CaWRKY30 Positively Regulates Pepper Immunity by Targeting CaWRKY40 against Ralstonia solanacearum Inoculation through Modulating Defense-Related Genes
Abstract
:1. Introduction
2. Results
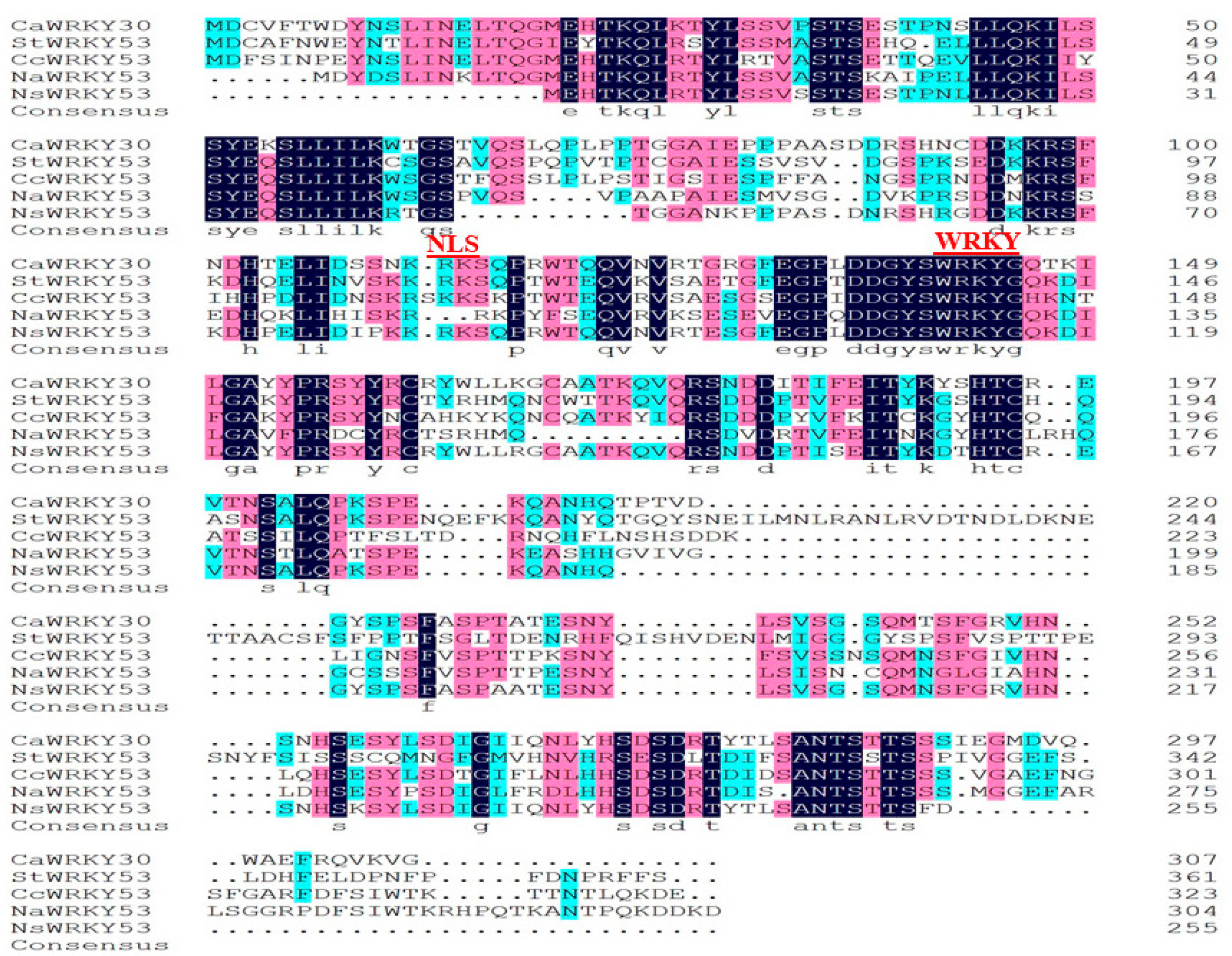
2.1. Cloning and Sequence Analysis of CaWRKY30
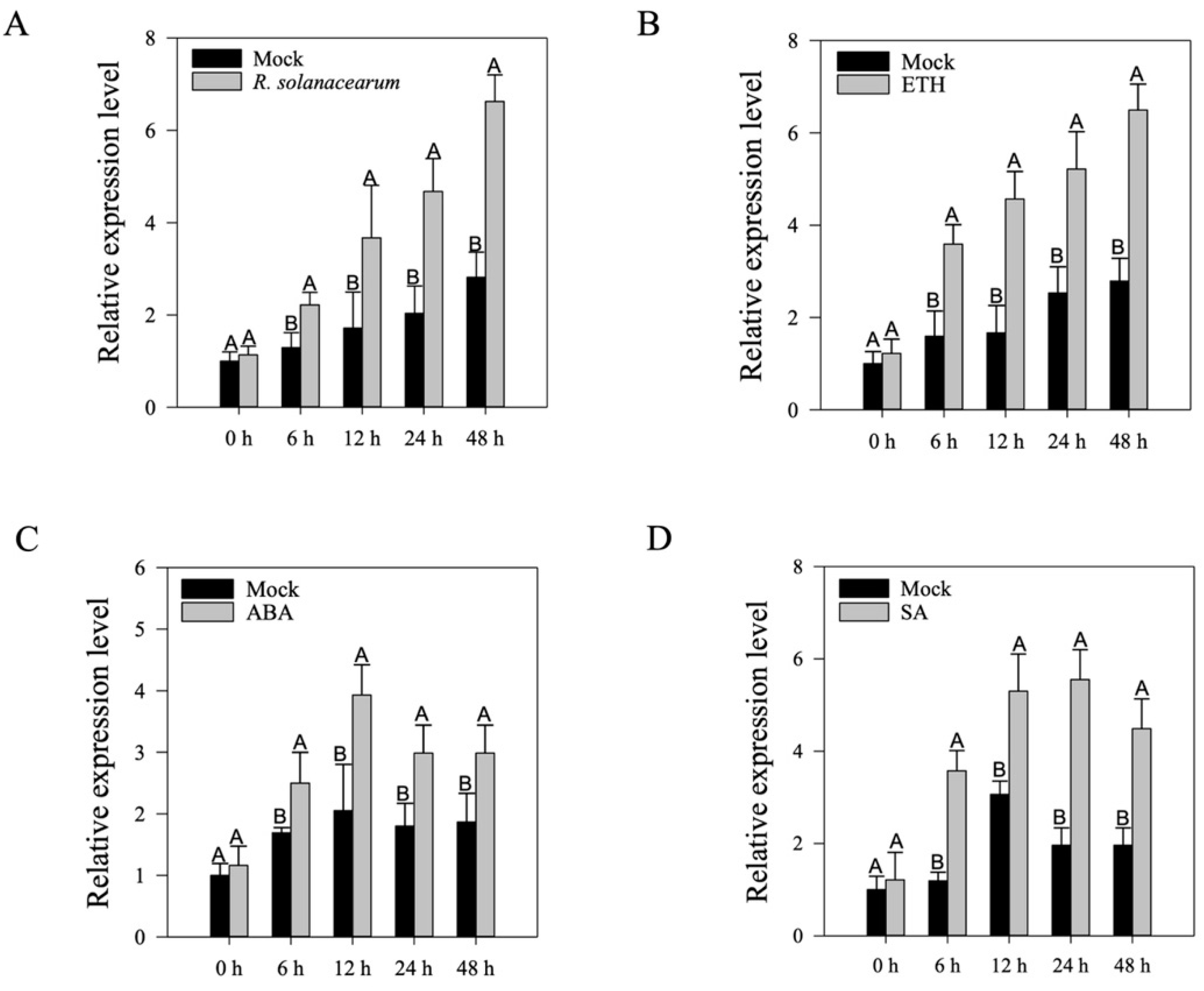
2.2. Transcriptional Expression of CaWRKY30 under RSI and Treatment with Various Phytohormones
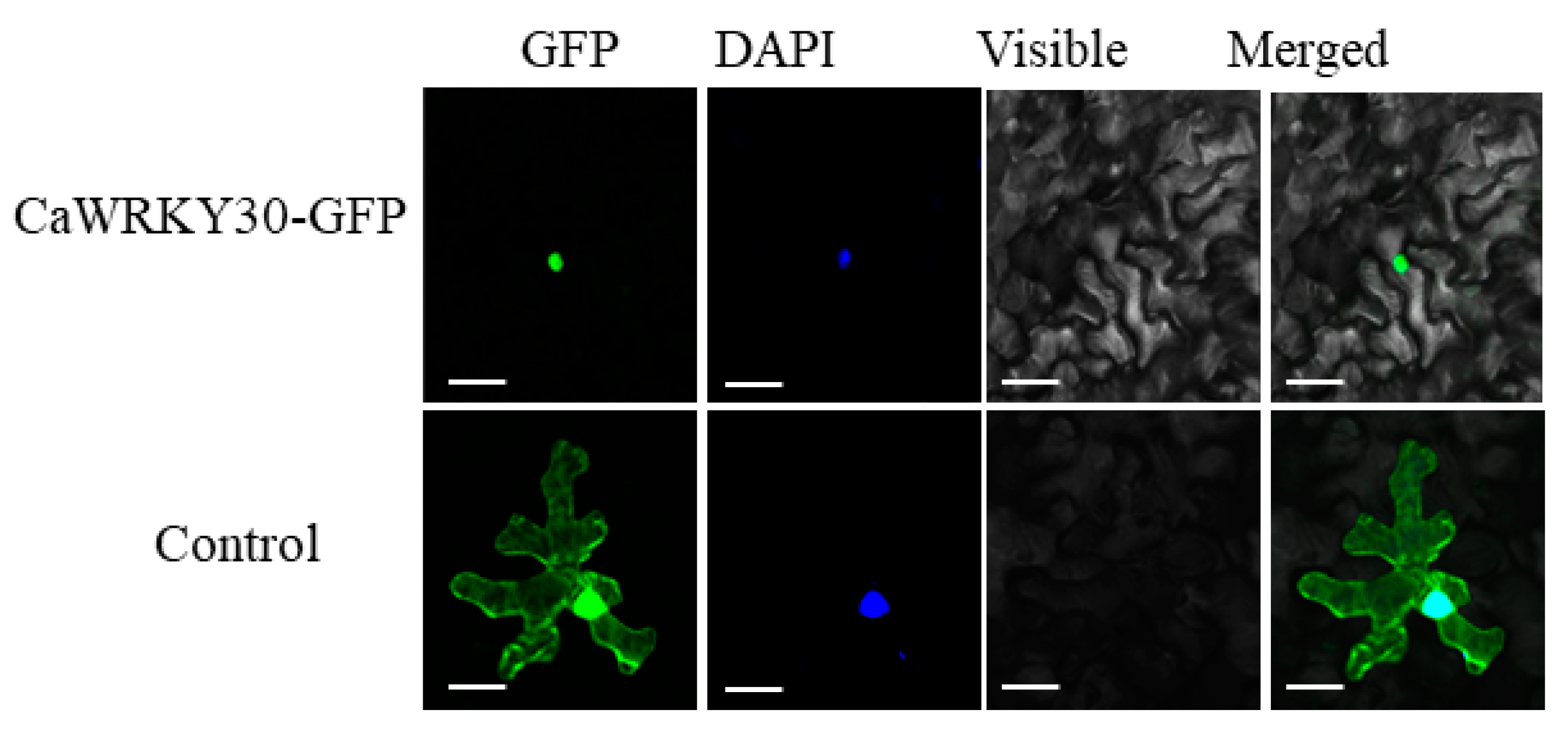
2.3. CaWRKY30 Was Found to Be Localized in Nucleus
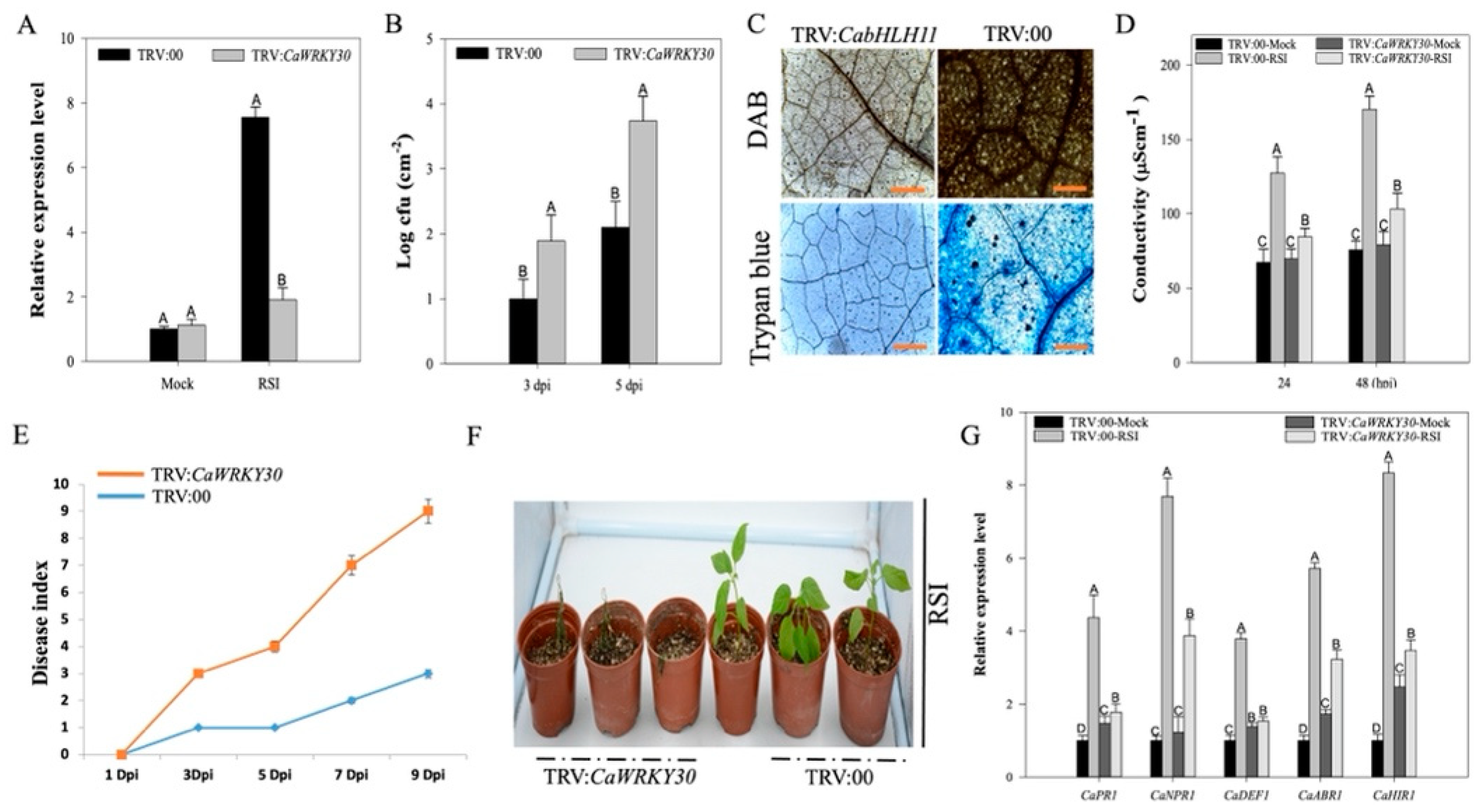
2.4. Silencing of CaWRKY30by VIGS Curtailed Pepper’s Immunity to RSI and Downregulated Immunity Associated Marker Genes
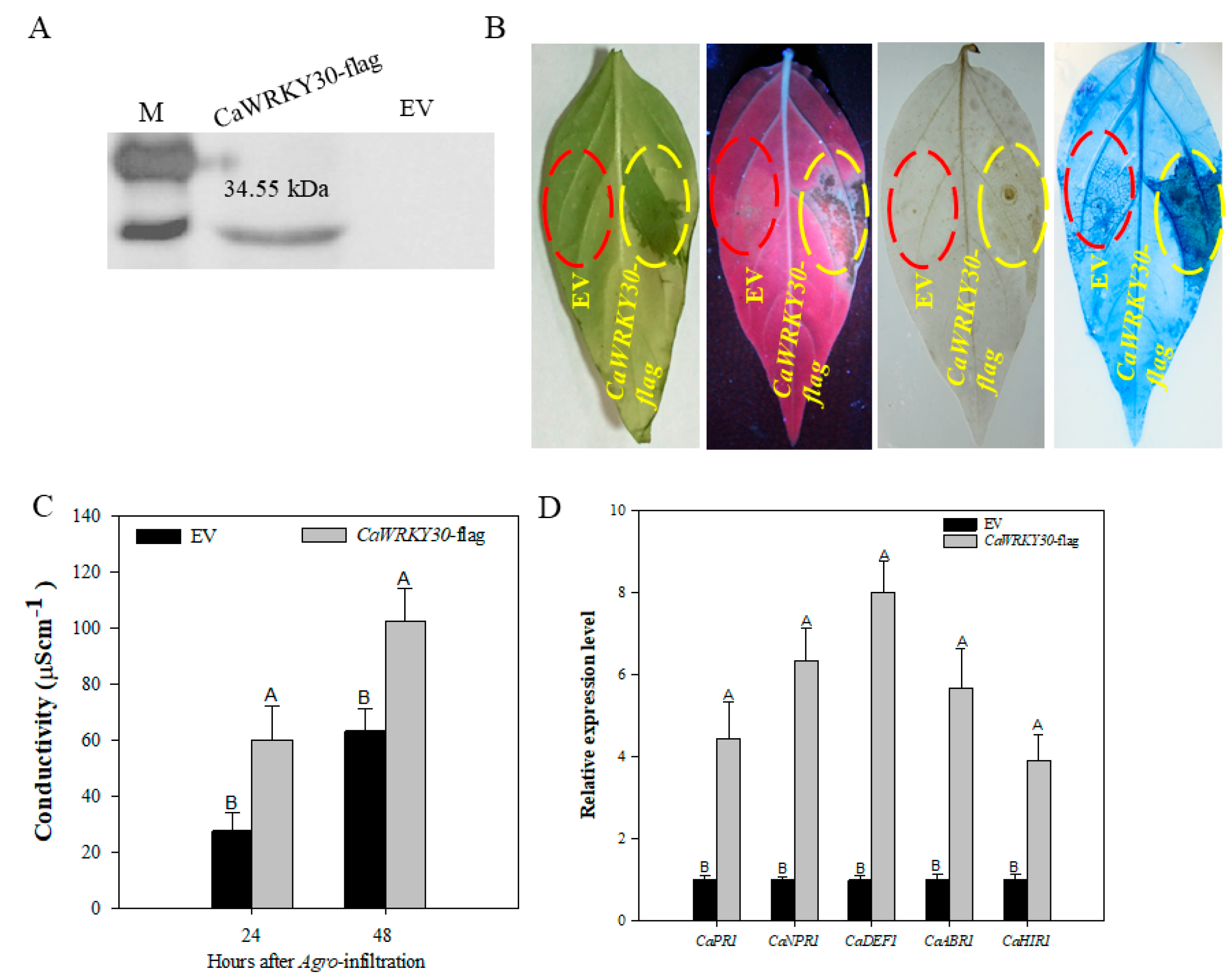
2.5. CaWRKY30 Transient Over-Expression Stimulated HR-Like Cell Death, Production of H2O2, and Transcriptional Upregulation of Immunity Related Marker Genes
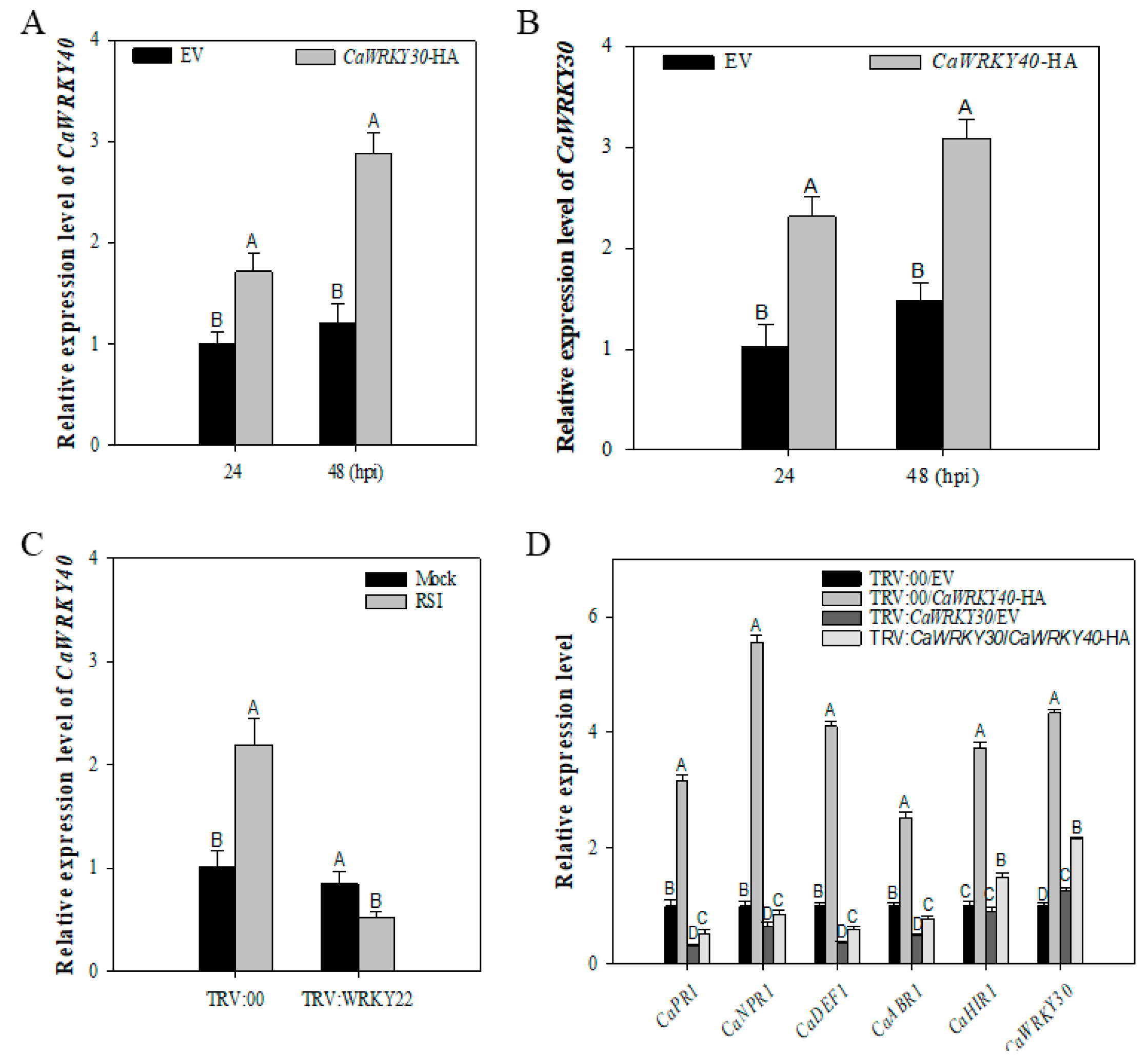
2.6. Inter-Relationship between CaWRKY30 and CaWRKY40
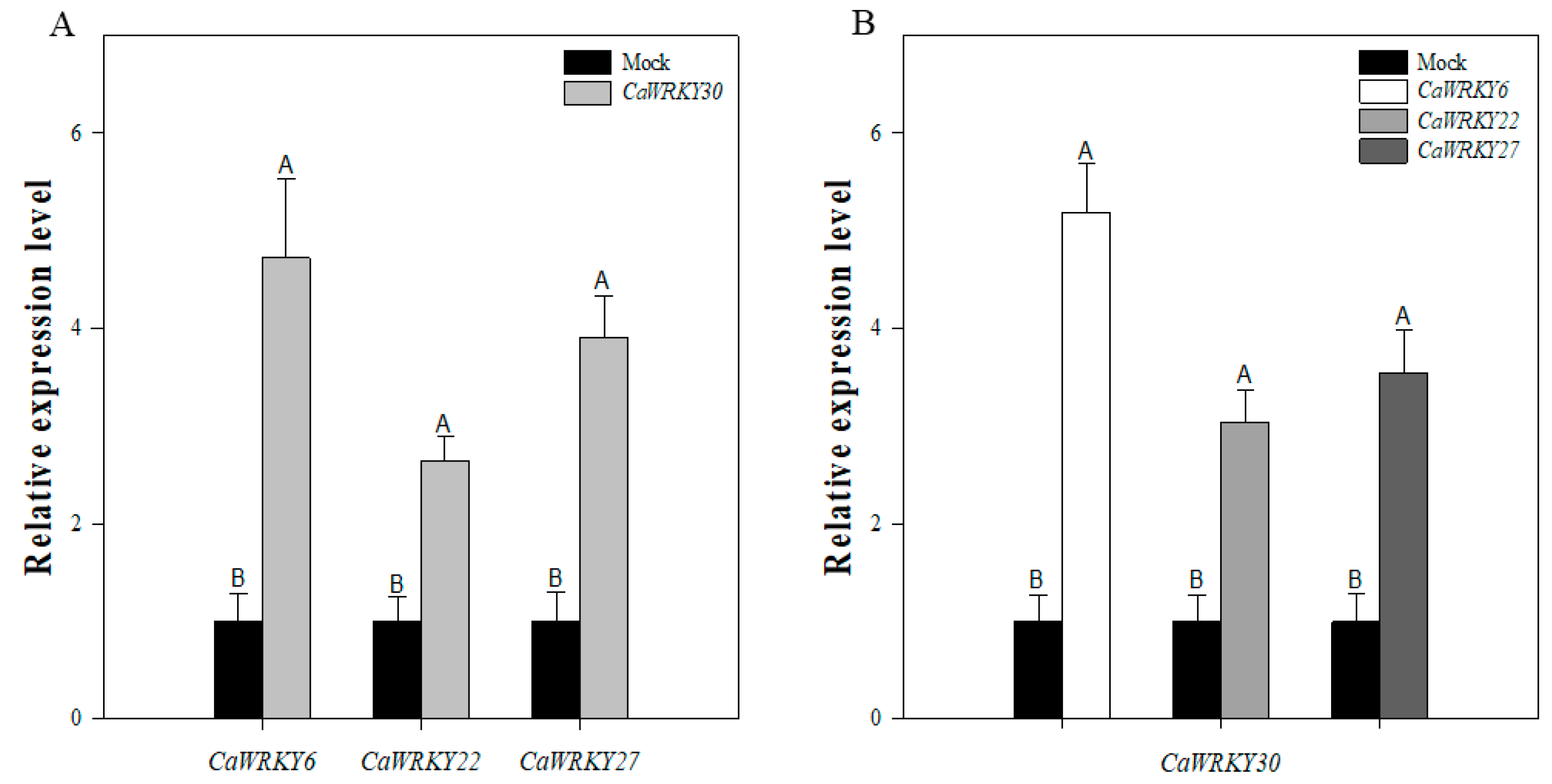
2.7. Inter-Relationship among CaWRY30 and CaWRKY6, CaWRKY22, and CaWRKY27
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Generation of Vectors
4.3. Pathogen’s Growth and Inoculation Procedures
4.4. Foliar Application of Phytohormones
4.5. Examination of CaWRKY30 Sub-Cellular Localization
4.6. Histochemical Staining
4.7. Silencing of CaWRKY30 through Virus-Induced Gene Silencing (VIGS)
4.8. CaWRKY30 Transient Over-Expression in Pepper Leaves
4.9. RNA Extraction and Quantitative Real Time RT-PCR
4.10. Estimation of ELECTRICAL Conductivity
4.11. Immuno Blotting
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arif, M.; Atta, S.; Bashir, M.A.; Khan, M.I.; Hussain, A.; Shahjahan, M.; Alwahibi, M.S.; Elshikh, M.S. The impact of Fosetyl-Aluminium application timing on Karnal bunt suppression and economic returns of bread wheat (Triticum aestivum L.). PLoS ONE 2021, 16, e0244931. [Google Scholar]
- Khalofah, A.; Khan, M.I.; Arif, M.; Hussain, A.; Ullah, R.; Irfan, M.; Mahpara, S.; Shah, R.U.; Ansari, M.J.; Kintl, A. Deep placement of nitrogen fertilizer improves yield, nitrogen use efficiency and economic returns of transplanted fine rice. PLoS ONE 2021, 16, e0247529. [Google Scholar] [CrossRef] [PubMed]
- Noman, A.; Hussain, A.; Adnan, M.; Khan, M.I.; Ashraf, M.F.; Zainab, M.; Khan, K.A.; Ghramh, H.A.; He, S. A novel MYB transcription factor CaPHL8 provide clues about evolution of pepper immunity against soil borne pathogen. Microb. Pathog. 2019, 137, 103758. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Noman, A.; Khan, M.I.; Zaynab, M.; Aqeel, M.; Anwar, M.; Ashraf, M.F.; Liu, Z.; Raza, A.; Mahpara, S. Molecular regulation of pepper innate immunity and stress tolerance: An overview of WRKY TFs. Microb. Pathog. 2019, 135, 103610. [Google Scholar] [CrossRef] [PubMed]
