Global Transcriptomic Analysis Reveals Differentially Expressed Genes Involved in Embryogenic Callus Induction in Drumstick (Moringa oleifera Lam.)
Abstract
:1. Introduction
2. Results
2.1. Morphological, Histological, and Biochemical Features
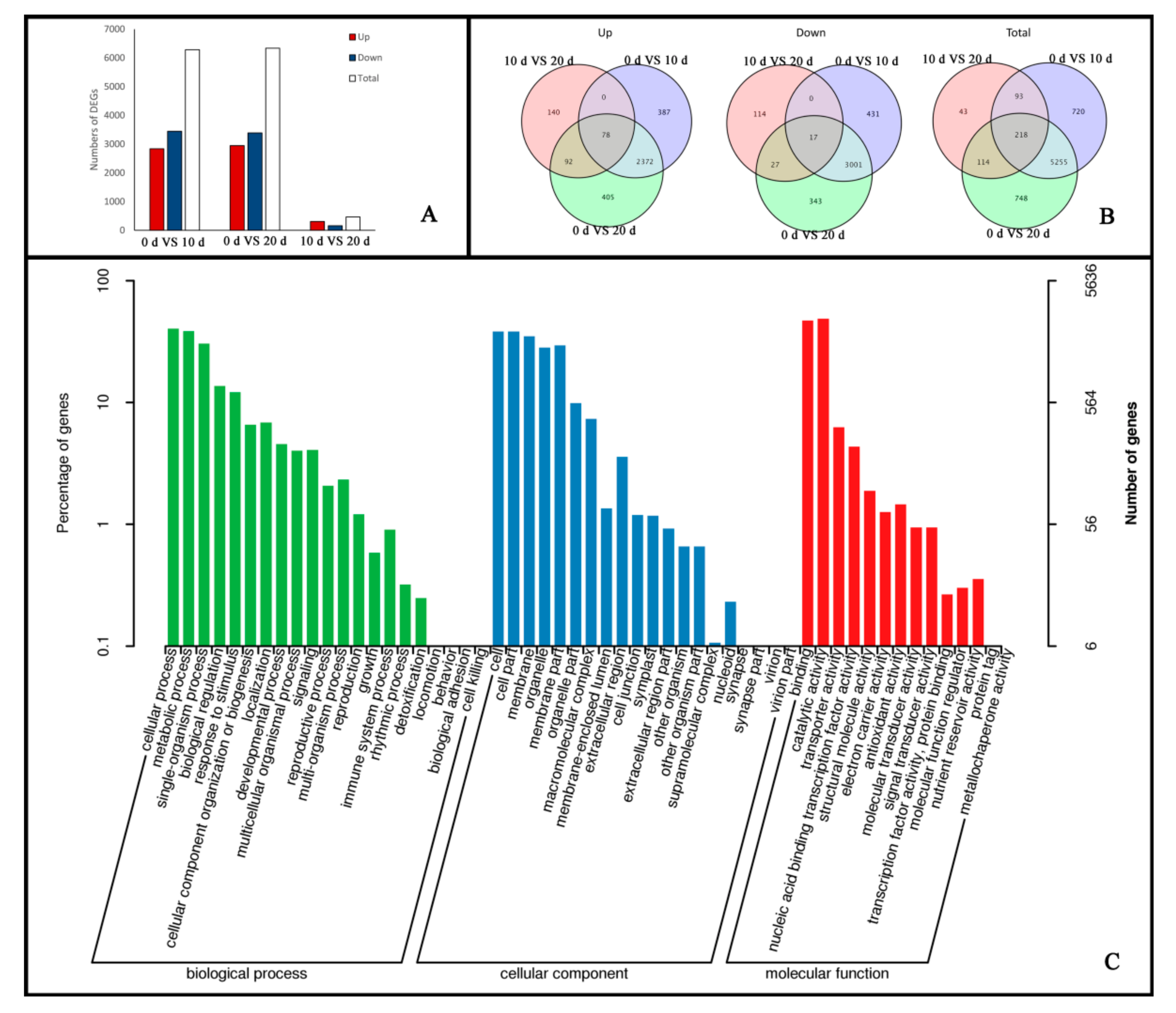
2.2. Global Transcriptomic Changes at Different Stages of Embryogenic Callus Induction Revealed by Transcriptome Sequencing Analysis
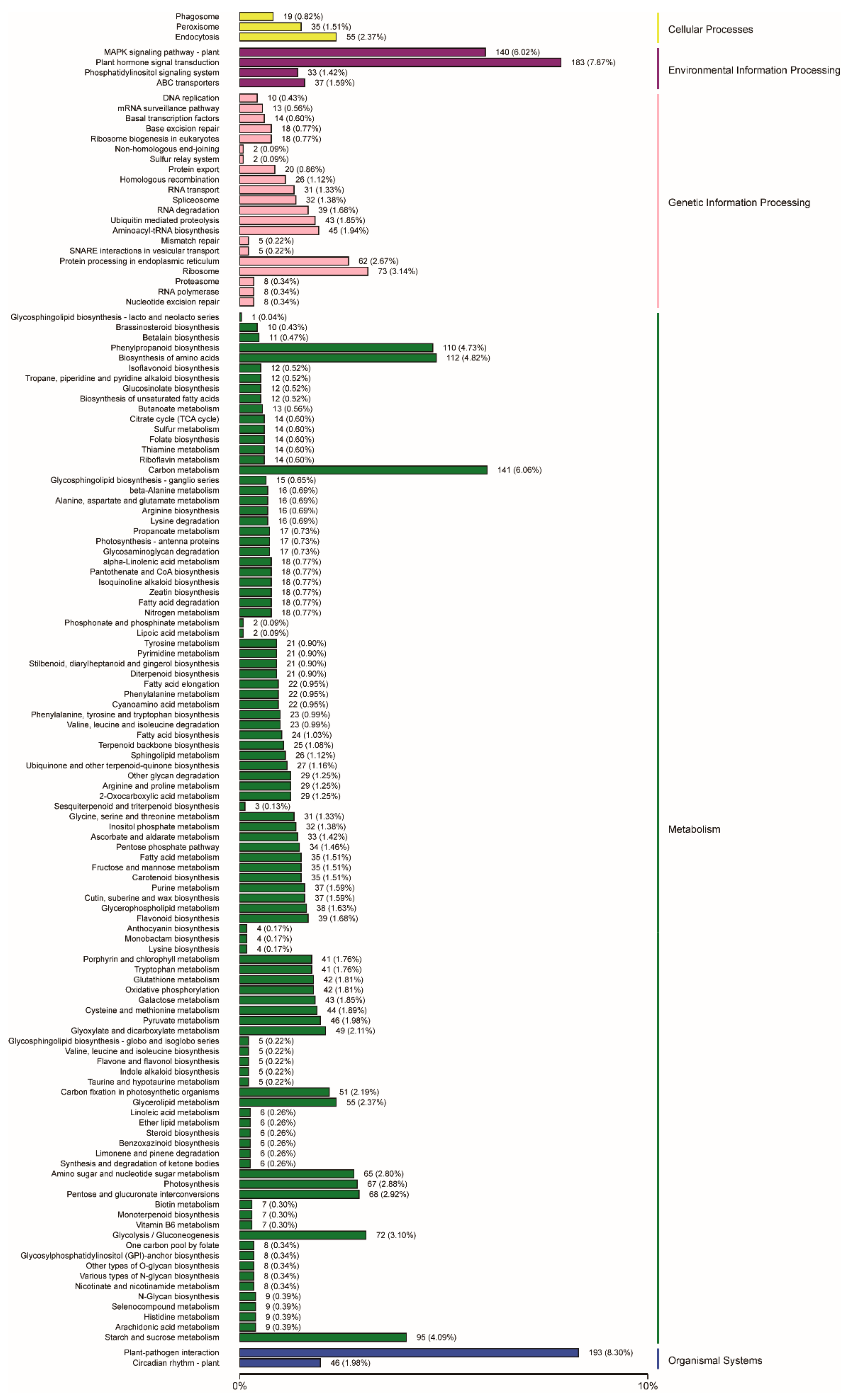
2.3. Functional Annotation and Classification of Genes Related to Embryogenic Callus Induction
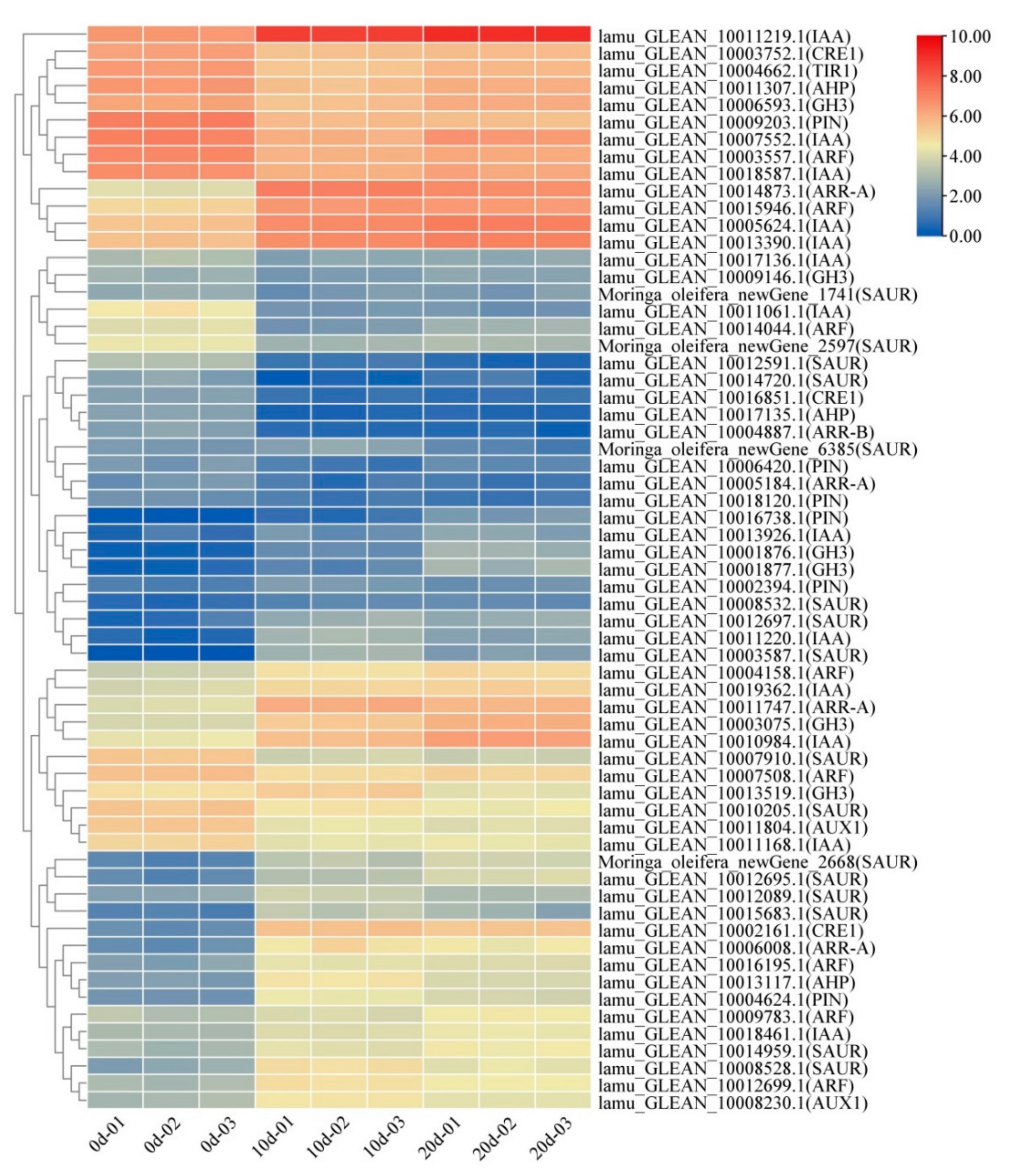
2.4. Auxin and Cytokinin Metabolism Involved in Embryogenic Callus Induction
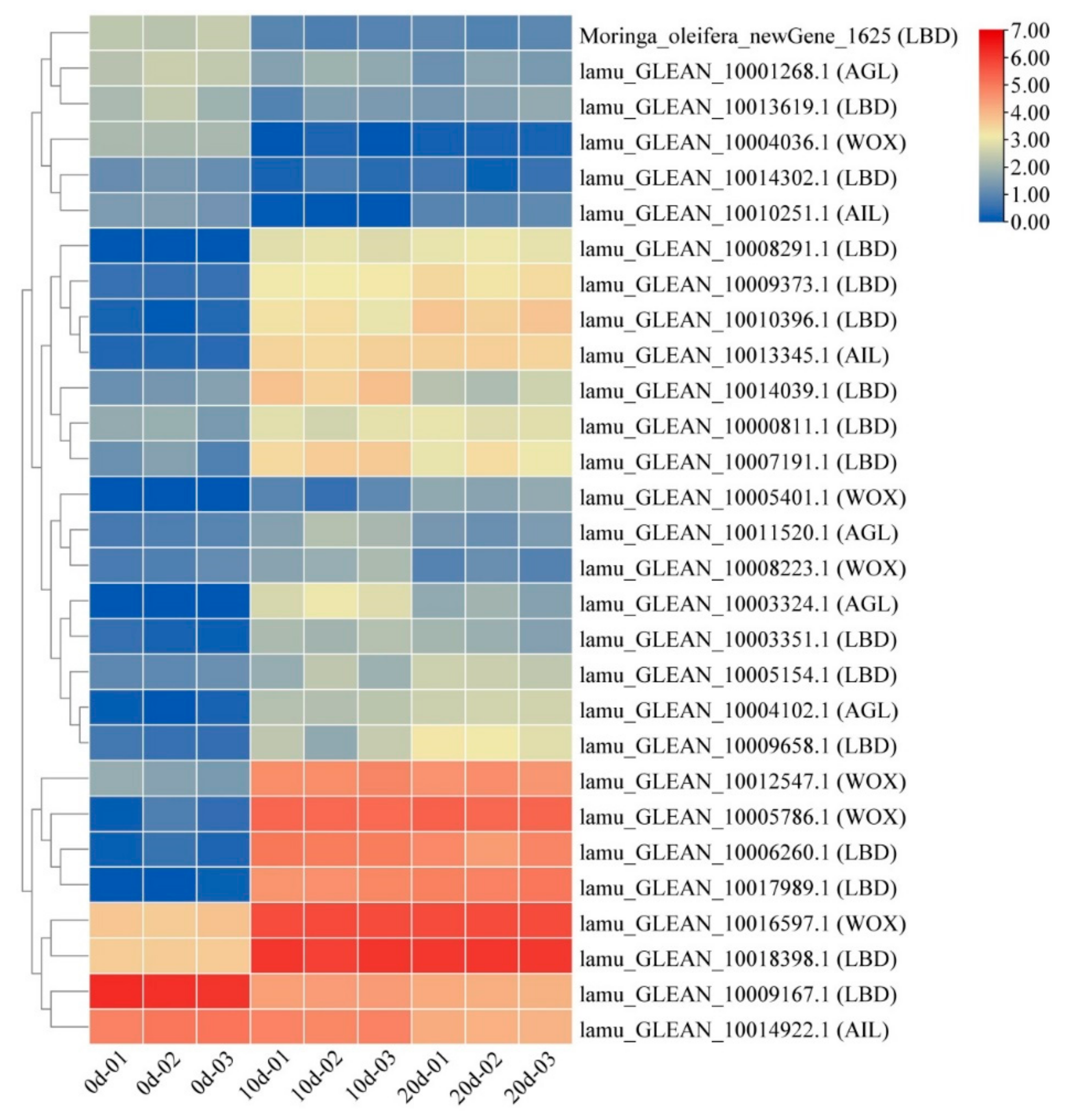
2.5. Embryogenesis-Labeled Genes Involved in Embryogenic Callus Induction
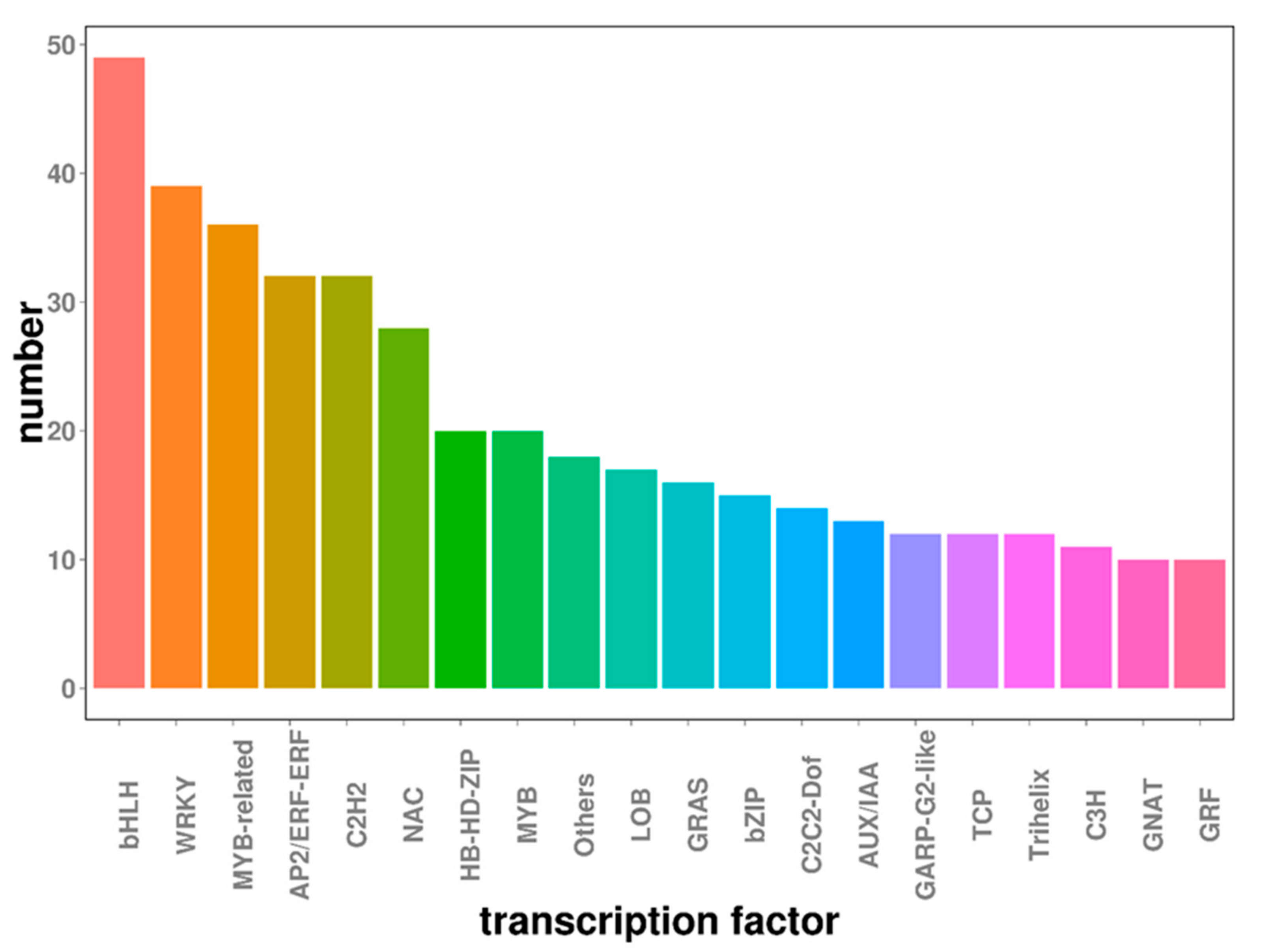
2.6. Transcription Factors Involved in Embryogenic Callus Induction
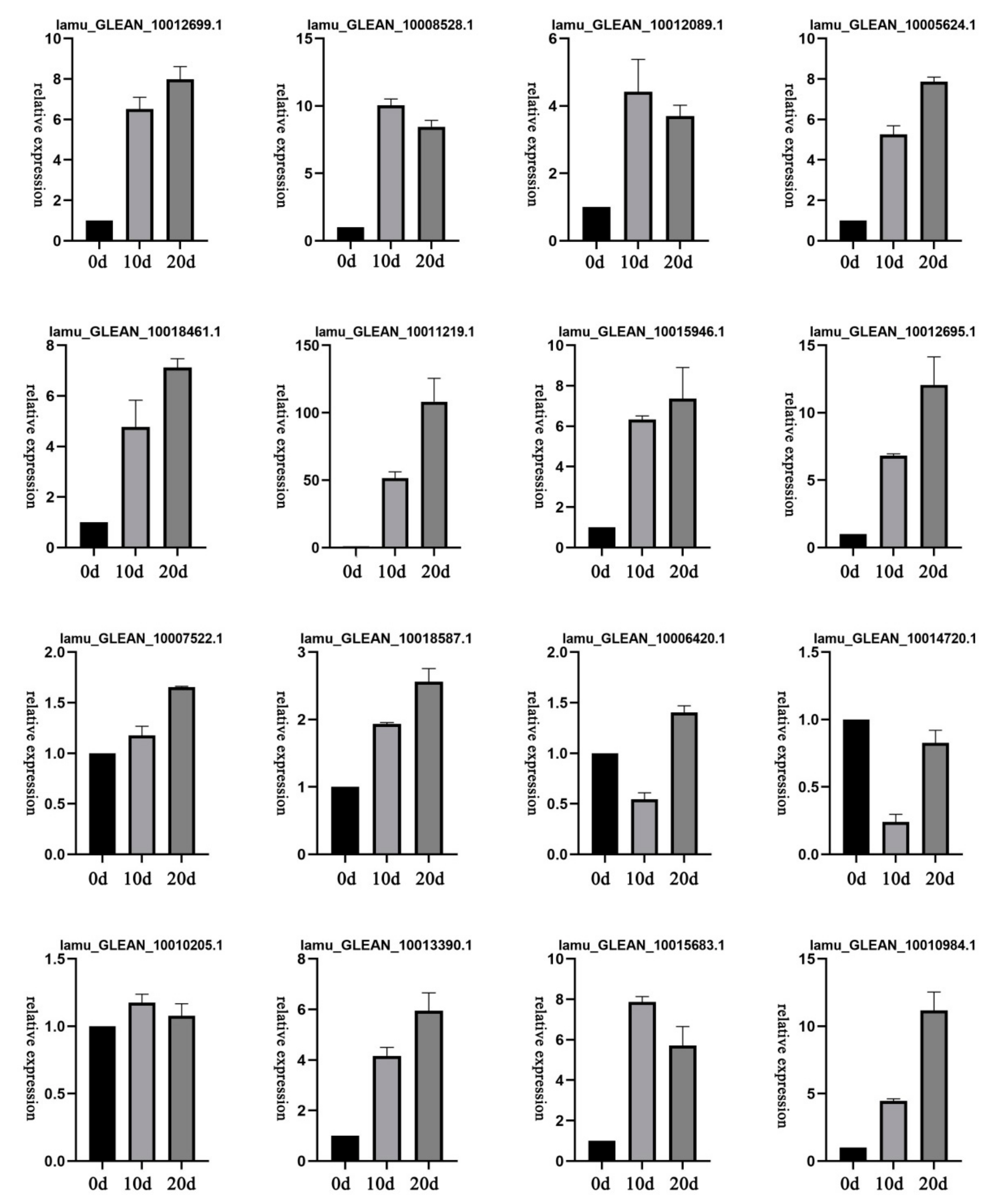
2.7. Expression Patterns of Selected DEGs
3. Discussion
3.1. Auxin and Cytokinin Play an Important Role in Drumstick EC Formation
3.2. Embryogenesis-Labeled Genes Play Important Roles in Drumstick EC Formation
3.3. Stress-Related Transcription Factors Play an Important Role in Drumstick EC Formation
4. Material and Methods
4.1. Induction of Embryogenic Callus and Plant Regeneration
4.2. Histomorphological and Histochemical Analyses
4.3. Library Preparation and Transcriptome Sequencing
4.4. Real-Time Quantitative PCR
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zimmerman, J.L. Somatic embryogenesis: A model for early development in higher plants. Plant Cell 1993, 5, 1411–1423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Abdallat, A.M.; Sawwan, J.S.; Al Zoubi, B. Agrobacterium tumefaciens-mediated transformation of callus cells of Crataegus aronia. Plant Cell Tissue Organ Cult. 2011, 104. [Google Scholar] [CrossRef]
