Abstract
Proteins targeted to the secretory pathway start their intracellular journey by being transported across biological membranes such as the endoplasmic reticulum (ER). A central component in this protein translocation process across the ER is the Sec61 translocon complex, which is only intracellularly expressed and does not have any enzymatic activity. In addition, Sec61 translocon complexes are difficult to purify and to reconstitute. Screening for small molecule inhibitors impairing its function has thus been notoriously difficult. However, such translocation inhibitors may not only be valuable tools for cell biology, but may also represent novel anticancer drugs, given that cancer cells heavily depend on efficient protein translocation into the ER to support their fast growth. In this review, different inhibitors of protein translocation will be discussed, and their specific mode of action will be compared. In addition, recently published screening strategies for small molecule inhibitors targeting the whole SRP-Sec61 targeting/translocation pathway will be summarized. Of note, slightly modified assays may be used in the future to screen for substances affecting SecYEG, the bacterial ortholog of the Sec61 complex, in order to identify novel antibiotic drugs.
1. Introduction
With the evolution of simple cellular structures to multi organelle compartmentalized cells, the transport of proteins across biological membranes has become an unavoidable challenge. Extracellular and integral membrane proteins—synthesized in the cytosol—need to be translocated either across or integrated into bilipid membranes, in order to reach their final destination. Since the discovery of the secretory pathway [1,2,3,4,5], numerous studies have shed light on the different targeting signals, translocation modes, and pathways used by proteins to cross the endoplasmic reticulum (ER) membrane, which is the first and decisive step in the secretory pathway for protein biogenesis (see Figure 1) [6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22]. After maturation in the ER lumen, the proteins are embedded in vesicles and travel through the Golgi apparatus to the cell membrane. Here, the vesicles fuse with the cell membrane, resulting in the expression of the membrane protein at the cell surface or in the secretion of the soluble protein into the extracellular environment.
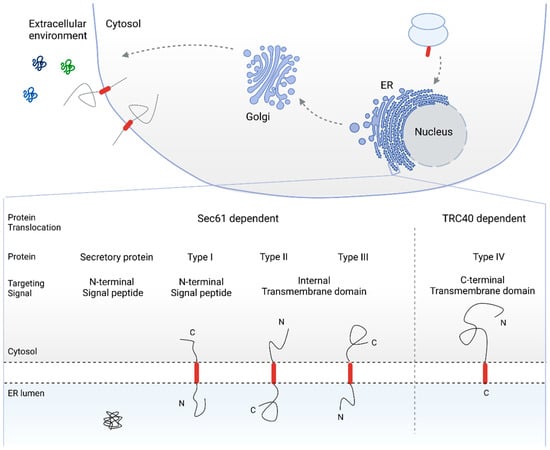
Figure 1.
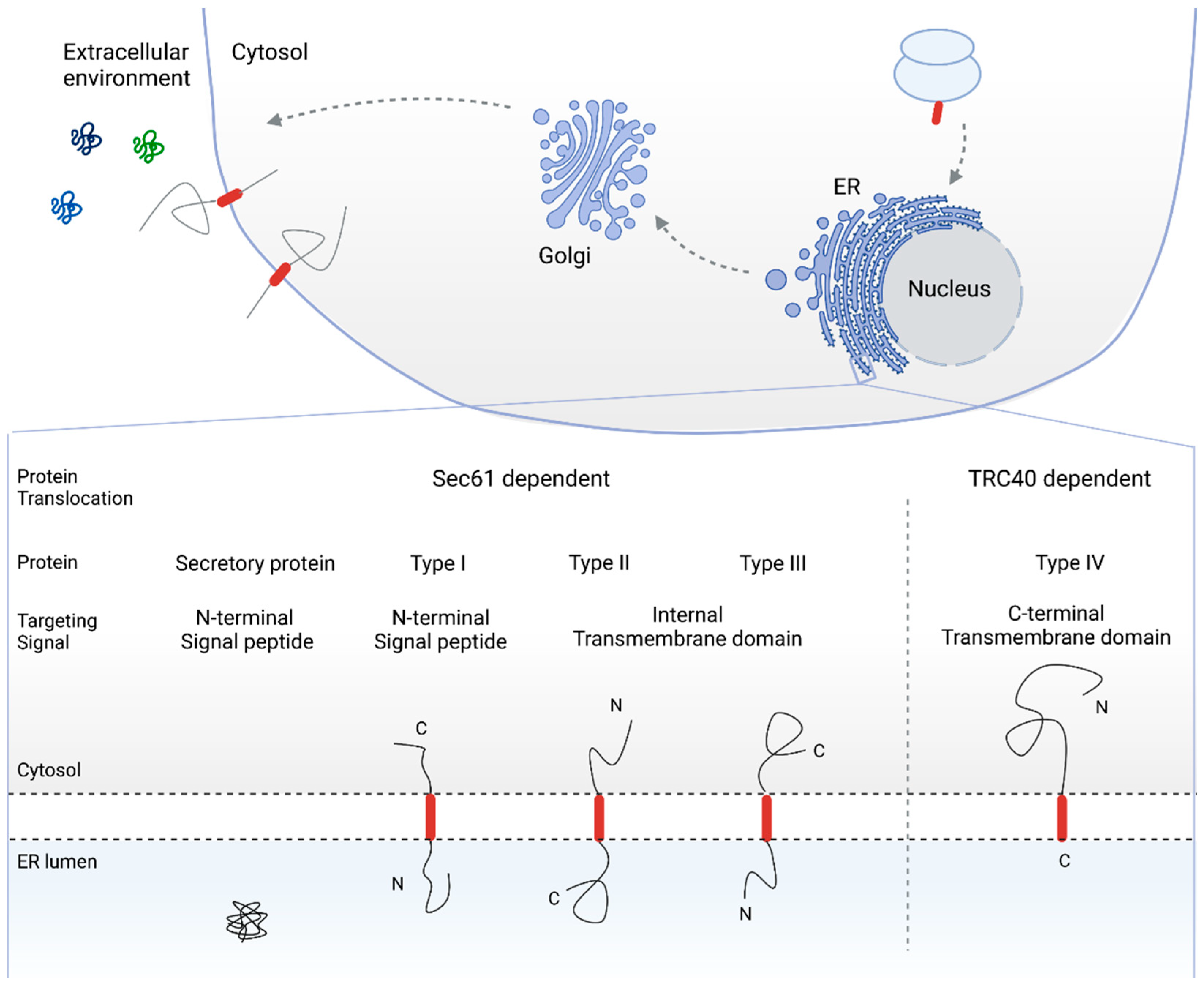
Overview of the secretory pathway for protein biogenesis and topology-based classification of secretory and single pass membrane proteins. Cleavable N-terminal SPs target secretory and type I membrane proteins to the ER membrane, resulting in an N-terminally translocated topology. In the case of type II and type III membrane proteins, the TMD functions as a targeting sequence. Depending on the overall hydrophobicity and charge of the topological sequences adjacent to the TMD, the C- or N-terminal end of the protein is translocated into the ER lumen (type II and type III single pass membrane protein, respectively). Type IV, or TA proteins, are targeted to the ER membrane via the C-terminal TMD. As a result, TA proteins are post-translationally translocated via the TRC-40 pathway. SP: signal peptide, ER: endoplasmic reticulum, TMD: transmembrane domain, TA: tail-anchored, TRC-40: transmembrane recognition complex subunit of 40 kDa.
The targeting signals that drive proteins toward the secretory pathway include N-terminal (cleavable) signal peptides (SPs) as well as transmembrane domains (TMDs). As SPs and TMDs are intrinsic targeting signals for the Sec61 dependent pathway for protein co- and post-translational translocation [8,14,23,24,25,26,27], C-terminally located targeting signals route the respective protein to a different translocation pathway. In tail-anchored (TA) proteins, for instance, the TMD serves as the ER membrane targeting signal. The specific C-terminal location of the targeting TMD, however, restrict TA proteins to the transmembrane recognition complex subunit of the 40 kDa (TRC40) pathway for post-translational translocation as the targeting TMD emerges from the ribosome only when translation is completed [22,28,29,30,31,32,33,34]. In fact, single pass membrane proteins are often classified based on their targeting signal and topology after ER translocation (see Figure 1).
Evidenced by the evolutionary conservation of the translocation pathways over the different domains of life, correct translocation of proteins across the ER membrane is essential for the proper functioning of cells [21,35,36,37].
Small molecule inhibitors have therefore become an attractive tool to gain insights into the complex multistep nature of the different translocation routes known today [38,39,40,41,42]. However, new insights in this domain have quickly arisen. In this review, we update the previous knowledge of Sec61 dependent protein translocation in higher eukaryotes, and discuss the newest insights and mode of action of specific translocation inhibitors. Additional focus is placed on the interaction sites of each inhibitor within the translocation complex and screening methods to identify novel signal recognition particle (SRP)-Sec61 specific pathway inhibitors.
2. The Sec61 Dependent Pathway for Co- and Post-Translational Protein Translocation
The Sec61 dependent translocation can occur in two modes (i.e., co- or post-translational translocation). Intrinsic to the terms, they refer to the translocation process occurring simultaneous with or after completion of protein translation, respectively. Post-translational translocation is most commonly used by small secretory proteins (SSP) (less than approximately 100 amino acid residues) and is best understood in fungi and bacteria. Co-translational translocation of proteins over the ER membrane, on the other hand, is a complex multistep process, as shown in Figure 2, which is mostly employed by higher eukaryotes [16,21,25,43,44,45].
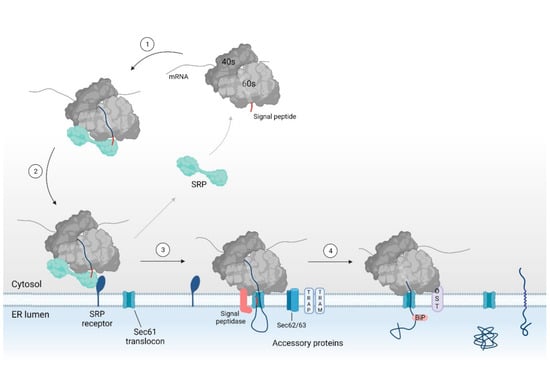
Figure 2.
