Fukutin Protein Participates in Cell Proliferation by Enhancing Cyclin D1 Expression through Binding to the Transcription Factor Activator Protein-1: An In Vitro Study
Abstract
1. Introduction
2. Results
2.1. Fukutin Is Localized in Both the Nucleus and Cytoplasm
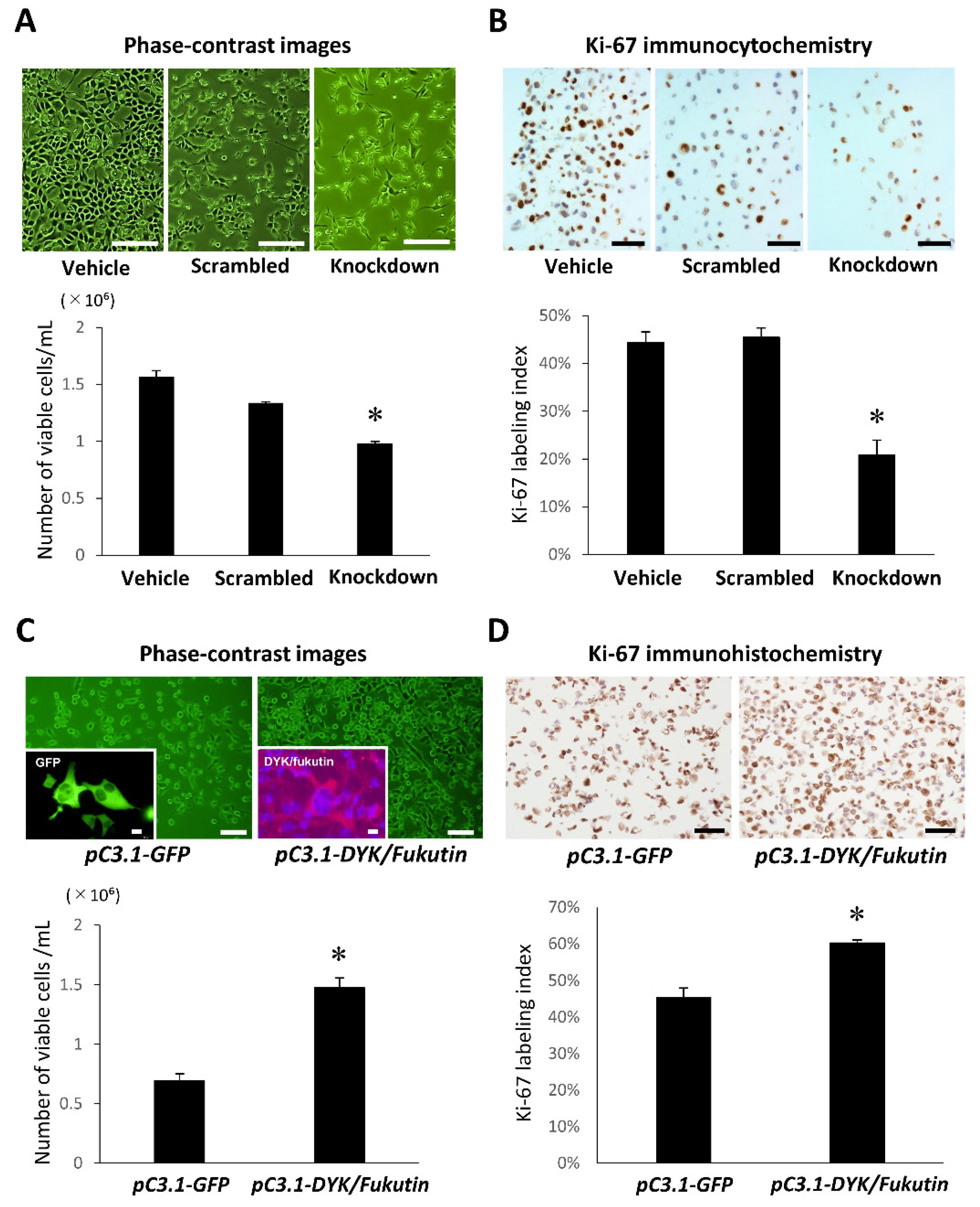
2.2. Knockdown and Overexpression of Fukutin Alter Proliferation Ability
2.3. Knockdown and Overexpression of Fukutin Alter Cyclin D1 Transcript and Protein Levels
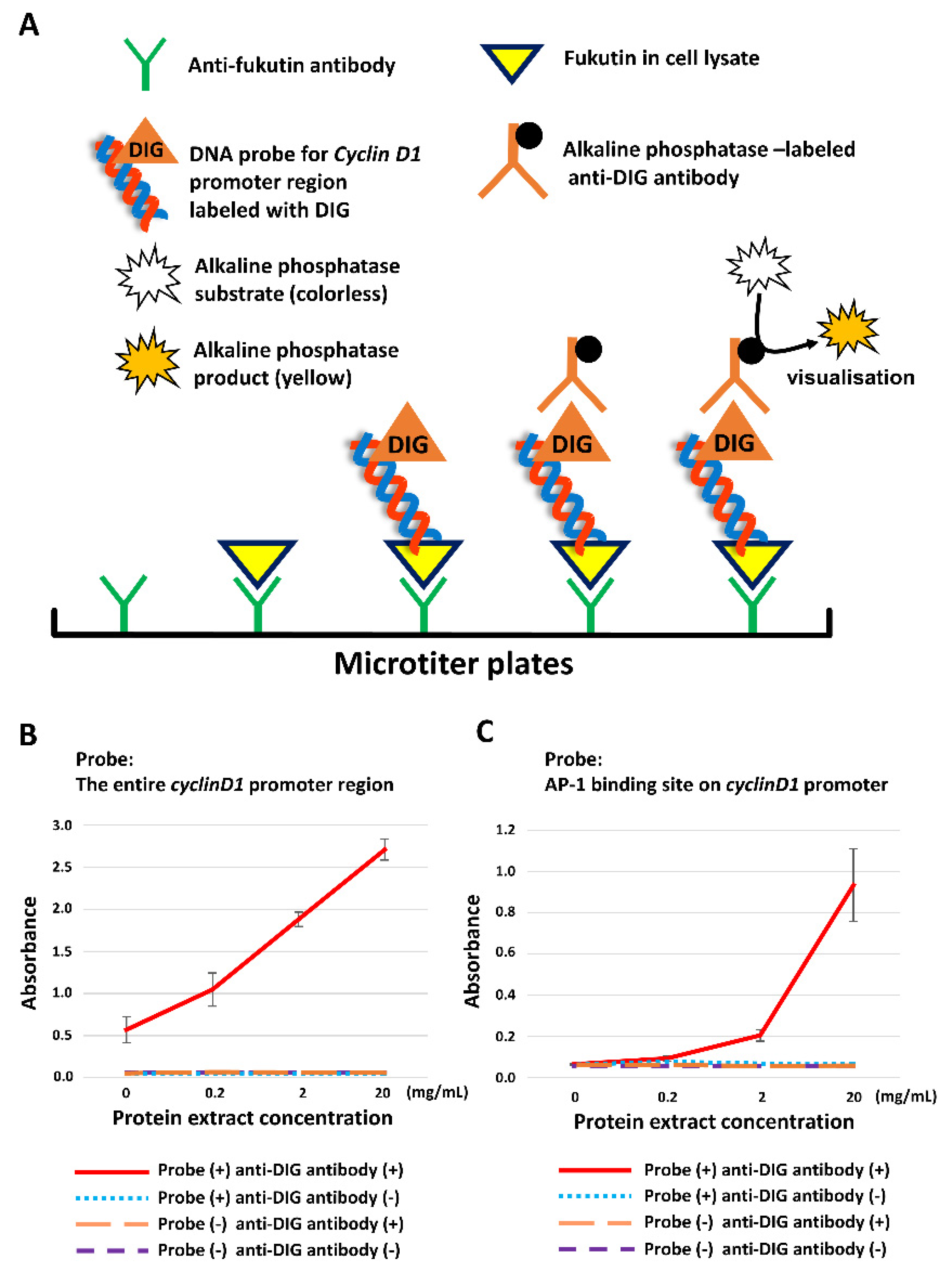
2.4. Fukutin Protein Binds to the Promoter Region of Cyclin D1
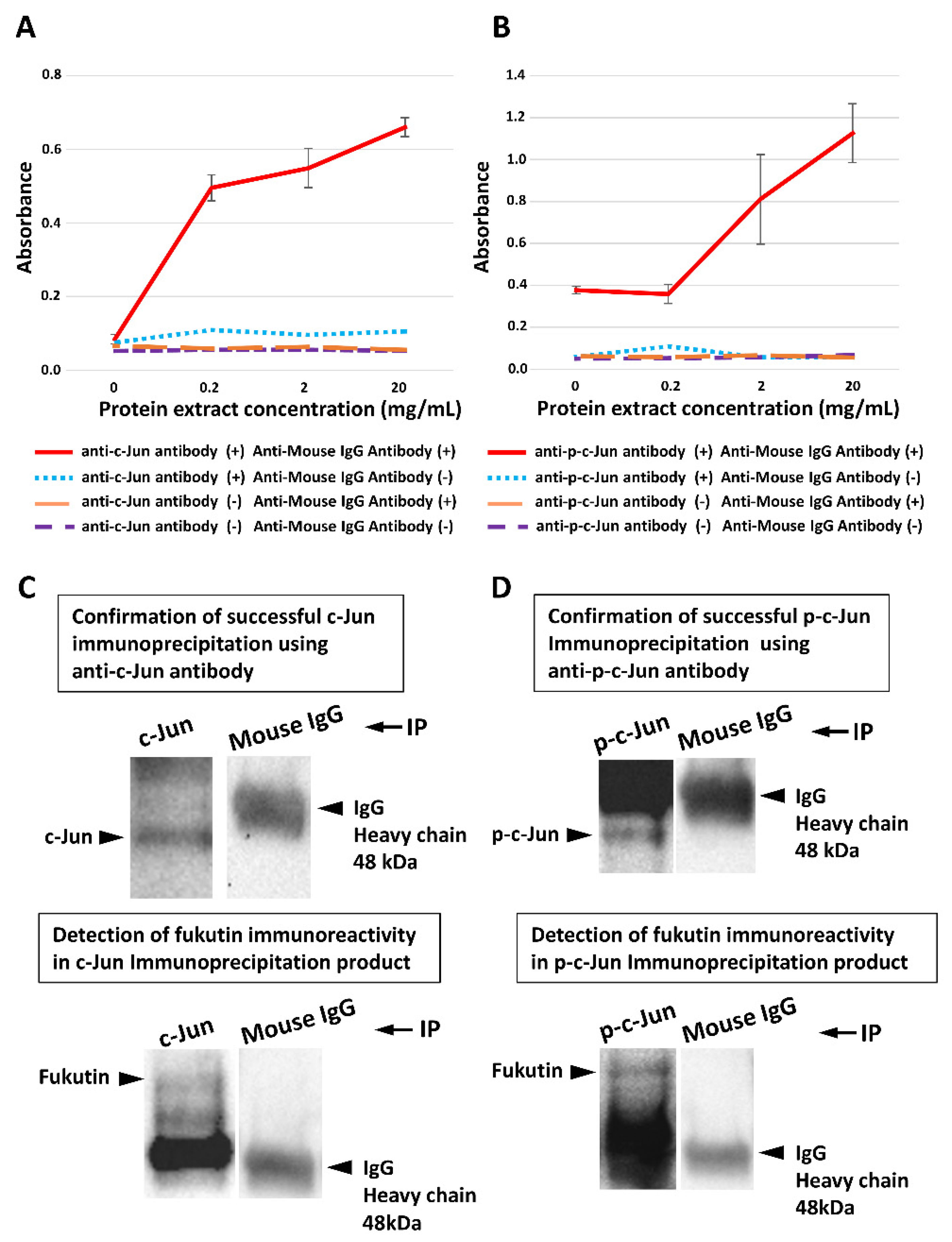
2.5. Fukutin Protein Binds to the AP-1 Component c-Jun/p-c-Jun
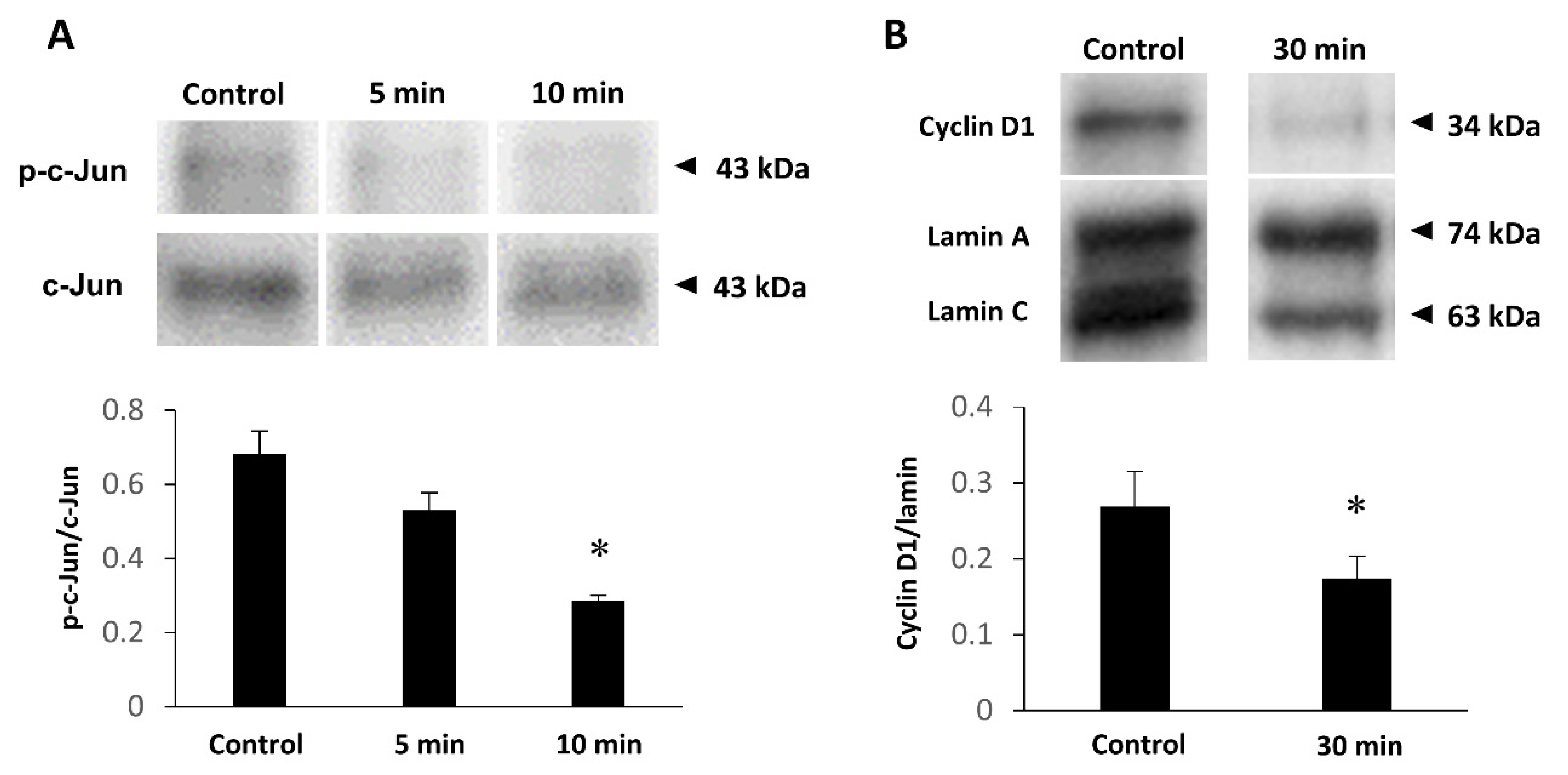
2.6. JNK Inhibition Downregulates Cyclin D1 Expression
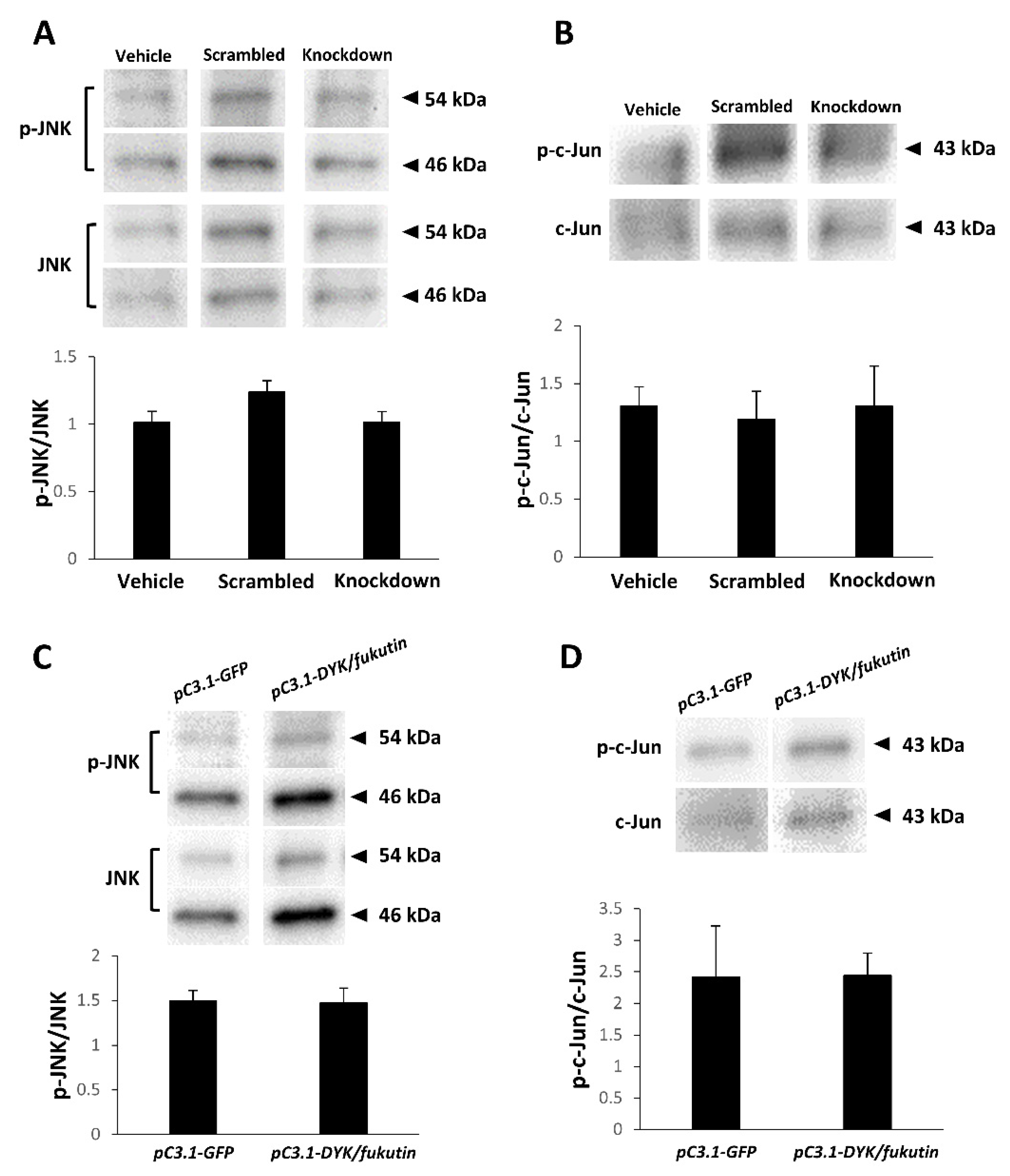
2.7. Knockdown or Overexpression of Fukutin Does Not Affect Phosphorylation of JNK or c-Jun
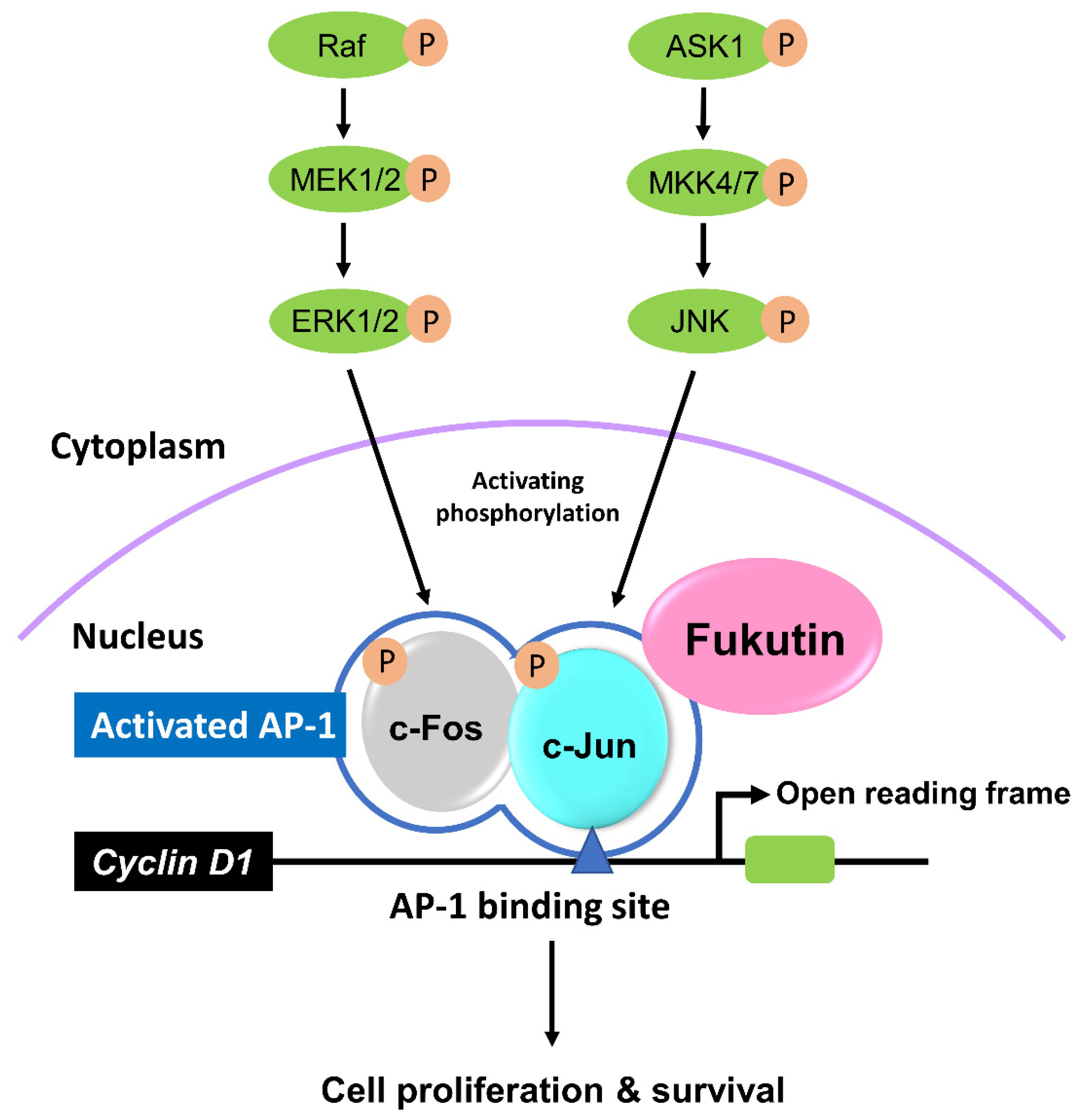
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Experimental Design
4.2. Knockdown Fukutin
4.3. Overexpression of Fukutin
4.4. JNK Inhibition
4.5. Immunocytochemistry on Cell Block Sections
4.6. Immunocytochemistry on Culture Slides
4.7. Semiquantitative RT-PCR Analysis
4.8. Protein Extraction
4.9. Immunoprecipitation
4.10. Western Blot Analysis
4.11. Preparation of DNA Probes for ELISA-Based Assay
4.12. ELISA
4.13. Sandwich ELISA
4.14. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fukuyama, Y.; Kawazura, M.; Haruna, H. A peculiar form of congenital progressive muscular dystrophy. Paediatr. Univ. Tokyo 1960, 4, 5–8. [Google Scholar]
