Cinobufagin Exerts Anticancer Activity in Oral Squamous Cell Carcinoma Cells through Downregulation of ANO1
Abstract
1. Introduction
2. Results
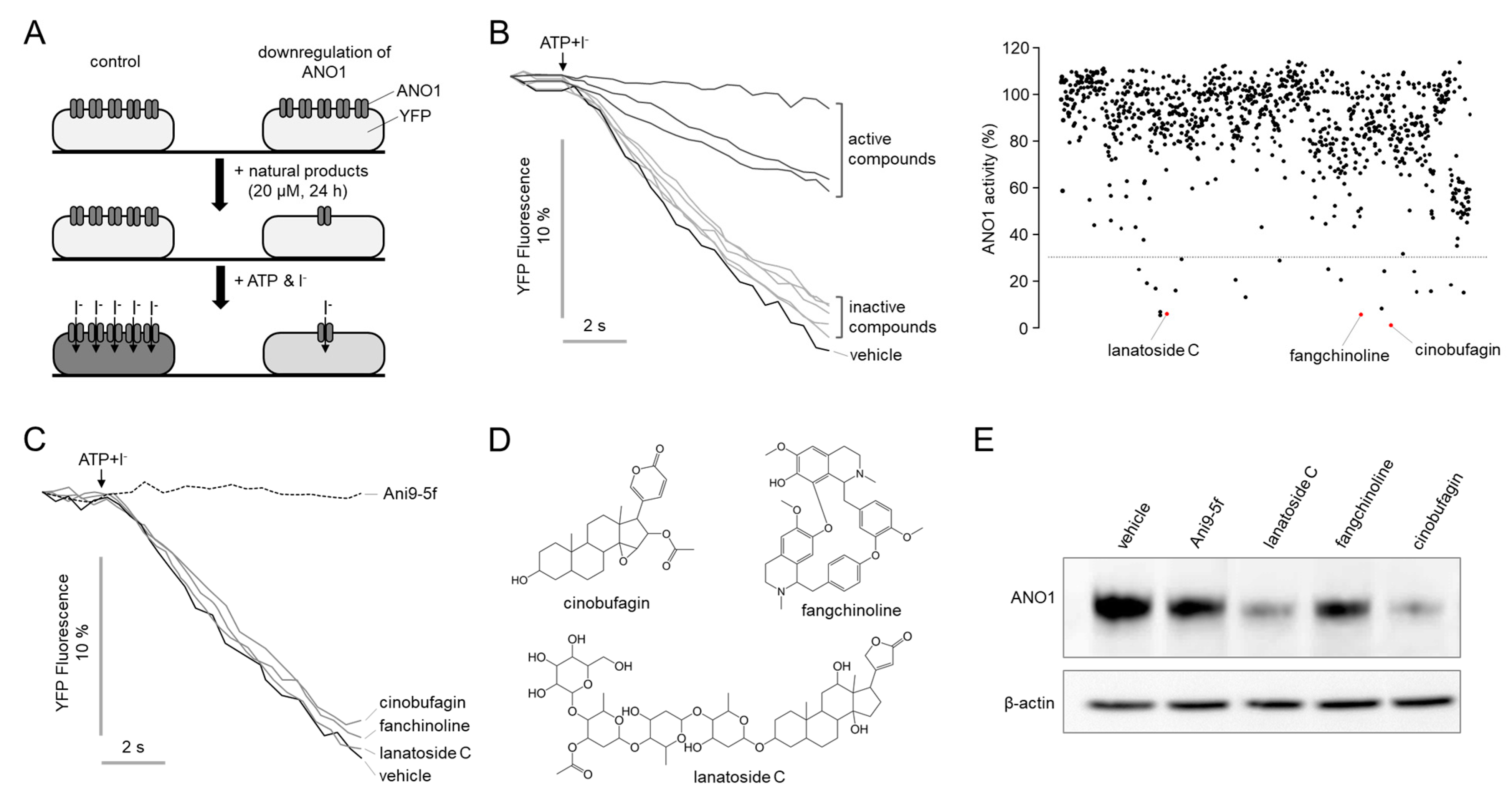
2.1. Identification of Novel Compounds That Downregulate ANO1
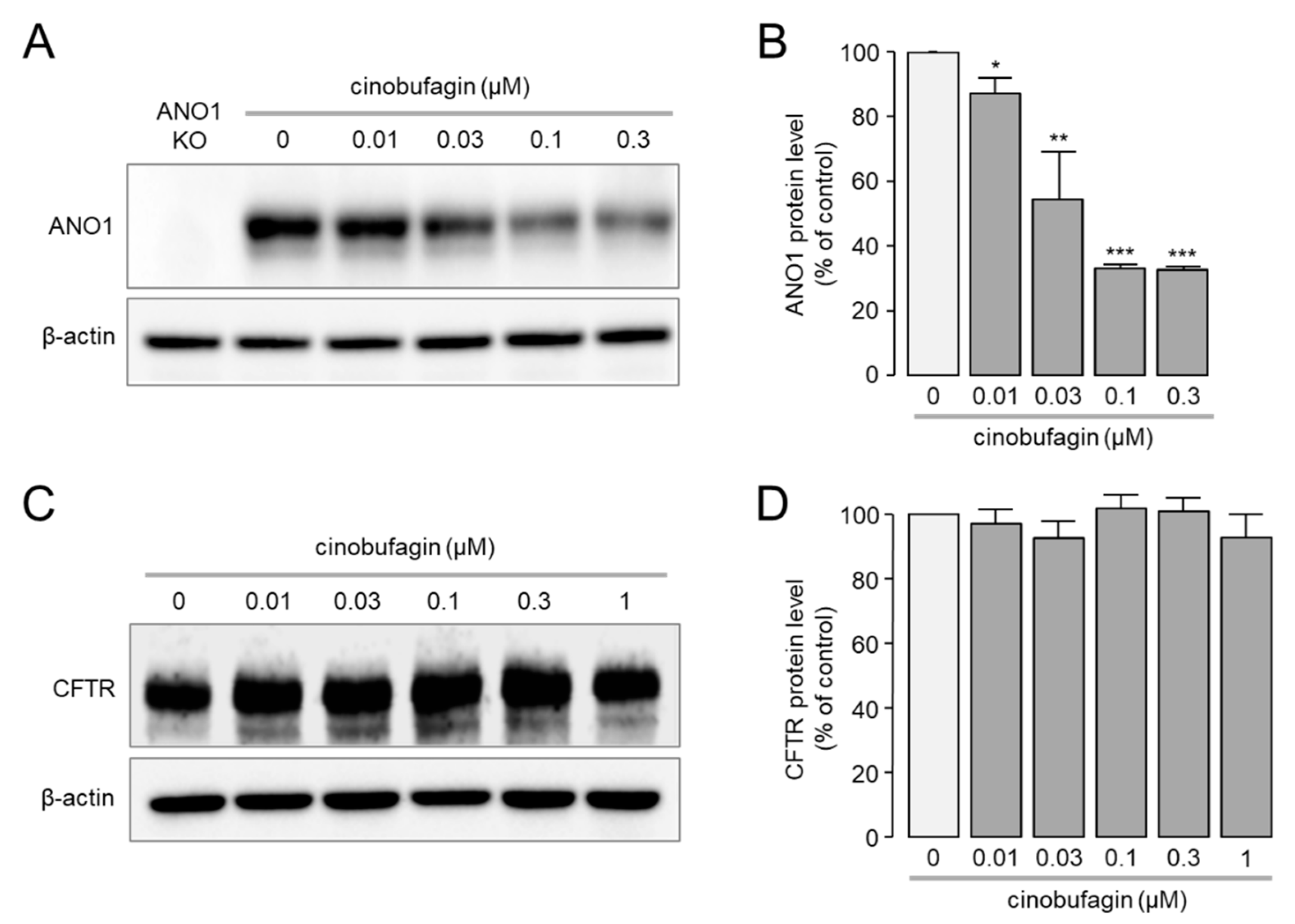
2.2. Effect of Cinobufagin on Protein Expression Levels of ANO1 and CFTR
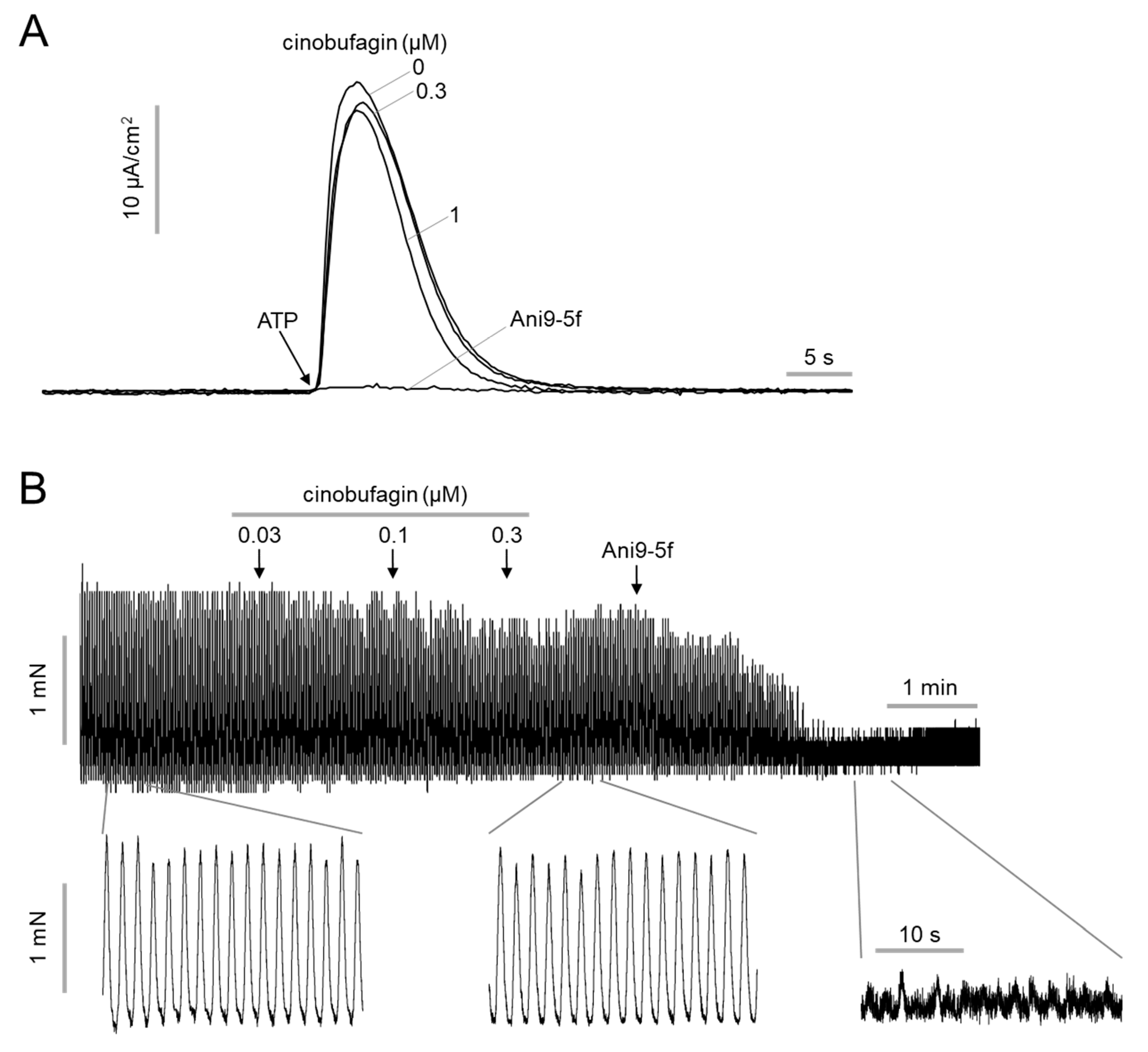
2.3. Effect of Cinobufagin on ANO1 Acitivy and Intestinal Smooth Muscle Contration
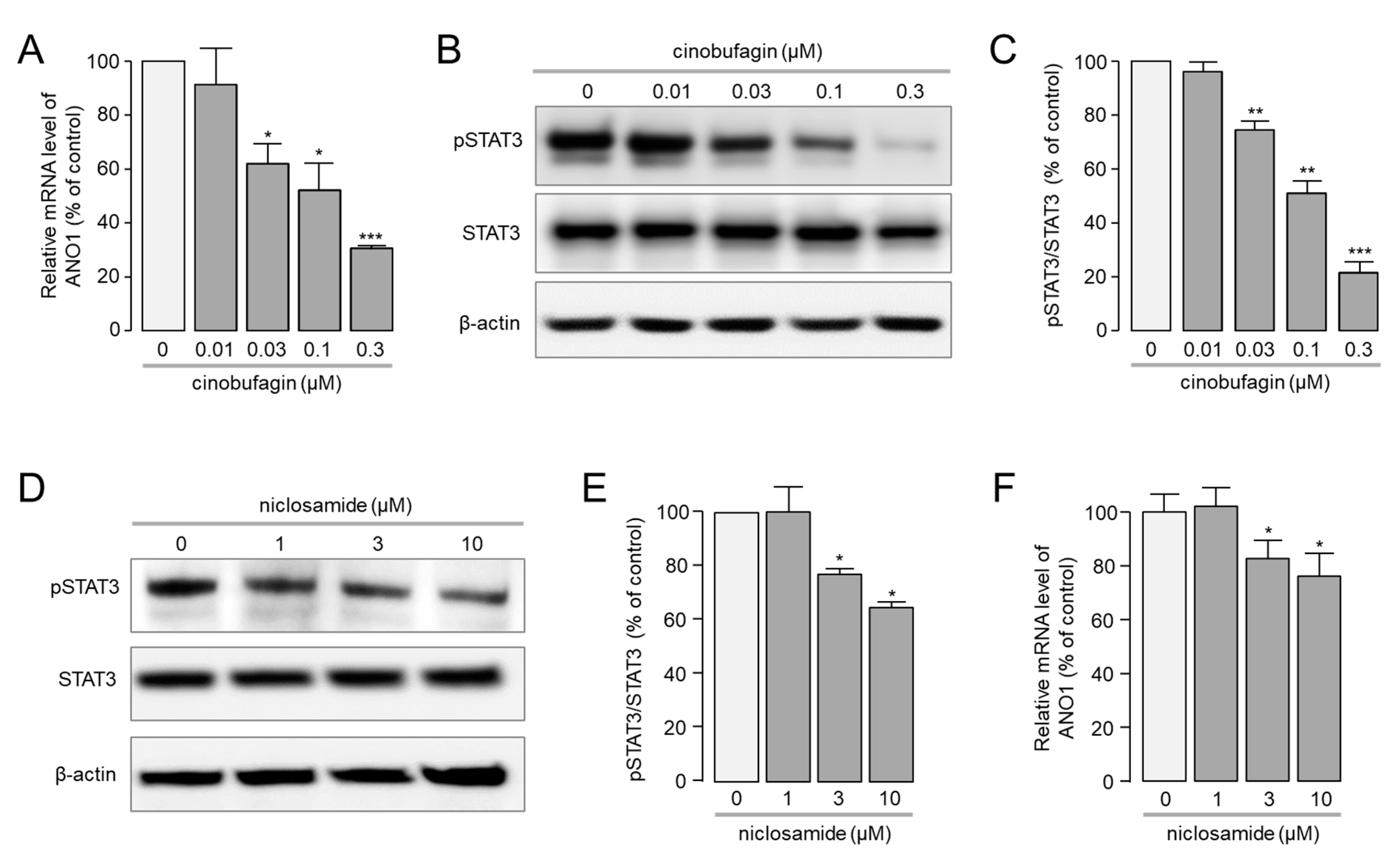
2.4. Cinobufagin Reduced the Phosphorylation of STAT3 and ANO1 Gene Transcription
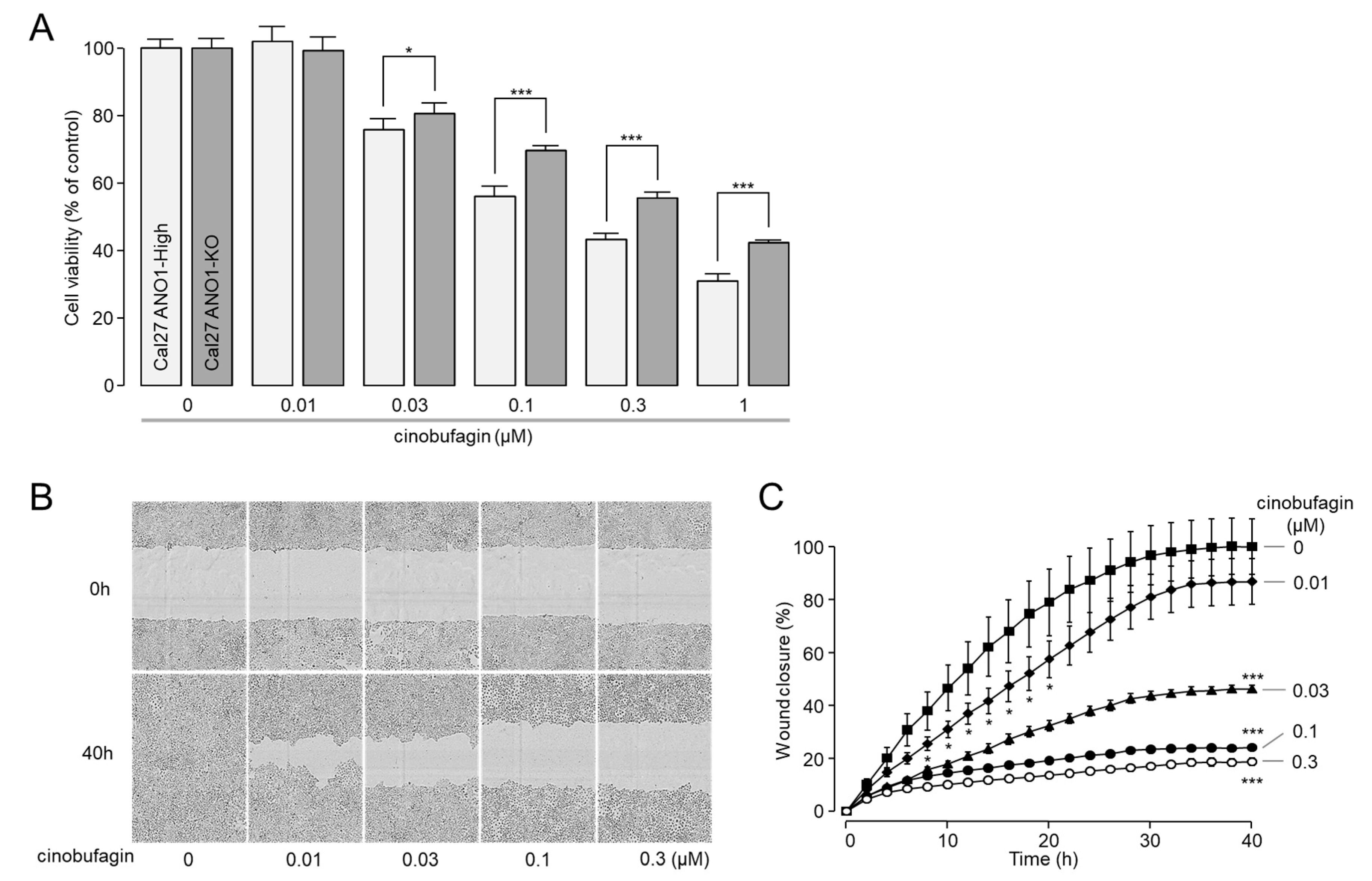
2.5. Cinobufagin Reduced Cell Proliferation and Migration in CAL-27 Cells
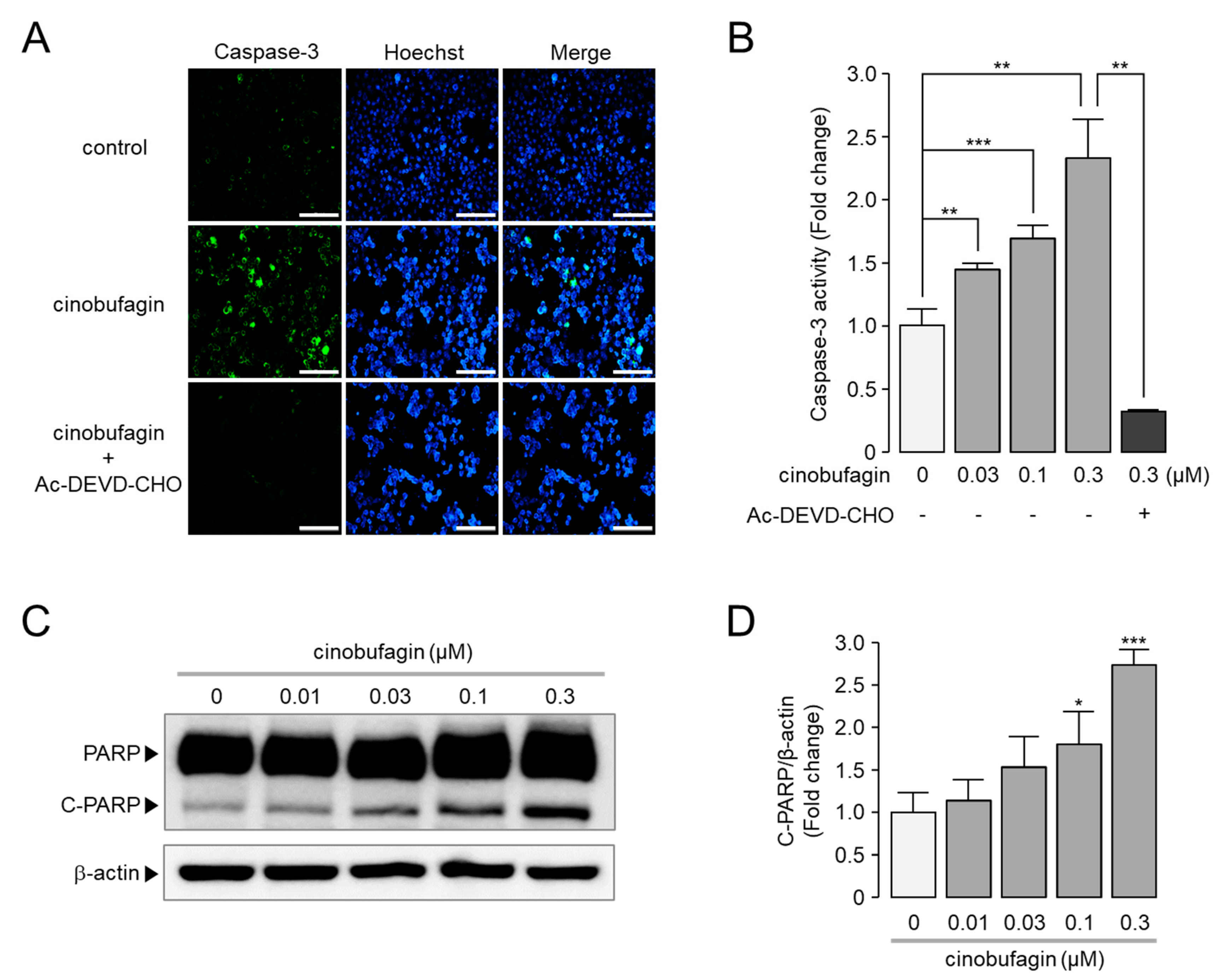
2.6. Cinobufagin-Induced Activation of Caspase-3 and Cleavage of PARP
3. Discussion
4. Materials and Methods
4.1. Material and Solutions
4.2. Cell Culture
4.3. Cell Based YFP Fluorescence Quenching Assay
4.4. Apical Membrane Circuit Measurement
4.5. Immunoblot Analysis
4.6. Real-Time RT-PCR Analysis
4.7. Cell Viability Assay
4.8. Wound Healing Assay
4.9. Caspase-3 Activity Assay
4.10. Intestinal Smooth Muscle Contraction
4.11. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ANO1 | Anoctamin1 |
| OSCC | Oral squamous cell carcinoma |
| HPB242 | 2,4-bis (p-hydroxyphenyl)-2-butenal |
| Sp1 | Specificity protein 1 |
| G3BP1 | GAP SH3 Binding Protein 1 |
| TMEM16A | Transmembrane member 16A |
| CFTR | Cystic fibrosis transmembrane conductance regulator |
| HNSCC | Head and neck squamous cell carcinoma |
References
- Markopoulos, A.K. Current aspects on oral squamous cell carcinoma. Open Dent. J. 2012, 6, 126–130. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Bennardo, L.; Bennardo, F.; Giudice, A.; Passante, M.; Dastoli, S.; Morrone, P.; Provenzano, E.; Patruno, C.; Nisticò, S.P. Local Chemotherapy as an Adjuvant Treatment in Unresectable Squamous Cell Carcinoma: What Do We Know So Far? Curr. Oncol. 2021, 28, 2317–2325. [Google Scholar] [CrossRef] [PubMed]
