Abstract
SHORT VEGETATIVE PHASE (SVP) genes are members of the well-known MADS-box gene family that play a key role in regulating vital developmental processes in plants. Hemerocallis are perennial herbs that exhibit continuous flowering development and have been extensively used in landscaping. However, there are few reports on the regulatory mechanism of flowering in Hemerocallis. To better understand the molecular basis of floral formation of Hemerocallis, we identified and characterized the SVP-like gene HkSVP from the Hemerocallis cultivar ‘Kanai Sensei’. Quantitative RT-PCR (qRT-PCR) indicated that HkSVP transcript was mainly expressed in the vegetative growth stage and had the highest expression in leaves, low expression in petals, pedicels and fruits, and no expression in pistils. The HkSVP encoded protein was localized in the nucleus of Arabidopsis protoplasts and the nucleus of onion epidermal cells. Yeast two hybrid assay revealed that HKSVP interacted with Hemerocallis AP1 and TFL1. Moreover, overexpression of HkSVP in Arabidopsis resulted in delayed flowering and abnormal phenotypes, including enriched trichomes, increased basal inflorescence branches and inhibition of inflorescence formation. These observations suggest that the HkSVP gene may play an important role in maintaining vegetative growth by participating in the construction of inflorescence structure and the development of flower organs.
1. Introduction
The transition from vegetative to reproductive growth is a major developmental switch in the life cycle of plants and is required for successful sexual reproduction of flowering plants. This process is controlled by environmental stimuli signals and the complex internal genetic network [1,2]. Molecular genetic analyses have identified three major genetic pathways FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), and LEAFY (LFY), that promote the floral transition in Arabidopsis [3,4]. The sequential interactions mediated by the above integrators constitute a conserved master regulatory pathway that controls vegetative phase changes in plants [5].
SHORT VEGETATIVE PHASE (SVP), which encodes a MADS-box transcription factor that represses the FT and SOC1 floral pathways to suppress the transition of reproductive development, could be involved in maintenance of the plant vegetative growth period and determination of the specificity of the flower meristem [6,7,8,9,10]. In Arabidopsis thaliana, SVP is expressed broadly during vegetative development including in leaves and shoot apices [11]. It was reported that two paralogous genes, SVP and AGAMOUS-LIKE 24 (AGL24), competitively bind APETALA1 (AP1) to regulate the expression of AGAMOUS (AG), thereby affecting floral transition during the early stages of flower development [8,12,13,14]. Furthermore, SVP acts redundantly and directly to interact with TERMINAL FLOWER1 (TFL1) in the establishment of floral meristems derived from the inflorescence meristem [15]. In addition, SVP mediates ambient temperature signalling by regulating FT expression [6] and responds to endogenous signals from the autonomous and gibberellin (GA) pathways to directly control SOC1 transcription [16].
To date, several homologues of SVP have been characterized from different plant species including Lycopersicon esculentum [17], Eucalyptus grandis [10], Medicago L. [18], Actinidia chinensis [19], Poncirus trifoliata [9], Hordeum vulgare [20,21], Brassica L. [22], Pharbitis nil [23], Paeonia suffruticosa [24], Narcissus tazetta [25], Antirrhinum majus [26], Prunus mume [27] and Mangifera indica [28]. It has been reported that ectopic overexpression of SVP in transgenic Arabidopsis svp mutants leads to the formation of bud-like structures and leaf-like sepals instead of flowers [10,18], causes floral defects including delayed flowering in wild type (WT) Arabidopsis [18], and inhibits early transition and prolonged co-florescence development in tobacco [9]. These results suggest that plants have evolved diversified SVPs to address changes in species and environmental stress in terms of their roles in determining the vegetative phase and floral transition [12].
Hemerocallis fulva, commonly known as day lily, is an economically important landscape plant with a long history of cultivation in China [29]. The flowering and floral setting of Hemerocallis are valuable ornamental characteristics [30,31]. Little is known however about the floral transition of Hemerocallis, and the only flowering-related gene reported is HkTFL1 [32]. In this study, we report the isolation and characterization of an SVP-like gene, named HkSVP, from Hemerocallis fulva. HkSVP was used for amino acid sequence phylogenetic analysis, expression profiling, protein integrates flowering signals, and functional analysis of overexpression in Arabidopsis. The data presented herein enrich our understanding of floral pathway integrators in Hemerocallis.
2. Results
2.1. Sequence Alignment and Phylogenetic Analysis
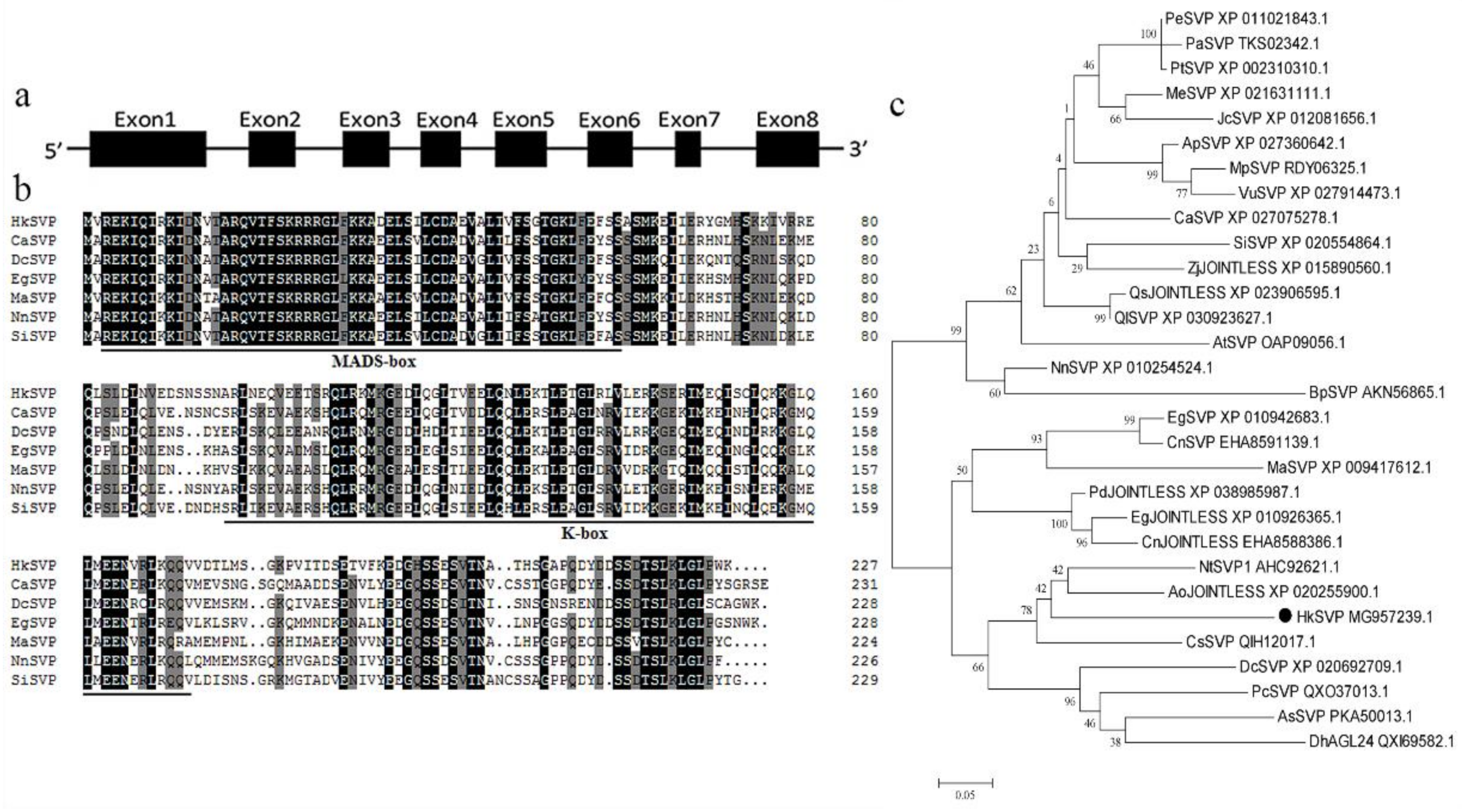
The full-length HkSVP cDNA sequence was obtained from the total RNA of young Hemerocallis ‘Kanai Sensei’ leaves by RT-PCR. Oligo d(T)18 was used for mRNA reverse transcription, and the specific primers HkSVP-F/R for the conserved 5′ and 3′ UTRs were used separately for PCR amplification (GenBank accession No. MG957239.1). The open reading frame of HkSVP consisted of 684 bp and was predicted to encode a protein of 227 amino acids. The theoretical isoelectric point (pI) of HkSVP was 6.38, and the calculated molecular weight of HkSVP was 25.83 kDa. Analysis of the full-length cDNA and genomic DNA sequences of HkSVP indicated that there were eight exons in HkSVP (Figure 1a). The putative HkSVP protein contains a MIKC-II type MADS-box motif in the N-terminus and a K-box motif in the middle domains and shares high similarity with other SVP-related proteins (Figure 1b).