- Osawa, M.; Sumida, S.; Suzuki, N.; Arai, Y.; Ikenaka, H.; Murasugi, H.; Shishikura, K.; Suzuki, H.; Saito, K.; Fukuyama, Y. Fukuyama type congenital progressive muscular dystrophy. In Congenital Muscular Dystrophies; Fukuyama, Y., Osawa, M., Saito, K., Eds.; Elsevier: Amsterdam, The Netherlands, 1997; pp. 31–68. [Google Scholar]
- Kobayashi, K.; Nakahori, Y.; Miyake, M.; Matsumura, K.; Kondo-Iida, E.; Nomura, Y.; Segawa, M.; Yoshioka, M.; Saito, K.; Osawa, M.; et al. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature 1998, 394, 388–392. [Google Scholar] [CrossRef] [PubMed]
- Kanagawa, M.; Kobayashi, K.; Tajiri, M.; Manya, H.; Kuga, A.; Yamaguchi, Y.; Akasaka, M.K.; Furukawa, J.; Mizuno, M.; Kawakami, H.; et al. Identification of a Post-translational Modification with Ribitol-Phosphate and Its Defect in Muscular Dystrophy. Cell Rep. 2016, 14, 2209–2223. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.K.; Ogawa, M.; Tagawa, K.; Noguchi, S.; Ishihara, T.; Nonaka, I.; Arahata, K. Selective deficiency of α-dystroglycan in Fukuyama-type congenital muscular dystrophy. Neurology 2001, 57, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Toyoda, C.; Kobayashi, M.; Kondo, E.; Saito, K.; Osawa, M. Pial-glial barrier abnormalities in fetuses with Fukuyama congenital muscular dystrophy. Brain Dev. 1997, 19, 35–42. [Google Scholar] [CrossRef]
