Aging, Bone Marrow and Next-Generation Sequencing (NGS): Recent Advances and Future Perspectives
Abstract
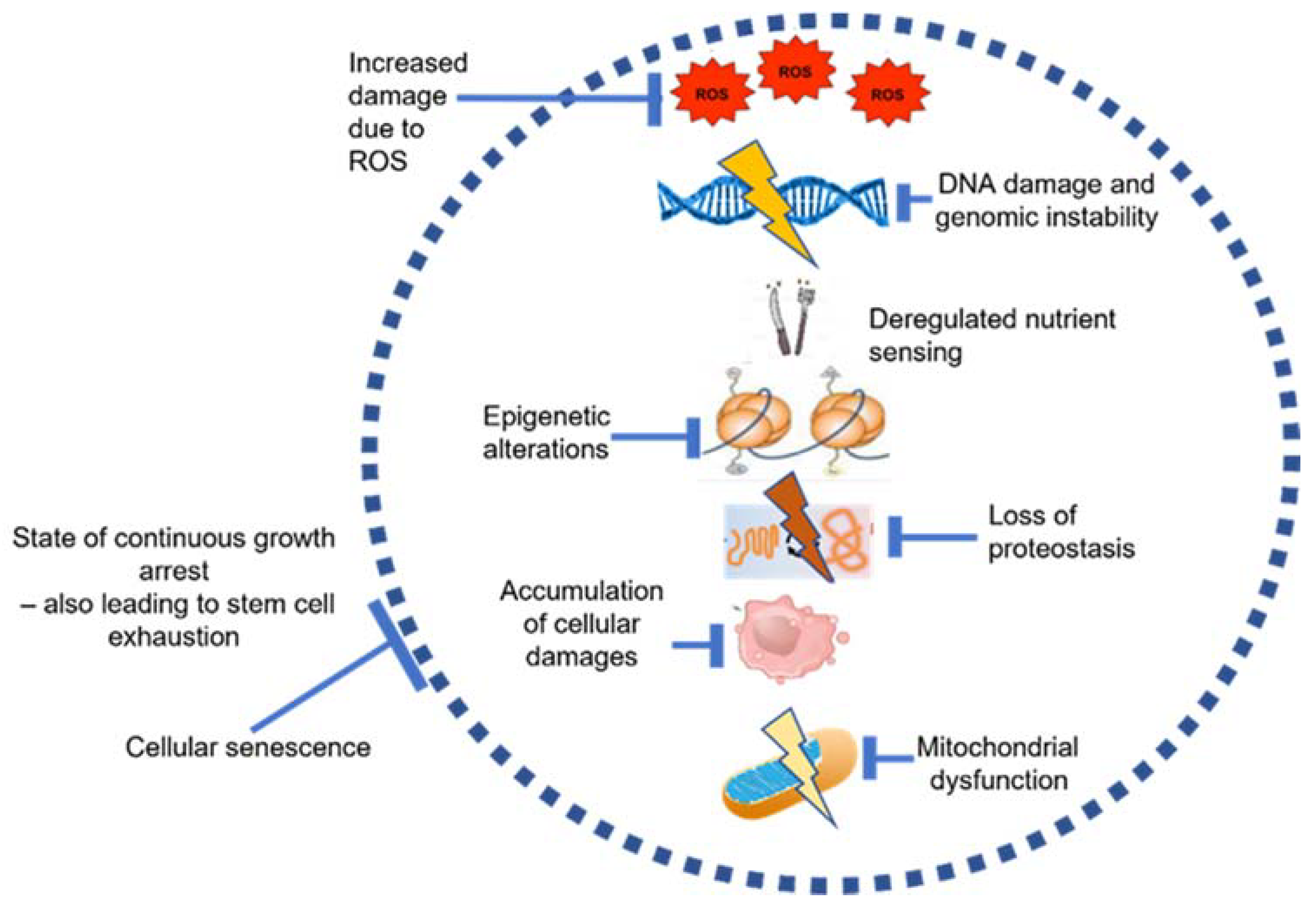
1. Introduction
2. Aging in the Bone Marrow
2.1. The Aging Bone Marrow (BM) Niche
2.2. Age-Related Changes in Haematopoietic Stem Cells (HSCs)
2.3. Age-Related Changes in Mesenchymal Stem Cells (MSCs)
2.4. Age-Related Changes in B Cells and T Cells: Immune Aging and Immunosenescence
2.4.1. B Cells
2.4.2. T Cells
3. NGS and Its Applications in Aging Studies
3.1. Targeted Sequencing Approaches
3.1.1. DNA Panels
3.1.2. ScRNA and Bulk RNA-Seq
4. The Future of NGS in Aging Studies—Potential, Perspectives and Limitations
4.1. Potential and Perspectives of NGS in Aging Studies
4.2. Limitations of NGS in Aging Studies
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Ageing and Health. Available online: https://www.who.int/news-room/fact-sheets/detail/ageing-and-health (accessed on 15 November 2020).