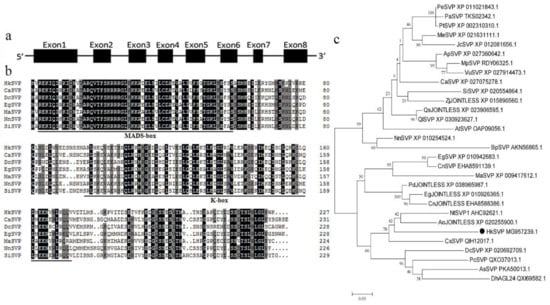
Figure 1.
Schematic of the HkSVP gene structure and systemic phylogenetic relationship. (a) HkSVP CDS gene structure. Lines indicate introns, and black boxes indicate exons from the open reading frame. Numbers represent the exon or intron lengths in base pairs. The ATG translation initiation site and TAA translation stop site are marked. (b) Comparison of SVP amino acid sequences from selected species. Identical or conserved residues are indicated in black, similar residues are indicated in grey, and similar or weakly similar residues are indicated in white. The MADS-domain and K-domain are indicated by black lines. (c) Phylogenetic tree analysis of the amino acid sequence alignment of the Hemerocallis SVP protein with SVP proteins from other plant species. The trees were constructed by MEGA 7.0 software, using the minimum evolution phylogeny tested with 1000 bootstrap replicates, and the cut-off value was set to 50%. PeSVP (Populus euphratica, XP_011021843.1), PaSVP (Populus alba, TKS02342.1), PtSVP (Populus trichocarpa, XP_002310310.1), MeSVP (Manihot esculenta, XP_021631111.1), JcSVP (Jatropha curcas, XP_012081656.1), ApSVP (Abrus precatorius, XP_027360642.1), MpSVP (Mucuna pruriens, RDY06325.1), VuSVP (Vigna unguiculata, XP_027914473.1), CaSVP (Coffea arabica, XP_027075278.1), SiSVP (Sesamum indicum, XP_020554864.1), ZjJOINTLESS (Ziziphus jujuba, XP_015890560.1), QsJOINTLESS (Quercus suber, XP_023906595.1), QlSVP (Quercus lobata, XP_030923627.1), AtSVP (Arabidopsis thaliana, OAP09056.1), NnSVP (Nelumbo nucifera, XP_010254524.1), BpSVP (Betula platyphylla, AKN56865.1), EgSVP (Elaeis guineensis, XP_010942683.1), CnSVP (Cocos nucifera, EHA8591139.1), MaSVP (Musa acuminata, XP_009417 612.1), PdJOINTLESS (Phoenix dactylifera, XP_038985987.1), EgJOINTLESS (Elaeis guineensis, XP_010926365.1), CnJOINTLESS (Cocos nucifera, EHA8588386.1), NtSVP1 (Narcissus tazetta, AHC92621.1), AoJOINTLESS (Asparagus officinalis, XP_020255900.1), HkSVP (Hemerocallis ‘Kanai Sensei’, MG957239.1), CsSVP (Crocus sativus, QIH12017.1), DcSVP (Dendrobium catenatum, XP_020692709.1), PcSVP (Paphiopedilum callosum, QXO37013.1), AsSVP (Apostasia shenzhenica, PKA50013.1), DhAGL24 (Dendrobium hybrid, QXI69582.1).
We performed phylogenetic tree analysis to better understand the genetic relationship between HkSVP and other reported SVP-like genes. As shown in Figure 1c, the evolutionary tree is divided into two different evolutionary groups. The SVP homologues of monocotyledons and dicotyledons are clustered into different branches, indicating that the SVP gene was highly conserved during evolution. HkSVP is clustered with Narcissus tazetta, Asparagus officinalis, Crocus sativus and Dendrobium catenatum, indicating that it is more closely related to monocotyledonous plants and shows high homology with genes from these plants. It is genetically distant from and shows low homology with genes from woody plants such as Populus trichocarpa, Populus euphratica and Populus alba. suggesting a conserved function with these evolutionarily closer SVP homologues.
2.2. Subcellular Localization of HkSVP
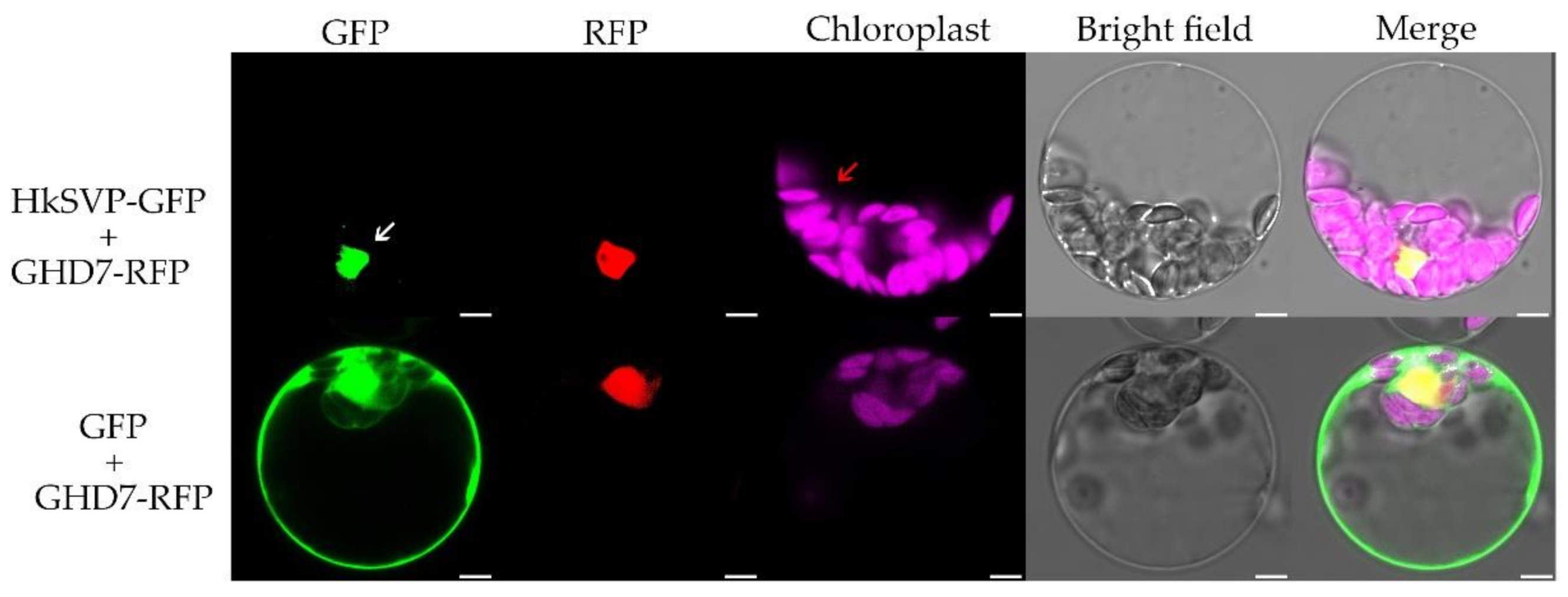
To investigate the subcellular localization of HkSVP, we co-transformed the vector PAN580-HkSVP-GFP and nuclear marker gene pRFP-OsGHD7 [33] into Arabidopsis protoplasts derived from the WT. The colocalized fluorescent signals from GFP and RFP merged in the nuclear region, suggesting that HkSVP is a nucleus-localized protein (Figure 2). Furthermore, the pROKII-HkSVP-GFP fusion construct was transformed into onion epidermal cells using a gene gun. The green fluorescence signal of HkSVP-GFP was also distributed in the nuclear region (Figure S1). To confirm the confocal microscopy results and explore the subcellular location of HkSVP in more detail, we carried out analysis of the localization signals in the HkSVP amino acid sequence by using the WoLF PSORT tool (http://wolfpsort.org, accessed on 1 August 2019) [34]. The results showed that there was a predicted nuclear localization signal peptide in the N-terminal region with the PERIPHERAL motif, and that there was an XXRR-like membrane retention signal motif in the N-terminus along with a VREK motif, but other signal motifs for subcellular localization of the HkSVP protein were unexpectedly detected in the C-terminus region. The subcellular localization pattern indicated that the MADS-box protein HkSVP functions as a transcription factor in the nucleus.