- Yamamoto, T.; Kato, Y.; Kawaguchi, M.; Shibata, N.; Kobayashi, M. Expression and localization of fukutin, POMGnT1, and POMT1 in the central nervous system: Consideration for functions of fukutin. Med. Electron. Microsc. 2004, 37, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Shibata, N.; Saito, Y.; Osawa, M.; Kobayashi, M. Functions of fukutin, a gene responsible for Fukuyama type congenital muscular dystrophy, in neuromuscular system and other somatic organs. Cent. Nerv. Syst. Agents Med. Chem. 2010, 10, 169–179. [Google Scholar] [CrossRef]
- Saito, Y.; Yamamoto, T.; Ohtsuka-Tsurumi, E.; Oka, A.; Mizuguchi, M.; Itoh, M.; Voit, T.; Kato, Y.; Kobayashi, M.; Saito, Y.; et al. Fukutin expression in mouse non-muscle somatic organs: Its relationship to the hypoglycosylation of alpha -dystroglycan in Fukuyama-type congenital muscular dystrophy. Brain Dev. 2004, 26, 469–479. [Google Scholar] [CrossRef]
- Yamamoto, T.; Kato, Y.; Shibata, N.; Sawada, T.; Osawa, M.; Kobayashi, M. A role of fukutin, a gene responsible for Fukuyama type congenital muscular dystrophy, in cancer cells: A possible role to suppress cell proliferation. Int. J. Exp. Pathol. 2008, 89, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, J.; Ishikwa, K.; Kobayashi, K.; Kondo-lida, E.; Fukuyama, M.; Mizusawa, H.; Takashima, S.; Sakakihara, Y.; Nakamura, Y.; Toda, T. Neuronal expression of the fukutin gene. Hum. Mol. Genet. 2000, 9, 3083–3090. [Google Scholar]
- Oo, H.-Z.; Sentani, K.; Mukai, S.; Hattori, T.; Shinmei, S.; Goto, K.; Sakamoto, N.; Naito, Y.; Anami, K.; Trang, P.T.B.; et al. Fukutin, identified by the Escherichia coli ampicillin secretion trap (CAST) method, participates in tumor progression in gastric cancer. Gastric Cancer. 2016, 19, 443–452. [Google Scholar] [CrossRef][Green Version]