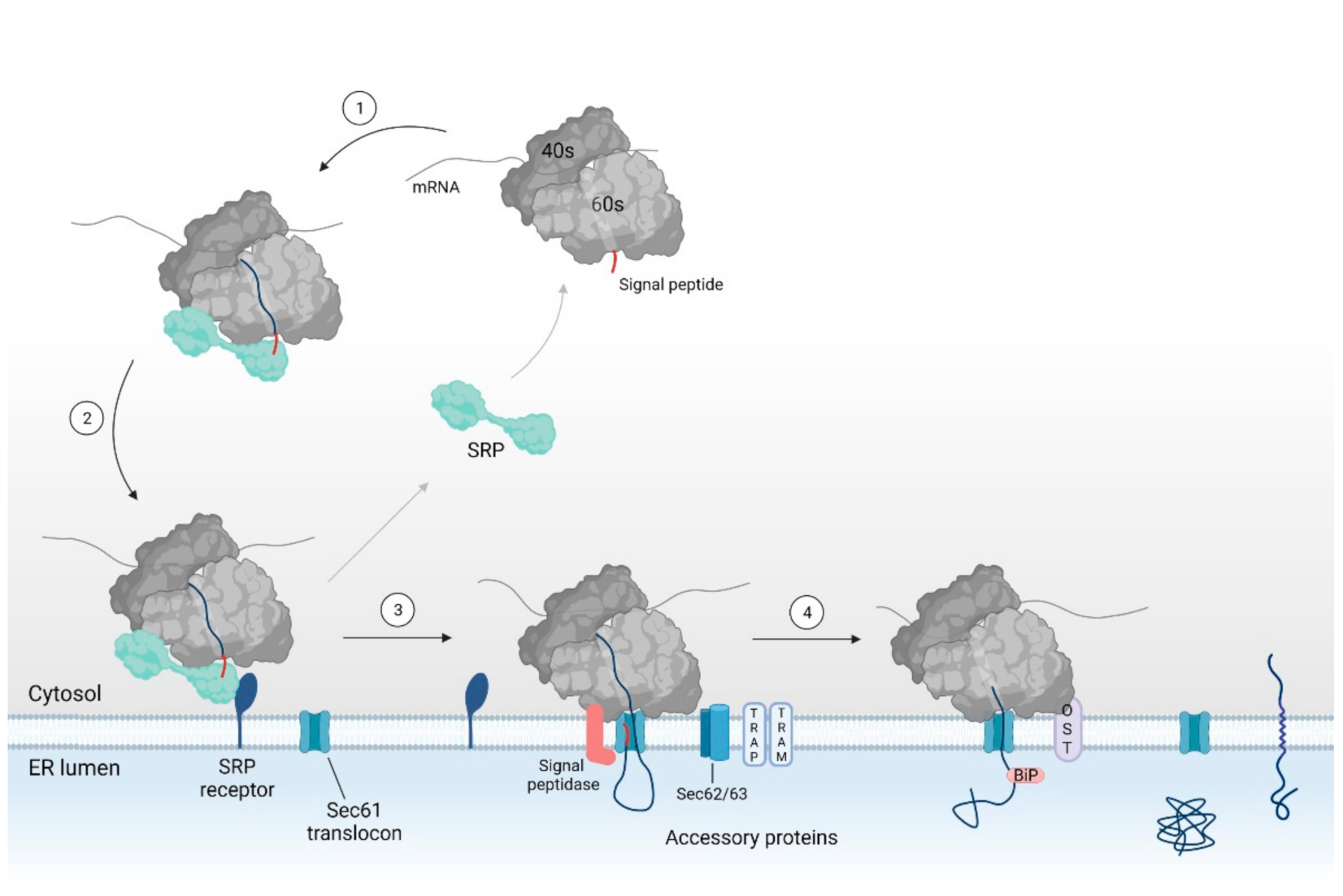
Overview of the SRP dependent pathway for co-translational translocation via the Sec61 translocon. A secretory or integral membrane protein is targeted toward the ER membrane by means of SRP binding to the signal sequence (i.e., the SP or TMD (steps 1–2)). SRP binding stalls protein translation to keep the nascent chain in a translocation competent state. At the ER membrane, SRP interacts with the SRP receptor. The RNC complex is then transferred to the Sec61 translocon (step 3). Interaction of the ribosome with the translocon reinitiates translation and induces conformational changes within Sec61α, eventually leading to protein translocation. In the case of a weak hydrophobic SP or TMD, the protein requires help from accessory proteins such as TRAP, TRAM, Sec62, and/or Sec63 for protein translocation. In the ER lumen, the SP is cleaved by the signal peptidase complex and the protein is glycosylated by the OST complex (step 4). SRP: signal recognition particle, ER: endoplasmic reticulum, SP: signal peptide, TMD: transmembrane domain, RNC: ribosomal nascent chain, TRAP: translocon-associated protein, TRAM: translocating chain-associating membrane protein, OST: oligosaccharyl transferase.
In short, when the targeting signal emerges from the ribosome, it is recognized and bound to by SRP. Upon binding of SRP, protein translation is stalled and the ribosome-nascent chain (RNC) complex is targeted to the ER membrane through binding of SRP to its receptor [46,47,48,49,50]. Next, the targeting sequence interacts with the Sec61 translocon (i.e., the protein conducting channel). Binding of the ribosome to the translocon reinitiates translation of the nascent chain and induces translocation of the preprotein into the ER lumen [27,49,50,51,52,53,54]. In the lumen, the signal peptidase and oligosaccharyl transferase (OST) complex allow for further maturation of the translocated preprotein by cleaving the protein’s signal peptide and by glycosylation of the mature protein part, respectively [55,56,57,58,59].
SRP mediated protein targeting to the ER membrane is the most common in eukaryotes and therefore forms the focus of this review. However, proteins can also be SRP independently targeted to the ER membrane, in which case specific chaperone activity is required. For an overview of SRP-independent pathways for protein targeting to the ER, the reader is referred to other publications [11,12,16,43,60,61,62,63].
2.1. SRP Dependent Protein Targeting to the ER Membrane Keeps the Protein in a Translocation Competent State
When a secretory or integral membrane protein is translated in the cytosol and the targeting signal (i.e., SP or TMD) emerges from the ribosomal exit tunnel, it is recognized and bound to by SRP [46,47,64,65,66,67,68] (see Figure 2). SRP is a ribonucleoprotein complex consisting of six subunits (SRP9, 14, 19, 54, 68 and 72 m) and a 7S RNA molecule, which assemble into two SRP domains [22,46,48]. SRP 19, SRP54, SRP68, and SRP72 as well as the majority of the SRP RNA make up the S domain of SRP, which holds the recognition and binding site for the emerging SP. The remaining two proteins SRP9 and SRP14 as well as the 5′ and 3′ end of the RNA molecule form the Alu domain of SRP [46]. The Alu domain interacts with the ribosome elongation site, resulting in the transient retardation of protein translation [46,47,48]. Hence, SRP binding to the RNC complex locks the nascent chain in a translocation competent (i.e., unfolded) state by inducing a translational arrest.
Next, the RNC complex is targeted toward the ER membrane. Here, SRP interacts with the SRP receptor (SR) [46,47,64,65,66,67,68]. The SR then mediates the transfer of the RNC complex to the Sec61 translocon, the central component, and protein-conducting channel of the Sec61 dependent pathway for protein translocation [22,46,47,48].
2.2. Binding of the RNC Complex Induces Dynamic Conformational Changes in the Translocon
The Sec61 translocon is a heterotrimeric complex that consists of Sec61α, β, and γ monomers (see Figure 3). The Sec61α subunit, composed of ten transmembrane helices (TMH), forms the central pore of the translocon [27,51,52,53,54,69,70]. In the quiescent, or native state, the translocon is axially closed by a lumenal plug domain in the central pore of the complex (see Figure 3, depicted as a single helix in red). In addition, the translocon is also laterally sealed by the lateral gate formed by the interhelical interactions between TMH2 and TMH3 (blue helices in Figure 3) and TMH7 and TMH8 (green helices in Figure 3) [51,52,53]. The interface between TMH2 and TMH7 near the cytosolic side of the translocon also serves as the recognition site for the targeting sequence of the protein nascent chain [27].
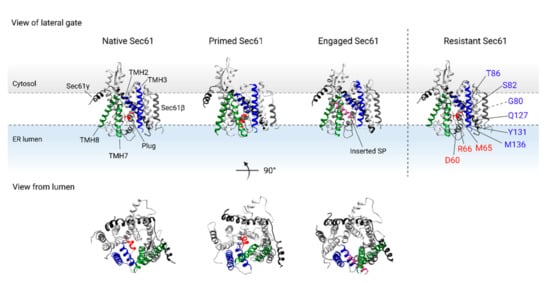
Figure 3.