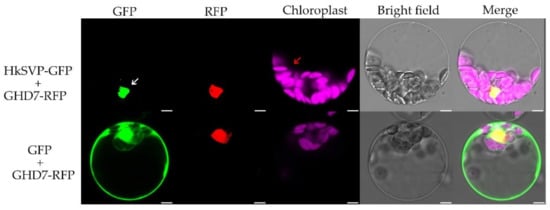
Figure 2.
Subcellular localization of the HkSVP protein. PAN580-HkSVP-GFP and nuclear marker were transiently coexpressed in Arabidopsis protoplasts, and overlapping GFP and RFP signals were observed in the nucleus. White and red arrowheads indicate the nucleus and chloroplast, respectively. Scale bar, 5 µm.
2.3. HkSVP Interacts with the HkTFL1 and HkAP1 Proteins from Hemerocallis
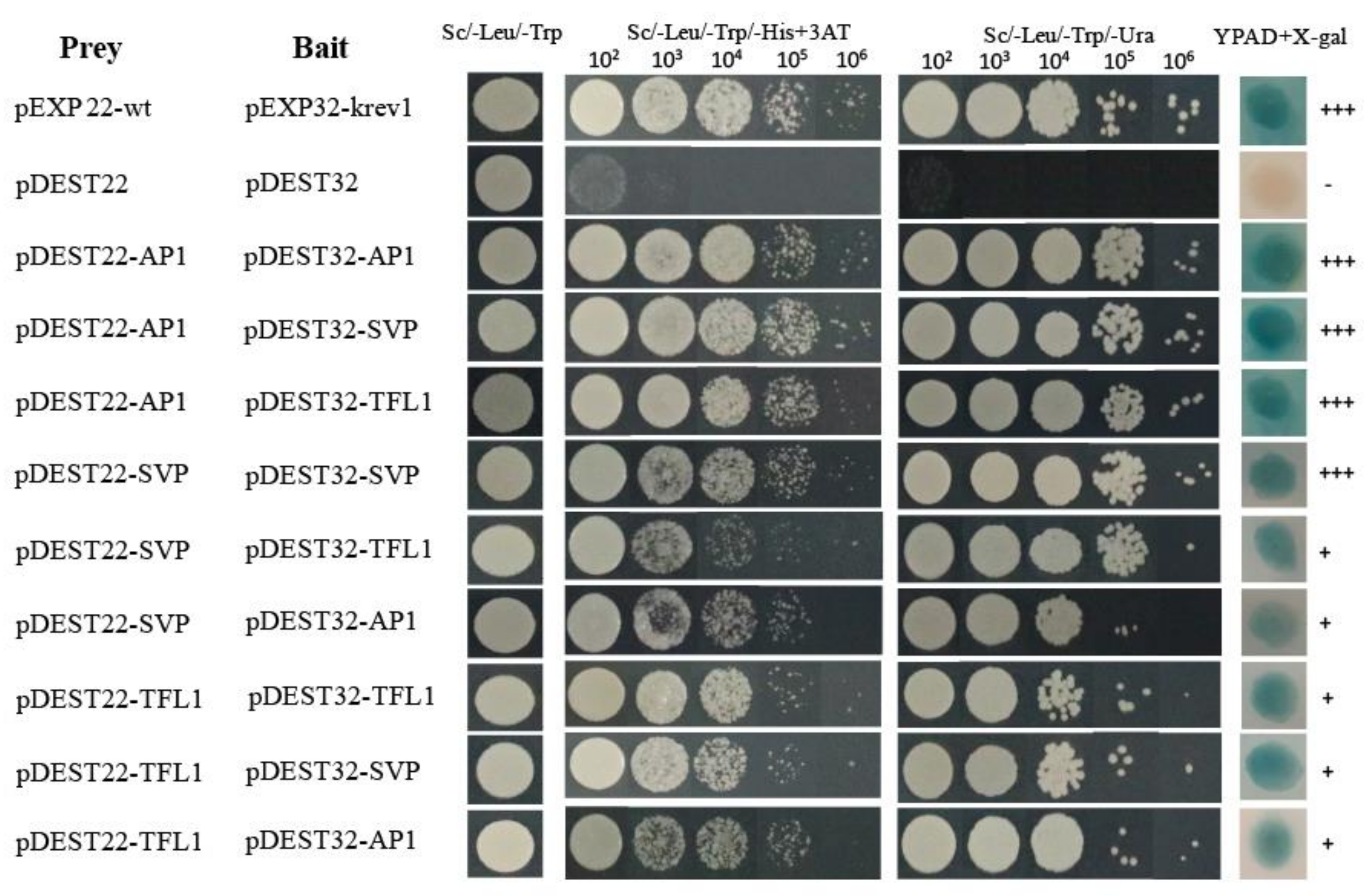
To further understand the functional interactions of HkSVP with other endogenous flowering-related proteins of Hemerocallis for floral development, the coding sequences of HkSVP, HkAP1, and HkTFL1 from Hemerocallis were fused downstream of the GAL4 activation domain (AD) in the bait vector pDEST22, and other sequences were fused downstream of the GAL4 DNA-binding domain (BD) in the prey vector pDEST32. The BD and AD, cloned in the prey vector pDEST32 and bait vector pDEST22, respectively, were tested for their ability to interact with HkSVP. Analysis of the interactions of HkSVP with HkAP1 and HkTFL1 yielded positive results, with viable colonies on double drop out (Leu-Trp) medium (Figure 3). Through the self-activation experiment with the bait plasmid, it was concluded that 25 mM 3-aminotriazole (3AT) could be used as the screening concentration to inhibit the growth of false-positive yeast transformants (Figure S2). The interactions between these proteins and their interaction partners were further confirmed with the development of viable blue colonies on YPDA medium with the β-galactosidase induction assay (X-gal assay). The strength of interactions between the proteins was visually interpreted from the intensity of the colonies on triple drop out (Leu-Trp-His) and (Leu-Trp-Ura) medium, which was found to be the strongest for the interaction of AP1 with SVP and the weakest for the interaction of SVP with AP1. The interaction between the AP1 and SVP proteins has been reported in Arabidopsis [35], and AP1 was also shown to interact with TFL1 [36]. Therefore, we also performed a yeast two-hybrid assay using SVP and TFL1 from Hemerocallis to obtain insight into the functional similarity of the SVP proteins from Hemerocallis and Arabidopsis. The results of this analysis indicate that HkSVP can interact with both HkAP1 and HkTFL1. This suggested that the protein-protein interaction domains of MADS-box and PEBP proteins from Hemerocallis may interact specifically with related partners and might be conserved during floral induction and development among different species. Based on the data obtained in this study and that already available in the literature regarding protein-protein interactions among different species MADS-box genes [37], an interaction of the current state of the protein-protein interaction network is presented in Figure 3.