- Ahn, M.Y.; Yoon, J.H. Histone deacetylase 8 as a novel therapeutic target in oral squamous cell carcinoma. Oncol. Rep. 2017, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Chae, J.I.; Lee, R.; Cho, J.; Hong, J.; Shim, J.H. Specificity protein 1 is a novel target of 2,4-bis (p-hydroxyphenyl)-2-butenal for the suppression of human oral squamous cell carcinoma cell growth. J. Biomed. Sci. 2014, 21, 4. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, P.; Liu, Y.; Zhang, Y.; Fan, W.; Upton, M.P.; Lohavanichbutr, P.; Houck, J.R.; Doody, D.R.; Futran, N.D.; et al. Integrative genomics in combination with RNA interference identifies prognostic and functionally relevant gene targets for oral squamous cell carcinoma. PLoS Genet. 2013, 9, e1003169. [Google Scholar] [CrossRef]
- Liu, J.; Lian, X.; Liu, F.; Yan, X.; Cheng, C.; Cheng, L.; Sun, X.; Shi, Z. Identification of Novel Key Targets and Candidate Drugs in Oral Squamous Cell Carcinoma. Curr. Bioinform. 2020, 15, 328–337. [Google Scholar] [CrossRef]
- Caputo, A.; Caci, E.; Ferrera, L.; Pedemonte, N.; Barsanti, C.; Sondo, E.; Pfeffer, U.; Ravazzolo, R.; Zegarra-Moran, O.; Galietta, L.J. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science 2008, 322, 590–594. [Google Scholar] [CrossRef]
- Yang, Y.D.; Cho, H.; Koo, J.Y.; Tak, M.H.; Cho, Y.; Shim, W.S.; Park, S.P.; Lee, J.; Lee, B.; Kim, B.M.; et al. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature 2008, 455, 1210–1215. [Google Scholar] [CrossRef]
- Schroeder, B.C.; Cheng, T.; Jan, Y.N.; Jan, L.Y. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell 2008, 134, 1019–1029. [Google Scholar] [CrossRef]
- Rock, J.R.; O’Neal, W.K.; Gabriel, S.E.; Randell, S.H.; Harfe, B.D.; Boucher, R.C.; Grubb, B.R. Transmembrane protein 16A (TMEM16A) is a Ca2+-regulated Cl- secretory channel in mouse airways. J. Biol. Chem. 2009, 284, 14875–14880. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Rock, J.R.; Harfe, B.D.; Cheng, T.; Huang, X.; Jan, Y.N.; Jan, L.Y. Studies on expression and function of the TMEM16A calcium-activated chloride channel. Proc. Natl. Acad. Sci. USA 2009, 106, 21413–21418. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.J.; Blair, P.J.; Britton, F.C.; O’Driscoll, K.E.; Hennig, G.; Bayguinov, Y.R.; Rock, J.R.; Harfe, B.D.; Sanders, K.M.; Ward, S.M. Expression of anoctamin 1/TMEM16A by interstitial cells of Cajal is fundamental for slow wave activity in gastrointestinal muscles. J. Physiol. 2009, 587 Pt 20, 4887–4904. [Google Scholar] [CrossRef]
- Namkung, W.; Phuan, P.W.; Verkman, A.S. TMEM16A inhibitors reveal TMEM16A as a minor component of calcium-activated chloride channel conductance in airway and intestinal epithelial cells. J. Biol. Chem. 2011, 286, 2365–2374. [Google Scholar] [CrossRef] [PubMed]
- Lammie, G.A.; Peters, G. Chromosome 11q13 abnormalities in human cancer. Cancer Cells 1991, 3, 413–420. [Google Scholar]
- Peters, G.; Fantl, V.; Smith, R.; Brookes, S.; Dickson, C. Chromosome 11q13 markers and D-type cyclins in breast cancer. Breast Cancer Res. Treat. 1995, 33, 125–135. [Google Scholar] [CrossRef]
- Schuuring, E.; Verhoeven, E.; van Tinteren, H.; Peterse, J.L.; Nunnink, B.; Thunnissen, F.B.; Devilee, P.; Cornelisse, C.J.; van de Vijver, M.J.; Mooi, W.J.; et al. Amplification of genes within the chromosome 11q13 region is indicative of poor prognosis in patients with operable breast cancer. Cancer Res. 1992, 52, 5229–5234. [Google Scholar] [PubMed]
- Akervall, J.A.; Jin, Y.; Wennerberg, J.P.; Zätterström, U.K.; Kjellén, E.; Mertens, F.; Willén, R.; Mandahl, N.; Heim, S.; Mitelman, F. Chromosomal abnormalities involving 11q13 are associated with poor prognosis in patients with squamous cell carcinoma of the head and neck. Cancer 1995, 76, 853–859. [Google Scholar] [CrossRef]
- Huang, X.; Godfrey, T.E.; Gooding, W.E.; McCarty, K.S., Jr.; Gollin, S.M. Comprehensive genome and transcriptome analysis of the 11q13 amplicon in human oral cancer and synteny to the 7F5 amplicon in murine oral carcinoma. Genes Chromosom. Cancer 2006, 45, 1058–1069. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, C.; Wasylyk, C.; Li, Y.; Thomas, E.; Marisa, L.; Robé, A.; Roux, M.; Abecassis, J.; de Reyniès, A.; Wasylyk, B. ANO1 amplification and expression in HNSCC with a high propensity for future distant metastasis and its functions in HNSCC cell lines. Br. J. Cancer 2010, 103, 715–726. [Google Scholar] [CrossRef]