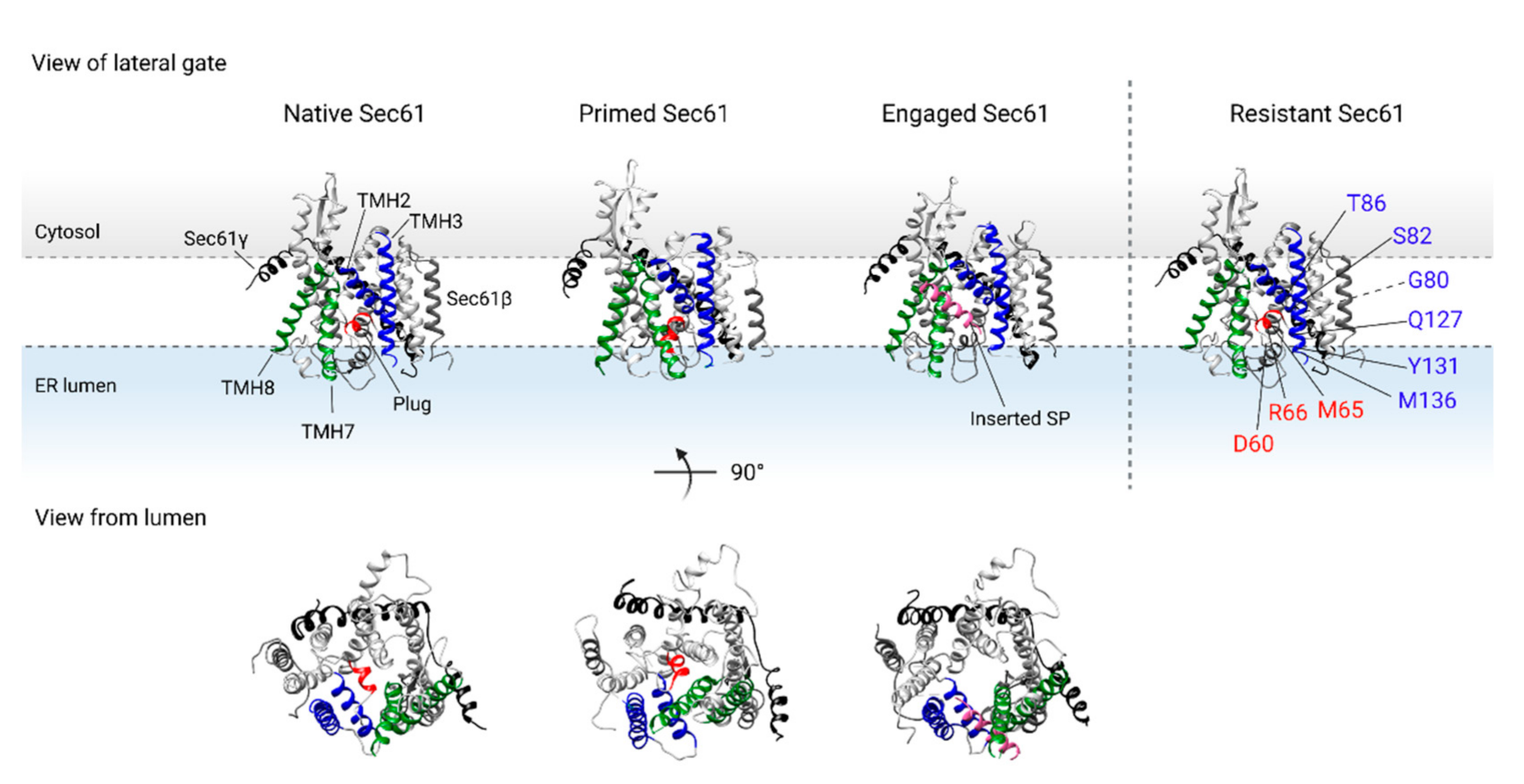
Dynamics of the TMHs of the Sec61 translocon (PDB 5A6U [69]) upon binding of the ribosome (primed state, PDB 3J7Q [52]) and insertion of the SP (engaged state, PDB 3JC2 [27]). Sec61α is shown in grey, Sec61β is shown in dark grey, and Sec61γ is shown in black. The interhelical interaction between TMH2 and TMH3 (shown in blue) on one half of the translocon, and TMH7 and TMH8 (shown in green) on the other half of the translocon form the lateral gate of Sec61α. Additionally, the translocon is closed axially by the lumenal plug domain of TMH2 (shown in red). Binding of the ribosome disrupts the interaction of TMH3 and TMH8 of the lateral gate and primes the translocon for insertion of the nascent protein chain, while the plug domain remains in place. The SP of the nascent chain (shown in pink) interacts with the lateral gate of the translocon, resulting in lateral escape from the translocon and insertion of the TMD into the ER membrane. In addition, the plug domain is displaced to allow for protein translocation into the ER lumen. Resistance conferring mutations located in the lateral gate or plug domain of Sec61α are shown in ‘Resistant Sec61’. TMH: transmembrane helix, SP: signal peptide, TMD: transmembrane domain, ER: endoplasmic reticulum.
Structural studies have shown that binding of the RNC complex to the translocon triggers dynamic conformational changes within Sec61α, resulting in the interrupted interhelical contact between the lateral gate TMH3 and TMH8 (see Figure 3) ‘primed Sec61’ [35,55,56,57,58,59]. Interestingly, the position of the plug domain, which seals the translocon on the lumenal side of the ER membrane, is almost unaltered upon ribosome binding [51,70]. Hence, ribosome binding to the Sec61 translocon reinitiates protein translation by the release of SRP, and primes the translocon to accept an incoming nascent chain.
The inserting nascent chain can then interact with the recognition site in the lateral gate, which further opens the lateral gate, and displaces the plug domain so that the translocon is opened toward the lipid bilayer for TMD insertion, and toward the lumen for protein translocation [14,27,51,53,54,71].
2.3. Assisted Opening of the Sec61 Translocon
With the rise in structural models explaining the dynamic interactions of the Sec61 translocon upon protein insertion, it has become clear that the hydrophobic strength of the targeting signal is crucial for protein translocation. After all, the SP and/or TMD needs to be sufficiently hydrophobic to disrupt the interhelical hydrophobic interaction between the TMHs of the lateral gate to open the translocon for lateral escape into the ER membrane [27,51,52,69]. In addition, the SP and/or TMD need to displace the plug domain in order for the protein to translocate over the ER membrane.
Hence, proteins with a—so-called—weak hydrophobic SP and/or TMD require additional accessory components such as the translocon-associated protein (TRAP), translocating chain-associated membrane protein (TRAM), Sec62, and/or Sec63 for the translocation into the ER lumen. The specific accessory translocation machinery that is required, is thought to be protein, and thus SP/TMD specific [14,54,71,72,73,74,75,76,77,78,79].
2.4. Chaperone Mediated Completion of Protein Translocation and Post-Translational Modifications in the ER Lumen
For the translocation of the last amino acid residues that remain in the ribosomal exit tunnel when translation is completed, proteins rely on the binding immunoglobin protein (BiP), a lumenal translocation chaperone. BiP acts as a molecular ratchet by binding to the preprotein and pulling it toward the ER lumen to complete translocation in an ATP dependent manner [80,81,82,83]. Once translocated, the proteins are post-translationally modified in the ER lumen. For instance, the SP is cleaved from the preprotein by the signal peptidase complex and the preprotein is glycosylated by the oligosaccharyl-transferase (OST) complex [55,57,58,59].
3. Translocation Inhibitors of the Sec61 Dependent Protein Translocation Pathway
Being a multistep process, ER protein transport provides many pitfalls for protein mis-translocation that are mostly corrected by cellular control systems and specialized clean-up pathways such as ER associated protein degradation (ERAD) [84,85,86,87,88,89]. The correct translocation of proteins is crucial for the proper functioning of cells. In fact, inefficient protein translocation has been linked to many liver, kidney, and metabolic diseases [36,90]. Cancer cells, on the other hand, depend heavily on efficient protein translocation into the ER to support their fast growth. As such, correct protein translocation is key for many fast-growing cancers [37,91,92,93,94,95,96,97]. In addition, viruses exploit the host ER protein translocation machinery for the synthesis of viral proteins and host related entry receptors [98,99,100,101,102,103]. It is therefore no surprise that different inhibitors have been identified that interact with the Sec61 dependent protein translocation process.
The inhibitors known today are natural products and synthetic small molecules that inhibit Sec61 dependent protein translocation with differential substrate selectivity. Evidenced by the fact that many inhibitors originate from therapeutic screening programs, the Sec61 translocon forms a promising target for therapeutic intervention (e.g., for anticancer, immunosuppressive, and/or antiviral treatment).
In the following section, we present an overview of the Sec61 inhibitors of protein translocation known today, with a focus on the discovery, structure–activity relationship (SAR), therapeutic activity, and (putative) interaction sites within the Sec61 translocon.
3.1. Sec61 Inhibitors of Natural Origin
3.1.1. HUN7293, CAM741, and Cotransin
Cell adhesion molecules play a critical role in the immune response by regulating leucocyte migration and cell-to-cell interaction at the site of inflammation. Therefore, the expression of cell adhesion molecules has become an interesting therapeutic target in a variety of inflammatory and autoimmune diseases that are characterized by the overexpression of cell adhesion molecules [104]. With this rationale, a screening program for the inhibition of cell adhesion molecule expression was set up and led to the identification of HUN-7293 [105]. HUN-7293 is a fungal cyclic heptadepsipeptide that selectively inhibits the expression of three cell adhesion molecules (i.e., vascular cell adhesion molecule 1 (VCAM-1), intracellular adhesion molecule 1 (ICAM-1), and E-selectin) [104,105]. With the HUN-7293 compound as the lead molecule of this new class of therapeutic agents, a complete library of analogs was synthesized to study the SAR [105,106,107]. This led to the identification of new inhibitors of cell adhesion molecule expression that have eventually also paved the way to study Sec61 dependent protein translocation [105,108,109,110].
A first HUN7293 analog is CAM741, a cyclopeptolide that selectively inhibits the expression of VCAM-1 by inhibition of the VCAM-1 co-translational translocation in the ER lumen [111,112]. By means of chemical cross-linking experiments, the authors showed that in the presence of CAM741, the VCAM-1 SP adopts an altered positioning relative to the Sec61α subunit of the translocon [108]. Later, vascular endothelial growth factor (VEGF) was identified as a second substrate for CAM741 [113]. As seen for VCAM-1, the VEGF SP is also diverted to a different position within Sec61α [113]. Hence, it was suggested that CAM741 interferes with the interaction between the SP and the SP recognition site in the lateral gate of the translocon, resulting in the incorrect insertion of the nascent chain and subsequent inhibition of protein translocation.
Around the same time as the identification of CAM741, another HUN-7293 analog, cotransin, was identified [109]. Cotransin selectively inhibits the expression of VCAM-1 and p-selectin via the inhibition of the co-translational translocation of these proteins across the ER membrane. In parallel to the CAM741 study, Garrison et al. showed that the orientation of the VCAM-1 nascent chain, with regard to the different translocon subunits, was altered in the presence of cotransin [109]. Photoaffinity labelling of cotransins confirmed earlier experiments that were performed on minimal liposomes (containing only the components that are crucial for translocation, i.e., Sec61 and SR), namely that the Sec61α subunit serves as target site for cotransin activity [109]. Later, a small subset of secretory and membrane proteins were identified as additional substrates for cotransin. Among these were angiotensinogen, β-lactamase, corticotropin releasing factor 1, endothelin B receptor, and aquaporin 2 (see Table 1) [105,109,110,114,115,116]. Initially, researchers believed that cotransin acts in a SP discriminatory manner, as so far, only secretory and type I membrane proteins with a SP targeting signal have been identified as cotransin substrates. CT08 and CT09, two cotransin analogs, however, showed activity against tumor necrosis factor α (TNFα), a single pass type II membrane protein with a non-cleavable TMD as a targeting signal [115]. From a proteomics study on cotrasin, Klein et al. concluded that the biosynthesis of almost all secreted proteins was cotransin-sensitive at a saturating concentration, whereas only a small subset of integral membrane proteins was affected at this concentration. Interestingly, for the integral membrane protein fraction, a conformational TMD consensus motif mediating cotransin sensitivity could be identified [110]. Hence, a cleavable SP is not a strict requirement for cotransin activity, leading to an unanticipated breadth of additional cotransin substrates.