- Pham, T.T.B.; Oue, N.; Yamamoto, M.; Fujihara, M.; Ishida, T.; Mukai, S.; Sakamoto, N.; Sentani, K.; Yasui, W. Characteristic expression of fukutin in gastric cancer among atomic bomb survivors. Oncol. Lett. 2017, 13, 937–941. [Google Scholar] [CrossRef]
- Rauscher, F.J.D.; Cohen, D.R.; Curran, T.; Bos, T.J.; Vogt, P.K.; Bohmann, D.; Tjian, R.; Franza, B.R., Jr. Fos-associated protein p39 is the product of the jun proto-oncogene. Science 1988, 240, 1010–1016. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, M.; Kolbus, A.; Piu, F.; Szabowski, A.; MohleSteinlein, U.; Tian, J.; Karin, M.; Angel, P.; Wagner, E.F. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999, 13, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Hao, X.; Tan, F.-F.; Pei, X.; Shang, L.-M.; Jiang, X.-L.; Yang, F. The elements of human cyclin D1 promoter and regulation involved. Clin. Epigenet. 2011, 2, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Angel, P.; Allegretto, E.A.; Okino, S.T.; Hattori, K.; Boyle, W.J.; Hunter, T.; Karin, M. Oncogene jun encodes a sequence-specific trans-activator similar to AP-1. Nature 1988, 332, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Bohmann, D.; Bos, T.J.; Admo, A.; Nishimura, T.; Vogt, P.K.; Tjian, R. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science 1987, 238, 1386–1392. [Google Scholar] [CrossRef]
- Chiu, R.; Boyle, W.J.; Meek, J.; Smeal, T.; Hunter, T.; Karin, M. The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell 1988, 54, 541–552. [Google Scholar] [CrossRef]