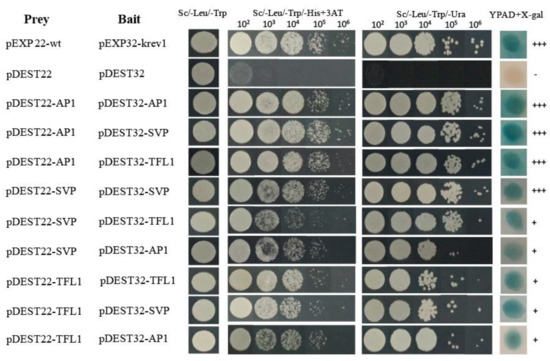
Figure 3.
Interactions between HkSVP, HkTFL1 and HkAP1 proteins determined by a yeast two-hybrid assay. The positive and negative controls were pEXP22-wt/pEXP32-krev1 and pDEST22/ pDEST32 (Thermo, PQ10001), respectively. All hybrid yeast strains were spotted on selection plates. 102–106, fold dilution of the yeast liquid culture. +++, very strong interaction; +, strong interaction.
2.4. Expression Analysis of HkSVP in Hemerocallis
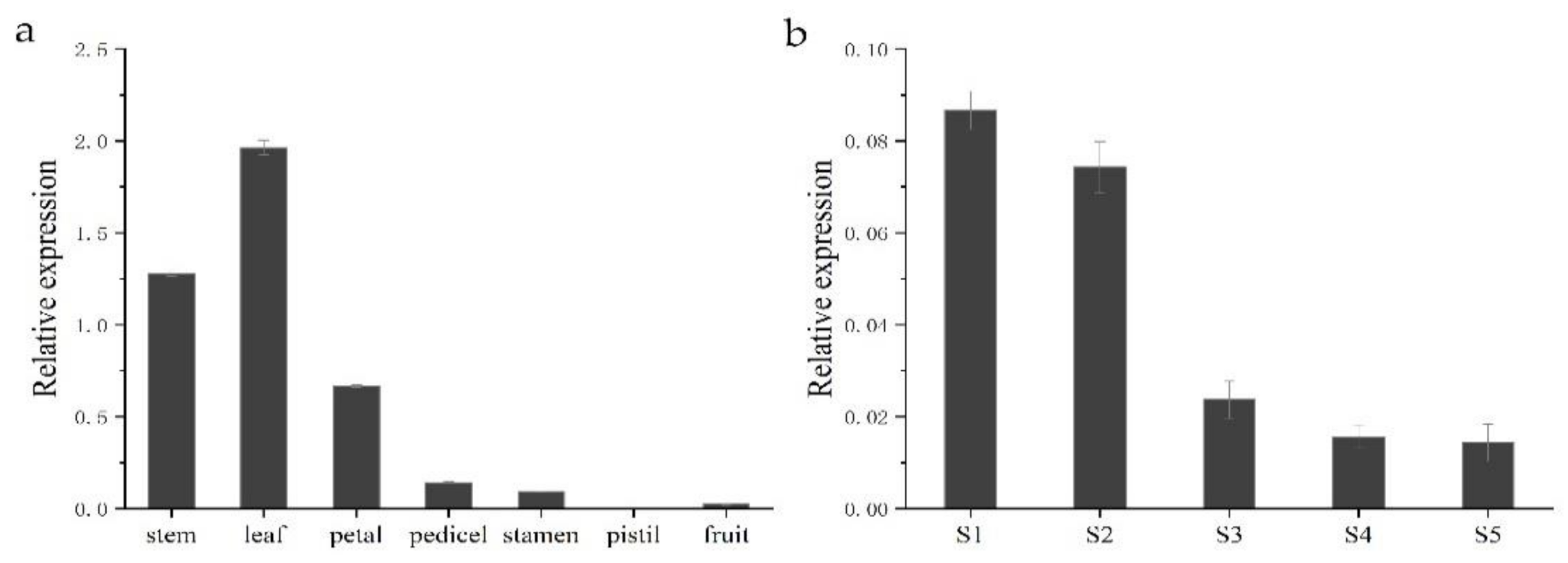
To gain insight into the functional role of HkSVP in Hemerocallis development and the floral formation stage, we performed spatial analysis of the expression of the HkSVP gene using qRT-PCR in different organs and at different developmental stages. As shown in Figure 4, different expression patterns were found in these six organs. HkSVP mRNA transcript was principally found in stem and leaf tissues, with the highest expression in leaf tissues, but it was significantly expressed at low levels in the petal, pedicel and fruit and was almost undetectable in the pistil. Therefore, it appears that HkSVP was expressed during the early vegetative phase and increased towards the transition from the vegetative phase to reproductive phase. These observations were consistent with the results presented for AtSVP and showed that HkSVP played an important role in determining the duration of the vegetative phase [12].
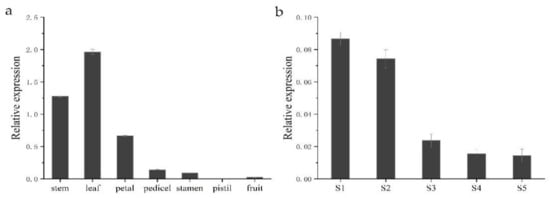
Figure 4.
HkSVP expression analysis in Hemerocallis by qRT-PCR. (a) Stem, leaf, petal, pedicel, stamen, pistil and fruit. (b) S1–S5 stages of flower development. Error bars represent standard deviations calculated from three replications. The Actin gene was used as an internal control (Table S1).
To fully characterize the expression patterns of the HkSVP gene during the floral formation stage, we analysed the expression of HkSVP during the floral organ morphological structure stages from S1 to S5. The highest HkSVP expression was observed in the S1 petal differentiation stage, and then the expression began to decrease gradually until the lowest expression in the S5 stage, which was shown as S1 > S2 > S3 > S4 > S5. Based on these results, we conclude that a high level of HkSVP expression occurs in the early floral development stage, which might contribute to inducing floral formation and repressing flowering [12].