Table 1.
Overview of Sec61 translocon inhibitors, substrate specificity, active concentration, and resistance conferring mutations.
Resistance studies on cotransin and analogs have shown that the lumenal region between the plug domain and lateral gate of the translocon serves as the active site of cotransin (see Table 1 and Figure 3) [115,116]. MacKinnon et al. showed that cotransin binding nearby the plug domain stabilizes the partially opened gate of Sec61α. In this model, the SP is prevented from entering the translocon, and TMD integration is hampered by blocking displacement of the plug domain [112,116,117].
Given the prominent role of VCAM-1, ICAM-1, and TNFα in the cellular immune response, HUN7293 as well as the related molecules CAM741 and cotransins might also be interesting as immunosuppressive agents [115,118]. A more recently identified cotransin substrate is the oncoprotein human epidermal growth factor receptor 3 (HER3), suggesting a potential anticancer activity for cotransin [119]. In addition, by blocking the Sec61 translocon with cotransin, researchers were able to show the importance of the translocon to support viral replication of the influenza A virus (IAV), the human immunodeficiency virus (HIV), and Dengue virus, implicating ER protein transport as a potential antiviral strategy [100].
3.1.2. Decatransin
In contrast to the earlier described inhibitors, fungal cyclic decadepsipeptide decatransin inhibits protein translocation independent of the targeting sequence, and translocation mode, suggesting a broad-spectrum activity. Resistance profiling studies indicate that decatransin binds to Sec61α in a similar, yet distinct manner than cotransin (see Table 1) [120]. Interestingly, cotransin and decatransin also showed cross inhibitory activity with the prokaryotic SecYEG translocon [120].
3.1.3. Apratoxin A and Coibamide A
Apratoxin A and Coibamide A are small molecules isolated from marine cyanobacteria that were originally investigated for their anticancer activity [121,122,123,124,125,126,127,128]. Natural products from marine organisms have a track record of antiproliferative activity in a variety of cancer cells that has led to the development of several clinical candidates [129]. Examples of such candidates from marine cyanobacteria are anti-tubulin agents, the cryptophycins, dolastatins 10 and 15, and curacin A [129,130]. Marine cyanobacteria have been shown to be an inexhaustible source of cytotoxic depsipeptides applicable to cancer research and potential pharmaceutical development [131,132].
Of the five naturally occurring apratoxins, apratoxin A exhibits the highest potency in various cancer cell lines, as the antiproliferative activity was found to be in the low nanomolar range. The antiproliferative activity was later assigned to the apratoxin A induced G1-phase cell cycle arrest and apoptosis [124]. Proteomics revealed that apratoxin A has a broad-spectrum activity as it reversibly downmodulates the expression of numerous ER resident proteins and cancer associated receptors via the inhibition of the co-translational translocation process [125]. Substrates of apratoxin A include gp130, c-MET, HER-2, PDGFR-β, insulin-like growth factor 1β, FGFR, and VEGFR2 [125]. The biological activity and structure have prompted researchers to study the total synthesis of apratoxins [125,133]. Hence, SAR studies have further investigated the selectivity profile of apratoxins, giving rise to apratoxin S4, a synthetic analog, with a more favorable cytotoxicity profile in vivo [121].
Based on the knowledge of other translocation inhibitors (i.e., CAM741 and cotransin), Sec61α was the suggested target candidate of apratoxins. In fact, via a radioactively labelled analog, the Sec61 complex was indeed identified as the molecular target of apratoxins [121]. A competitive binding assay with HUN7293 showed that apratoxins and HUN7293 likely have different binding sites within the translocon [121]. These results were confirmed by an additional study: mutagenesis and competitive photocrosslinking indicate that apratoxin A binds to the Sec61α lateral gate in a distinct manner as was seen for cotransins [122]. In fact, a mutagenesis study revealed that T86 and Y131, two residues located near the lumenal end of TMH2 and TMH3, respectively, are important for apratoxin A activity (see Table 1 and Figure 3).
A recent study suggests an antiviral potential of apratoxins, namely against the SARS-CoV-2 virus [134]. Since many of the apratoxin substrates are receptors that are validated targets for anticancer therapy [125], apratoxin A was thought to be the first anticancer agent to act through the mechanism of co-translational translocation inhibition. Around the same time, however, coibamide A has prompted scientists to investigate it for its unprecedented anticancer activity in vitro [127].
Coibamide A inhibits the migration, invasion, and cell cycle progression of glioblastoma cells [123] and has a broad-spectrum activity that shows substrate overlap with apratoxin A [128,135]. The anticancer activity of coibamide A was also shown in in vivo murine models, however, medicinal chemistry approaches are required to limit the observed dose induced toxicity. SAR analysis showed that the cyclization of the coibamide peptide is crucial for the biological activity, as two linear analogs no longer showed antiproliferative activity against glio- and neuroblastoma cancer cells [123].
By means of a photoaffinity labelled coibamide analog, researchers were able to identify the Sec61 translocon as the main target for coibamide A [135]. Later, resistance profiling suggested a distinct binding mode of coibamide A to Sec61α compared to the other known inhibitors [135]. In fact, the S71 residue that conferred coibamide A resistance upon mutation is located near the plug domain, and is shared only with decatransin, in contrast to the binding site of other inhibitors that are located in the area of the lateral gate (see Table 1 and Figure 3).
Interestingly, a recent study showed impaired autophagy to underly the anticancer activity of coibamide A [136].
3.1.4. Mycolactone
Mycolactone is a virulence factor produced by the Mycobacterium ulcerans and is responsible for the pathogenesis of Buruli ulcers, predominantly seen in West Africa, Australia, Asia, and South America. The immunosuppressive effect caused by mycolactone upon infection of Mycobacterium ulcerans was later assigned to a broad-spectrum inhibition of Sec61 dependent co-translational translocation of secretory proteins that are important in the innate and adaptive immune response such as cytokines, chemokines, and homing receptors into the ER [137,138,139,140,141,142,143,144]. Mycolactone has a complex chemical structure consisting of a 12-membered lactone ring and two polyketide-derived chains that branch from the core in a north and south position [144]. In fact, SAR studies on mycolactone show that the northern chain of the structure is crucial for the biological activity of mycolactone [144].
Competitive binding assays with cotransin showed that mycolactone dose dependently competes with cotransin for binding to the Sec61 translocon. Resistance studies later confirmed the Sec61 translocon, specifically residues near the plug domain of the translocon as the binding partner of mycolactone [145,146]. As summarized in Table 1, these binding sites overlap with other Sec61 inhibitors, suggesting a shared mechanism of action.
A proteomics study conducted on T-cells later confirmed the broad-spectrum activity of mycolactone, as 52 proteins were significantly downregulated in the presence of mycolactone. In fact, mycolactone substrates predominantly consist of single pass type I and type II membrane proteins, containing either a SP or TMD to target the proteins to the ER membrane [138,145,147]. Hence, mycolactone is indiscriminatory for the targeting signal. Later, a more selective effect of mycolactone was seen on the expression of SSPs, which translocate in a post-translational manner. At this point, mycolactone was hypothesized to stabilize the closed conformation of the Sec61 translocon. SSPs with a sufficiently hydrophobic SP were hypothesized to overcome mycolactone activity because of the short nature of their mature protein. The large mature protein part of substrates that are co-translationally translocated, however, retains them in the translocon, independent of the strength of their SP.
This hypothesis, however, was questioned with the determination of the 3D structure of a mycolactone inhibited Sec61 translocon. To date, mycolactone is the only known inhibitor for which a high-resolution structure of the inhibited Sec61 translocon exists [148]. Here, Gérard et al. showed that the conformation of Sec61 in the presence of mycolactone is actually favorable for SP engagement as mycolactone binding induces conformational changes that open the cytoplasmic end of TMH2 and TMH3 of the translocon [148]. The broad-spectrum activity of mycolactone, however, implies that the compound occupies a site in the translocon that is important for SP or TMD binding. In fact, structural analysis confirmed the mycolactone binding site in the cytosolic entrance of the translocon, normally occupied by the SP. Mycolactone is therefore thought to prevent the SP mediated opening of the translocon and subsequent dislocation of the plug domain [148]. Strikingly, Sec61α mutations that confer resistance to mycolactone activity are not located within the mycolactone binding pocket of Sec61α (see Table 1 and Figure 3). These resistance mutations, in fact, induce conformational changes in Sec61α that reduce the formation of the mycolactone binding site near the cytoplasmic end of the translocon [148]. As a consequence, mycolactone binds less efficiently to the mutant Sec61 translocons and no longer inhibits protein translocation into the ER lumen.
3.1.5. Ipomoeassin F
Ipomoeassin F (IpoF) is a natural plant derived resin glycoside cytotoxin that showed a high anticancer potency in different cell lines [149,150,151]. Later, IpoF was shown to be a non-selective inhibitor of protein secretion via the co-translational translocation process [151,152,153]. In vitro translocation assays showed that IpoF is specific for the inhibition of Sec61 dependent protein translocation as tail-anchored proteins, but also type III single pass membrane proteins were resistant to IpoF activity [151]. Furthermore, SAR studies showed that the ring size of the IpoF structure is correlated to the biological activity, as ring expansion enhanced IpoF cytotoxicity in cells and its potency to reduce in vitro protein translocation [154].
Resistance profiling shows that IpoF competes with other known inhibitors such as cotransin, apratoxin A, and mycolactone for Sec61α binding, suggesting that these inhibitors have at least partially overlapping interaction sites within the translocon [151].
In addition to the anticancer potency of IpoF, an in vitro SARS-CoV-2 antiviral activity was recently reported for IpoF through the inhibition of the co-translational translocation process of the SARS-CoV-2 spike proteins and the host cell membrane receptor ACE2 [152].
