F.; Puertas-Umbert, L.; Jiménez-Xarrie, E.; Vila, E.; Giménez-Llort, L. Sex-Dependent End-of-Life Mental and Vascular Scenarios for Compensatory Mechanisms in Mice with Normal and AD-Neurodegenerative Aging. Biomedicines 2021, 9, 111. [Google Scholar] [CrossRef] [PubMed]
- Whitehouse, P.J.; Price, D.L.; Struble, R.G.; Clark, A.W.; Coyle, J.T.; Delon, M.R. Alzheimer’s Disease and Senile Dementia: Loss of Neurons in the Basal Forebrain. Science 1982, 215, 1237–1239. [Google Scholar] [CrossRef] [PubMed]
- Vazin, T.; Ball, K.A.; Lu, H.; Park, H.; Ataeijannati, Y.; Head-Gordon, T.; Poo, M.; Schaffer, D. V Efficient Derivation of Cortical Glutamatergic Neurons from Human Pluripotent Stem Cells: A Model System to Study Neurotoxicity in Alzheimer’s Disease. Neurobiol. Dis. 2014, 62, 62–72. [Google Scholar] [CrossRef] [Green Version]
- Palmer, A.M.; Francis, P.T.; Benton, J.S.; Sims, N.R.; Mann, D.M.; Neary, D.; Snowden, J.S.; Bowen, D.M. Presynaptic Serotonergic Dysfunction in Patients with Alzheimer’s Disease. J. Neurochem. 1987, 48, 8–15. [Google Scholar] [CrossRef]
- Bondareff, W.; Mountjoy, C.Q.; Roth, M. Selective Loss of Neurones of Origin of Adrenergic Projection to Cerebral Cortex (Nucleus Locus Coeruleus) in Senile Dementia. Lancet 1981, 1, 783–784. [Google Scholar] [CrossRef]
- Rodríguez-Puertas, R.; Nilsson, S.; Pascual, J.; Pazos, A.; Hökfelt, T. 125I-Galanin Binding Sites in Alzheimer’s Disease: Increases in Hippocampal Subfields and a Decrease in the Caudate Nucleus. J. Neurochem. 1997, 68, 1106–1113. [Google Scholar] [CrossRef]
- Manuel, I.; Lombardero, L.; Llorente-Ovejero, A.; Rodríguez-Puertas, R. Chapter 27—Neuropeptides and neurolipids: What they are and how they relate to Alzheimer’s disease. In Genetics, Neurology, Behavior, and Diet in Dementia; Martin, C.R., Preedy, V.R., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 423–439. ISBN 978-0-12-815868-5. [Google Scholar]
- Llorente-Ovejero, A.; Martínez-Gardeazabal, J.; Moreno-Rodríguez, M.; Lombardero, L.; González de San Román, E.; Manuel, I.; Giralt, M.T.; Rodríguez-Puertas, R. Specific Phospholipid Modulation by Muscarinic Signaling in a Rat Lesion Model of Alzheimer’s Disease. ACS Chem. Neurosci. 2021, 12, 2167–2181. [Google Scholar] [CrossRef]
- Astarita, G.; Jung, K.-M.; Vasilevko, V.; Dipatrizio, N.V.; Martin, S.K.; Cribbs, D.H.; Head, E.; Cotman, C.W.; Piomelli, D. Elevated Stearoyl-CoA Desaturase in Brains of Patients with Alzheimer’s Disease. PLoS ONE 2011, 6, e24777. [Google Scholar] [CrossRef] [Green Version]
- Martin, M.; Ledent, C.; Parmentier, M.; Maldonado, R.; Valverde, O. Involvement of CB1 Cannabinoid Receptors in Emotional Behaviour. Psychopharmacology 2002, 159, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Cunnane, S.C.; Schneider, J.A.; Tangney, C.; Tremblay-Mercier, J.; Fortier, M.; Bennett, D.A.; Morris, M.C. Plasma and Brain Fatty Acid Profiles in Mild Cognitive Impairment and Alzheimer’s Disease. J. Alzheimer’s Dis. 2012, 29, 691–697. [Google Scholar] [CrossRef]
- Torres, M.; Price, S.L.; Fiol-Deroque, M.A.; Marcilla-Etxenike, A.; Ahyayauch, H.; Barceló-Coblijn, G.; Terés, S.; Katsouri, L.; Ordinas, M.; López, D.J.; et al. Membrane Lipid Modifications and Therapeutic Effects Mediated by Hydroxydocosahexaenoic Acid on Alzheimer’s Disease. Biochim. Biophys. Acta 2014, 1838, 1680–1692. [Google Scholar] [CrossRef] [Green Version]
- González-Domínguez, R.; García-Barrera, T.; Gómez-Ariza, J.L. Combination of Metabolomic and Phospholipid-Profiling Approaches for the Study of Alzheimer’s Disease. J. Proteom. 2014, 104, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Farooqui, A.A.; Horrocks, L.A.; Farooqui, T. Interactions between Neural Membrane Glycerophospholipid and Sphingolipid Mediators: A Recipe for Neural Cell Survival or Suicide. J. Neurosci. Res. 2007, 85, 1834–1850. [Google Scholar] [CrossRef] [PubMed]