- WHO NIoH. Global Health and Aging. 2011. Available online: https://www.who.int/news-room/fact-sheets/detail/ageing-and-health (accessed on 7 July 2021).
- Franceschi, C.; Garagnani, P.; Morsiani, C.; Conte, M.; Santoro, A.; Grignolio, A.; Monti, D.; Capri, M.; Salvioli, S. The Continuum of Aging and Age-Related Diseases: Common Mechanisms but Different Rates. Front. Med. 2018, 5, 61. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Schultz, M.B.; Sinclair, D.A. When stem cells grow old: Phenotypes and mechanisms of stem cell aging. Development 2016, 143, 3–14. [Google Scholar] [CrossRef]
- Harman, D. The Free Radical Theory of Aging: Effect of Age on Serum Copper Levels. J. Gerontol. 1965, 20, 151–153. [Google Scholar] [CrossRef]
- Blackburn, E.H.; Epel, E.S.; Lin, J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science 2015, 350, 1193–1198. [Google Scholar] [CrossRef]
- Fukada, S.-I.; Ma, Y.; Uezumi, A. Adult stem cell and mesenchymal progenitor theories of aging. Front. Cell Dev. Biol. 2014, 2, 10. [Google Scholar] [CrossRef] [PubMed]
- Powers, E.T.; Morimoto, R.I.; Dillin, A.; Kelly, J.W.; Balch, W.E. Biological and Chemical Approaches to Diseases of Proteostasis Deficiency. Annu. Rev. Biochem. 2009, 78, 959–991. [Google Scholar] [CrossRef]
- Fraga, M.; Esteller, M. Epigenetics and aging: The targets and the marks. Trends Genet. 2007, 23, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, C.; Campisi, J. Chronic Inflammation (Inflammaging) and Its Potential Contribution to Age-Associated Diseases. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2014, 69, S4–S9. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Zhang, X.; Zheng, S.; Khanabdali, R.; Kalionis, B.; Wu, J.; Wan, W.; Tai, X. An Update on Inflamm-Aging: Mechanisms, Prevention, and Treatment. J. Immunol. Res. 2016, 2016, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, P.; El-Jawhari, J.J.; Giannoudis, P.V.; Burska, A.N.; Ponchel, F.; Jones, E.A. Age Related Changes in Bone Marrow Mesenchymal Stromal Cells: A Potential Impact on Osteoporosis and Osteoarthritis Development. Cell Transplant. 2017, 26. [Google Scholar] [CrossRef] [PubMed]
- Loeser, R.F. Age-Related Changes in the Musculoskeletal System and the Development of Osteoarthritis. Clin. Geriatr. Med. 2010, 26, 371–386. [Google Scholar] [CrossRef] [PubMed]
- Ahamad, N.; Rath, P. Bone Marrow Stem Cells, Aging, and Age-Related Diseases. In Models, Molecules and Mechanisms in Biogerontology; Springer: Singapore, 2021; pp. 321–352. [Google Scholar]
- Kovtonyuk, L.V.; Fritsch, K.; Feng, X.; Manz, M.; Takizawa, H. Inflamm-Aging of Hematopoiesis, Hematopoietic Stem Cells, and the Bone Marrow Microenvironment. Front. Immunol. 2016, 7, 502. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- de Magalhães, J.P.; Finch, C.E.; Janssens, G. Next-generation sequencing in aging research: Emerging applications, problems, pitfalls and possible solutions. Ageing Res. Rev. 2010, 9, 315–323. [Google Scholar] [CrossRef]