2.5. Ectopic Expression of HkSVP in Arabidopsis
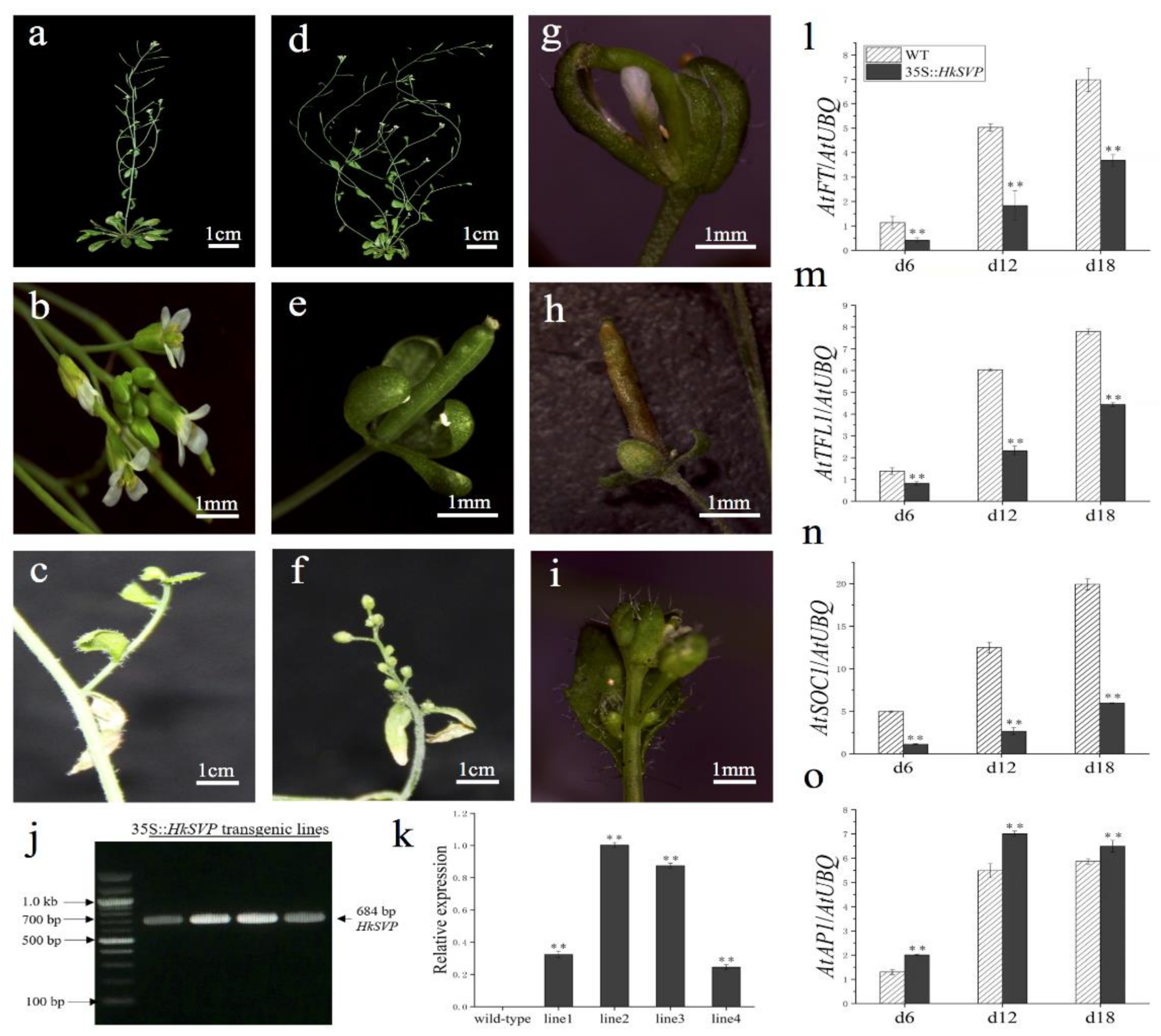
To assess the potential roles of HkSVP in the control of flowering time and regulation of flower development, the HkSVP protein was overexpressed in transgenic Arabidopsis. Four independent T1 generation transgenic lines were examined for kanamycin resistance, and the results were subsequently confirmed with RT-PCR and qRT-PCR analyses (Figure 5j,k). In the qRT-PCR data, line 1 and line 4 represent low level overexpression, line 2 and line 3 represent the highest level of overexpression of HkSVP in Arabidopsis. Lines 1, 2, 3, and 4 (L1, L2, L3, and L4) were screened using the selection method described above until T3 generation transgenic seeds were obtained. Four of the transformed Arabidopsis T3 lines had significantly delayed blooms under long-day growth conditions compared to WT plants during floral development (Table 1). At the onset of flower development, the transgenic plants were easily distinguished from WT Col, and flowers appeared at 35 days. However, the transgenic plants flowered after approximately 53 days (Table 1).
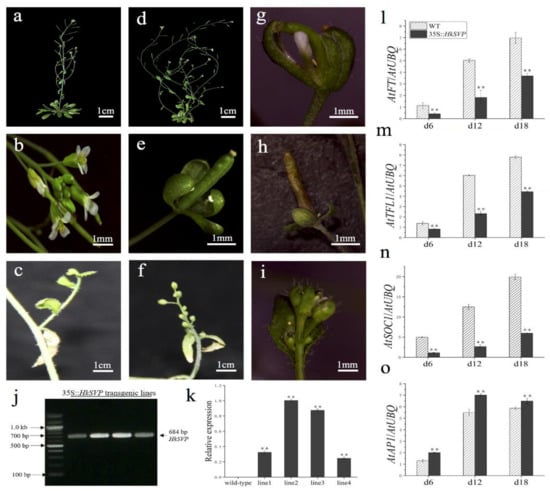
Figure 5.
Phenotypes of p35S::HkSVP transgenic Arabidopsis thaliana. (a–c) Wild-type Arabidopsis. (d) Basal inflorescence branches increased. (e,h) Leafy sepals persistent. (f) The number of flower buds on the inflorescence of secondary branches increased. (g) Sepals abnormally enlarged. (i) Increased density of trichomes. (j) Results of an RT-PCR analysis of HkSVP expression in transgenic Arabidopsis. M, DL 2000 Marker; 1-4, independent 35S::HkSVP transgenic lines. (k) Results of qRT-PCR analysis of HkSVP transcript in transgenic Arabidopsis lines. Wild-type plants served as controls. (l–o) AtFT expression, AtTFL1 expression, AtSOC1 expression, and AtAP1 expression. The error bars represent the standard deviation of the data obtained from three biological replicates. The expression levels of all the genes were normalized against AtUBQ-F and AtUBQ-R expression (Table S1). Values are presented as the mean ± standard deviation of three biological replicates. ** indicates significant differences (p < 0.01) according to Student’s test.

Table 1.
Flowering phenotypes of 35S::HkSVP in Arabidopsis under long-day conditions.
HkSVP affects the inflorescence structure of Arabidopsis. Compared with the WT (Figure 5a,b), transgenic lines 2 and 3 showed a significant increase in the number of typical basal branches (Figure 5d). At the same time, the plants also showed different degrees of abnormality in controlling the development of flower meristems. Leafy sepals are leathery in flower organs and reside at the base of developing pod (Figure 5e,h). The line 2 flower organ showed that the volume of sepals increased, and the abnormal flower structure with missing petals and stamens was wrapped in it (Figure 5g). Among the inflorescences formed on some secondary branches, WT Arabidopsis produced only a small number of flower buds (Figure 5c). In contrast, the number of flower buds on the inflorescence attached to the secondary branches of the transformed line increased significantly and grow alternately on the whole extended internode (Figure 5f). The density of trichome on flower buds and leaves of individual transformed lines was significantly higher than that of the WT (Figure 5i). In summary, HkSVP delayed flowering time in transgenic lines and as a floral repressor plays an important role in controlling inflorescence structure and flower organ morphology.
Furthermore, qRT-PCR was used to analyse the expression of flowering related genes (AtFT, AtTFL1, AtSOC1, AtAP1) in transgenic plants at different growth stages (6, 12, 18 days after seed germination) (Figure 5l–o). The results showed that the expression levels of AtFT, AtTFL1, and AtSOC1 genes in transgenic plants were significantly lower than those in the WT, especially the AtSOC1 gene was only weakly expressed in 35S::HkSVP. In contrast, the expression level of flower meristem determining gene AtAP1 was consistent with that in the WT, and both lines showed a similar trend with the gradual increase in plant growth and development.
3. Discussion
We isolated the HkSVP gene from Hemerocallis ‘Kanai Sensei’ and examined its expression patterns and functions. Sequence analysis and multiple sequence alignment confirmed that HkSVP gene is similar to other SVP genes from different species. These genes contain MADS-box and K-box domains, which play a key role in DNA binding and protein complex formation [38]. Phylogenetic tree analysis showed that HkSVP is highly homologous to the SVP genes of monocotyledon plant species, and with the highest sequence similarity with SVP gene of Narcissus tazetta. The localization of HkSVP-GFP fusion protein in the nucleus also indicates that HkSVP may have transcription factor activity.