- Geraldo, L.H.M.; de Sampaio Spohr, T.C.L.; do Amaral, R.F.; da Fonseca, A.C.C.; Garcia, C.; de Almeida Mendes, F.; Freitas, C.; dosSantos, M.F.; Lima, F.R.S. Role of Lysophosphatidic Acid and Its Receptors in Health and Disease: Novel Therapeutic Strategies. Signal Transduct. Target. Ther. 2021, 6, 45. [Google Scholar] [CrossRef]
- Hao, Y.; Guo, M.; Feng, Y.; Dong, Q.; Cui, M. Lysophospholipids and Their G-Coupled Protein Signaling in Alzheimer’s Disease: From Physiological Performance to Pathological Impairment. Front. Mol. Neurosci. 2020, 13, 58. [Google Scholar] [CrossRef] [Green Version]
- Leong, W.I.; Saba, J.D. S1P Metabolism in Cancer and Other Pathological Conditions. Biochimie 2010, 92, 716–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manuel, I.; González de San Román, E.; Giralt, M.T.; Ferrer, I.; Rodríguez-Puertas, R. Type-1 Cannabinoid Receptor Activity during Alzheimer’s Disease Progression. J. Alzheimer’s Dis. 2014, 42, 761–766. [Google Scholar] [CrossRef] [Green Version]
- Lombardero, L.; Llorente-Ovejero, A.; Manuel, I.; Rodríguez-Puertas, R. Chapter 28—Neurotransmitter receptors in Alzheimer’s disease: From glutamatergic to cholinergic receptors. In Genetics, Neurology, Behavior, and Diet in Dementia; Martin, C.R., Preedy, V.R., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 441–456. ISBN 978-0-12-815868-5. [Google Scholar]
- Katona, I.; Freund, T.F. Endocannabinoid Signaling as a Synaptic Circuit Breaker in Neurological Disease. Nat. Med. 2008, 14, 923–930. [Google Scholar] [CrossRef]
- Choi, J.W.; Chun, J. Lysophospholipids and Their Receptors in the Central Nervous System. Biochim. Biophys. Acta 2013, 1831, 20–32. [Google Scholar] [CrossRef] [Green Version]
- Westlake, T.M.; Howlett, A.C.; Bonner, T.I.; Matsuda, L.A.; Herkenham, M. Cannabinoid Receptor Binding and Messenger RNA Expression in Human Brain: An in Vitro Receptor Autoradiography and in Situ Hybridization Histochemistry Study of Normal Aged and Alzheimer’s Brains. Neuroscience 1994, 63, 637–652. [Google Scholar] [CrossRef]
- Ramírez, B.G.; Blázquez, C.; Gómez del Pulgar, T.; Guzmán, M.; de Ceballos, M.L. Prevention of Alzheimer’s Disease Pathology by Cannabinoids: Neuroprotection Mediated by Blockade of Microglial Activation. J. Neurosci. 2005, 25, 1904–1913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farkas, S.; Nagy, K.; Palkovits, M.; Kovács, G.G.; Jia, Z.; Donohue, S.; Pike, V.; Halldin, C.; Máthé, D.; Harkany, T.; et al. [125I]SD-7015 Reveals Fine Modalities of CB₁ Cannabinoid Receptor Density in the Prefrontal Cortex during Progression of Alzheimer’s Disease. Neurochem. Int. 2012, 60, 286–291. [Google Scholar] [CrossRef] [Green Version]
- Benito, C.; Núñez, E.; Tolón, R.M.; Carrier, E.J.; Rábano, A.; Hillard, C.J.; Romero, J. Cannabinoid CB2 Receptors and Fatty Acid Amide Hydrolase Are Selectively Overexpressed in Neuritic Plaque-Associated Glia in Alzheimer’s Disease Brains. J. Neurosci. 2003, 23, 11136–11141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Agacinski, G.; Williams, J.H.; Wilcock, G.K.; Esiri, M.M.; Francis, P.T.; Wong, P.T.-H.; Chen, C.P.; Lai, M.K.P. Intact Cannabinoid CB1 Receptors in the Alzheimer’s Disease Cortex. Neurochem. Int. 2010, 57, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Mechoulam, R.; Lichtman, A.H. Neuroscience. Stout Guards of the Central Nervous System. Science 2003, 302, 65–67. [Google Scholar] [CrossRef] [PubMed]
- Ehrhart, J.; Obregon, D.; Mori, T.; Hou, H.; Sun, N.; Bai, Y.; Klein, T.; Fernandez, F.; Tan, J.; Shytle, R.D. Stimulation of Cannabinoid Receptor 2 (CB2) Suppresses Microglial Activation. J. Neuroinflamm. 2005, 2, 29. [Google Scholar] [CrossRef] [Green Version]