- Chen, Y.-J.; Chang, W.-A.; Huang, M.-S.; Chen, C.-H.; Wang, K.-Y.; Hsu, Y.-L.; Kuo, P.-L. Identification of novel genes in aging osteoblasts using next-generation sequencing and bioinformatics. Oncotarget 2017, 8, 113598–113613. [Google Scholar] [CrossRef] [PubMed]
- Biolabs NE. NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® | NEB. 2016. Available online: https://international.neb.com/products/e7645-nebnext-ultra-ii-dna-library-prep-kit-for-illumina#Product%20Information (accessed on 10 July 2021).
- Illumina. Nextera XT DNA Library Prep Kit. 2016. Available online: https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/nextera-xt-dna.html (accessed on 10 July 2021).
- Buermans, H.; Dunnen, J.D. Next generation sequencing technology: Advances and applications. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2014, 1842, 1932–1941. [Google Scholar] [CrossRef] [PubMed]
- Logsdon, G.A.; Vollger, M.R.; Eichler, E.E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 2020, 21. [Google Scholar] [CrossRef]
- Oikonomopoulos, S.; Bayega, A.; Fahiminiya, S.; Djambazian, H.; Berube, P.; Ragoussis, J. Methodologies for Transcript Profiling Using Long-Read Technologies. Front. Genet. 2020, 11, 606. [Google Scholar] [CrossRef]
- Petric, R.C.; Pop, L.-A.; Jurj, A.; Raduly, L.; Dumitrascu, D.; Dragos, N.; Neagoe, I.B. Next generation sequencing applications for breast cancer research. Clujul. Med. 2015, 88, 278–287. [Google Scholar] [CrossRef]
- Reagan, M.R.; Rosen, C.J. Navigating the bone marrow niche: Translational insights and cancer-driven dysfunction. Nat. Rev. Rheumatol. 2016, 12, 154–168. [Google Scholar] [CrossRef]
- Wang, H.; Leng, Y.; Gong, Y. Bone Marrow Fat and Hematopoiesis. Front. Endocrinol. 2018, 9, 694. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Ferrer, S.; Michurina, T.V.; Ferraro, F.; Mazloom, A.R.; MacArthur, B.; Lira, S.A.; Scadden, D.T.; Ma’Ayan, A.; Enikolopov, G.; Frenette, P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nat. Cell Biol. 2010, 466, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.-Y.; Jeong, S.-Y.; Lee, H.-R.; Oh, I.-H. Age-related differences in the bone marrow stem cell niche generate specialized microenvironments for the distinct regulation of normal hematopoietic and leukemia stem cells. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Li, J.; Liu, X.; Zuo, B.; Zhang, L. The Role of Bone Marrow Microenvironment in Governing the Balance between Osteoblastogenesis and Adipogenesis. Aging Dis. 2016, 7, 514–525. [Google Scholar] [CrossRef] [PubMed]