- Elegba, W.; McCallum, E.; Gruissem, W. Efficient genetic transformation and regeneration of a farmer-preferred Cassava cultivar from Ghana. Front. Plant Sci. 2021, 12, 668042. [Google Scholar] [CrossRef] [PubMed]
- Saika, H.; Toki, S. Mature seed-derived callus of the model indica rice variety Kasalath is highly competent in Agrobacterium-mediated transformation. Plant Cell Rep. 2010, 29, 1351–1364. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhang, D.; Que, Q.; Chen, X.; Ouyang, K. Plant regeneration and Agrobacterium-mediated transformation of the miracle tree Neolamarckia cadamba. Ind. Crop. Prod. 2019, 130, 443–449. [Google Scholar] [CrossRef]
- Passamani, L.Z.; Reis, R.S.; Vale, E.M.; Sousa, K.R.; Aragao, V.P.M.; Santa-Catarina, C.; Silveira, V. Long-term culture with 2,4-dichlorophenoxyacetic acid affects embryogenic competence in sugarcane callus via changes in starch, polyamine and protein profiles. Plant Cell Tissue Organ Cult. 2019, 140, 415–429. [Google Scholar] [CrossRef]
- Wang, Y.; Ruemmele, B.A.; Chandlee, J.M.; Sullivan, W.M.; Knapp, J.E.; Kausch, A.P. Embryogenic callus induction and plant regeneration media for bentgrasses and annual bluegrass. In Vitro Cell. Dev. Biol.-Plant 2002, 38, 460–467. [Google Scholar] [CrossRef]
- Jabeen, N.; Chaudhry, Z.; Mirza, B.; Rashid, H. Effect of genotype and explant type on in vitro shoot regeneration of tomato (Lycopersicon esculentum Mill.). Pak. J. Bot. 2005, 37, 899–903. [Google Scholar]
- Kadhimi, A.A.; Alhasnawi, A.N.; Isahak, A.; Ashraf, M.F.; Mohamad, A.; Doni, F.; Yusoffi, W.M.W. Effect of Genotype and Growth Regulators in Induction of Embryogenic Callus in Rice. Pure Appl. Microbiol. 2014, 8, 4573–4578. [Google Scholar]