- Reboledo, G.; Agorio, A.d.; Vignale, L.; Batista-García, R.A.; de León, I.P. Transcriptional profiling reveals conserved and species-specific plant defense responses during the interaction of Physcomitrium patens with Botrytis cinerea. Plant Mol. Biol. 2021, 1–21. [Google Scholar] [CrossRef]
- Aerts, N.; Mendes, M.P.; van Wees, S.C. Multiple levels of crosstalk in hormone networks regulating plant defense. Plant J. 2021, 105, 489–504. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Shi, L.; Weng, Y.; Zou, H.; Li, X.; Yang, S.; Qiu, S.; Huang, X.; Huang, J.; Hussain, A. ChiIV3 acts as a novel target of WRKY40 to mediate pepper immunity against Ralstonia solanacearum infection. Mol. Plant-Microbe Interact. 2019, 32, 1121–1133. [Google Scholar] [CrossRef] [PubMed]
- ul Haq, M.I.; Maqbool, M.M.; Ali, A.; Farooq, S.; Khan, S.; Saddiq, M.S.; Khan, K.A.; Ali, S.; Khan, M.I.; Hussain, A. Optimizing planting geometry for barley-Egyptian clover intercropping system in semi-arid sub-tropical climate. PLoS ONE 2020, 15, e0233171. [Google Scholar]
- Hussain, A.; Li, X.; Weng, Y.; Liu, Z.; Ashraf, M.F.; Noman, A.; Yang, S.; Ifnan, M.; Qiu, S.; Yang, Y. CaWRKY22 acts as a positive regulator in pepper response to Ralstonia solanacearum by constituting networks with CaWRKY6, CaWRKY27, CaWRKY40, and CaWRKY58. Int. J. Mol. Sci. 2018, 19, 1426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukhtar, M.S.; Liu, X.; Somssich, I.E. Elucidating the role of WRKY27 in male sterility in Arabidopsis. Plant Signal. Behav. 2018, 13, e1363945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.I.; Zhang, Y.; Liu, Z.; Hu, J.; Liu, C.; Yang, S.; Hussain, A.; Ashraf, M.F.; Noman, A.; Shen, L. CaWRKY40b in pepper acts as a negative regulator in response to Ralstonia solanacearum by directly modulating defense genes including CaWRKY40. Int. J. Mol. Sci. 2018, 19, 1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakshi, M.; Oelmüller, R. WRKY transcription factors: Jack of many trades in plants. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.-P.; Xu, H.; Wang, B.; Su, X.-D. Crystal structures of N-terminal WRKY transcription factors and DNA complexes. Protein Cell 2020, 11, 208–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Li, C.; Wang, H.; Guo, Z. WRKY transcription factors: Evolution, binding, and action. Phytopathol. Res. 2019, 1, 13. [Google Scholar] [CrossRef]
- Shen, L.; Liu, Z.; Yang, S.; Yang, T.; Liang, J.; Wen, J.; Liu, Y.; Li, J.; Shi, L.; Tang, Q. Pepper CabZIP63 acts as a positive regulator during Ralstonia solanacearum or high temperature–high humidity challenge in a positive feedback loop with CaWRKY40. J. Exp. Bot. 2016, 67, 2439–2451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schluttenhofer, C.; Yuan, L. Regulation of Specialized Metabolism by WRKY Transcription Factors. Plant Physiol. 2015, 167, 295–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Yu, K.; Pieterse, C.M.; Bakker, P.A.; Berendsen, R.L. Beneficial microbes going underground of root immunity. Plant Cell Environ. 2019, 42, 2860–2870. [Google Scholar] [CrossRef] [Green Version]