- Bakiri, L.; Lallemand, D.; Wetzel, E.B.; Yaniv, M. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: A role in the control of cyclin D1 expression. EMBO J. 2000, 19, 2056–2068. [Google Scholar] [CrossRef]
- Shaulian, E.; Karin, M. AP-1 in cell proliferation and survival. Oncogene 2001, 20, 2390–2400. [Google Scholar] [CrossRef]
- Trop-Steinberg, S.; Azar, Y. AP-1 Expression and its Clinical Relevance in Immune Disorders and Cancer. Am. J. Med. Sci. 2017, 353, 474–483. [Google Scholar] [CrossRef] [PubMed]
- Oak, S.A.; Zhou, Y.W.; Jarrett, H.W. Skeletal muscle signaling pathway through the dystrophin glycoprotein complex and Rac1. J. Biol. Chem. 2003, 278, 39287–39295. [Google Scholar] [CrossRef] [PubMed]
- Tachikawa, M.; Kanagawa, M.; Yu, C.-C.; Kobayashi, K.; Toda, T. Mislocalization of fukutin protein by disease-causing missense mutations can be rescued with treatments directed at folding amelioration. J. Biol. Chem. 2012, 287, 8398–8406. [Google Scholar] [CrossRef]
- Haro, C.; Uribe, M.L.; Quereda, C.; Cruces, J.; Martín-Nieto, J. Expression in retinal neurons of fukutin and FKRP, the protein products of two dystroglycanopathy-causative genes. Mol. Vis. 2018, 24, 43–58. [Google Scholar]
- Kobayashi, K.; Sasaki, J.; Kondo-Iida, E.; Fukuda, Y.; Kinoshita, M.; Sunada, Y.; Nakamura, Y.; Toda, T. Structural organization, complete genomic sequences and mutational analyses of the Fukuyama-type congenital muscular dystrophy gene, fukutin. FEBS Lett. 2001, 489, 192–196. [Google Scholar] [CrossRef]