- Umemura, K.; Yamashita, N.; Yu, X.; Arima, K.; Asada, T.; Makifuchi, T.; Murayama, S.; Saito, Y.; Kanamaru, K.; Goto, Y.; et al. Autotaxin Expression Is Enhanced in Frontal Cortex of Alzheimer-Type Dementia Patients. Neurosci. Lett. 2006, 400, 97–100. [Google Scholar] [CrossRef]
- Awada, R.; Rondeau, P.; Grès, S.; Saulnier-Blache, J.S.; Lefebvre d’Hellencourt, C.; Bourdon, E. Autotaxin Protects Microglial Cells against Oxidative Stress. Free Radic. Biol. Med. 2012, 52, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Dong, Y.; Cui, M.-Z.; Xu, X. Lysophosphatidic Acid Induces Increased BACE1 Expression and Aβ Formation. Biochim. Biophys. Acta 2013, 1832, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Sayas, C.L.; Moreno-Flores, M.T.; Avila, J.; Wandosell, F. The Neurite Retraction Induced by Lysophosphatidic Acid Increases Alzheimer’s Disease-like Tau Phosphorylation. J. Biol. Chem. 1999, 274, 37046–37052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayas, C.L.; Avila, J.; Wandosell, F. Regulation of Neuronal Cytoskeleton by Lysophosphatidic Acid: Role of GSK-3. Biochim. Biophys. Acta 2002, 1582, 144–153. [Google Scholar] [CrossRef]
- Weiner, J.A.; Hecht, J.H.; Chun, J. Lysophosphatidic Acid Receptor Gene Vzg-1/LpA1/Edg-2 Is Expressed by Mature Oligodendrocytes during Myelination in the Postnatal Murine Brain. J. Comp. Neurol. 1998, 398, 587–598. [Google Scholar] [CrossRef]
- Zheng, Z.-Q.; Fang, X.-J.; Zhang, Y.; Qiao, J.-T. Neuroprotective Effect of Lysophosphatidic Acid on AbetaP31-35-Induced Apoptosis in Cultured Cortical Neurons. Sheng Li Xue Bao 2005, 57, 289–294. [Google Scholar] [PubMed]
- Ceccom, J.; Loukh, N.; Lauwers-Cances, V.; Touriol, C.; Nicaise, Y.; Gentil, C.; Uro-Coste, E.; Pitson, S.; Maurage, C.A.; Duyckaerts, C.; et al. Reduced Sphingosine Kinase-1 and Enhanced Sphingosine 1-Phosphate Lyase Expression Demonstrate Deregulated Sphingosine 1-Phosphate Signaling in Alzheimer’s Disease. Acta Neuropathol. Commun. 2014, 2, 12. [Google Scholar] [CrossRef]
- Couttas, T.A.; Kain, N.; Daniels, B.; Lim, X.Y.; Shepherd, C.; Kril, J.; Pickford, R.; Li, H.; Garner, B.; Don, A.S. Loss of the Neuroprotective Factor Sphingosine 1-Phosphate Early in Alzheimer’s Disease Pathogenesis. Acta Neuropathol. Commun. 2014, 2, 9. [Google Scholar] [CrossRef] [Green Version]
- Healy, L.M.; Antel, J.P. Sphingosine-1-Phosphate Receptors in the Central Nervous and Immune Systems. Curr. Drug Targets 2016, 17, 1841–1850. [Google Scholar] [CrossRef]
- Rubino, T.; Guidali, C.; Vigano, D.; Realini, N.; Valenti, M.; Massi, P.; Parolaro, D. CB1 Receptor Stimulation in Specific Brain Areas Differently Modulate Anxiety-Related Behaviour. Neuropharmacology 2008, 54, 151–160. [Google Scholar] [CrossRef]
- Laviolette, S.R.; Grace, A.A. Cannabinoids Potentiate Emotional Learning Plasticity in Neurons of the Medial Prefrontal Cortex through Basolateral Amygdala Inputs. J. Neurosci. 2006, 26, 6458–6468. [Google Scholar] [CrossRef] [Green Version]
- España, J.; Giménez-Llort, L.; Valero, J.; Miñano, A.; Rábano, A.; Rodriguez-Alvarez, J.; LaFerla, F.M.; Saura, C.A. Intraneuronal Beta-Amyloid Accumulation in the Amygdala Enhances Fear and Anxiety in Alzheimer’s Disease Transgenic Mice. Biol. Psychiatry 2010, 67, 513–521. [Google Scholar] [CrossRef]
- Kalifa, S.; Polston, E.K.; Allard, J.S.; Manaye, K.F. Distribution Patterns of Cannabinoid CB1 Receptors in the Hippocampus of APPswe/PS1ΔE9 Double Transgenic Mice. Brain Res. 2011, 1376, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Aso, E.; Palomer, E.; Juvés, S.; Maldonado, R.; Muñoz, F.J.; Ferrer, I. CB1 Agonist ACEA Protects Neurons and Reduces the Cognitive Impairment of AβPP/PS1 Mice. J. Alzheimer’s Dis. 2012, 30, 439–459. [Google Scholar] [CrossRef] [Green Version]