- Noman, A.; Liu, Z.; Yang, S.; Shen, L.; Hussain, A.; Ashraf, M.F.; Khan, M.I.; He, S. Expression and functional evaluation of CaZNF830 during pepper response to Ralstonia solanacearum or high temperature and humidity. Microb. Pathog. 2018, 118, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Noman, A.; Hussain, A.; Ashraf, M.F.; Khan, M.I.; Liu, Z.; He, S. CabZIP53 is targeted by CaWRKY40 and act as positive regulator in pepper defense against Ralstonia solanacearum and thermotolerance. Environ. Exp. Bot. 2019, 159, 138–148. [Google Scholar] [CrossRef]
- Ashraf, M.F.; Yang, S.; Wu, R.; Wang, Y.; Hussain, A.; Noman, A.; Khan, M.I.; Liu, Z.; Qiu, A.; Guan, D.; et al. Capsicum annuum HsfB2a Positively Regulates the Response to Ralstonia solanacearum Infection or High Temperature and High Humidity Forming Transcriptional Cascade with CaWRKY6 and CaWRKY40. Plant Cell Physiol. 2018, 59, 2608–2623. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Dossa, K.; You, J.; Zhang, Y.; Li, D.; Zhou, R.; Yu, J.; Wei, X.; Zhu, X.; Jiang, S.; et al. High-resolution temporal transcriptome sequencing unravels ERF and WRKY as the master players in the regulatory networks underlying sesame responses to waterlogging and recovery. Genomics 2021, 113, 276–290. [Google Scholar] [CrossRef]
- Yang, X.; Zhou, Z.; Fu, M.; Han, M.; Liu, Z.; Zhu, C.; Wang, L.; Zheng, J.; Liao, Y.; Zhang, W.; et al. Transcriptome-wide identification of WRKY family genes and their expression profiling toward salicylic acid in Camellia japonica. Plant Signal. Behav. 2021, 16, 1844508. [Google Scholar] [CrossRef] [PubMed]
- Pervez, M.; Babar, M.; Iqbal, J.; Hasnain, M.; Abbas, S.; Shah, S.; Ashraf, M. In Silico Structural and Functional Analysis of Wrky1 and Wrky3 Genes in The Selected Cereal Crops. J. Anim. Plant Sci.-JAPS 2021, 31, 322–341. [Google Scholar]
- Liu, Z.-Q.; Liu, Y.-Y.; Shi, L.-P.; Yang, S.; Shen, L.; Yu, H.-X.; Wang, R.-Z.; Wen, J.-Y.; Tang, Q.; Hussain, A.; et al. SGT1 is required in PcINF1/SRC2-1 induced pepper defense response by interacting with SRC2-1. Sci. Rep. 2016, 6, 21651. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, K. Biological Activities of Red Pepper (Capsicum annuum) and Its Pungent Principle Capsaicin: A Review. Crit. Rev. Food Sci. Nutr. 2016, 56, 1488–1500. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Shi, L.; Yang, S.; Lin, Y.; Weng, Y.; Li, X.; Hussain, A.; Noman, A.; He, S. Functional and Promoter Analysis of ChiIV3, a Chitinase of Pepper Plant, in Response to Phytophthora capsici Infection. Int. J. Mol. Sci. 2017, 18, 1661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, M.; Luo, D.-X.; Khan, A.; Haq, S.U.; Gai, W.-X.; Zhang, H.-X.; Cheng, G.-X.; Muhammad, I.; Gong, Z.