- Kang, H.; Lee, C.; Kwon, S.; Park, J.; Kang, K.; Shim, D. Comparative transcriptome analysis during developmental stages of direct somatic embryogenesis in Tilia amurensis Rupr. Sci. Rep. 2021, 11, 6359. [Google Scholar] [CrossRef]
- Lai, Z.; Lin, Y. Analysis of the global transcriptome of longan (Dimocarpus longan Lour.) embryogenic callus using Illumina paired-end sequencing. BMC Genom. 2013, 14, 561. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Yang, Y.; Lin, M.; Li, S.; Tang, Y.; Chen, H.; Chen, X. An efficient micropropagation protocol for direct organogenesis from leaf explants of an economically valuable plant, drumstick (Moringa oleifera Lam.). Ind. Crop. Prod. 2017, 103, 59–63. [Google Scholar] [CrossRef]
- Lin, M.; Zhang, J.; Chen, X. Bioactive flavonoids in Moringa oleifera and their health-promoting properties. J. Funct. Foods 2018, 47, 469–479. [Google Scholar] [CrossRef]
- Leone, A.; Spada, A.; Battezzati, A.; Schiraldi, A.; Aristil, J.; Bertoli, S. Cultivation, genetic, ethnopharmacology, phytochemistry and pharmacology of Moringa oleifera leaves: An overview. Int. J. Mol. Sci. 2015, 16, 12791–12835. [Google Scholar] [CrossRef]
- Sengupta, M.E.; Keraita, B.; Olsen, A.; Boateng, O.K.; Thamsborg, S.M.; Pálsdóttir, G.R.; Dalsgaard, A. Use of Moringa oleifera seed extracts to reduce helminth egg numbers and turbidity in irrigation water. Water Res. 2012, 46, 3646–3656. [Google Scholar] [CrossRef]
- Bakre, A.G.; Aderibigbe, A.O.; Ademowo, O.G. Studies on neuropharmacological profile of ethanol extract of Moringa oleifera leaves in mice. J. Ethnopharmacol. 2013, 149, 783–789. [Google Scholar] [CrossRef]
- Kituyi, J.L.; Foulkes, M.; Worsfold, P.; Ongulu, R.A.; Kiplagat, A.; Gachanja, A. Efficiency of pre-treated Moringa oleifera for the removal of Cd2+ and Zn2+ ions from waste waters. Ecohydrol. Hydrobiol. 2013, 13, 267–271. [Google Scholar] [CrossRef]