- González-Gualda, E.; Baker, A.G.; Fruk, L.; Muñoz-Espín, D. A guide to assessing cellular senescence in vitro and in vivo. FEBS J. 2021, 288, 56–80. [Google Scholar] [CrossRef]
- Guidi, N.; Marka, G.; Sakk, V.; Zheng, Y.; Florian, M.C.; Geiger, H. An aged bone marrow niche restrains rejuvenated hematopoietic stem cells. STEM Cells 2021, 39, 1101–1106. [Google Scholar] [CrossRef] [PubMed]
- Connor, K.M.; Hsu, Y.; Aggarwal, P.K.; Capone, S.; Colombo, A.R.; Ramsingh, G. Understanding metabolic changes in aging bone marrow. Exp. Hematol. Oncol. 2018, 7, 13. [Google Scholar] [CrossRef]
- Groarke, E.M.; Young, N.S. Aging and Hematopoiesis. Clin. Geriatr. Med. 2019, 35, 285–293. [Google Scholar] [CrossRef]
- Moehrle, B.M.; Geiger, H. Aging of hematopoietic stem cells: DNA damage and mutations? Exp. Hematol. 2016, 44, 895–901. [Google Scholar] [CrossRef]
- Lee, J.; Yoon, S.R.; Choi, I.; Jung, H. Causes and Mechanisms of Hematopoietic Stem Cell Aging. Int. J. Mol. Sci. 2019, 20, 1272. [Google Scholar] [CrossRef]
- Strzelecka, P.M.; Damm, F. Haematopoietic ageing through the lens of single-cell technologies. Dis. Model. Mech. 2021, 14. [Google Scholar] [CrossRef]
- Nakamura-Ishizu, A.; Ito, K.; Suda, T. Hematopoietic Stem Cell Metabolism during Development and Aging. Dev. Cell 2020, 54, 239–255. [Google Scholar] [CrossRef]
- Ho, Y.-H.; Méndez-Ferrer, S. Microenvironmental contributions to hematopoietic stem cell aging. Haematologica 2020, 105, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Akunuru, S.; Geiger, H. Aging, Clonality, and Rejuvenation of Hematopoietic Stem Cells. Trends Mol. Med. 2016, 22, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Mejia-Ramirez, E.; Florian, M.C. Understanding intrinsic hematopoietic stem cell aging. Haematologica 2019, 105, 22–37. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; MacDougald, O.A. Stem cell factor: The bridge between bone marrow adipocytes and hematopoietic cells. Haematologica 2019, 104, 1689–1691. [Google Scholar] [CrossRef] [PubMed]
- Ambrosi, T.H.; Scialdone, A.; Graja, A.; Gohlke, S.; Jank, A.-M.; Bocian, C.; Woelk, L.; Fan, H.; Logan, D.W.; Schurmann, A.; et al. Adipocyte Accumulation in the Bone Marrow during Obesity and Aging Impairs Stem Cell-Based Hematopoietic and Bone Regeneration. Cell Stem Cell 2017, 20, 771–784.e6. [Google Scholar] [CrossRef]
- Zhang, Z.; Huang, Z.; Ong, B.; Sahu, C.; Zeng, H.; Ruan, H.-B. Bone marrow adipose tissue-derived stem cell factor mediates metabolic regulation of hematopoiesis. Haematologica 2019, 104, 1731–1743. [Google Scholar] [CrossRef] [PubMed]
- Ghannam, S.; Bouffi, C.; Djouad, F.; Jorgensen, C.; Noël, D. Immunosuppression by mesenchymal stem cells: Mechanisms and clinical applications. Stem Cell Res. Ther. 2010, 1, 1–7. [Google Scholar] [CrossRef]