- Holcomb, L.; Gordon, M.N.; McGowan, E.; Yu, X.; Benkovic, S.; Jantzen, P.; Wright, K.; Saad, I.; Mueller, R.; Morgan, D.; et al. Accelerated Alzheimer-Type Phenotype in Transgenic Mice Carrying Both Mutant Amyloid Precursor Protein and Presenilin 1 Transgenes. Nat. Med. 1998, 4, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Varo, R.; Trujillo-Estrada, L.; Sanchez-Mejias, E.; Torres, M.; Baglietto-Vargas, D.; Moreno-Gonzalez, I.; De Castro, V.; Jimenez, S.; Ruano, D.; Vizuete, M.; et al. Abnormal Accumulation of Autophagic Vesicles Correlates with Axonal and Synaptic Pathology in Young Alzheimer’s Mice Hippocampus. Acta Neuropathol. 2012, 123, 53–70. [Google Scholar] [CrossRef] [Green Version]
- Bedse, G.; Romano, A.; Cianci, S.; Lavecchia, A.M.; Lorenzo, P.; Elphick, M.R.; Laferla, F.M.; Vendemiale, G.; Grillo, C.; Altieri, F.; et al. Altered Expression of the CB1 Cannabinoid Receptor in the Triple Transgenic Mouse Model of Alzheimer’s Disease. J. Alzheimer’s Dis. 2014, 40, 701–712. [Google Scholar] [CrossRef]
- Llorente-Ovejero, A.; Manuel, I.; Lombardero, L.; Giralt, M.T.; Ledent, C.; Giménez-Llort, L.; Rodríguez-Puertas, R. Endocannabinoid and Muscarinic Signaling Crosstalk in the 3xTg-AD Mouse Model of Alzheimer’s Disease. J. Alzheimer’s Dis. 2018, 64, 117–136. [Google Scholar] [CrossRef] [PubMed]
- Lauckner, J.E.; Hille, B.; Mackie, K. The Cannabinoid Agonist WIN55,212-2 Increases Intracellular Calcium via CB1 Receptor Coupling to Gq/11 G Proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 19144–19149. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, R.; Goffin, K.; Van den Stock, J.; De Winter, F.-L.; Cleeren, E.; Bormans, G.; Tournoy, J.; Persoons, P.; Van Laere, K.; Vandenbulcke, M. In Vivo Type 1 Cannabinoid Receptor Availability in Alzheimer’s Disease. Eur. Neuropsychopharmacol. J. Eur. Coll. Neuropsychopharmacol. 2014, 24, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Blankman, J.L.; Cravatt, B.F. Chemical Probes of Endocannabinoid Metabolism. Pharmacol. Rev. 2013, 65, 849–871. [Google Scholar] [CrossRef] [Green Version]
- Hecht, J.H.; Weiner, J.A.; Post, S.R.; Chun, J. Ventricular Zone Gene-1 (Vzg-1) Encodes a Lysophosphatidic Acid Receptor Expressed in Neurogenic Regions of the Developing Cerebral Cortex. J. Cell Biol. 1996, 135, 1071–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shano, S.; Moriyama, R.; Chun, J.; Fukushima, N. Lysophosphatidic Acid Stimulates Astrocyte Proliferation through LPA1. Neurochem. Int. 2008, 52, 216–220. [Google Scholar] [CrossRef]
- Möller, T.; Contos, J.J.; Musante, D.B.; Chun, J.; Ransom, B.R. Expression and Function of Lysophosphatidic Acid Receptors in Cultured Rodent Microglial Cells. J. Biol. Chem. 2001, 276, 25946–25952. [Google Scholar] [CrossRef] [Green Version]
- Tham, C.-S.; Lin, F.-F.; Rao, T.S.; Yu, N.; Webb, M. Microglial Activation State and Lysophospholipid Acid Receptor Expression. Int. J. Dev. Neurosci. Off. J. Int. Soc. Dev. Neurosci. 2003, 21, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Allard, J.; Barron, S.; Trottier, S.; Cervera, P.; Daumas-Duport, C.; Leguern, E.; Brice, A.; Schwartz, J.C.; Sokoloff, P. Edg-2 in Myelin-Forming Cells: Isoforms, Genomic Mapping, and Exclusion in Charcot-Marie-Tooth Disease. Glia 1999, 26, 176–185. [Google Scholar] [CrossRef]
- Desai, M.K.; Sudol, K.L.; Janelsins, M.C.; Mastrangelo, M.A.; Frazer, M.E.; Bowers, W.J. Triple-Transgenic Alzheimer’s Disease Mice Exhibit Region-Specific Abnormalities in Brain Myelination Patterns Prior to Appearance of Amyloid and Tau Pathology. Glia 2009, 57, 54–65. [Google Scholar] [CrossRef] [Green Version]
- Harms, M.P.; Kotyk, J.J.; Merchant, K.M. Evaluation of White Matter Integrity in Ex Vivo Brains of Amyloid Plaque-Bearing APPsw Transgenic Mice Using Magnetic Resonance Diffusion Tensor Imaging. Exp. Neurol. 2006, 199, 408–415. [Google Scholar] [CrossRef]