3.2. Synthetic Sec61 Inhibitors
3.2.1. Cyclotriazadisulfonamide
In contrast to the previously discussed Sec61 inhibitors, cyclotriazadisulfonamide (CADA) is a synthetic small molecule translocation inhibitor that was first discovered during a human immunodeficiency virus (HIV)-screening program [103]. It was shown that CADA downmodulates huCD4 expression on a wide range of cells [99,102,103]. Since huCD4 is the main entry receptor for HIV, the reduced expression of huCD4 in the presence of CADA explains the observed antiviral effect of the compound [102,103,162]. In addition to the reported CD4-mediated antiviral effect for CADA, recently, a CD8+ T-cell mediated immunosuppressive effect was described that is related to the CADA-induced suppression of CD137 upregulation [158]. Furthermore, partial downmodulation of the sortilin protein by CADA [159] has recently been linked to a reduction in progranulin-induced breast cancer stem cell propagation [163], thus, suggesting an additional anticancer effect for CADA.
The relatively small size of CADA stimulated the synthesis of numerous analogs that could be implemented in SAR studies [102,163,164,165,166,167,168,169]. These SAR studies were all based on the biological effect of CADA analogs on the cellular expression of the huCD4 receptor, and structure optimization resulted in improved activity going from µM to the nM range [165]. An important condition for the preservation of activity is the closed 12-membered ring structure of the compound, given that open ring analogs did not exert any activity on huCD4 [166]. A first quantitative SAR study pointed to the importance of a relatively large, hydrophobic tail group for high impact on huCD4 [164]. In contrast to the symmetrical nature of the lead compound CADA, a subsequent SAR study revealed that unsymmetrical CADA analogs with two different side arms exerted the highest activity [168].
Mechanistic studies showed that CADA directly interacts with huCD4 SP and its reorientation within the Sec61 translocon during the co-translational translocation process of the human CD4 preprotein [170]. In fact, CADA was the first translocation inhibitor for which a direct binding to a SP was shown [170], which distinguishes it from the group of Sec61 translocon binding inhibitors described above. Specific residues in the vicinity of the hydrophobic h-region of the huCD4 SP were identified as being critical for the sensitivity to CADA [171]. Furthermore, a proteomics study on T-cells was performed and identified only five substrates for CADA (see Table 1), suggesting a selective nature of the compound [103,157,158,159]. Importantly, all substrates carried a cleavable SP as a targeting sequence, implicating that these proteins are Sec61 selective proteins for co-translational translocation. One can thus speculate that the common factor, Sec61α, is a target for CADA binding, however, the importance of direct interaction of CADA with the protein SP cannot be ruled out [170]. Evidence to confirm these hypotheses is awaited as well as the analysis of more potent CADA analogs on substrate selectivity and translocation inhibition.
3.2.2. Eeyarestatin
The ER to cytosol degradation pathway for the disposal of misfolded proteins is an attractive target of intervention for diseases characterized by impaired protein degradation such as Alzheimer’s, Parkinson’s, prion, and Huntington’s disease [172,173,174]. It was in this regard that Eeyarestatin (ES) I and II, two structurally related chemical molecules, were identified from a library to screen for ERAD inhibitors [172,173]. ESI and ESII were shown to bind with the ER membrane bound p97 complex of ERAD, finally resulting in hampered deubiquitination of misfolded proteins, an essential step for proper proteasomal degradation [175]. As a result, misfolded proteins accumulate and rapidly induce ER stress [175,176]. However, it became clear that ESI and ESII also interfere at a step prior to proteasomal degradation. In fact, studies on ESR35, an ESI analog, showed a broad-spectrum inhibition of protein translocation [160]. Further analysis of the ES compounds suggests that ES targets a component in the Sec61 translocon and thereby sterically prevents the transfer of the RNC complex from the SRP targeting machinery to the Sec61 translocation machinery [160].
Since ES, and other inhibitors for that matter, interacts with the Sec61 translocon to prevent protein translocation into the ER lumen, they may indirectly induce Ca2+ leakage from the ER lumen, the major intracellular Ca2+ storage [177]. In fact, it was shown that ES, via its 5-NF moiety, induces Ca2+ leakage from cells [178,179]. ESI, ESII, and ES24, a minimal analog that closely resembles the 5-NF moiety of ES [179], have been shown to prevent protein translocation, however, while keeping Sec61 in a Ca2+ permeable state [178]. This apparent contradiction was reconciled in a mechanistic model for the action of ES compounds on Sec61 complexes [178]. Docking analysis of the Sec61 translocon structure revealed that ES, and particularly the 5-NF group, putatively interacts with the cytosolic end of the Sec61α lateral gate. The binding of ES in the space between TMH2 and TMH7 hampers conformational changes of Sec61α that are required for protein translocation [178]. Hence, this model suggests that ES binding stabilizes the primed, Ca2+ permeable, state of the Sec61 translocon by preventing the lateral gate to close [178].
Since the Sec61 translocon is tightly linked to the ERAD pathway for protein degradation, it is suggested that the inhibition of the ER translocation machinery might simultaneously also block both the retrotranslocation of misfolded proteins to the cytosol [160]. The resulting accumulation of cytosolic and misfolded proteins subsequently induces ER stress, which explains the cytotoxic effect of ESI in cellula, and even suggests an anticancer activity of ES [160,180].
In fact, induced tumor cell death upon ES treatment was reported in vitro, and appeared to be enhanced upon co-treatment with proteasomal inhibitors such as bortezomib [160,173,175,176,181,182,183]. Recently, an antibacterial activity of ES24, a smaller analog of ESI, was described. ES24 impairs protein translocation in E. coli via interaction with the SecYEG translocon, the prokaryotic orthologue of the Sec61 translocon [184].
3.2.3. KZR-261 and KZR-834
The most recent Sec61 dependent protein translocation inhibitors are KZR-261 and KZR-834, two structural analogs that were identified in an anticancer medicinal chemistry screening program [161]. A proteomics study that assayed KZR-261 and KZR-834 activity on different tumor cell lines showed that both compounds had a broad-spectrum activity in vitro in the nanomolar range, with a preference for secreted and type I membrane proteins [161]. In fact, in vivo studies have been performed and have even led to the selection of KZR-261 for clinical development to profile the safety and early efficacy of this novel compound [161]. Among the other substrates are more therapeutic targets such as VEGF, VEGFR, and EGFR, suggesting, besides the anticancer activity, also an immunosuppressive potency for KZR-261 and KZR-834 [161].
4. High Throughput Screening Assays to Define Novel Inhibitors of the Sec61 Complex
Novel small molecule inhibitors of protein translocation at the Sec61 complex may be important innovations in pharmacology and may have several medicinal indications in the future. From the previous section, it is clear that Sec61 inhibitors act by a selective or non-selective mechanism of action [39,90].
Broad-spectrum or non-selective inhibitors impair translocation of many or even all proteins targeted to the Sec61 complex [39,90]. They may represent novel cancer drugs slowing down tumor cell growth. The advantage of Sec61 complex inhibitors over the still widely used cancer drugs affecting nucleic acid biosynthesis would be that they should not be mutagenic by themselves. Non-selective compounds may also be used to decelerate the production of viruses in infected cells, in particular, that of the enveloped viruses that possess integral membrane proteins at the surface using the Sec pathway. It is conceivable that such non-selective substances should target the Sec61 complex alone, rather than the signal sequences of the substrate proteins.
Selective inhibitors, instead, target translocation of a small subset of proteins at the Sec61 complex in a signal sequence discriminatory manner [39,90]. Selective inhibitors may be used in the future to downregulate the biosynthesis of proteins of interest. While a number of selective compounds have been described in the past decade, a specific substance inhibiting translocation of only one protein is not known thus far. Of note, the term selective inhibitor should only be used when proteomic experiments are performed, supporting this classification.
The inhibitors of translocation outlined above were described for the eukaryotic Sec61 complex present in the ER membrane. The orthologous bacterial SecYEG complex translocates proteins across the prokaryotic plasma membrane, therefore, inhibitors of the SecYEG complex may represent novel antibiotics that are urgently needed. A proof of principle for SecYEG inhibition was published, for example, decatransin [99] and eeyarestatin [81], although these compounds inhibit both the eukaryotic Sec61 complex and prokaryotic SecYEG.
The setup of high throughput screening assays for small molecule inhibitors of the Sec61 complex is notoriously difficult. This is essentially due to three properties of the Sec61 complex. (i) Most importantly, the heterotrimeric Sec61 core complex (Sec61αβγ) has no enzymatic activity by itself. Sec61α gating (i.e., the switch from the closed to the open state) is facilitated by ribosomes, signal sequences, and auxiliary factors such as TRAP and/or Sec62/Sec63 (recent reviews: [131,133]). Translocation itself is facilitated by the BiP chaperone ratchet mechanism at the ER lumenal side [83]. (ii) The accessibility of the complex for compounds in live cells is limited because it is only expressed intracellularly in the ER membrane. (iii) Finally, the complex is difficult to isolate and to reconstitute functionally in larger amounts, making in vitro assays unfavorable.
Although a lot of structural information has been published for the Sec61 complex in the past decade, no attempts have been made to define inhibitors by in silico screening. The main obstacle for in silico screening is the Sec61α architecture itself: it forms a huge and highly dynamic aqueous pore with diameters ranging from 12 to 22 Angstrom. The fact that the channel handles a multitude of different proteins implies that it has slightly different interaction sites for protein substrates. It is consequently very difficult to define specific inhibitor binding sites by in silico approaches.