-H. Classification and Genome-Wide Analysis of Chitin-Binding Proteins Gene Family in Pepper (Capsicum annuum L.) and Transcriptional Regulation to Phytophthora capsici, Abiotic Stresses and Hormonal Applications. Int. J. Mol. Sci. 2018, 19, 2216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noman, A.; Liu, Z.; Aqeel, M.; Zainab, M.; Khan, M.I.; Hussain, A.; Ashraf, M.F.; Li, X.; Weng, Y.; He, S. Basic leucine zipper domain transcription factors: The vanguards in plant immunity. Biotechnol. Lett. 2017, 39, 1779–1791. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Shi, Y.; Zou, L.; Huang, J.; Shen, L.; Wang, Y.; Guan, D.; He, S. Pepper CaMLO6 Negatively Regulates Ralstonia solanacearum Resistance and Positively Regulates High Temperature and High Humidity Responses. Plant Cell Physiol. 2020, 61, 1223–1238. [Google Scholar] [CrossRef]
- Arif, M.; Atta, S.; Bashir, M.A.; Hussain, A.; Khan, M.I.; Farooq, S.; Hannan, A.; Islam, S.U.; Umar, U.U.D.; Khan, M.; et al. Molecular characterization and RSV Co-infection of Nicotiana benthamiana with three distinct begomoviruses. Methods 2020, 183, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.G.; Allen, C. Ralstonia solanacearum genes induced during growth in tomato: An inside view of bacterial wilt. Mol. Microbiol. 2004, 53, 1641–1660. [Google Scholar] [CrossRef] [PubMed]
- Elphinstone, J.G. The Current Bacterial Wilt Situation: A Global Overview, Bacterial Wilt Disease and the Ralstonia Solanacearum Species Complex; APS Press: St. Paul, MN, USA, 2005; pp. 9–28. [Google Scholar]
- Argueso, C.T.; Ferreira, F.J.; Kieber, J.J. Environmental perception avenues: The interaction of cytokinin and environmental response pathways. Plant Cell Environ. 2009, 32, 1147–1160. [Google Scholar] [CrossRef]
- Choi, J.; Huh, S.U.; Kojima, M.; Sakakibara, H.; Paek, K.-H.; Hwang, I. The Cytokinin-Activated Transcription Factor ARR2 Promotes Plant Immunity via TGA3/NPR1-Dependent Salicylic Acid Signaling in Arabidopsis. Dev. Cell 2010, 19, 284–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shigenaga, A.M.; Berens, M.L.; Tsuda, K.; Argueso, C.T. Towards engineering of hormonal crosstalk in plant immunity. Curr. Opin. Plant Biol. 2017, 38, 164–172. [Google Scholar] [CrossRef]
- Silvar, C.; Merino, F.; Díaz, J. Differential activation of defense-related genes in susceptible and resistant pepper cultivars infected with Phytophthora capsici. J. Plant Physiol. 2008, 165, 1120–1124. [Google Scholar] [CrossRef]
- Cargnello, M.; Roux, P.P. Activation and Function of the MAPKs and Their Substrates, the MAPK-Activated Protein Kinases. Microbiol. Mol. Biol. Rev. 2011, 75, 50–83. [Google Scholar] [CrossRef] [Green Version]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef] [Green Version]
- Adnan, M.; Islam, W.; Noman, A.; Hussain, A.; Anwar, M.; Khan, M.U.; Akram, W.; Ashraf, M.F.; Raza, M.F. Q-SNARE protein FgSyn8 plays important role in growth, DON production and pathogenicity of Fusarium graminearum. Microb. Pathog. 2019, 140, 103948. [Google Scholar] [CrossRef]