- Liu, W.; Lu, M.; Liu, B.; Huang, Y.; Wang, K. Inhibition of Ca(2+)-activated Cl(−) channel ANO1/TMEM16A expression suppresses tumor growth and invasiveness in human prostate carcinoma. Cancer Lett. 2012, 326, 41–51. [Google Scholar] [CrossRef]
- Britschgi, A.; Bill, A.; Brinkhaus, H.; Rothwell, C.; Clay, I.; Duss, S.; Rebhan, M.; Raman, P.; Guy, C.T.; Wetzel, K.; et al. Calcium-activated chloride channel ANO1 promotes breast cancer progression by activating EGFR and CAMK signaling. Proc. Natl. Acad. Sci. USA 2013, 110, E1026–E1034. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, L.; Ou, Y.; Gao, Z.; Li, E.; Li, X.; Zhang, W.; Wang, J.; Xu, L.; Zhou, Y.; et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 2014, 509, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, C.; Martins, J.R.; Rudin, F.; Schneider, S.; Dietsche, T.; Fischer, C.A.; Tornillo, L.; Terracciano, L.M.; Schreiber, R.; Bubendorf, L.; et al. Enhanced expression of ANO1 in head and neck squamous cell carcinoma causes cell migration and correlates with poor prognosis. PLoS ONE 2012, 7, e43265. [Google Scholar] [CrossRef]
- Duvvuri, U.; Shiwarski, D.J.; Xiao, D.; Bertrand, C.; Huang, X.; Edinger, R.S.; Rock, J.R.; Harfe, B.D.; Henson, B.J.; Kunzelmann, K.; et al. TMEM16A induces MAPK and contributes directly to tumorigenesis and cancer progression. Cancer Res. 2012, 72, 3270–3281. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, Y.; Ren, Y.; Kang, L.; Zhang, L. Transmembrane protein with unknown function 16A overexpression promotes glioma formation through the nuclear factor-κB signaling pathway. Mol. Med. Rep. 2014, 9, 1068–1074. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Liu, F.; Ji, K.; Liu, N.; He, Y.; Zhang, W.; Wang, L. MicroRNA-381 inhibits the metastasis of gastric cancer by targeting TMEM16A expression. J. Exp. Clin. Cancer Res. 2017, 36, 29. [Google Scholar] [CrossRef]