- Sreelatha, S.; Jeyachitra, A.; Padma, P.R. Antiproliferation and induction of apoptosis by Moringa oleifera leaf extract on human cancer cells. Food Chem. Toxicol. 2011, 49, 1270–1275. [Google Scholar] [CrossRef]
- Muhl, Q.E.; du Toit, E.S.; Steyn, J.M.; Apostolides, Z. Bud development, flowering and fruit set of Moringa oleifera Lam. (Horseradish Tree) as affected by various irrigation levels. J. Agric. Rural. Dev. Trop. Subtrop. 2013, 114, 79–87. [Google Scholar]
- Batra, S.; Kumar, S. Agrobacterium-mediated transient GUS gene expression in buffel grass (Cenchrus ciliaris L.). J. Appl. Genet. 2003, 44, 449–458. [Google Scholar]
- Takamizo, T.; Sato, H. Protocol for Agrobacterium-mediated transformation of tall fescue and future perspective on the application of genome editing. Plant Biotechnol. 2020, 37, 157–161. [Google Scholar] [CrossRef]
- Zhang, J.; Lin, M.; Chen, H.; Chen, X. Agrobacterium tumefaciens-mediated transformation of drumstick (Moringa oleifera Lam.). Biotechnol. Biotechnol. Equip. 2017, 31, 1126–1131. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Sugimoto, K.; Iwase, A. Plant callus: Mechanisms of induction and repression. Plant Cell 2013, 25, 3159–3173. [Google Scholar] [CrossRef] [Green Version]
- Che, P.; Gingerich, D.J.; Lall, S.; Howell, S.H. Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell 2002, 14, 2771–2785. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, X.; Guo, X.; Cui, Y.; Yang, H. The roles of Aux/IAA gene family in development of Dendrocalamus sinicus (Poaceae: Bambusoideae) inferred by comprehensive analysis and expression profiling. Mol. Biol. Rep. 2019, 46, 1625–1634. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Gray, W.M. SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kant, S.; Bi, Y.M.; Zhu, T.; Rothstein, S.J. SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol. 2009, 151, 691–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roosjen, M.; Paque, S.; Weijers, D. Auxin response factors: Output control in auxin biology. J. Exp. Bot. 2018, 69, 179–188. [Google Scholar] [CrossRef]