- Ross, C.A.; Liu, Y.; Shen, Q.J. The WRKY Gene Family in Rice (Oryza sativa). J. Integr. Plant Biol. 2007, 49, 827–842. [Google Scholar] [CrossRef]
- Imran, Q.M.; Hussain, A.; Mun, B.-G.; Lee, S.-U.; Asaf, S.; Ali, M.A.; Lee, I.-J.; Yun, B.-W. Transcriptome wide identification and characterization of NO-responsive WRKY transcription factors in Arabidopsis thaliana L. Environ. Exp. Bot. 2018, 148, 128–143. [Google Scholar] [CrossRef]
- Cai, H.; Yang, S.; Yan, Y.; Xiao, Z.; Cheng, J.; Wu, J.; Qiu, A.; Lai, Y.; Mou, S.; Guan, D.; et al. CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 2015, 66, 3163–3174. [Google Scholar] [CrossRef] [Green Version]
- Dang, F.F.; Wang, Y.N.; Yu, L.; Eulgem, T.; Lai, Y.; Liu, Z.Q.; Wang, X.; Qiu, A.L.; Zhang, T.X.; Lin, J. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 2013, 36, 757–774. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dang, F.; Liu, Z.; Wang, X.; Eulgem, T.; Lai, Y.; Yu, L.; She, J.; Shi, Y.; Lin, J. C a WRKY 58, encoding a group I WRKY transcription factor of Capsicum annuum, negatively regulates resistance to Ralstonia solanacearum infection. Mol. Plant Pathol. 2013, 14, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Yang, S.; Yang, T.; Liang, J.; Cheng, W.; Wen, J.; Liu, Y.; Li, J.; Shi, L.; Tang, Q.; et al. CaCDPK15 positively regulates pepper responses to Ralstonia solanacearum inoculation and forms a positive-feedback loop with CaWRKY40 to amplify defense signaling. Sci. Rep. 2016, 6, 22439. [Google Scholar] [CrossRef] [Green Version]
- Chanwala, J.; Satpati, S.; Dixit, A.; Parida, A.; Giri, M.K.; Dey, N. Genome-wide identification and expression analysis of WRKY transcription factors in pearl millet (Pennisetum glaucum) under dehydration and salinity stress. BMC Genom. 2020, 21, 231. [Google Scholar] [CrossRef] [Green Version]
- Mine, A.; Seyfferth, C.; Kracher, B.; Berens, M.L.; Becker, D.; Tsuda, K. The Defense Phytohormone Signaling Network Enables Rapid, High-Amplitude Transcriptional Reprogramming during Effector-Triggered Immunity. Plant Cell 2018, 30, 1199–1219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pieterse, C.M.A.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S.C. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.-M.; Rhee, J.-S.; Jeong, C.-B.; Seo, J.S.; Park, G.S.; Lee, Y.-M.; Lee, J.-S. Heavy metals induce oxidative stress and trigger oxidative stress-mediated heat shock protein (hsp) modulation in the intertidal copepod Tigriopus japonicus. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2014, 166, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Sabater-Jara, A.B.; Almagro, L.; Belchí-Navarro, S.; Ferrer, M.Á.; Barceló, A.R.; Pedreño, M.Á. Induction of sesquiterpenes, phytoesterols and extracellular pathogenesis-related proteins in elicited cell cultures of Capsicum annuum. J. Plant Physiol. 2010, 167, 1273–1281. [Google Scholar] [CrossRef]