The interactions between the proteins revealed an important mode underlying the regulatory function. MADS-box transcription factors play a key role in plant flowering and its gene regulatory network [39]. As a flowering inhibitor, the TFL1-like gene in the PEBP family plays a key role in prolonging plant vegetative growth and delaying plant flowering [32]. Therefore, the study of the protein interaction between PEBP and MADS-box can lay a foundation for clarifying the molecular mechanism of the flowering regulation network. The results of this study show that the two-way hybridization between HkAP1, HkSVP, and HkTFL1 proteins can interact directly. It is speculated that they are jointly involved in various ways of regulating target genes, including the transformation from vegetative growth to reproductive growth, controlling the morphological structure of plants and the growth and development state of flower sequence.
Previous studies have indicated that the continuous flowering phenotype was related to the number of plant vegetative branches during hybridization of Hemerocallis in the field [29,32]. HkSVP expression has a broad expression pattern throughout various tissues of plants. The accumulation of HkSVP was higher in full leaves and stems, consistent with previous reports on SVP in Arabidopsis [12]. However, the expression levels of HkSVP decreased significantly during floral bud differentiation development. It is speculated that this gene plays an important regulatory role in controlling vegetative growth and transforming from vegetative growth to reproductive growth. A similar expression pattern was reported in an SVP homologue from another perennial plant, Rubus idaeus L. [40]. This pattern was consistent with the annual phase transition in woody plants, which does not occur in annual herbaceous plants.
SVP is a flowering repressor in Arabidopsis and reporting functionally delays flowering time. The overexpression of SVP genes from different species has been confirmed to cause alterations in flowering time and floral morphology [41,42,43]. In this research, HkSVP was found to improve the basal inflorescence branch of overexpressed transgenic Arabidopsis. Therefore, it was speculated that the HkSVP gene was primarily related to the control of the inflorescence branches of continuously flowering Hemerocallis. This result coincides with reports in transgenic EgrSVP and PmSVP2 genes in Arabidopsis causing additional inflorescences [10,27], and the overexpression of Narcissus tazetta NtSVP1 in Arabidopsis increased basal inflorescences [25]. However, 35S::HkSVP Arabidopsis showed abnormal floral development and a range of abnormal floral morphologies, such as abnormally enlarged sepals, persistent leafy sepals and an increased density of trichomes, which are similar to the phenotypes of KiSVP, PtSVP, and EgSVP overexpressing Arabidopsis plants [9,10,44]. At the same time, the ectopic expression of HkSVP enhanced the number of flower buds of every single inflorescence on the basal inflorescence. It is speculated that the HkSVP gene plays an important role in prolonging the single flower period and maintaining the vegetative growth of inflorescences. As the upstream regulator of the TFL1 gene, SVP cooperates with the SOC1, AGL24, and SEP4 genes to inhibit the expression of the TFL1 gene to regulate the inflorescence branching pattern of Arabidopsis and further affect the expression of the AP1 and LFY genes. This determines reproductive success, results from coordinated regulatory events mediated by both positive and negative regulators that consistently fine-tune the timing of flowering, and subsequent floral meristem development. Ultimately, this ensures that plants develop appropriate inflorescence architecture [15].
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
The cultivar of Hemerocallis ‘Kanai Sensei’ was field grown at the Xiaotangshan Experimental Center of Beijing Forestry University (116.3°E,40.0°N, Beijing, China). The plant tissues, including stem, leaf, petal, pedicel, stamen, pistil and fruit were collected from the whole mature plant, as well as flower organs at different developmental stages and frozen in liquid nitrogen immediately and then stored at −80 °C before use. The five flower development stages S1–S5 were petal differentiation (S1), stamen differentiation (S2), pistil differentiation (S3), ovule and anther formation (S4), ovule and anthers maturity (S5) based on current report [32]. Arabidopsis ecotype Columbia (Col) and transgenic Arabidopsis plants were grown in a climate incubator (4000 lx) at 22 °C under long day conditions (16 h light/8 h dark).
4.2. Isolation of HkSVP
The cDNA sequence of HkSVP gene was identified from transcriptome of Hemerocallis. The full-length cDNA of the HkSVP was cloned from total RNA of Hemerocallis ‘Kanai Sensei’ young leaves by RT-PCR. The olig d(T)18 was reverse primer, and specific primers HkSVP-F and HkSVP-R (Table S1) were used to PCR amplification by FastPfu Fly DNA Polymerase (TransGen Biotech, Beijing, China), full-length sequences of the gene were obtained. The PCR products were purified and cloned into pEASY-Blunt Cloning Kit (TransGen Biotech) to do DNA sequencing.
4.3. Sequence Alignment and Phylogenetic Analysis of HkSVP
Alignment of cDNA and amino acid sequences were proceeded by BLAST (http://blast.ncbi.nlm.nih.gov/Blast, accessed on 11 June 2019). The conserved domains of HkSVP was predicted by Pfam version 29.0 software (http://pfam.xfam.org/, accessed on 11 June 2019). The full-length amino acid sequence of HkSVP protein in Hemerocallis was aligned with orthologs protein sequences from various plants through ClustalW2 software. The physical and chemical parameters of HkSVP were predicted by ProtParam (https://web.expasy.org/protparam/, accessed on 12 June 2019). The phylogenetic trees were constructed by MEGA 7.0 software, and bootstrap values were derived from 1000 replicates, and the cutoff value was set to 50%.
4.4. Quantitative Real-Time PCR (qRT-PCR) Analysis
The tissue specific expression patterns of HkSVP from different developmental periods was analyzed by quantitative real-time PCR (qRT-PCR). For the spatial expression patterns of HkSVP, floral organs (petal, stamen and pistil), leaf, pedicel, stem and fruit were sampled. For the expression pattern of HkSVP in different flowering stages, floral bud were sampled at S1–S5 stage [32].
A one mg aliquot of total RNA treated with 5U DNase I (Invitrogen, Carlsbad, CA, USA) was used for reverse-transcription by PrimeScript RT Reagent Kit (TaKaRa, Beijing, China). Quantitative real-time RT-PCR (qRT-PCR) was carried out using CFX Connect System (Bio-Rad, Hercules, CA, USA) with SYBR Premix EX TaqTM II (TaKaRa). The specific primers HkSVP-QF and HkSVP-QR (Table S1) to detect the gene expression levels were designed by Primer Premier 5.0 software. Hemerocallis Actin-F and Actin-R gene were used to as an internal control [32]. To estimate the transcript levels of flowering-related genes in transgenic and wild-type Arabidopsis by qRT-PCR, aerial parts of different growth cycles Arabidopsis plants were harvested before inflorescences were formed. Specific primers for flowering-related genes were used for qRT-PCR, with the AtUBQ gene as an internal reference. All the primers used for qRT-PCR in the present study are listed in Table S1. Each sample had three biological replicates, while had three technical replicates for each biological replicate. The cycle threshold 2−△△Ct based method was used to calculate the relative gene expression [45].
4.5. Subcellular Localization of the HkSVP
The pAN580-GFP and pROKII-GFP expression vector carrying the CaMV 35S promoter were digested with restriction enzymes XbaI and BamHI. The full-length HkSVP ORF (without a stop codon) amplified with primers HkSVP-SF and HkSVP-SR (Table S1) was ligated into the linearized pAN580-35S::HkSVP:GFP and pROKII-35S::HkSVP:GFP vector according to the homologous recombination method of the In-Fusion HD Cloning Kit (Takara). The construct without HkSVP (35S::GFP) was used as a control. The constructs were transformed into the Arabidopsis protoplasts according to established protocol [46]. The fusion of pAN580-HkSVP fluorescence and nuclear marker fluorescence was observed under laser confocal microscope (Zeiss LSM 710, Zeiss, Oberkoche, Germany). The tests of subcellular localization were repeated in onion epidermal cells. The pROKII-HkSVP-GFP fusion plasmid was transferred into onion epidermal cells by GJ-1000 Gene Gun System (Scientz, Beijing, China). The fluorescence signal was observed with a laser confocal microscope (Zeiss LSM 710).