- Buechel, S.; Leibfried, A.; To, J.P.C.; Zhao, Z.; Andersen, S.U.; Kieber, J.J.; Lohmann, J.U. Role of A-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. Eur. J. Cell Biol. 2010, 89, 279–284. [Google Scholar] [CrossRef]
- Gordon-Kamm, B.; Sardesai, N.; Arling, M.; Lowe, K.; Hoerster, G.; Betts, S.; Jones, T. Using morphogenic genes to improve recovery and regeneration of transgenic plants. Plant 2019, 8, 38. [Google Scholar] [CrossRef] [Green Version]
- Rocha, D.I.; Monte-Bello, C.C.; Aizza, L.C.B.; Dornelas, M.C. A passion fruit putative ortholog of the SOMATIC EMBRYOGENESIS RECEPTOR KINASE1 gene is expressed throughout the in vitro de novo shoot organogenesis developmental program. Plant Cell Tissue Organ Cult. 2016, 125, 107–117. [Google Scholar] [CrossRef]
- Xu, C.; Cao, H.F.; Zhang, Q.Q.; Wang, H.Z.; Xin, W.; Xu, E.J.; Zhang, S.Q.; Yu, R.X.; Ye, D.X.; Hu, Y.X. Control of auxin-induced callus formation by bZIP59–LBD complex in Arabidopsis regeneration. Nat. Plants 2018, 4, 108–115. [Google Scholar] [CrossRef]
- Zheng, Q.; Zheng, Y.; Perry, S.E. AGAMOUS-Like15 promotes somatic embryogenesis in Arabidopsis and soybean in part by the control of ethylene biosynthesis and response. Plant Physiol. 2013, 161, 2113–2127. [Google Scholar] [CrossRef] [Green Version]
- Haecker, A.; Gross-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef] [Green Version]