- Barsalobres-Cavallari, C.; Petitot, A.; Severino, F.; Maia, I.; Fernandez, D. Host response profiling to fungal infection: Molecular cloning, characterization and expression analysis of NPR1 gene from coffee (Coffea arabica). Microb. Pathog. Strateg. Combat. Sci. Technol. Educ. 2013, 4, 411–418. [Google Scholar]
- Wang, Q.; Qiu, B.; Li, S.; Zhang, Y.; Cui, X.; Ge, F.; Liu, D. A methyl jasmonate induced defensin like protein from Panax notoginseng confers resistance against Fusarium solani in transgenic tobacco. Biol. Plant. 2019, 63, 797–807. [Google Scholar] [CrossRef] [Green Version]
- Choi, D.S.; Hwang, B.K. Proteomics and Functional Analyses of Pepper Abscisic Acid–Responsive 1 (ABR1), Which Is Involved in Cell Death and Defense Signaling. Plant Cell 2011, 23, 823–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.W.; Kim, Y.J.; Hwang, B.K. The Hypersensitive Induced Reaction and Leucine-Rich Repeat Proteins Regulate Plant Cell Death Associated with Disease and Plant Immunity. Mol. Plant-Microbe Interact. 2011, 24, 68–78. [Google Scholar] [CrossRef] [Green Version]
- Scarpeci, T.E.; Zanor, M.I.; Mueller-Roeber, B.; Valle, E.M. Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol. Biol. 2013, 83, 265–277. [Google Scholar] [CrossRef]
- El-Esawi, M.A.; Al-Ghamdi, A.A.; Ali, H.M.; Ahmad, M. Overexpression of AtWRKY30 Transcription Factor Enhances Heat and Drought Stress Tolerance in Wheat (Triticum aestivum L.). Genes 2019, 10, 163. [Google Scholar] [CrossRef] [Green Version]
- Berens, M.L.; Berry, H.M.; Mine, A.; Argueso, C.T.; Tsuda, K. Evolution of Hormone Signaling Networks in Plant Defense. Annu. Rev. Phytopathol. 2017, 55, 401–425. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, B.N.; Brooks, D.M. Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 2002, 5, 325–331. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D. Hormone Crosstalk in Plant Disease and Defense: More than Just Jasmonate-Salicylate Antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Ngou, B.P.M.; Ding, P.; Xin, X.-F. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 2021, 62, 102030. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.P.; Somssich, I.E. The Role of WRKY Transcription Factors in Plant Immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [Green Version]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.; Cai, H.; Qin, Y.; Mullis, A.; Lin, Z.; Zhang, L. The WRKY Transcription Factor Family in Model Plants and Crops. Crit. Rev. Plant Sci. 2017, 36, 311–335. [Google Scholar] [CrossRef]
- Qiao, Z.; Li, C.-L.; Zhang, W. WRKY1 regulates stomatal movement in drought-stressed Arabidopsis thaliana. Plant Mol. Biol. 2016, 91, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Azeem, F.; Nawaz, M.A.; Acet, T.; Abbas, A.; Imran, Q.M.; Shah, K.H.; Rehman, H.M.; Chung, G.; Yang, S.H.; et al. Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. J. Plant Physiol. 2018, 226, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-Q.; Yan, L.; Wu, Z.; Mei, C.; Lu, K.; Yu, Y.