4.6. Yeast Two-Hybrid Analyses
Protein interactions of HkSVP, HkAP1 (GenBank accession No. MG957240) and HkTFL1 (GenBank accession No. KY569646) were screened using the ProQuest two hybrid system (Invitrogen). Briefly, the coding sequence region of HkSVP, HkAP1 and HkTFL1 were constructed into pDEST22 as the prey vector, and the coding sequence region of HkSVP, HkAP1 and HkTFL1 were constructed into pDEST32 as the bait vector. pDEST33-SVP/AP1/TFL1 plasmid and pDEST22-SVP/AP1/TFL1 were co-transformed into MaV203 yeast cells. All protocols were carried out strictly according to the manufacturer’s user manual. Primers used are listed in Supplemental Table S1.
4.7. Generation of Transgenic Arabidopsis Plants
The coding sequencing reigon of HkSVP gene amplified with primers HkSVP-OE-F and HkSVP-OE-R was introduced into binary T-DNA vector pROKII by BamHI and KpnI, that was driven by the 35S promoter. The construct was transformed into Agrobacterium GV3101 competent cells using a freeze-thaw technique [47]. The Arabidopsis thaliana (Col-0) was used to transformation by the floral dipping method [48,49]. The resistant seedlings werer screened by 50 mg/L kanamycin-labeled and identified by PCR of HkSVP specific primers. The transgenic plants were transplanted into sterilized soil until harvest T3 generation seeds. Flowering time was measured by scoring the number of rosette leaves when the height of scape was 1 cm. All transgenic lines included 10 plants for statistics.
5. Conclusions
In this study, HkSVP gene was isolated and identified from Hemerocallis. The temporal and tissue expression analysis showed that the HkSVP was plays an important regulatory role in controlling vegetative growth and transforming from vegetative growth to reproductive growth. A yeast two hybrid assays showed that HkSVP protein could interact with the flowering-related proteins HkAP1, HkTFL1. Furthermore, HkSVP was found to improve the basal inflorescence branch of overexpressed transgenic Arabidopsis, and showed abnormal floral development and a range of abnormal floral morphologies. The data presented herein may provide new insights into the regulatory mechanisms of flower organ development and inflorescence structure of Hemerocallis and may be useful for comprehensive analyses of flower opening.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms222112010/s1.
Author Contributions
Y.G., Y.L. and Q.Z. designed and conceived the experiment. Y.L. conducted vector construction, transformation and molecular characterization of mutants. Y.L., Y.G. and L.Y. analyzed the data. Y.L. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31770736), Natural Science Foundation of Heilongjiang Province of China (YQ2021C014), the “Young Talents” Project of Northeast Agricultural University, China (20QC04).
Institutional Review Board Statement
No applicable.
Informed Consent Statement
No applicable.
Data Availability Statement
No applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wellmer, F.; Riechmann, J.L. Gene networks controlling the initiation of flower development. Trends Genet. 2010, 26, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.R.G.; Ai, X.-Y.; Zhang, J.-Z. Genetic regulation of flowering time in annual and perennial plants. Wiley Interdiscip. Rev. RNA 2013, 5, 347–359. [Google Scholar] [CrossRef]
- Samach, A.; Onouchi, H.; Gold, S.E.; Ditta, G.S.; Schwarz-Sommer, Z.; Yanofsky, M.F.; Coupland, G. Distinct Roles of CONSTANS Target Genes in Reproductive Development of Arabidopsis. Science 2000, 288, 1613–1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blazquez, M.; Weigel, D. Integration of floral inductive signals in Arabidopsis. Nature 2000, 404, 889–892. [Google Scholar] [CrossRef]
- Haghighi, R.; Tabatabaei, B.E.S.; Maibody, S.A.M.M.; Talebi, M.; Molina, R.V.; Nebauer, S.G.; Renau-Morata, B. A flowering inhibitor of the temperature-dependent pathway in Crocus sativus L. Mol. Biol. Rep. 2020, 47, 2171–2179. [Google Scholar] [CrossRef]
- Lee, J.H.; Yoo, S.J.; Park, S.H.; Hwang, I.; Ahn, J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007, 21, 397–402. [Google Scholar] [CrossRef] [Green Version]
- Gregis, V.; Andrés, F.; Sessa, A.; Guerra, R.F.; Simonini, S.; Mateos, J.L.; Torti, S.; Zambelli, F.; Prazzoli, G.M.; Bjerkan, K.N.; et al. Identification of pathways directly regulated by short vegetative phase during vegetative and reproductive development in Arabidopsis. Genome Biol. 2013, 14, 26. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Xu, Y.; Tan, E.L.; Kumar, P.P. AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc. Natl. Acad. Sci. USA 2002, 99, 16336–16341. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.-M.; Zhang, J.-Z.; Mei, L.; Deng, X.-X.; Hu, C.-G.; Yao, J.-L. PtSVP, an SVP homolog from trifoliate orange (Poncirus trifoliata L. Raf.), shows seasonal periodicity of meristem determination and affects flower development in transgenic Arabidopsis and tobacco plants. Plant Mol. Biol. 2010, 74, 129–142. [Google Scholar] [CrossRef]
- Brill, E.M.; Watson, J.M. Ectopic expression of a Eucalyptus grandis SVP orthologue alters the flowering time of Arabidopsis thaliana. Funct. Plant Biol. 2004, 31, 217–224. [Google Scholar] [CrossRef]
- Yu, H.; Ito, T.; Wellmer, F.; Meyerowitz, E.M. Repression of agamous-like 24 is a crucial step in promoting flower development. Nat. Genet. 2004, 36, 157–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartmann, U.; Hohmann, S.; Nettesheim, K.; Wisman, E.; Saedler, H.; Huijser, P. Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J. 2000, 21, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Gregis, V.; Sessa, A.; Colombo, L.; Kater, M.M. AGL24, Short vegetative phase, and apetala1 redundantly Control agamous during Early Stages of Flower Development in Arabidopsis. Plant Cell 2006, 18, 1373–1382. [Google Scholar] [CrossRef] [Green Version]
- Gregis, V.; Sessa, A.; Colombo, L.; Kater, M.M. Agamous-like24 and short vegetative phase determine floral meristem identity in Arabidopsis. Plant J. 2008, 56, 891–902. [Google Scholar] [CrossRef]
- Liu, C.; Teo, Z.W.N.; Bi, Y.; Song, S.; Xi, W.; Yang, X.; Yin, Z.; Yu, H. A Conserved Genetic Pathway Determines Inflorescence Architecture in Arabidopsis and Rice. Dev. Cell 2013, 24, 612–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Liu, C.; Shen, L.; Wu, Y.; Chen, H.; Robertson, M.; Helliwell, C.; Ito, T.; Meyerowitz, E.; Yu, H. A Repressor Complex Governs the Integration of Flowering Signals in Arabidopsis. Dev. Cell 2008, 15, 110–120. [Google Scholar] [CrossRef] [Green Version]
- Mao, L.; Begum, D.; Chuang, H.-W.; Budiman, M.A.; Szymkowiak, E.J.; Irish, E.E.; Wing, R. Jointless is a MADS-box gene controlling tomato flower abscissionzone development. Nature 2000, 406, 910–913. [Google Scholar] [CrossRef]
- Jaudal, M.; Monash, J.; Zhang, L.; Wen, J.; Mysore, K.S.; Macknight, R.; Putterill, J. Overexpression of Medicago SVP genes causes floral defects and delayed flowering in Arabidopsis but only affects floral development in Medicago. J. Exp. Bot. 2013, 65, 429–442. [Google Scholar] [CrossRef] [Green Version]