- Kandel, E.R.; Squire, L.R. Neuroscience: Breaking down Scientific Barriers to the Study of Brain and Mind. Science 2000, 290, 1113–1120. [Google Scholar] [CrossRef]
- García-Díaz, B.; Riquelme, R.; Varela-Nieto, I.; Jiménez, A.J.; de Diego, I.; Gómez-Conde, A.I.; Matas-Rico, E.; Aguirre, J.Á.; Chun, J.; Pedraza, C.; et al. Loss of Lysophosphatidic Acid Receptor LPA1 Alters Oligodendrocyte Differentiation and Myelination in the Mouse Cerebral Cortex. Brain Struct. Funct. 2015, 220, 3701–3720. [Google Scholar] [CrossRef]
- Wu, Y.; Ma, Y.; Liu, Z.; Geng, Q.; Chen, Z.; Zhang, Y. Alterations of Myelin Morphology and Oligodendrocyte Development in Early Stage of Alzheimer’s Disease Mouse Model. Neurosci. Lett. 2017, 642, 102–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, A.K.; Hajra, A.K. Quantification, Characterization and Fatty Acid Composition of Lysophosphatidic Acid in Different Rat Tissues. Lipids 1989, 24, 329–333. [Google Scholar] [CrossRef] [Green Version]
- Mihara, Y.; Horikawa, M.; Sato, S.; Eto, F.; Hanada, M.; Banno, T.; Arima, H.; Ushirozako, H.; Yamada, T.; Xu, D.; et al. Lysophosphatidic Acid Precursor Levels Decrease and an Arachidonic Acid-Containing Phosphatidylcholine Level Increases in the Dorsal Root Ganglion of Mice after Peripheral Nerve Injury. Neurosci. Lett. 2019, 698, 69–75. [Google Scholar] [CrossRef]
- Aytan, N.; Choi, J.-K.; Carreras, I.; Brinkmann, V.; Kowall, N.W.; Jenkins, B.G.; Dedeoglu, A. Fingolimod Modulates Multiple Neuroinflammatory Markers in a Mouse Model of Alzheimer’s Disease. Sci. Rep. 2016, 6, 24939. [Google Scholar] [CrossRef] [Green Version]
- Yeh, C.-Y.; Vadhwana, B.; Verkhratsky, A.; Rodríguez, J.J. Early Astrocytic Atrophy in the Entorhinal Cortex of a Triple Transgenic Animal Model of Alzheimer’s Disease. ASN Neuro 2011, 3, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Rozen, S.; Boyle, S.H.; Hellegers, C.; Cheng, H.; Burke, J.R.; Welsh-Bohmer, K.A.; Doraiswamy, P.M.; Kaddurah-Daouk, R. Metabolomics in Early Alzheimer’s Disease: Identification of Altered Plasma Sphingolipidome Using Shotgun Lipidomics. PLoS ONE 2011, 6, e21643. [Google Scholar] [CrossRef]
- Mendis, L.H.S.; Grey, A.C.; Faull, R.L.M.; Curtis, M.A. Hippocampal Lipid Differences in Alzheimer’s Disease: A Human Brain Study Using Matrix-Assisted Laser Desorption/Ionization-Imaging Mass Spectrometry. Brain Behav. 2016, 6, e00517. [Google Scholar] [CrossRef]
- Koal, T.; Klavins, K.; Seppi, D.; Kemmler, G.; Humpel, C. Sphingomyelin SM(D18:1/18:0) Is Significantly Enhanced in Cerebrospinal Fluid Samples Dichotomized by Pathological Amyloid-Β42, Tau, and Phospho-Tau-181 Levels. J. Alzheimer’s Dis. 2015, 44, 1193–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, J.-N.; Zhu, Z.; Itokazu, Y.; Wang, G.; Dinkins, M.B.; Zhong, L.; Lin, H.-P.; Elsherbini, A.; Leanhart, S.; Jiang, X.; et al. Novel Function of Ceramide for Regulation of Mitochondrial ATP Release in Astrocytes. J. Lipid Res. 2018, 59, 488–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottfries, C.G.; Jungbjer, B.; Karlsson, I.; Svennerholm, L. Reductions in Membrane Proteins and Lipids in Basal Ganglia of Classic Alzheimer Disease Patients. Alzheimer Dis. Assoc. Disord. 1996, 10, 77–81. [Google Scholar] [CrossRef]
- Orešič, M.; Hyötyläinen, T.; Herukka, S.-K.; Sysi-Aho, M.; Mattila, I.; Seppänan-Laakso, T.; Julkunen, V.; Gopalacharyulu, P.V.; Hallikainen, M.; Koikkalainen, J.; et al. Metabolome in Progression to Alzheimer’s Disease. Transl. Psychiatry 2011, 1, e57. [Google Scholar] [CrossRef]