-T.; Liang, S.; Zhang, X.-F.; Wang, X.-F.; Zhang, D.-P. Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. J. Exp. Bot. 2012, 63, 6371–6392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, F.-C.; Chou, M.-Y.; Chou, S.-J.; Li, Y.-R.; Peng, H.-P.; Shih, M.-C. Submergence Confers Immunity Mediated by the WRKY22 Transcription Factor in Arabidopsis. Plant Cell 2013, 25, 2699–2713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doll, J.; Muth, M.; Riester, L.; Nebel, S.; Bresson, J.; Lee, H.-C.; Zentgraf, U. Arabidopsis thaliana WRKY25 Transcription Factor Mediates Oxidative Stress Tolerance and Regulates Senescence in a Redox-Dependent Manner. Front. Plant Sci. 2020, 10, 1734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Cai, H.; Su, Z.; Wang, L.; Huang, X.; Zhang, M.; Chen, P.; Dai, X.; Zhao, H.; Palanivelu, R.; et al. KLU suppresses megasporocyte cell fate through SWR1-mediated activation of WRKY28 expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E526–E535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birkenbihl, R.P.; Diezel, C.; Somssich, I.E. Arabidopsis WRKY33 Is a Key Transcriptional Regulator of Hormonal and Metabolic Responses toward Botrytis cinerea Infection. Plant Physiol. 2012, 159, 266–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalachova, T.; Leontovyčová, H.; Iakovenko, O.; Pospíchalová, R.; Maršík, P.; Klouček, P.; Janda, M.; Valentová, O.; Kocourková, D.; Martinec, J. Interplay between phosphoinositides and actin cytoskeleton in the regulation of immunity related responses in Arabidopsis thaliana seedlings. Environ. Exp. Bot. 2019, 167, 103867. [Google Scholar] [CrossRef]
- Chen, L.; Xiang, S.; Chen, Y.; Li, D.; Yu, D. Arabidopsis WRKY45 Interacts with the DELLA Protein RGL1 to Positively Regulate Age-Triggered Leaf Senescence. Mol. Plant 2017, 10, 1174–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Nolan, T.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54 and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Response. Plant Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Huhn, K.; Brandt, R.; Potschin, M.; Bieker, S.; Straub, D.; Doll, J.; Drechsler, T.; Zentgraf, U.; Wenkel, S. REVOLUTA and WRKY53 connect early and late leaf development in Arabidopsis. Development 2014, 141, 4772–4783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukushima, S.; Mori, M.; Sugano, S.; Takatsuji, H. Transcription Factor WRKY62 Plays a Role in Pathogen Defense and Hypoxia-Responsive Gene Expression in Rice. Plant Cell Physiol. 2016, 57, 2541–2551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rishmawi, L.; Pesch, M.; Jüngst, C.; Schauss, A.C.; Schrader, A.; Hülskamp, M. Non-Cell-Autonomous Regulation of Root Hair Patterning Genes by WRKY75 in Arabidopsis. Plant Physiol. 