- Duan, P.J.; Zhao, J.H.; Xie, L.L. Cul4B promotes the progression of ovarian cancer by upregulating the expression of CDK2 and CyclinD1. J. Ovarian Res. 2020, 13, 76. [Google Scholar] [CrossRef]
- Sallinen, S.L.; Sallinen, P.K.; Kononen, J.T.; Syrja Koski, K.M.; Nupponen, N.N.; Rantala, I.S.; Helen, P.T.; Helin, H.J.; Haapasalo, H.K. Cyclin D1 expression in astrocytomas is associated with cell proliferation activity and patient prognosis. J. Pathol. 1999, 188, 289–293. [Google Scholar] [CrossRef]
- Li, T.; Song, T.; Ni, L.; Yang, G.; Song, X.; Wu, L.; Liu, B.; Liu, C. The p-ERK–p-c-Jun–cyclinD1 pathway is involved in proliferation of smooth muscle cells after exposure to cigarette smoke extract. Biochem. Biophys. Res. Commun. 2014, 453, 316–320. [Google Scholar] [CrossRef]
- Lavoie, J.N.; L’Allemain, G.; Brunet, A.; Müller, R.; Pouysse’gur, J. Cyclin D1 Expression Is Regulated Positively by the p42/p44MAPK and Negatively by the p38/HOGMAPK Pathway. J. Biol. Chem. 1996, 271, 20608–20616. [Google Scholar] [CrossRef]
- Hsia, E.Y.; Goodson, M.L.; Zou, J.X.; Privaisky, M.L.; Chen, H.-W. Nuclear receptor coregulators as a new paradigm for therapeutic targeting. Adv. Drug Deliv. Rev. 2010, 62, 1227–1237. [Google Scholar] [CrossRef]
- Albanese, C.; D’Amico, M.; Reutens, A.T.; Fu, M.; Watanabe, G.; Lee, R.J.; Kitsis, R.N.; Henglein, B.; Avantaggiati, M.; Somasundaram, K.; et al. Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J. Biol. Chem. 1999, 274, 34186–34195. [Google Scholar] [CrossRef] [PubMed]