- Whiley, L.; Sen, A.; Heaton, J.; Proitsi, P.; García-Gómez, D.; Leung, R.; Smith, N.; Thambisetty, M.; Kloszewska, I.; Mecocci, P.; et al. Evidence of Altered Phosphatidylcholine Metabolism in Alzheimer’s Disease. Neurobiol. Aging 2014, 35, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Holtzman, D.M.; McKeel, D.W.J. Plasmalogen Deficiency in Early Alzheimer’s Disease Subjects and in Animal Models: Molecular Characterization Using Electrospray Ionization Mass Spectrometry. J. Neurochem. 2001, 77, 1168–1180. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.K.; Wengenack, T.M.; Curran, G.L.; Poduslo, J.F. Reduced Membrane Lipids in the Cortex of Alzheimer’s Disease Transgenic Mice. Neurochem. Res. 2009, 34, 102–108. [Google Scholar] [CrossRef]
- Cheng, H.; Zhou, Y.; Holtzman, D.M.; Han, X. Apolipoprotein E Mediates Sulfatide Depletion in Animal Models of Alzheimer’s Disease. Neurobiol. Aging 2010, 31, 1188–1196. [Google Scholar] [CrossRef] [Green Version]
- Chan, R.B.; Oliveira, T.G.; Cortes, E.P.; Honig, L.S.; Duff, K.E.; Small, S.A.; Wenk, M.R.; Shui, G.; Di Paolo, G. Comparative Lipidomic Analysis of Mouse and Human Brain with Alzheimer Disease. J. Biol. Chem. 2012, 287, 2678–2688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tajima, Y.; Ishikawa, M.; Maekawa, K.; Murayama, M.; Senoo, Y.; Nishimaki-Mogami, T.; Nakanishi, H.; Ikeda, K.; Arita, M.; Taguchi, R.; et al. Lipidomic Analysis of Brain Tissues and Plasma in a Mouse Model Expressing Mutated Human Amyloid Precursor Protein/Tau for Alzheimer’s Disease. Lipids Health Dis. 2013, 12, 68. [Google Scholar] [CrossRef] [Green Version]
- Emre, C.; Do, K.V.; Jun, B.; Hjorth, E.; Alcalde, S.G.; Kautzmann, M.-A.I.; Gordon, W.C.; Nilsson, P.; Bazan, N.G.; Schultzberg, M. Age-Related Changes in Brain Phospholipids and Bioactive Lipids in the APP Knock-in Mouse Model of Alzheimer’s Disease. Acta Neuropathol. Commun. 2021, 9, 116. [Google Scholar] [CrossRef] [PubMed]
- Urano, S.; Sato, Y.; Otonari, T.; Makabe, S.; Suzuki, S.; Ogata, M.; Endo, T. Aging and Oxidative Stress in Neurodegeneration. Biofactors 1998, 7, 103–112. [Google Scholar] [CrossRef]
- Conquer, J.A.; Tierney, M.C.; Zecevic, J.; Bettger, W.J.; Fisher, R.H. Fatty Acid Analysis of Blood Plasma of Patients with Alzheimer’s Disease, Other Types of Dementia, and Cognitive Impairment. Lipids 2000, 35, 1305–1312. [Google Scholar] [CrossRef]
- Youdim, K.A.; Martin, A.; Joseph, J.A. Essential Fatty Acids and the Brain: Possible Health Implications. Int. J. Dev. Neurosci. Off. J. Int. Soc. Dev. Neurosci. 2000, 18, 383–399. [Google Scholar] [CrossRef] [Green Version]
- Kanfer, J.N.; Singh, I.N.; Pettegrew, J.W.; McCartney, D.G.; Sorrentino, G. Phospholipid Metabolism in Alzheimer’s Disease and in a Human Cholinergic Cell. J. Lipid Mediat. Cell Signal. 1996, 14, 361–363. [Google Scholar] [CrossRef]
- Mapstone, M.; Cheema, A.K.; Fiandaca, M.S.; Zhong, X.; Mhyre, T.R.; MacArthur, L.H.; Hall, W.J.; Fisher, S.G.; Peterson, D.R.; Haley, J.M.; et al. Plasma Phospholipids Identify Antecedent Memory Impairment in Older Adults. Nat. Med. 2014, 20, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, D.T.; Lemere, C.A.; Selkoe, D.J.; Clemens, J.A. Cytosolic Phospholipase A2 (CPLA2) Immunoreactivity Is Elevated in Alzheimer’s Disease Brain. Neurobiol. Dis. 1996, 3, 51–63. [Google Scholar] [CrossRef] [Green Version]
- Colangelo, V.; Schurr, J.; Ball, M.J.; Pelaez, R.P.; Bazan, N.G.; Lukiw, W.J. Gene Expression Profiling of 12633 Genes in Alzheimer Hippocampal CA1: Transcription and Neurotrophic Factor down-Regulation and up-Regulation of Apoptotic and pro-Inflammatory Signaling. J. Neurosci. Res. 2002, 70, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Gónzalez de San Román, E.; Manuel, I.; Giralt, M.T.; Ferrer, I.; Rodríguez-Puertas, R. Imaging Mass Spectrometry (IMS) of Cortical Lipids from Preclinical to Severe Stages of Alzheimer’s Disease. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1604–1614. [Google Scholar] [CrossRef]