2014, 165, 186–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, D.S.; Hwang, I.S.; Hwang, B.K. Requirement of the Cytosolic Interaction between Pathogenesis-Related Protein10 and Leucine-Rich Repeat Protein1 for Cell Death and Defense Signaling in Pepper. Plant Cell 2012, 24, 1675–1690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korasick, D.; McMichael, C.; Walker, K.A.; Anderson, J.C.; Bednarek, S.Y.; Heese, A. Novel Functions of Stomatal Cytokinesis-Defective 1 (SCD1) in Innate Immune Responses against Bacteria. J. Biol. Chem. 2010, 285, 23342–23350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azevedo, C.; Sadanandom, A.; Kitagawa, K.; Freialdenhoven, A.; Shirasu, K.; Schulze-Lefert, P. The RAR1 Interactor SGT1, an Essential Component of R Gene-Triggered Disease Resistance. Science 2002, 295, 2073–2076. [Google Scholar] [CrossRef]
- Liu, Z.; Shi, L.; Liu, Y.; Tang, Q.; Shen, L.; Yang, S.; Cai, J.; Yu, H.; Wang, R.; Wen, J.; et al. Genome-wide identification and transcriptional expression analysis of mitogen-activated protein kinase and mitogen-activated protein kinase kinase genes in Capsicum annuum. Front. Plant Sci. 2015, 6, 780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, I.S.; Hwang, B.K. The Pepper Mannose-Binding Lectin Gene CaMBL1 Is Required to Regulate Cell Death and Defense Responses to Microbial Pathogens. Plant Physiol. 2011, 155, 447–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, A.; Khan, M.I.; Albaqami, M.; Mahpara, S.; Noorka, I.R.; Ahmed, M.A.A.; Aljuaid, B.S.; El-Shehawi, A.M.; Liu, Z.; Farooq, S.; et al. CaWRKY30 Positively Regulates Pepper Immunity by Targeting CaWRKY40 against Ralstonia solanacearum Inoculation through Modulating Defense-Related Genes. Int. J. Mol. Sci. 2021, 22, 12091. https://doi.org/10.3390/ijms222112091
Hussain A, Khan MI, Albaqami M, Mahpara S, Noorka IR, Ahmed MAA, Aljuaid BS, El-Shehawi AM, Liu Z, Farooq S, et al. CaWRKY30 Positively Regulates Pepper Immunity by Targeting CaWRKY40 against Ralstonia solanacearum Inoculation through Modulating Defense-Related Genes. International Journal of Molecular Sciences. 2021; 22(21):12091. https://doi.org/10.3390/ijms222112091
Chicago/Turabian StyleHussain, Ansar, Muhammad Ifnan Khan, Mohammed Albaqami, Shahzadi Mahpara, Ijaz Rasool Noorka, Mohamed A. A. Ahmed, Bandar S. Aljuaid, Ahmed M. El-Shehawi, Zhiqin Liu, Shahid Farooq, and et al. 2021. "CaWRKY30 Positively Regulates Pepper Immunity by Targeting CaWRKY40 against Ralstonia solanacearum Inoculation through Modulating Defense-Related Genes" International Journal of Molecular Sciences 22, no. 21: 12091. https://doi.org/10.3390/ijms222112091
APA StyleHussain, A., Khan, M. I., Albaqami, M., Mahpara, S., Noorka, I. R., Ahmed, M. A. A., Aljuaid, B. S., El-Shehawi, A. M., Liu, Z., Farooq, S., & Zuan, A. T. K. (2021). CaWRKY30 Positively Regulates Pepper Immunity by Targeting CaWRKY40 against Ralstonia solanacearum Inoculation through Modulating Defense-Related Genes. International Journal of Molecular Sciences, 22(21), 12091. https://doi.org/10.3390/ijms222112091