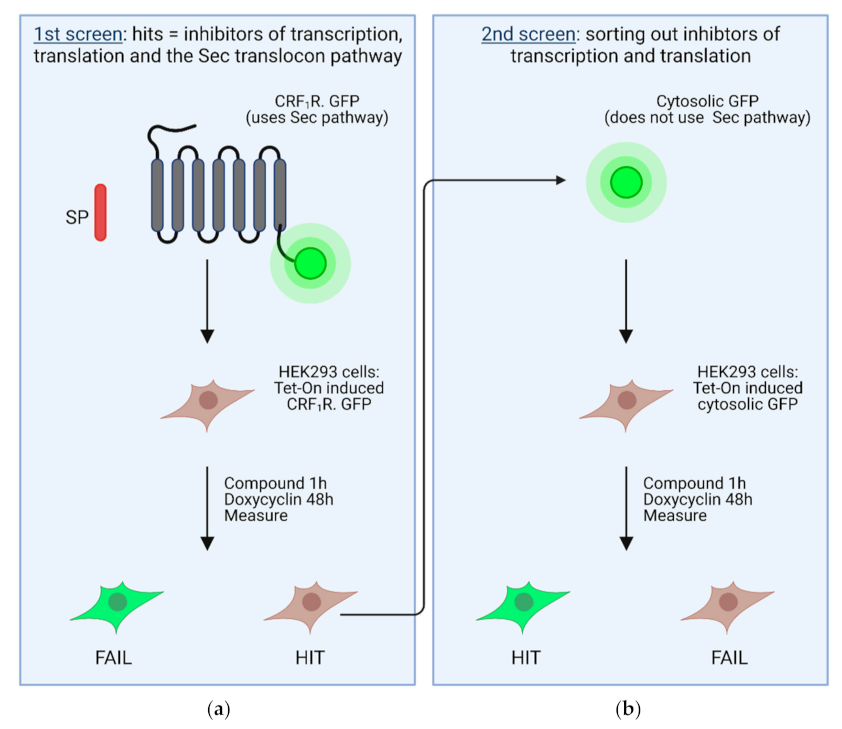
Despite all of these potential obstacles, a whole-cell screening approach was recently published for inhibitors of the Sec61 complex using two succeeding screening steps [67]. In a primary screen, inhibitors for transcription, translation, and the SRP-Sec61 targeting/translocation pathway were selected (Figure 4a). To this end, a heptahelical G protein-coupled receptor was used as the target, which was C-terminally tagged with GPP (CRF1R.GFP; Figure 4a). The CRF1R.GFP possesses a cleavable signal peptide and thus uses the SRP-Sec61 targeting/translocation pathway [134]. SRP binding to the signal peptide of this construct should decelerate or even arrest translation. The idea is that inhibitors of the SRP-Sec61 targeting/translocation pathway from a compound library should decrease or even prevent CRF1R.GFP biosynthesis, and consequently expression of the C-terminal GFP tag, which could be measured fluorimetrically [77]. A decreased GFP fluorescence, however, may also be observed when inhibitors of the transcription/translation machinery are present. Taking all hit compounds of the primary screen, the latter were deselected with a secondary screen using unfused, cytosolic GFP protein as a target (Figure 4b) [77]. GFP alone does not use the SRP-Sec61 targeting/translocation pathway and its expression depends only on transcription and translation. Compounds were considered as inhibitors of the SRP-Sec61 targeting/translocation pathway when they behaved as hits in the primary screen, but not in the secondary screen [77].
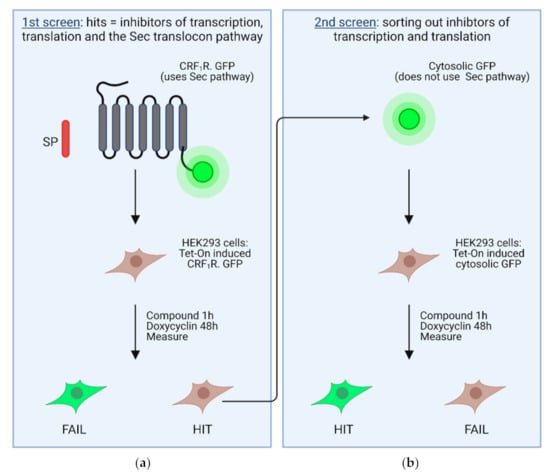
Figure 4.
Scheme of the primary and the secondary screen in stably transfected HEK 293 cells. In the primary screen (a), Tet-On-controlled CRF1R.GFP was used, a GFP-tagged GPCR possessing a cleavable signal peptide that uses the SRP-Sec61 targeting/translocation pathway. The secondary screen (b) was performed with Tet-On-controlled, unfused, soluble GFP, which does not use the SRP-Sec61 targeting/translocation pathway. Hits of the primary screen were used in the secondary screen to deselect inhibitors of the transcription/translation machinery. Figure modified from [67].
Using this screening setup and a library of 37,312 substances, 1052 compounds were identified in the primary screen [77]. This number was reduced to 28 compounds following the secondary screen. Following an in vitro biosynthesis assay in live cells, five compounds were considered to represent real hits with a potential to inhibit the SRP-Sec61 targeting/translocation pathway. For one of them, namely FMP-401319-3, it could be shown by an in vitro transcription/translation/translocation assay that it acts indeed in a post targeting step at the level of the Sec61 complex. The potency of compound FMP-401319-3, however, was only in the low micromolar range. It remains to be determined whether it could be optimized by medicinal chemistry methods in the future. Using a much larger library might also be helpful to identify compounds with higher potency.
Of note, a slightly modified methodology may be used in the future to screen for substances affecting SecYEG, the bacterial ortholog of the Sec61 complex in order to derive novel antibiotic drugs. No such specific inhibitor for the prokaryotic SecYEG complex has been reported thus far, except for decatransin [120] and eeyarestatin [184], which both inhibit the Sec61 and SecYEG complexes.
5. Summary
Protein translocation is by far the most crucial process for the overall protein biogenesis and correct functioning of proteins in cellular processes, and homeostasis in general. This is evidenced by the fact that incorrect protein translocation is linked to numerous metabolic and protein folding diseases.
Today, different inhibitors of the Sec61 dependent protein translocation pathway have been identified. The chemical structure, compound concentration, and substrate targeting sequence are factors that ultimately contribute to the substrate specificity and selectivity of these compounds. As many inhibitors share binding regions within the Sec61α subunit, the translocon shows great potential as a molecular target in different therapeutic areas such as anticancer, immunosuppressive, and antiviral treatment. An intriguing fact of the inhibitors discussed in this review, is that they share a certain level of structural resemblance: they belong to the family of macrocyclic depsipeptides. The macrocyclic nature of the compounds, however, is associated with challenges regarding the synthesis, plasma stability, and/or stereochemical complexity.
By means of two-step whole cell screening approaches, researchers therefore aim to discover novel inhibitors specific to the SRP-Sec61 translocation pathway. A methodology that might also be expanded to screen for molecules that affect SecYEG, the bacterial ortholog of the Sec61 complex, in order to discover new antibiotic drugs. Undoubtedly, inhibitors of protein translocation will find their way into the clinic as promising therapeutics to treat various diseases.
Author Contributions
E.P., R.S. and K.V. jointly wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Siekevitz, P.; Palade, G.E. A cytochemical study on the pancreas of the guinea pig. 5. In vivo incorporation of leucine-1-C14 into the chymotrypsinogen of various cell fractions. J. Cell Biol. 1958, 7, 619–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caro, L.G.; Palade, G.E. Protein synthesis, storage, and discharge in the pancreatic exocrine cell. An autoradiographic study. J. Cell Biol. 1964, 20, 473–495. [Google Scholar] [PubMed]
- Jamieson, J.D.; Palade, G.E. Intracellular transport of secretory proteins in the pancreatic endocrine cell: II. Transport to Condensing Vacuoles and Zymogen Granules. J. Cell Biol. 1967, 34, 597–615. [Google Scholar] [CrossRef] [Green Version]
- Jamieson, J.D.; Palade, G.E. Intracellular transport of secretory proteins in the pancreatic endocrine cell: I. Role of the Peripheral Elements of the Golgi Complex. J. Cell Biol. 1967, 34, 577–596. [Google Scholar] [CrossRef] [Green Version]
- Matlin, K.S.; Caplan, M.J. The secretory pathway at 50: A golden anniversary for some momentous grains of silver. Mol. Biol. Cell 2017, 28, 229–232. [Google Scholar] [CrossRef] [Green Version]
- Walter, P.; Gilmore, R.; Müller, M.; Blobel, G. The protein translocation machinery of the endoplasmic reticulum. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1982, 300, 225–228. [Google Scholar] [CrossRef]
- Walter, P.; Gilmore, R.; Blobel, G. Protein translocation across the endoplasmic reticulum. Cell 1984, 38, 5–8. [Google Scholar] [CrossRef]
- Von Heijne, G. Signal sequences. The limits of variation. J. Mol. Biol. 1985, 184, 99–105. [Google Scholar] [CrossRef]
- Cross, B.C.; Sinning, I.; Luirink, J.; High, S. Delivering proteins for export from the cytosol. Nat. Rev. Mol. Cell Biol. 2009, 10, 255–264. [Google Scholar] [CrossRef]
- Mateja, A.; Keenan, R.J. A structural perspective on tail-anchored protein biogenesis by the GET pathway. Curr. Opin. Struct. Biol. 2018, 51, 195–202. [Google Scholar] [CrossRef]
- Ast, T.; Cohen, G.; Schuldiner, M. A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell 2013, 152, 1134–1145. [Google Scholar] [CrossRef] [Green Version]
- Aviram, N.; Ast, T.; Costa, E.A.; Arakel, E.C.; Chuartzman, S.G.; Jan, C.H.; Hassdenteufel, S.; Dudek, J.; Jung, M.; Schorr, S.; et al. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature 2016, 540, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, J.J.; High, S. The Sec61 complex is located in both the ER and the ER-Golgi intermediate compartment. J. Cell Sci. 1999, 112 Pt 10, 1477–1486. [Google Scholar] [PubMed]
- Kim, S.J.; Mitra, D.; Salerno, J.R.; Hegde, R.S. Signal sequences control gating of the protein translocation channel in a substrate-specific manner. Dev. Cell 2002, 2, 207–217. [Google Scholar] [CrossRef] [Green Version]
- Kraut-Cohen, J.; Afanasieva, E.; Haim-Vilmovsky, L.; Slobodin, B.; Yosef, I.; Bibi, E.; Gerst, J.E. Translation- and SRP-independent mRNA targeting to the endoplasmic reticulum in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 2013, 24, 3069–3084. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.; Powis, K.; High, S. Post-translational translocation into the endoplasmic reticulum. Biochim. Biophys. Acta 2013, 1833, 2403–2409. [Google Scholar] [CrossRef]
- Mandon, E.C.; Trueman, S.F.; Gilmore, R. Translocation of proteins through the Sec61 and SecYEG channels. Curr. Opin. Cell Biol. 2009, 21, 501–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borgese, N.; Coy-Vergara, J.; Colombo, S.F.; Schwappach, B. The Ways of Tails: The GET Pathway and more. Protein J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Denic, V. A portrait of the GET pathway as a surprisingly complicated young man. Trends Biochem. Sci. 2012, 37, 411–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chartron, J.W.; Clemons, W.M., Jr.; Suloway, C.J. The complex process of GETting tail-anchored membrane proteins to the ER. Curr. Opin. Struct. Biol. 2012, 22, 217–224. [Google Scholar] [CrossRef] [Green Version]