- Billings, L.M.; Oddo, S.; Green, K.N.; McGaugh, J.L.; LaFerla, F.M. Intraneuronal Abeta Causes the Onset of Early Alzheimer’s Disease-Related Cognitive Deficits in Transgenic Mice. Neuron 2005, 45, 675–688. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, S.A.; Reyzer, M.L.; Caprioli, R.M. Direct Tissue Analysis Using Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry: Practical Aspects of Sample Preparation. J. Mass Spectrom. 2003, 38, 699–708. [Google Scholar] [CrossRef]
- Yang, J.; Caprioli, R.M. Matrix Sublimation/Recrystallization for Imaging Proteins by Mass Spectrometry at High Spatial Resolution. Anal. Chem. 2011, 83, 5728–5734. [Google Scholar] [CrossRef] [Green Version]

| Basal Binding (nCi/g t.e.) | WIN55,212-2 Stimulation (%) | |||
|---|---|---|---|---|
| Brain Region | WT | 3xTg-AD | WT | 3xTg-AD |
| Amygdala | ||||
| Anterior | 234.9 ± 18.3 | 218.2 ± 20.9 | 204.5 ± 30.1 | 185.8 ± 37.7 |
| Posterior | 216.6 ± 14.6 | 266.4 ± 23.5 | 295.5 ± 41.7 | 112.8 ± 28.9 ** |
| Internal capsule | 143.1 ± 19.0 | 120.8 ± 9.8 | 52.9 ± 12.5 | 36.2 ± 11.1 |
| Striatum | 178.8 ± 8.4 | 187.4 ± 12.8 | 250.2 ± 27.7 | 226.1 ± 26.4 |
| Cerebellum | ||||
| White matter | 60.2 ± 6.5 | 51.7 ± 5.7 | 66.1 ± 34.8 | 93.2 ± 19.6 |
| Gray matter | 54.7 ± 4.7 | 66.7 ± 7.3 | 814.8 ± 56.3 | 658.2 ± 49.2 |
| Cortex | ||||
| Cingular | 164.2 ± 13.0 | 166.0 ± 12.4 | 308.4 ± 45.5 | 267.3 ± 38.2 |
| Motor Layers I-VI | 152.4 ± 7.9 | 178.4 ± 13.1 | 267.3 ± 25.6 | 228.6 ± 29.7 |
| Layer VI | 175.3 ± 55.4 | 190.8 ± 52.8 | 435.4 ± 58.2 | 238.4 ± 22.9 ** |
| Corpus callosum | 146.6 ± 11.8 | 127.2 ± 10.2 | 72.1 ± 13.1 | 84.6 ± 14.5 |
| Globus pallidus | 200.6 ± 12.3 | 204.7 ± 17.7 | 870.2 ± 69.4 | 872.5 ± 52.2 |
| Hippocampus | ||||
| CA1 | 132.3 ± 10.8 | 143.4 ± 9.9 | 314.8 ± 47.1 | 238.4 ± 27.1 |
| Dentate gyrus | 121.1 ± 15.4 | 143.5 ± 8.8 | 209.8 ± 38.9 | 202.5 ± 21.5 |
| Hypothalamus | 217.6 ± 16.6 | 243.3 ± 22.8 | 162.5 ± 20.8 | 169.3 ± 16.3 |
| Thalamic nuclei | ||||
| Anteroventral | 113.7 ± 11.7 | 138.6 ± 10.2 | 128.5 ± 35.4 | 86.1 ± 30.3 |
| Thalamus | 89.8 ± 7.8 | 112.7 ± 9.9 | 109.3 ± 29.9 | 92.5 ± 15.6 |
| Basal Nucleus | 215.6 ± 14.7 | 227.3 ± 15.9 | 271.3 ± 39.1 | 274.9 ± 39.9 |
| Substantia Nigra | 190.1 ± 16.9 | 172.1 ± 8.1 | 1062.7 ± 79.8 | 979.3 ± 65.7 |
| LPA Stimulation (%) | ||
|---|---|---|
| Brain Region | WT | 3xTg-AD |
| Amygdala | ||
| Anterior | 20.5 ± 11.7 | 13.0 ± 10.3 |
| Posterior | 28.8 ± 12.8 | 8.1 ± 15.9 |
| Internal capsule | 30.2 ± 9.6 | 32.8 ± 6.2 |
| Striatum | 3.3 ± 7.2 | 23.1 ± 3.8 * |
| Cerebellum | ||
| White matter | 76.6 ± 18.4 | 111.3 ± 19.7 |
| Gray matter | 62.0 ± 15.7 | 82.9 ± 19.7 |
| Cortex | ||
| Cingular | 13.2 ± 7.2 | 20.8 ± 6.4 |
| Motor | 6.2 ± 12.4 | 26.7 ± 7.6 * |
| Corpus Callosum | 90.8 ± 12.3 | 189.6 ± 17.4 * |
| Globus pallidus | 22.1 ± 9.3 | 29.8 ± 6.9 |
| Hippocampus | ||
| CA1 | −18.7 ± 7.8 | 22.7 ± 4.4 * |
| Dentate gyrus | 54.7 ± 11.0 | 49.5 ± 23.2 |
| Hypothalamus | 34.5 ± 19.9 | 22.6 ± 10.2 |
| Thalamic nuclei | ||
| Anteroventral | 22.6 ± 14.2 | 20.6 ± 8.2 |
| Thalamus | 20.6 ± 13.2 | 24.6 ± 11.1 |
| Basal Nucleus | 22.9 ± 10.7 | 39.8 ± 8.5 |
| Substantia Nigra | 23.1 ± 13.7 | 25.4 ± 7.8 |
| CYM5442 Stimulation (%) | ||
| Brain Region | WT | 3xTg-AD |
| Amygdala | ||
| Anterior | 487 ± 91.4 | 515 ± 91.7 |
| Posterior | 334 ± 33.9 | 397 ± 49.5 |
| Internal capsule | 193 ± 45.0 | 131 ± 16.0 |
| Striatum | 446 ± 58.9 | 375 ± 29.1 |
| Cerebellum | ||
| White matter | 183 ± 37.1 | 166 ± 19.5 |
| Gray matter | 329 ± 41.9 | 343 ± 52.3 |
| Cortex | ||
| Cingular | 789 ± 131.0 | 997 ± 173.0 |
| Motor | 690 ± 94.0 | 677 ± 79.0 |
| Entorhinal | 542 ± 104.0 | 387 ± 52.0 |
| Frontal | 475 ± 46.3 | 483 ± 56.0 |
| Corpus callosum | 243 ± 41.0 | 196 ± 29.0 |
| Globus pallidus | 468 ± 61.9 | 365 ± 41.3 |
| Hippocampus | ||
| CA1 | 542 ± 58.7 | 328 ± 29.2 ** |
| CA3 | 328 ± 33.4 | 221 ± 11.6 ** |
| Dentate gyrus | 606 ± 58.8 | 439 ± 34.9 * |
| Hypothalamus | 178 ± 50.16 | 188 ± 37.2 |
| Thalamic nuclei | ||