- Horwitz, E.M.; Le Blanc, K.; Dominici, M.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.C.; Deans, R.J.; Krause, D.S.; Keating, A. Clarification of the nomenclature for MSC: The International Society for Cellular Therapy position statement. Cytotherapy 2005, 7, 393–395. [Google Scholar] [CrossRef]
- Boxall, S.A.; Jones, E. Markers for Characterization of Bone Marrow Multipotential Stromal Cells. Stem Cells Int. 2012, 2012, 1–12. [Google Scholar] [CrossRef]
- Siegel, G.; Kluba, T.; Hermanutz-Klein, U.; Bieback, K.; Northoff, H.; Schäfer, R. Phenotype, donor age and gender affect function of human bone marrow-derived mesenchymal stromal cells. BMC Med. 2013, 11, 146. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.; Boyette, L.B.; Tuan, R.S. Characterization of bone marrow-derived mesenchymal stem cells in aging. Bone 2015, 70, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, P.; El-Jawhari, J.J.; Burska, A.N.; Ponchel, F.; Giannoudis, P.V.; Jones, E.A. The Analysis of In Vivo Aging in Human Bone Marrow Mesenchymal Stromal Cells Using Colony-Forming Unit-Fibroblast Assay and the CD45lowCD271+ Phenotype. Stem Cells Int. 2019, 2019, 1–14. [Google Scholar] [CrossRef]
- Josephson, A.M.; Bradaschia-Correa, V.; Lee, S.; Leclerc, K.; Patel, K.S.; Lopez, E.M.; Litwa, H.; Neibart, S.S.; Kadiyala, M.; Wong, M.Z.; et al. Age-related inflammation triggers skeletal stem/progenitor cell dysfunction. Proc. Natl. Acad. Sci. USA 2019, 116, 6995–7004. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kim, C.; Choi, Y.S.; Kim, M.; Park, C.; Suh, Y. Age-related alterations in mesenchymal stem cells related to shift in differentiation from osteogenic to adipogenic potential: Implication to age-associated bone diseases and defects. Mech. Ageing Dev. 2012, 133, 215–225. [Google Scholar] [CrossRef]
- Yang, Y.-M.; Li, P.; Cui, D.-C.; Dang, R.-J.; Zhang, L.; Wen, N.; Jiang, X.-X. Effect of aged bone marrow microenvironment on mesenchymal stem cell migration. AGE 2015, 37, 1–9. [Google Scholar] [CrossRef][Green Version]
- Ganguly, P.; Burska, A.N.; Davis, C.L.M.; El-Jawhari, J.J.; Giannoudis, P.V.; Jones, E.A. Intrinsic Type 1 Interferon (IFN1) Profile of Uncultured Human Bone Marrow CD45lowCD271+ Multipotential Stromal Cells (BM-MSCs): The Impact of Donor Age, Culture Expansion and IFNα and IFNβ Stimulation. Biomedicines 2020, 8, 214. [Google Scholar] [CrossRef] [PubMed]
- Fulop, T.; Larbi, A.; Dupuis, G.; Le Page, A.; Frost, E.H.; Cohen, A.A.; Witkowski, J.M.; Franceschi, C. Immunosenescence and Inflamm-Aging as Two Sides of the Same Coin: Friends or Foes? Front. Immunol. 2018, 8, 1960. [Google Scholar] [CrossRef]