- Spiegelman, B.M.; Heinrich, R.; Farber, D. Biological Contorol through Regulated Transcriptional Coactivators. Cell 2004, 119, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Arias, J.; Alberts, A.S.; Brindle, P.; Claret, F.-X.; Smeal, T.; Karin, M.; Fremisco, J.; Montminy, M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature 1994, 370, 226–228. [Google Scholar] [CrossRef] [PubMed]
- Kwok, R.P.; Lundblad, J.R.; Chrivia, J.C.; Richards, J.P.; Bachinger, H.P.; Brannan, R.G.; Roberts, S.G.E.; Green, M.R.; Goodman, R.H. Nuclear protein CBP is a coactivetor for the transcription factor CREB. Nature 1994, 370, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Oehler, T.; Wilhelm, D.; Angel, P.; Kouzarides, T. Stimulation of c-Jun activity by CBP: C-Jun residues Ser63/73 are required for CBP induced stimulation in vivo end CBP binding in vitro. Oncogene 1995, 11, 2509–2514. [Google Scholar] [PubMed]
- Smith, M.J.; Prochownik, E.V. Inhibition of c-jun Causes Reversible Proliferative Arrest and Withdrawal from the Cell Cycle. Blood 1992, 79, 2107–2115. [Google Scholar] [CrossRef]
- Claret, F.X.; Hibi, M.; Dhut, S.; Toda, T.; Karin, M. A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature 1996, 383, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Kato, Y.; Karita, M.; Takeiri, H.; Muramatsu, F.; Kobayashi, M.; Saito, K.; Osawa, M. Fukutin expression in glial cells and neurons: Implication in the brain lesions of Fukuyama congenital muscular dystrophy. Acta Neuropathol. 2002, 104, 217–224. [Google Scholar] [CrossRef]