- Palovaara, J.; Hallberg, H.; Stasolla, C.; Hakman, I. Comparative expression pattern analysis of WUSCHEL-related homeobox 2 (WOX2) and WOX8/9 in developing seeds and somatic embryos of the gymnosperm Picea abies. New Phytol. 2010, 188, 122–135. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, Z.; Lyu, M.; Yuan, Y.; Wu, B. Characterization of JsWOX1 and JsWOX4 during Callus and Root Induction in the Shrub Species Jasminum sambac. Plants 2019, 8, 79. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.H.; Guo, H.X.; Zhang, L.; Fan, Y.J.; Wu, J.F.; Tang, Z.M.; Zhang, Y.; Fan, Y.P.; Zeng, F.C. Dynamic transcriptome analysis reveals uncharacterized complex regulatory pathway underlying genotype-recalcitrant somatic embryogenesis transdifferentiation in Cotton. Genes 2020, 11, 519. [Google Scholar] [CrossRef]
- Guo, J.R.; Sun, B.X.; He, H.R.; Zhang, Y.F.; Tian, H.Y.; Wang, B.S. Current understanding of bHLH transcription factors in plant abiotic stress tolerance. Int. J. Mol. Sci. 2021, 22, 4921. [Google Scholar] [CrossRef]
- Li, W.X.; Pang, S.Y.; Lu, Z.G.; Jin, B. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plant 2020, 9, 1515. [Google Scholar] [CrossRef]
- Zhao, P.C.; Hou, S.L.; Guo, X.F.; Jia, J.T.; Yang, W.G.; Liu, Z.J.; Chen, S.; Li, X.; Qi, D.; Liu, G.; et al. A MYB-related transcription factor from sheepgrass, LcMYB2, promotes seed germination and root growth under drought stress. BMC Plant Biol. 2019, 19, 564. [Google Scholar] [CrossRef] [Green Version]
- Iwase, A.; Mitsuda, N.; Koyama, T.; Hiratsu, K.; Kojima, M.; Arai, T.; Inoue, Y.; Seki, M.; Sakakibara, H.; Sugimoto, K.; et al. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr. Biol. 2011, 21, 508–514. [Google Scholar] [CrossRef] [Green Version]
- An, H.; Zhang, J.; Xu, F.; Jiang, S.; Zhang, X. Transcriptomic profiling and discovery of key genes involved in adventitious root formation from green cuttings of highbush blueberry (Vaccinium corymbosum L.). BMC Plant Biol. 2020, 20, 182. [Google Scholar] [CrossRef] [Green Version]