- Hassdenteufel, S.; Nguyen, D.; Helms, V.; Lang, S.; Zimmermann, R. ER import of small human presecretory proteins: Components and mechanisms. FEBS Lett. 2019, 593, 2506–2524. [Google Scholar] [CrossRef]
- Jadhav, B.; McKenna, M.; Johnson, N.; High, S.; Sinning, I.; Pool, M.R. Mammalian SRP receptor switches the Sec61 translocase from Sec62 to SRP-dependent translocation. Nat. Commun. 2015, 6, 10133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rapoport, T.A. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 2007, 450, 663–669. [Google Scholar] [CrossRef]
- Aviram, N.; Schuldiner, M. Embracing the void—How much do we really know about targeting and translocation to the endoplasmic reticulum? Curr. Opin. Cell Biol. 2014, 29, 8–17. [Google Scholar] [CrossRef]
- Johnson, N.; Hassdenteufel, S.; Theis, M.; Paton, A.W.; Paton, J.C.; Zimmermann, R.; High, S. The signal sequence influences post-translational ER translocation at distinct stages. PLoS ONE 2013, 8, e75394. [Google Scholar] [CrossRef] [Green Version]
- Kriegler, T.; Magoulopoulou, A.; Amate Marchal, R.; Hessa, T. Measuring Endoplasmic Reticulum Signal Sequences Translocation Efficiency Using the Xbp1 Arrest Peptide. Cell Chem. Biol. 2018, 25, 880–890. [Google Scholar] [CrossRef] [PubMed]
- Voorhees, R.M.; Hegde, R.S. Structure of the Sec61 channel opened by a signal sequence. Science 2016, 351, 88–91. [Google Scholar] [CrossRef] [Green Version]
- Hassdenteufel, S.; Schauble, N.; Cassella, P.; Leznicki, P.; Muller, A.; High, S.; Jung, M.; Zimmermann, R. Ca2+-calmodulin inhibits tail-anchored protein insertion into the mammalian endoplasmic reticulum membrane. FEBS Lett. 2011, 585, 3485–3490. [Google Scholar] [CrossRef] [Green Version]
- Casson, J.; McKenna, M.; Hassdenteufel, S.; Aviram, N.; Zimmerman, R.; High, S. Multiple pathways facilitate the biogenesis of mammalian tail-anchored proteins. J. Cell Sci. 2017, 130, 3851–3861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colombo, S.F.; Cardani, S.; Maroli, A.; Vitiello, A.; Soffientini, P.; Crespi, A.; Bram, R.F.; Benfante, R.; Borgese, N. Tail-anchored Protein Insertion in Mammals: Function and reciprocal interactions of the two subunits of the TRC40 receptor. J. Biol. Chem. 2016, 291, 15292–15306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coy-Vergara, J.; Rivera-Monroy, J.; Urlaub, H.; Lenz, C.; Schwappach, B. A trap mutant reveals the physiological client spectrum of TRC40. J. Cell Sci. 2019, 132, jcs230094. [Google Scholar] [CrossRef] [Green Version]
- Borgese, N.; Fasana, E. Targeting pathways of C-tail-anchored proteins. Biochim. Biophys. Acta (BBA) Biomembr. 2011, 1808, 937–946. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, Y.; Sakisaka, T. Molecular Machinery for Insertion of Tail-Anchored Membrane Proteins into the Endoplasmic Reticulum Membrane in Mammalian Cells. Mol. Cell 2012, 48, 387–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ott, M.; Marques, D.; Funk, C.; Bailer, S.M. Asna1/TRC40 that mediates membrane insertion of tail-anchored proteins is required for efficient release of Herpes simplex virus 1 virions. Virol. J. 2016, 13, 175. [Google Scholar] [CrossRef]
- Lang, S.; Benedix, J.; Fedeles, S.V.; Schorr, S.; Schirra, C.; Schauble, N.; Jalal, C.; Greiner, M.; Hassdenteufel, S.; Tatzelt, J.; et al. Different effects of Sec61alpha, Sec62 and Sec63 depletion on transport of polypeptides into the endoplasmic reticulum of mammalian cells. J. Cell Sci. 2012, 125, 1958–1969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, S.; Pfeffer, S.; Lee, P.H.; Cavalie, A.; Helms, V.; Forster, F.; Zimmermann, R. An Update on Sec61 Channel Functions, Mechanisms, and Related Diseases. Front. Physiol. 2017, 8, 887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linxweiler, M.; Schick, B.; Zimmermann, R. Let’s talk about Secs: Sec61, Sec62 and Sec63 in signal transduction, oncology and personalized medicine. Signal Transduct. Target. Ther. 2017, 2, 17002. [Google Scholar] [CrossRef] [PubMed]
- Kalies, K.U.; Romisch, K. Inhibitors of Protein Translocation across the ER Membrane. Traffic 2015, 16, 1027–1038. [Google Scholar] [CrossRef] [Green Version]
- Van Puyenbroeck, V.; Vermeire, K. Inhibitors of protein translocation across membranes of the secretory pathway: Novel antimicrobial and anticancer agents. Cell. Mol. Life Sci. CMLS 2018, 75, 1541–1558. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, R.; Eyrisch, S.; Ahmad, M.; Helms, V. Protein translocation across the ER membrane. Biochim. Biophys. Acta 2011, 1808, 912–924. [Google Scholar] [CrossRef] [Green Version]
- Matlack, K.E.S.; Mothes, W.; Rapoport, T.A. Protein Translocation: Tunnel Vision. Cell 1998, 92, 381–390. [Google Scholar] [CrossRef] [Green Version]
- Luesch, H.; Paavilainen, V.O. Natural products as modulators of eukaryotic protein secretion. Nat. Prod. Rep. 2020, 37, 717–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakkaraju, A.K.; Thankappan, R.; Mary, C.; Garrison, J.L.; Taunton, J.; Strub, K. Efficient secretion of small proteins in mammalian cells relies on Sec62-dependent posttranslational translocation. Mol. Biol. Cell 2012, 23, 2712–2722. [Google Scholar] [CrossRef] [PubMed]
- Denks, K.; Vogt, A.; Sachelaru, I.; Petriman, N.A.; Kudva, R.; Koch, H.G. The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Mol. Membr. Biol. 2014, 31, 58–84. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Dudek, J.; Zimmermann, R.; Forster, F. Organization of the native ribosome-translocon complex at the mammalian endoplasmic reticulum membrane. Biochim. Biophys. Acta 2016, 1860, 2122–2129. [Google Scholar] [CrossRef]
- Luirink, J.; Sinning, I. SRP-mediated protein targeting: Structure and function revisited. Biochim. Biophys. Acta 2004, 1694, 17–35. [Google Scholar] [CrossRef] [PubMed]
- Lakkaraju, A.K.; Mary, C.; Scherrer, A.; Johnson, A.E.; Strub, K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell 2008, 133, 440–451. [Google Scholar] [CrossRef] [Green Version]
- Wild, K.; Juaire, K.D.; Soni, K.; Shanmuganathan, V.; Hendricks, A.; Segnitz, B.; Beckmann, R.; Sinning, I. Reconstitution of the human SRP system and quantitative and systematic analysis of its ribosome interactions. Nucleic Acids Res. 2019, 47, 3184–3196. [Google Scholar] [CrossRef]
- Dudek, J.; Pfeffer, S.; Lee, P.H.; Jung, M.; Cavalie, A.; Helms, V.; Forster, F.; Zimmermann, R. Protein transport into the human endoplasmic reticulum. J. Mol. Biol. 2015, 427, 1159–1175. [Google Scholar] [CrossRef]
- Nyathi, Y.; Wilkinson, B.M.; Pool, M.R. Co-translational targeting and translocation of proteins to the endoplasmic reticulum. Biochim. Biophys. Acta 2013, 1833, 2392–2402. [Google Scholar] [CrossRef]
- Voorhees, R.M.; Hegde, R.S. Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. eLife 2015, 4, e07975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voorhees, R.M.; Fernandez, I.S.; Scheres, S.H.; Hegde, R.S. Structure of the mammalian ribosome-Sec61 complex to 3.4 A resolution. Cell 2014, 157, 1632–1643. [Google Scholar] [CrossRef] [Green Version]
- Voorhees, R.M.; Hegde, R.S. Toward a structural understanding of co-translational protein translocation. Curr. Opin. Cell Biol. 2016, 41, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Dudek, J.; Schaffer, M.; Ng, B.G.; Albert, S.; Plitzko, J.M.; Baumeister, W.; Zimmermann, R.; Freeze, H.H.; Engel, B.D.; et al. Dissecting the molecular organization of the translocon-associated protein complex. Nat. Commun. 2017, 8, 14516. [Google Scholar] [CrossRef]
- Braunger, K.; Pfeffer, S.; Shrimal, S.; Gilmore, R.; Berninghausen, O.; Mandon, E.C.; Becker, T.; Forster, F.; Beckmann, R. Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science 2018, 360, 215–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, L.; Wang, T.; Zhao, G.; Kovach, A.; Li, H. The atomic structure of a eukaryotic oligosaccharyltransferase complex. Nature 2018, 555, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Dudek, J.; Gogala, M.; Schorr, S.; Linxweiler, J.; Lang, S.; Becker, T.; Beckmann, R.; Zimmermann, R.; Forster, F. Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon. Nat. Commun. 2014, 5, 3072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liaci, A.M.; Steigenberger, B.; Telles de Souza, P.C.; Tamara, S.; Grollers-Mulderij, M.; Ogrissek, P.; Marrink, S.J.; Scheltema, R.A.; Forster, F. Structure of the human signal peptidase complex reveals the determinants for signal peptide cleavage. Mol. Cell 2021, 81, 3934–3948. [Google Scholar] [CrossRef] [PubMed]
- Auclair, S.M.; Bhanu, M.K.; Kendall, D.A. Signal peptidase I: Cleaving the way to mature proteins. Protein Sci. 2012, 21, 13–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Jomaa, A.; Jaskolowski, M.; Yang, C.I.; Ban, N.; Shan, S.O. The molecular mechanism of cotranslational membrane protein recognition and targeting by SecA. Nat. Struct. Mol. Biol. 2019, 26, 919–929. [Google Scholar] [CrossRef]