- Macintyre, E.H.; Pontén, J.; Vatter, A.E. The ultrastructure of human and murine astrocytes and of human fibroblasts in culture. Acta Pathol. Microbiol. Scand. A 1972, 80, 267–283. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okamura, Y.; Yamamoto, T.; Tsukui, R.; Kato, Y.; Shibata, N. Fukutin Protein Participates in Cell Proliferation by Enhancing Cyclin D1 Expression through Binding to the Transcription Factor Activator Protein-1: An In Vitro Study. Int. J. Mol. Sci. 2021, 22, 12153. https://doi.org/10.3390/ijms222212153
Okamura Y, Yamamoto T, Tsukui R, Kato Y, Shibata N. Fukutin Protein Participates in Cell Proliferation by Enhancing Cyclin D1 Expression through Binding to the Transcription Factor Activator Protein-1: An In Vitro Study. International Journal of Molecular Sciences. 2021; 22(22):12153. https://doi.org/10.3390/ijms222212153
Chicago/Turabian StyleOkamura, Yukinori, Tomoko Yamamoto, Ryota Tsukui, Yoichiro Kato, and Noriyuki Shibata. 2021. "Fukutin Protein Participates in Cell Proliferation by Enhancing Cyclin D1 Expression through Binding to the Transcription Factor Activator Protein-1: An In Vitro Study" International Journal of Molecular Sciences 22, no. 22: 12153. https://doi.org/10.3390/ijms222212153
APA StyleOkamura, Y., Yamamoto, T., Tsukui, R., Kato, Y., & Shibata, N. (2021). Fukutin Protein Participates in Cell Proliferation by Enhancing Cyclin D1 Expression through Binding to the Transcription Factor Activator Protein-1: An In Vitro Study. International Journal of Molecular Sciences, 22(22), 12153. https://doi.org/10.3390/ijms222212153