- Deng, L.; Wu, Y.; Li, J.; OuYang, K.; Ding, M.; Zhang, J.; Li, S.Q.; Lin, M.F.; Chen, H.B.; Hu, X.S.; et al. Screening Reliable Reference Genes for RT-qPCR Analysis of Gene Expression in Moringa oleifera. PLoS ONE 2006, 11, e0159458. [Google Scholar] [CrossRef]

| Samples | Clean Reads | Mapped Reads | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|
| 0 d-1 | 44,952,482 | 42,277,829 (94.05%) | 98.3 | 95.04 | 45.62 |
| 0 d-2 | 38,824,772 | 36,488,916 (93.98%) | 98.24 | 94.98 | 45.56 |
| 0 d-3 | 43,843,258 | 41,495,852 (94.65%) | 98.31 | 95.04 | 45.55 |
| 10 d-1 | 43,791,194 | 41,857,113 (95.58%) | 98.31 | 95.08 | 45.05 |
| 10 d-2 | 43,102,782 | 40,697,612 (94.42%) | 98.22 | 94.86 | 44.92 |
| 10 d-3 | 42,518,186 | 40,566,753 (95.41%) | 98.25 | 94.92 | 45.01 |
| 20 d-1 | 41,091,584 | 38,900,779 (94.67%) | 98.2 | 94.94 | 45.14 |
| 20 d-2 | 43,883,936 | 40,957,664 (93.33%) | 98.21 | 94.93 | 45.31 |
| 20 d-3 | 44,517,654 | 41,830,480 (93.96%) | 98.25 | 95.02 | 45.17 |
| Annotated Databases | Annotated_Number | Percentage (%) | 300 ≤ Length < 1000 | Length ≥ 1000 |
|---|---|---|---|---|
| COG | 6337 | 26.86 | 1905 | 4359 |
| GO | 16,918 | 71.71 | 6809 | 9618 |
| KEGG | 13,097 | 55.52 | 4901 | 7915 |
| KOG | 11,384 | 48.26 | 4096 | 7034 |
| Pfam | 16,457 | 69.76 | 6321 | 9809 |
| Swiss-Prot | 15,255 | 64.66 | 5858 | 9011 |
| eggNOG | 2579 | 10.93 | 1156 | 1266 |
| NR | 21,382 | 90.64 | 9422 | 11,256 |
| At least one database | 21,439 | 90.88 | 9454 | 11,264 |
| Gene ID | Forward Primers | Reversed Primers |
|---|---|---|
| ACP2. | GAAACCAATGAGCACCCAGC | GATGAATACCAGTCCACCGCAAC |
| lamu_GLEAN_10018587.1 | ATCTCCTCCCTGCTGCTTG | CTTGACTTGTCGCCCTCTT |
| lamu_GLEAN_10010205.1 | GGAGCAAACGCCACTGTCA | TTCATCACCCTCGCCATCA |
| lamu_GLEAN_10012089.1 | CACCTGATTCCCTTCCATA | ATACTCTTCCTCGGCTTCC |
| lamu_GLEAN_10007552.1 | GCACCCTTCTTCCTTCTG | TCCTCGGAGCTTGTCTTT |