- Aiello, A.; Farzaneh, F.; Candore, G.; Caruso, C.; Davinelli, S.; Gambino, C.M.; Ligotti, M.E.; Zareian, N.; Accardi, G. Immunosenescence and Its Hallmarks: How to Oppose Aging Strategically? A Review of Potential Options for Therapeutic Intervention. Front. Immunol. 2019, 10, 2247. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.T.; Casciaro, M.; Gangemi, S.; Buquicchio, R. Immunosenescence in aging: Between immune cells depletion and cytokines up-regulation. Clin. Mol. Allergy 2017, 15, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Keren, Z.; Naor, S.; Nussbaum, S.; Golan, K.; Itkin, T.; Sasaki, Y.; Schmidt-Supprian, M.; Lapidot, T.; Melamed, D. B-cell depletion reactivates B lymphopoiesis in the BM and rejuvenates the B lineage in aging. Blood 2011, 117, 3104–3112. [Google Scholar] [CrossRef] [PubMed]
- Goronzy, J.J.; Weyand, C.M. Mechanisms underlying T cell ageing. Nat. Rev. Immunol. 2019, 19, 573–583. [Google Scholar] [CrossRef]
- Mittelbrunn, M.; Kroemer, G. Hallmarks of T cell aging. Nat. Immunol. 2021, 22, 687–698. [Google Scholar] [CrossRef]
- Yang, H.-J.; Ratnapriya, R.; Cogliati, T.; Kim, J.-W.; Swaroop, A. Vision from next generation sequencing: Multi-dimensional genome-wide analysis for producing gene regulatory networks underlying retinal development, aging and disease. Prog. Retin. Eye Res. 2015, 46, 1–30. [Google Scholar] [CrossRef]
- Mertens, J.; Paquola, A.C.; Ku, M.; Hatch, E.; Böhnke, L.; Ladjevardi, S.; McGrath, S.; Campbell, B.; Lee, H.; Herdy, J.R.; et al. Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell 2015, 17, 705–718. [Google Scholar] [CrossRef]
- Kowalczyk, M.S.; Tirosh, I.; Heckl, D.; Rao, T.N.; Dixit, A.; Haas, B.J.; Schneider, R.K.; Wagers, A.J.; Ebert, B.L.; Regev, A. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015, 25, 1860–1872. [Google Scholar] [CrossRef]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-Related Clonal Hematopoiesis Associated with Adverse Outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef]
- Genovese, G.; Kähler, A.K.; Handsaker, R.E.; Lindberg, J.; Rose, S.A.; Bakhoum, S.F.; Chambert, K.; Mick, E.; Neale, B.M.; Fromer, M.; et al. Clonal Hematopoiesis and Blood-Cancer Risk Inferred from Blood DNA Sequence. N. Engl. J. Med. 2014, 371, 2477–2487. [Google Scholar] [CrossRef]
- McKerrell, T.; Park, N.; Chi, J.; Collord, G.; Moreno, T.; Ponstingl, H.; Dias, J.; Gerasimou, P.; Melanthiou, K.; Prokopiou, C.; et al. JAK2 V617F hematopoietic clones are present several years prior to MPN diagnosis and follow different expansion kinetics. Blood Adv. 2017, 1, 968–971. [Google Scholar] [CrossRef]
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The cancer genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef]