- Hu, C.; Zhang, R.; Jiang, D. TMEM16A as a Potential Biomarker in the Diagnosis and Prognosis of Lung Cancer. Arch. Iran. Med. 2019, 22, 32–38. [Google Scholar]
- Seo, Y.; Lee, H.K.; Park, J.; Jeon, D.K.; Jo, S.; Jo, M.; Namkung, W. Ani9, A Novel Potent Small-Molecule ANO1 Inhibitor with Negligible Effect on ANO2. PLoS ONE 2016, 11, e0155771. [Google Scholar] [CrossRef]
- Seo, Y.; Kim, J.; Chang, J.; Kim, S.S.; Namkung, W.; Kim, I. Synthesis and biological evaluation of novel Ani9 derivatives as potent and selective ANO1 inhibitors. Eur. J. Med. Chem. 2018, 160, 245–255. [Google Scholar] [CrossRef]
- Seo, Y.; Ryu, K.; Park, J.; Jeon, D.K.; Jo, S.; Lee, H.K.; Namkung, W. Inhibition of ANO1 by luteolin and its cytotoxicity in human prostate cancer PC-3 cells. PLoS ONE 2017, 12, e0174935. [Google Scholar] [CrossRef]
- Seo, Y.; Park, J.; Kim, M.; Lee, H.K.; Kim, J.H.; Jeong, J.H.; Namkung, W. Inhibition of ANO1/TMEM16A Chloride Channel by Idebenone and Its Cytotoxicity to Cancer Cell Lines. PLoS ONE 2015, 10, e0133656. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.; Anh, N.H.; Heo, Y.; Park, S.H.; Kiem, P.V.; Lee, Y.; Yen, D.T.H.; Jo, S.; Jeon, D.; Tai, B.H.; et al. Novel ANO1 Inhibitor from Mallotus apelta Extract Exerts Anticancer Activity through Downregulation of ANO1. Int. J. Mol. Sci. 2020, 21, 6470. [Google Scholar] [CrossRef]
- Forrest, A.S.; Joyce, T.C.; Huebner, M.L.; Ayon, R.J.; Wiwchar, M.; Joyce, J.; Freitas, N.; Davis, A.J.; Ye, L.; Duan, D.D.; et al. Increased TMEM16A-encoded calcium-activated chloride channel activity is associated with pulmonary hypertension. Am. J. Physiol. Cell Physiol. 2012, 303, C1229–C1243. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.J.; Shi, J.; Pritchard, H.A.; Chadha, P.S.; Leblanc, N.; Vasilikostas, G.; Yao, Z.; Verkman, A.S.; Albert, A.P.; Greenwood, I.A. Potent vasorelaxant activity of the TMEM16A inhibitor T16A(inh)-A01. Br. J. Pharmacol. 2013, 168, 773–784. [Google Scholar] [CrossRef]