| Anteroventral | 177 ± 35.2 | 204 ± 62.8 |
| Thalamus | 188 ± 48.7 | 209 ± 50.1 |
| Basal Nucleus | 448 ± 46.2 | 373 ± 23.9 |
| Substantia Nigra | 855 ± 116.8 | 544 ± 74.3 * |
| Granular olfactory bulb | 1653 ± 156.9 | 950 ± 65.9 ** |
| Anterior olfactory Nucleus | 1255 ± 107.4 | 804 ± 168.3 * |
| Cortex | Hippocampus | Striatum | Amygdala | Cerebellum | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assignment | m/z | WT | 3xTg-AD | WT | 3xTg-AD | WT | 3xTg-AD | WT | 3xTg-AD | WT | 3xTg-AD |
| PA(34:1)+K+ | 713.4535 | 15.1 ± 1.5 | 25.0 ± 2.6 ** | 17.4 ± 1.1 | 25.8 ± 2.9 * | 13.7 ± 0.9 | 20.0 ± 2.4 * | 22.7 ± 1.8 | 25.7 ± 3.3 | 14.1 ± 2.4 | 15.3 ± 2.2 |
| PC(16:0/16:0)+ | 734.5721 | 57.2 ± 4.1 | 76.9 ± 7.6 * | 70.1 ± 4.3 | 71.8 ± 4.9 | 69.8 ± 3.4 | 69.2 ± 3.5 | 58.0 ± 0.5 | 75.3 ± 5.9 * | 55.5 ± 4.3 | 70.9 ± 3.1 * |
| PC(16:0/18:1)+ | 760.5658 | 82.3 ± 5.7 | 83.4 ± 4.8 | 86.1 ± 4.5 | 87.2 ± 4.8 | 88.7 ± 5.7 | 90.1 ± 6.9 | 65.6 ± 1.0 | 78.8 ± 5.1 * | 96.0 ± 2.1 | 96.1 ± 2.1 |
| SM(d18:1/18:0)+K+ | 769.5656 | 38.0 ± 3.5 | 36.1 ± 1.5 | 30.9 ± 0.9 | 35.9 ± 1.4 * | 23.3 ± 2.5 | 25.1 ± 1.6 | 34.8 ± 1.8 | 41.0 ± 1.5 * | 25.1 ± 3.0 | 26.5 ± 2.0 |
| PC(36:4)+ | 782.5654 | 33.1 ± 2.9 | 30.7 ± 2.1 | 37.3 ± 1.7 | 35.4 ± 1.1 | 34.6 ± 1.1 | 32.7 ± 1.3 | 43.9 ± 1.6 | 36.2 ± 2.9 * | 29.8 ± 0.8 | 24.9 ± 1.1 ** |
| PC(38:6)+ | 806.5711 | 15.0 ± 0.2 | 10.5 ± 0.7 ** | 9.8 ± 0.9 | 9.3 ± 0.5 | 11.3 ± 0.8 | 10.7 ± 1.3 | 5.3 ± 0.8 | 6.4 ± 0.8 | 19.9 ± 1.1 | 13.1 ± 1.2 ** |
| SM(d35:1)− | 715.5764 | 26.2 ± 1.4 | 37.4 ± 3.7 * | 50.2 ± 2.8 | 65.0 ± 5.9 * | 35.4 ± 2.1 | 38.1 ± 2.9 | 55.5 ± 2.8 | 67.5 ± 3.1 * | 41.2 ± 5.3 | 48.3 ± 6.0 |
| PI(16:0/20:4)− | 857.5190 | 22.7 ± 0.6 | 19.1 ± 0.3 ** | 14.5 ± 0.5 | 11.1 ± 0.3 ** | 12.1 ± 0.2 | 11.6 ± 0.2 | 10.5 ± 0.2 | 9.1 ± 0.6 | 10.8 ± 0.6 | 9.7 ± 1.1 |
| CPI(40:2)+MBT | 896.5779 | 11.2 ± 1.5 | 12.7 ± 2.8 | 23.8 ± 1.1 | 23.9 ± 2.1 | 15.3 ± 1.1 | 19.7 ± 3.1 | 25.5 ± 2.1 | 17.2 ± 2.0 * | 20.9 ± 3.3 | 17.6 ± 2.5 |
| PI(40:5)− | 911.5411 | 14.7 ± 0.7 | 8.8 ± 1.7 ** | 11.3 ± 0.7 | 9.9 ± 1.2 | 10.2 ± 0.6 | 10.3 ± 1.1 | 9.6 ± 1.1 | 8.7 ± 1.9 | 10.1 ± 0.8 | 8.2 ± 1.6 |
| 925.5556 | 14.8 ± 1.7 | 6.3 ± 2.8 * | 11.8 ± 0.9 | 3.7 ± 2.1 ** | 9.8 ± 1.5 | 7.5 ± 2.7 | 11.9 ± 0.9 | 5.7 ± 2.8 * | 10.5 ± 1.3 | 8.1 ± 2.7 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González de San Román, E.; Llorente-Ovejero, A.; Martínez-Gardeazabal, J.; Moreno-Rodríguez, M.; Giménez-Llort, L.; Manuel, I.; Rodríguez-Puertas, R. Modulation of Neurolipid Signaling and Specific Lipid Species in the Triple Transgenic Mouse Model of Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 12256. https://doi.org/10.3390/ijms222212256
González de San Román E, Llorente-Ovejero A, Martínez-Gardeazabal J, Moreno-Rodríguez M, Giménez-Llort L, Manuel I, Rodríguez-Puertas R. Modulation of Neurolipid Signaling and Specific Lipid Species in the Triple Transgenic Mouse Model of Alzheimer’s Disease. International Journal of Molecular Sciences. 2021; 22(22):12256. https://doi.org/10.3390/ijms222212256
Chicago/Turabian StyleGonzález de San Román, Estibaliz, Alberto Llorente-Ovejero, Jonatan Martínez-Gardeazabal, Marta Moreno-Rodríguez, Lydia Giménez-Llort, Iván Manuel, and Rafael Rodríguez-Puertas. 2021. "Modulation of Neurolipid Signaling and Specific Lipid Species in the Triple Transgenic Mouse Model of Alzheimer’s Disease" International Journal of Molecular Sciences 22, no. 22: 12256. https://doi.org/10.3390/ijms222212256
APA StyleGonzález de San Román, E., Llorente-Ovejero, A., Martínez-Gardeazabal, J., Moreno-Rodríguez, M., Giménez-Llort, L., Manuel, I., & Rodríguez-Puertas, R. (2021). Modulation of Neurolipid Signaling and Specific Lipid Species in the Triple Transgenic Mouse Model of Alzheimer’s Disease. International Journal of Molecular Sciences, 22(22), 12256. https://doi.org/10.3390/ijms222212256