- Lodato, M.A.; Woodworth, M.B.; Lee, S.; Evrony, G.D.; Mehta, B.K.; Karger, A.; Lee, S.; Chittenden, T.W.; D’Gama, A.M.; Cai, X.; et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 2015, 350, 94–98. [Google Scholar] [CrossRef]
- Knouse, K.A.; Wu, J.; Amon, A. Assessment of megabase-scale somatic copy number variation using single-cell sequencing. Genome Res. 2016, 26, 376–384. [Google Scholar] [CrossRef]
- Pathak, S.; Rowczenio, D.M.; Owen, R.G.; Doody, G.M.; Newton, D.J.; Taylor, C.; Taylor, J.; Cargo, C.; Hawkins, P.N.; Krause, K.; et al. Exploratory study of MYD88 L265P, rare NLRP3 variants and clonal hematopoiesis prevalence in patients with Schnitzler’s Syndrome. Arthritis Rheumatol. 2019, 71, 2121–2125. [Google Scholar] [CrossRef]
- Hennrich, M.L.; Romanov, N.; Horn, P.; Jaeger, S.; Eckstein, V.; Steeples, V.; Ye, F.; Ding, X.; Poisa-Beiro, L.; Lai, M.C.; et al. Cell-specific proteome analyses of human bone marrow reveal molecular features of age-dependent functional decline. Nat. Commun. 2018, 9, 1–18. [Google Scholar] [CrossRef]
- Picelli, S.; Faridani, O.R.; Björklund, A.K.; Winberg, G.; Sagasser, S.; Sandberg, R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protocols 2014, 9, 171–181. [Google Scholar] [CrossRef]
- Oetjen, K.A.; Lindblad, K.E.; Goswami, M.; Gui, G.; Dagur, P.K.; Lai, C.; Dillon, L.W.; McCoy, J.P.; Hourigan, C.S. Human bone marrow assessment by single-cell RNA sequencing, mass cytometry, and flow cytometry. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Shaw, B.E.; Ball, L.; Beksac, M.; Bengtsson, M.; Confer, D.; Diler, S.; Fechter, M.; Greinix, H.; Koh, M.; Lee, S.; et al. Donor safety: The role of the WMDA in ensuring the safety of volunteer unrelated donors: Clinical and ethical considerations. Bone Marrow Transplant. 2010, 45, 832–838. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, Y.; Zhang, Q.; Ren, X.; Zhang, Z. Direct Comparative Analyses of 10X Genomics Chromium and Smart-seq2. Genom. Proteom. Bioinform. 2021. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Milon, B.; Ott, S.; Zhao, X.; Sadzewicz, L.; Shetty, A.; Boger, E.T.; Tallon, L.J.; Morell, R.J.; Mahurkar, A.; et al. A comparative analysis of library prep approaches for sequencing low input translatome samples. BMC Genom. 2018, 19, 696. [Google Scholar] [CrossRef]
- Cheung, P.; Vallania, F.; Warsinske, H.C.; Donato, M.; Schaffert, S.; Chang, S.E.; Dvorak, M.; Dekker, C.L.; Davis, M.M.; Utz, P.J.; et al. Single-Cell Chromatin Modification Profiling Reveals Increased Epigenetic Variations with Aging. Cell 2018, 173, 1385–1397.e14. [Google Scholar] [CrossRef]