- Huang, F.; Zhang, H.; Wu, M.; Yang, H.; Kudo, M.; Peters, C.J.; Woodruff, P.G.; Solberg, O.D.; Donne, M.L.; Huang, X.; et al. Calcium-activated chloride channel TMEM16A modulates mucin secretion and airway smooth muscle contraction. Proc. Natl. Acad. Sci. USA 2012, 109, 16354–16359. [Google Scholar] [CrossRef]
- Malysz, J.; Gibbons, S.J.; Saravanaperumal, S.A.; Du, P.; Eisenman, S.T.; Cao, C.; Oh, U.; Saur, D.; Klein, S.; Ordog, T.; et al. Conditional genetic deletion of Ano1 in interstitial cells of Cajal impairs Ca(2+) transients and slow waves in adult mouse small intestine. Am. J. Physiol. Gastrointest Liver Physiol. 2017, 312, G228–G245. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta 2013, 1830, 3670–3695. [Google Scholar] [CrossRef]
- Guo, S.; Chen, Y.; Shi, S.; Wang, X.; Zhang, H.; Zhan, Y.; An, H. Arctigenin, a novel TMEM16A inhibitor for lung adenocarcinoma therapy. Pharmacol. Res. 2020, 155, 104721. [Google Scholar] [CrossRef]
- Wang, Q.; Bai, L.; Luo, S.; Wang, T.; Yang, F.; Xia, J.; Wang, H.; Ma, K.; Liu, M.; Wu, S.; et al. TMEM16A Ca(2+)-activated Cl(−) channel inhibition ameliorates acute pancreatitis via the IP(3)R/Ca(2+)/NFκB/IL-6 signaling pathway. J. Adv. Res. 2020, 23, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.V.; Gajowniczek, P.; Way, I.P.; Lee, D.Y.; Jiang, J.; Yuza, Y.; Classon, M.; Haber, D.A.; Settleman, J. A common signaling cascade may underlie "addiction" to the Src, BCR-ABL, and EGF receptor oncogenes. Cancer Cell 2006, 10, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Grandis, J.R.; Chakraborty, A.; Zeng, Q.; Melhem, M.F.; Tweardy, D.J. Downmodulation of TGF-alpha protein expression with antisense oligonucleotides inhibits proliferation of head and neck squamous carcinoma but not normal mucosal epithelial cells. J. Cell Biochem. 1998, 69, 55–62. [Google Scholar] [CrossRef]
- Redell, M.S.; Tweardy, D.J. Targeting transcription factors for cancer therapy. Curr. Pharm. Des. 2005, 11, 2873–2887. [Google Scholar] [CrossRef] [PubMed]