| lamu_GLEAN_10006420.1 | TGACAAGGATTTGGAGGGT | CAAGCGTAGGAGTTGGGAT |
| lamu_GLEAN_10008528.1 | AGGAGGAAGGCCAGGATTAC | AAGATTGGGTGGTTGAGGTG |
| lamu_GLEAN_10012699.1 | GCCCTCTTGTTTCTTTGCC | GTGATGGAAGATTCGGGTAGT |
| lamu_GLEAN_10012695.1 | ATGGCGATCAGGAAGTCA | TTGCTCGTCGTGGTAGGTC |
| lamu_GLEAN_10010984.1 | CACCAGACAGGGACGATG | ACCGCCACCACTACAACC |
| lamu_GLEAN_10018461.1 | TGGAGGCTACTGAGGCTAGT | TGACAGACCACAGAAGGGA |
| lamu_GLEAN_10011219.1 | GCTCTACGTGGGACGCTA | CACCTTCAGGCATTTACCG |
| lamu_GLEAN_10015946.1 | GCTTACCTCCACAACTTATC | TAGGCATCCTTCTGCTCTT |
| lamu_GLEAN_10013390.1 | CTACAGAACTGAGGCTTGG | AAGGAGTGGAAATAGTGGC |
| lamu_GLEAN_10015683.1 | GAAGCCGAGTCCGAGTTTG | CCTTGACGAATACGACCACC |
| lamu_GLEAN_10014720.1 | CTGCTGCTGTCAACAAGGT | GCCGCATCCAGTAATCTCA |
| lamu_GLEAN_10005624.1 | GGGCTTGCCTGTTATGGT | ACTCAGTTGCTGGCTTTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, E.; Zheng, M.; Zou, X.; Huang, X.; Yang, H.; Chen, X.; Zhang, J. Global Transcriptomic Analysis Reveals Differentially Expressed Genes Involved in Embryogenic Callus Induction in Drumstick (Moringa oleifera Lam.). Int. J. Mol. Sci. 2021, 22, 12130. https://doi.org/10.3390/ijms222212130
Yang E, Zheng M, Zou X, Huang X, Yang H, Chen X, Zhang J. Global Transcriptomic Analysis Reveals Differentially Expressed Genes Involved in Embryogenic Callus Induction in Drumstick (Moringa oleifera Lam.). International Journal of Molecular Sciences. 2021; 22(22):12130. https://doi.org/10.3390/ijms222212130
Chicago/Turabian StyleYang, Endian, Mingyang Zheng, Xuan Zou, Xiaoling Huang, Heyue Yang, Xiaoyang Chen, and Junjie Zhang. 2021. "Global Transcriptomic Analysis Reveals Differentially Expressed Genes Involved in Embryogenic Callus Induction in Drumstick (Moringa oleifera Lam.)" International Journal of Molecular Sciences 22, no. 22: 12130. https://doi.org/10.3390/ijms222212130