- Stolzing, A.; Jones, E.; McGonagle, D.; Scutt, A. Age-related changes in human bone marrow-derived mesenchymal stem cells: Consequences for cell therapies. Mech. Ageing Dev. 2008, 129, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Frasca, D.; Blomberg, B.B. Aging affects human B cell responses. J. Clin. Immunol. 2011, 31, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Chambers, S.M.; Shaw, C.; Gatza, C.; Fisk, C.J.; Donehower, L.A.; Goodell, M.A. Aging Hematopoietic Stem Cells Decline in Function and Exhibit Epigenetic Dysregulation. PLoS Biol. 2007, 5, e201. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zeng, X.; Xu, Y.; Wang, B.; Zhao, Y.; Lai, X.; Qian, P.; Huang, H. Mechanisms and rejuvenation strategies for aged hematopoietic stem cells. J. Hematol. Oncol. 2020, 13, 1–16. [Google Scholar] [CrossRef]
- Ambrosi, T.H.; Marecic, O.; McArdle, A.; Sinha, R.; Gulati, G.S.; Tong, X.; Wang, Y.; Steininger, H.M.; Hoover, M.Y.; Koepke, L.S.; et al. Aged skeletal stem cells generate an inflammatory degenerative niche. Nat. Cell Biol. 2021, 597, 256–262. [Google Scholar] [CrossRef]
- Wolock, S.L.; Krishnan, I.; Tenen, D.E.; Matkins, V.; Camacho, V.; Patel, S.; Agarwal, P.; Bhatia, R.; Tenen, D.G.; Klein, A.M.; et al. Mapping Distinct Bone Marrow Niche Populations and Their Differentiation Paths. Cell Rep. 2019, 28, 302–311.e5. [Google Scholar] [CrossRef]
- Zhong, L.; Yao, L.; Tower, R.J.; Wei, Y.; Miao, Z.; Park, J.; Shrestha, R.; Wang, L.; Yu, W.; Holdreith, N.; et al. Single cell transcriptomics identifies a unique adipose lineage cell population that regulates bone marrow environment. eLife 2020, 9. [Google Scholar] [CrossRef]
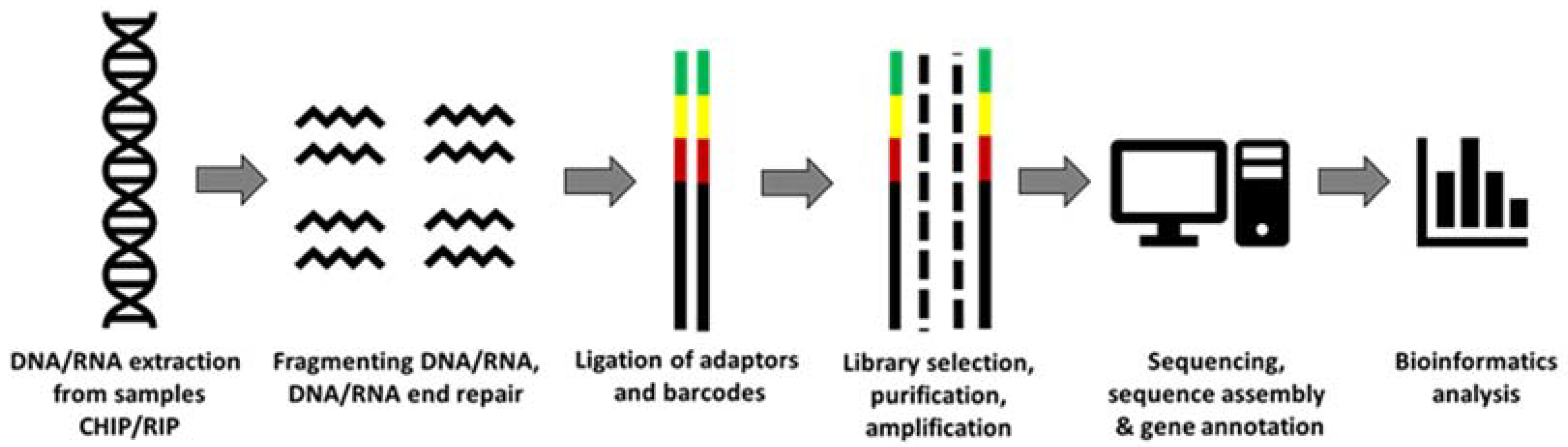

| Cells Explored | DNA/RNA | Key Findings | Reference |
|---|---|---|---|
| SSCs | RNA |
| [82] |
| Non-HSCs (MSCs) | RNA |
| [83] |
| MSC lineage cells | RNA |
| [84] |
| HSCs and MSCs | RNA |
| [71] |
| BM cells with focus on T cells | RNA |
| [73] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganguly, P.; Toghill, B.; Pathak, S. Aging, Bone Marrow and Next-Generation Sequencing (NGS): Recent Advances and Future Perspectives. Int. J. Mol. Sci. 2021, 22, 12225. https://doi.org/10.3390/ijms222212225
Ganguly P, Toghill B, Pathak S. Aging, Bone Marrow and Next-Generation Sequencing (NGS): Recent Advances and Future Perspectives. International Journal of Molecular Sciences. 2021; 22(22):12225. https://doi.org/10.3390/ijms222212225
Chicago/Turabian StyleGanguly, Payal, Bradley Toghill, and Shelly Pathak. 2021. "Aging, Bone Marrow and Next-Generation Sequencing (NGS): Recent Advances and Future Perspectives" International Journal of Molecular Sciences 22, no. 22: 12225. https://doi.org/10.3390/ijms222212225
APA StyleGanguly, P., Toghill, B., & Pathak, S. (2021). Aging, Bone Marrow and Next-Generation Sequencing (NGS): Recent Advances and Future Perspectives. International Journal of Molecular Sciences, 22(22), 12225. https://doi.org/10.3390/ijms222212225