- Wu, R.; Wang, T.; McGie, T.; Voogd, C.; Allan, A.C.; Hellens, R.P.; Varkonyi-Gasic, E. Overexpression of the kiwifruit SVP3 gene affects reproductive development and suppresses anthocyanin biosynthesis in petals, but has no effect on vegetative growth, dormancy, or flowering time. J. Exp. Bot. 2014, 65, 4985–4995. [Google Scholar] [CrossRef] [Green Version]
- Schmitz, J.; Franzen, R.; Ngyuen, T.H.; Garcia-Maroto, F.; Pozzi, C.; Salamini, F.; Rohde, W. Cloning, mapping and expression analysis of barley MADS-box genes. Plant Mol. Biol. 2000, 42, 899–913. [Google Scholar] [CrossRef]
- Trevaskis, B.; Tadege, M.; Hemming, M.N.; Peacock, W.J.; Dennis, E.S.; Sheldon, C. Short Vegetative Phase-Like MADS-Box Genes Inhibit Floral Meristem Identity in Barley. Plant Physiol. 2006, 143, 225–235. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Park, S.H.; Lee, J.S.; Ahn, J.H. A conserved role of short vegetative phase (SVP) in controlling flowering time of Brassica plants. Biochim. Biophys. Acta Gene Struct. Expr. 2007, 1769, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, R.; Sage-Ono, K.; Kamada, H.; Handa, H.; Ono, M. PnMADS1, encoding an StMADS11-clade protein, acts as a repressor of flowering in Pharbitis nil. Physiol. Plant. 2008, 133, 786–793. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xue, J.; Ahmadi, N.; Holloway, P.; Zhu, F.; Ren, X.; Zhang, X. Molecular characterization and expression patterns of PsSVP genes reveal distinct roles in flower bud abortion and flowering in tree peony (Paeonia suffruticosa). Can. J. Plant Sci. 2014, 94, 1181–1193. [Google Scholar] [CrossRef] [Green Version]
- Li, X.-F.; Wu, W.-T.; Zhang, X.-P.; Qiu, Y.; Zhang, W.; Li, R.; Xu, J.; Sun, Y.; Wang, Y.; Xu, L. Narcissus tazetta SVP-like gene NSVP1 affects flower development in Arabidopsis. J. Plant Physiol. 2015, 173, 89–96. [Google Scholar] [CrossRef]
- Masiero, S.; Li, M.; Will, I.; Hartmann, U.; Saedler, H.; Huijser, P.; Schwarz-Sommer, Z.; Sommer, H. Incomposita: A MADS-box gene controlling prophyll development and floral meristem identity in Antirrhinum. Development 2004, 131, 5981–5990. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhou, Y.; Yang, W.; Cheng, T.; Wang, J.; Zhang, Q. Isolation and functional characterization of SVP-like genes in Prunus mume. Sci. Hortic. 2017, 215, 91–101. [Google Scholar] [CrossRef]
- Mo, X.; Luo, C.; Yu, H.; Chen, J.; Liu, Y.; Xie, X.; Fan, Z.; He, X. Isolation and Functional Characterization of Two Short Vegetative Phase Homologous Genes from Mango. Int. J. Mol. Sci. 2021, 22, 9802. [Google Scholar] [CrossRef]
- Yuan, L.; Gao, Y.; Zhu, L.; Zhang, Q. Study on the cross breeding of continuous reblooming daylilies. J. China Agric. Univ. 2018, 23, 49–58. [Google Scholar]
- Danilevskaya, O.N.; Meng, X.; Ananiev, E.V. Concerted Modification of Flowering Time and Inflorescence Architecture by Ectopic Expression of TFL1-Like Genes in Maize. Plant. Physiol. 2010, 153, 238–251. [Google Scholar] [CrossRef] [Green Version]
- Hirota, S.K.; Nitta, K.; Suyama, Y.; Kawakubo, N.; Yasumoto, A.A. Pollinator-Mediated Selection on Flower Color, Flower Scent and Flower Morphology of Hemerocallis: Evidence from Genotyping Individual Pollen Grains on the Stigma. PLoS ONE 2015, 10, e0117885. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Gao, Y.; Yuan, L.; Zhang, Q. Functional characterization and spatial interaction of terminal flower 1 in Hemerocallis. Sci. Hortic. 2019, 253, 154–162. [Google Scholar] [CrossRef]
- Li, L.; He, Y.; Zhang, Z.; Shi, Y.; Zhang, X.; Xu, X.; Wu, J.-L.; Tang, S. OsNAC109 regulates senescence, growth and development by altering the expression of senescence- and phytohormone-associated genes in rice. Plant Mol. Biol. 2021, 105, 637–654. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teo, Z.W.N.; Song, S.; Wang, Y.-Q.; Liu, J.; Yu, H. New insights into the regulation of inflorescence architecture. Trends Plant Sci. 2014, 19, 158–165. [Google Scholar] [CrossRef] [PubMed]
- De Folter, S.; Immink, R.; Kieffer, M.; Pařenicová, L.; Henz, S.R.; Weigel, D.; Busscher, M.; Kooiker, M.; Colombo, L.; Kater, M.; et al. Comprehensive Interaction Map of the Arabidopsis MADS Box Transcription Factors. Plant Cell 2005, 17, 1424–1433. [Google Scholar] [CrossRef] [Green Version]
- Pelaz, S.; Gustafson-Brown, C.; Kohalmi, S.E.; Crosby, W.L.; Yanofsky, M.F. APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 2001, 26, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S.; Kennedy, A.; Pan, S.; Jermiin, L.S.; Melzer, R. Genome-wide analysis of MIKC -type MADS -box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. N. Phytol. 2019, 225, 511–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, G.G.; Dean, C. Arabidopsis, the Rosetta Stone of Flowering Time? Science 2002, 296, 285–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzitelli, L.; Hancock, R.D.; Haupt, S.; Walker, P.G.; Pont, S.D.A.; McNicol, J.; Cardle, L.; Morris, J.; Viola, R.; Brennan, R.; et al. Co-ordinated gene expression during phases of dormancy release in raspberry (Rubus idaeus L.) buds. J. Exp. Bot. 2007, 58, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Fornara, F.; Gregis, V.; Pelucchi, N.; Colombo, L.; Kater, M. The rice StMADS11-like genes OsMADS22 and OsMADS47 cause floral reversions in Arabidopsis without complementing the svp and agl24 mutants. J. Exp. Bot. 2008, 59, 2181–2190. [Google Scholar] [CrossRef] [Green Version]
- Ramamoorthy, R.; Phua, E.-E.K.; Lim, S.-H.; Tan, H.T.-W.; Kumar, P.P. Identification and Characterization of RcMADS1, an AGL24 Ortholog from the Holoparasitic Plant Rafflesia cantleyi Solms-Laubach (Rafflesiaceae). PLoS ONE 2013, 8, e67243. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhou, J.; Bracha-Drori, K.; Yalovsky, S.; Ito, T.; Yu, H. Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 2007, 134, 1901–1910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, R.-M.; Walton, E.F.; Richardson, A.; Wood, M.; Hellens, R.; Varkonyi-Gasic, E. Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J. Exp. Bot. 2011, 63, 797–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Sun, J.; Xia, H.; Zhao, C.; Hou, L.; Wang, B.; Li, A.; Chen, M.; Zhao, S.; Wang, X. Characterization of peanut phytochromes and their possible regulating roles in early peanut pod development. PLoS ONE 2018, 13, e0198041. [Google Scholar] [CrossRef] [PubMed]
- Sheen, J.; Hwang, S.; Niwa, Y.; Kobayashi, H.; Galbraith, D.W. Green-fluorescent protein as a new vital marker in plant cells. Plant J. 1995, 8, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Nelson, R.S.; Sherwood, J.L. Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. BioTechniques 1994, 16, 664–668. [Google Scholar] [PubMed]
- Clough, S.J.; Bent, A. Floral dip: A simplified method forAgrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Harrison, S.J.; Mott, E.K.; Parsley, K.; Aspinall, S.; Gray, J.C.; Cottage, A. A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2006, 2, 19. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).