- Grandis, J.R.; Drenning, S.D.; Zeng, Q.; Watkins, S.C.; Melhem, M.F.; Endo, S.; Johnson, D.E.; Huang, L.; He, Y.; Kim, J.D. Constitutive activation of Stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 4227–4232. [Google Scholar] [CrossRef]
- Song, Y.; Gao, J.; Guan, L.; Chen, X.; Gao, J.; Wang, K. Inhibition of ANO1/TMEM16A induces apoptosis in human prostate carcinoma cells by activating TNF-α signaling. Cell Death Dis. 2018, 9, 703. [Google Scholar] [CrossRef]
- Seo, Y.; Jeong, S.B.; Woo, J.H.; Kwon, O.B.; Lee, S.; Oh, H.I.; Jo, S.; Park, S.J.; Namkung, W.; Moon, U.Y.; et al. Diethylstilbestrol, a Novel ANO1 Inhibitor, Exerts an Anticancer Effect on Non-Small Cell Lung Cancer via Inhibition of ANO1. Int. J. Mol. Sci. 2021, 22, 7100. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Song, Y.; Gao, J.; Gao, J.; Wang, K. Inhibition of calcium-activated chloride channel ANO1 suppresses proliferation and induces apoptosis of epithelium originated cancer cells. Oncotarget 2016, 7, 78619–78630. [Google Scholar] [CrossRef]
- Yang, Z.; Luo, H.; Wang, H.; Hou, H. Preparative isolation of bufalin and cinobufagin from Chinese traditional medicine ChanSu. J. Chromatogr. Sci. 2008, 46, 81–85. [Google Scholar] [CrossRef][Green Version]
- De La Fuente, R.; Namkung, W.; Mills, A.; Verkman, A.S. Small-molecule screen identifies inhibitors of a human intestinal calcium-activated chloride channel. Mol. Pharmacol. 2008, 73, 758–768. [Google Scholar] [CrossRef]
- Kim, G.H.; Fang, X.Q.; Lim, W.J.; Park, J.; Kang, T.B.; Kim, J.H.; Lim, J.H. Cinobufagin Suppresses Melanoma Cell Growth by Inhibiting LEF1. Int. J. Mol. Sci. 2020, 21, 6706. [Google Scholar] [CrossRef]
- Zhang, G.; Wang, C.; Sun, M.; Li, J.; Wang, B.; Jin, C.; Hua, P.; Song, G.; Zhang, Y.; Nguyen, L.L.; et al. Cinobufagin inhibits tumor growth by inducing intrinsic apoptosis through AKT signaling pathway in human nonsmall cell lung cancer cells. Oncotarget 2016, 7, 28935–28946. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.; Lv, J.; Cai, C.; Lin, S.; Zhuo, E.; Wang, S. Cinobufagin, a bufadienolide, activates ROS-mediated pathways to trigger human lung cancer cell apoptosis in vivo. RSC Adv. 2017, 7, 25175–25181. [Google Scholar] [CrossRef]
- Bai, Y.; Wang, X.; Cai, M.; Ma, C.; Xiang, Y.; Hu, W.; Zhou, B.; Zhao, C.; Dai, X.; Li, X.; et al. Cinobufagin suppresses colorectal cancer growth via STAT3 pathway inhibition. Am. J. Cancer Res. 2021, 11, 200–214. [Google Scholar] [PubMed]
- Zhu, L.; Chen, Y.; Wei, C.; Yang, X.; Cheng, J.; Yang, Z.; Chen, C.; Ji, Z. Anti-proliferative and pro-apoptotic effects of cinobufagin on human breast cancer MCF-7 cells and its molecular mechanism. Nat. Prod. Res. 2018, 32, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.S.; Park, J.Y.; Park, S.H.; Ha, S.H.; An, A.R.; Noh, S.J.; Kwon, K.S.; Jung, S.H.; Park, H.S.; Kang, M.J.; et al. Expression of ANO1/DOG1 is associated with shorter survival and progression of breast carcinomas. Oncotarget 2018, 9, 607–621. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jo, S.; Yang, E.; Lee, Y.; Jeon, D.; Namkung, W. Cinobufagin Exerts Anticancer Activity in Oral Squamous Cell Carcinoma Cells through Downregulation of ANO1. Int. J. Mol. Sci. 2021, 22, 12037. https://doi.org/10.3390/ijms222112037
Jo S, Yang E, Lee Y, Jeon D, Namkung W. Cinobufagin Exerts Anticancer Activity in Oral Squamous Cell Carcinoma Cells through Downregulation of ANO1. International Journal of Molecular Sciences. 2021; 22(21):12037. https://doi.org/10.3390/ijms222112037
Chicago/Turabian StyleJo, Sungwoo, Eunhee Yang, Yechan Lee, Dongkyu Jeon, and Wan Namkung. 2021. "Cinobufagin Exerts Anticancer Activity in Oral Squamous Cell Carcinoma Cells through Downregulation of ANO1" International Journal of Molecular Sciences 22, no. 21: 12037. https://doi.org/10.3390/ijms222112037
APA StyleJo, S., Yang, E., Lee, Y., Jeon, D., & Namkung, W. (2021). Cinobufagin Exerts Anticancer Activity in Oral Squamous Cell Carcinoma Cells through Downregulation of ANO1. International Journal of Molecular Sciences, 22(21), 12037. https://doi.org/10.3390/ijms222112037