- Hassdenteufel, S.; Sicking, M.; Schorr, S.; Aviram, N.; Fecher-Trost, C.; Schuldiner, M.; Jung, M.; Zimmermann, R.; Lang, S. hSnd2 protein represents an alternative targeting factor to the endoplasmic reticulum in human cells. FEBS Lett. 2017, 591, 3211–3224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strzyz, P. The third route to the ER. Nat. Rev. Mol. Cell Biol. 2017, 18, 3. [Google Scholar] [CrossRef]
- Ast, T.; Schuldiner, M. All roads lead to Rome (but some may be harder to travel): SRP-independent translocation into the endoplasmic reticulum. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 273–288. [Google Scholar] [CrossRef]
- Egea, P.F.; Stroud, R.M.; Walter, P. Targeting proteins to membranes: Structure of the signal recognition particle. Curr. Opin. Struct. Biol. 2005, 15, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Halic, M.; Beckmann, R. The signal recognition particle and its interactions during protein targeting. Curr. Opin. Struct. Biol. 2005, 15, 116–125. [Google Scholar] [CrossRef]
- Hwang Fu, Y.H.; Chandrasekar, S.; Lee, J.H.; Shan, S.O. A molecular recognition feature mediates ribosome-induced SRP-receptor assembly during protein targeting. J. Cell Biol. 2019, 218, 3307–3319. [Google Scholar] [CrossRef] [Green Version]
- Kellogg, M.K.; Miller, S.C.; Tikhonova, E.B.; Karamyshev, A.L. SRPassing Co-translational Targeting: The Role of the Signal Recognition Particle in Protein Targeting and mRNA Protection. Int. J. Mol. Sci. 2021, 22, 6284. [Google Scholar] [CrossRef]
- Saraogi, I.; Shan, S.O. Molecular mechanism of co-translational protein targeting by the signal recognition particle. Traffic 2011, 12, 535–542. [Google Scholar] [CrossRef] [Green Version]
- Pfeffer, S.; Burbaum, L.; Unverdorben, P.; Pech, M.; Chen, Y.; Zimmermann, R.; Beckmann, R.; Forster, F. Structure of the native Sec61 protein-conducting channel. Nat. Commun. 2015, 6, 8403. [Google Scholar] [CrossRef] [Green Version]
- Pfeffer, S.; Brandt, F.; Hrabe, T.; Lang, S.; Eibauer, M.; Zimmermann, R.; Forster, F. Structure and 3D arrangement of endoplasmic reticulum membrane-associated ribosomes. Structure 2012, 20, 1508–1518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conti, B.J.; Devaraneni, P.K.; Yang, Z.; David, L.L.; Skach, W.R. Cotranslational stabilization of Sec62/63 within the ER Sec61 translocon is controlled by distinct substrate-driven translocation events. Mol. Cell 2015, 58, 269–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, D.; Stutz, R.; Schorr, S.; Lang, S.; Pfeffer, S.; Freeze, H.H.; Forster, F.; Helms, V.; Dudek, J.; Zimmermann, R. Proteomics reveals signal peptide features determining the client specificity in human TRAP-dependent ER protein import. Nat. Commun. 2018, 9, 3765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fons, R.D.; Bogert, B.A.; Hegde, R.S. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 2003, 160, 529–539. [Google Scholar] [CrossRef] [Green Version]
- Klein, M.C.; Lerner, M.; Nguyen, D.; Pfeffer, S.; Dudek, J.; Forster, F.; Helms, V.; Lang, S.; Zimmermann, R. TRAM1 protein may support ER protein import by modulating the phospholipid bilayer near the lateral gate of the Sec61-channel. Channels 2020, 14, 28–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamborero, S.; Vilar, M.; Martinez-Gil, L.; Johnson, A.E.; Mingarro, I. Membrane insertion and topology of the translocating chain-associating membrane protein (TRAM). J. Mol. Biol. 2011, 406, 571–582. [Google Scholar] [CrossRef] [Green Version]
- Russo, A. Understanding the mammalian TRAP complex function(s). Open Biol. 2020, 10, 190244. [Google Scholar] [CrossRef]
- Klein, W.; Rutz, C.; Eckhard, J.; Provinciael, B.; Specker, E.; Neuenschwander, M.; Kleinau, G.; Scheerer, P.; von Kries, J.P.; Nazare, M.; et al. Use of a sequential high throughput screening assay to identify novel inhibitors of the eukaryotic SRP-Sec61 targeting/translocation pathway. PLoS ONE 2018, 13, e0208641. [Google Scholar] [CrossRef]
- Hegde, R.S.; Voigt, S.; Rapoport, T.A.; Lingappa, V.R. TRAM Regulates the Exposure of Nascent Secretory Proteins to the Cytosol during Translocation into the Endoplasmic Reticulum. Cell 1998, 92, 621–631. [Google Scholar] [CrossRef] [Green Version]
- Voigt, S.; Jungnickel, B.; Hartmann, E.; Rapoport, T.A. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J. Cell Biol. 1996, 134, 25–35. [Google Scholar] [CrossRef] [Green Version]
- Alder, N.N.; Shen, Y.; Brodsky, J.L.; Hendershot, L.M.; Johnson, A.E. The molecular mechanisms underlying BiP-mediated gating of the Sec61 translocon of the endoplasmic reticulum. J. Cell Biol. 2005, 168, 389–399. [Google Scholar] [CrossRef]
- Alfaro-Valdés, H.; Burgos-Bravo, F.; Casanova-Morales, N.; Quiroga-Roger, D.; Wilson, C. Mechanical Properties of Chaperone BiP, the Master Regulator of the Endoplasmic Reticulum; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.H.; Law, D.T.; Williams, D.B. Binding protein BiP is required for translocation of secretory proteins into the endoplasmic reticulum in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1991, 88, 1565–1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matlack, K.E.; Misselwitz, B.; Plath, K.; Rapoport, T.A. BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell 1999, 97, 553–564. [Google Scholar] [CrossRef] [Green Version]
- Araki, K.; Nagata, K. Protein folding and quality control in the ER. Cold Spring Harb. Perspect. Biol. 2011, 3, a007526. [Google Scholar] [CrossRef] [Green Version]
- Comyn, S.A.; Chan, G.T.; Mayor, T. False start: Cotranslational protein ubiquitination and cytosolic protein quality control. J. Proteom. 2014, 100, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Ellgaard, L.; Helenius, A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003, 4, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Qi, L. Quality Control in the Endoplasmic Reticulum: Crosstalk between ERAD and UPR pathways. Trends Biochem. Sci. 2018, 43, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Kannan, K.; Vazquez-Laslop, N.; Mankin, A.S. Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell 2012, 151, 508–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verchot, J. The ER quality control and ER associated degradation machineries are vital for viral pathogenesis. Front. Plant Sci. 2014, 5, 66. [Google Scholar] [CrossRef] [Green Version]
- Sicking, M.; Lang, S.; Bochen, F.; Roos, A.; Drenth, J.P.H.; Zakaria, M.; Zimmermann, R.; Linxweiler, M. Complexity and Specificity of Sec61-Channelopathies: Human Diseases Affecting Gating of the Sec61 Complex. Cells 2021, 10, 1036. [Google Scholar] [CrossRef]
- Lu, Z.; Zhou, L.; Killela, P.; Rasheed, A.B.; Di, C.; Poe, W.E.; McLendon, R.E.; Bigner, D.D.; Nicchitta, C.; Yan, H. Glioblastoma proto-oncogene SEC61gamma is required for tumor cell survival and response to endoplasmic reticulum stress. Cancer Res. 2009, 69, 9105–9111. [Google Scholar] [CrossRef] [Green Version]
- Jung, V.; Kindich, R.; Kamradt, J.; Jung, M.; Müller, M.; Schulz, W.A.; Engers, R.; Unteregger, G.; Stöckle, M.; Zimmermann, R.; et al. Genomic and expression analysis of the 3q25-q26 amplification unit reveals TLOC1/SEC62 as a probable target gene in prostate cancer. Mol. Cancer Res. 2006, 4, 169–176. [Google Scholar] [CrossRef] [Green Version]
- Greiner, M.; Kreutzer, B.; Jung, V.; Grobholz, R.; Hasenfus, A.; Stöhr, R.F.; Tornillo, L.; Dudek, J.; Stöckle, M.; Unteregger, G.; et al. Silencing of the SEC62 gene inhibits migratory and invasive potential of various tumor cells. Int. J. Cancer 2011, 128, 2284–2295. [Google Scholar] [CrossRef]
- Linxweiler, M.; Linxweiler, J.; Barth, M.; Benedix, J.; Jung, V.; Kim, Y.J.; Bohle, R.M.; Zimmermann, R.; Greiner, M. Sec62 bridges the gap from 3q amplification to molecular cell biology in non-small cell lung cancer. Am. J. Pathol. 2012, 180, 473–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, C.S.L.; Pfohler, C.; Wahl, M.; Bochen, F.; Korner, S.; Kuhn, J.P.; Bozzato, A.; Schick, B.; Linxweiler, M. Expression of SEC62 Oncogene in Benign, Malignant and Borderline Melanocytic Tumors-Unmasking the Wolf in Sheep’s Clothing? Cancers 2021, 13, 1645. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.; Jiang, X.; Wang, J.; Sang, Z.; Guo, L.; Yin, G.; Wang, Y. SEC61G is upregulated and required for tumor progression in human kidney cancer. Mol. Med. Rep. 2021, 23, 427. [Google Scholar] [CrossRef] [PubMed]
- Witham, C.M.; Paxman, A.L.; Baklous, L.; Steuart, R.F.L.; Schulz, B.L.; Mousley, C.J. Cancer associated mutations in Sec61γ alter the permeability of the ER translocase. PLoS Genet. 2021, 17, e1009780. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, M.S.; Bagchi, P.; Cunningham, C.N.; Tsai, B. Opportunistic intruders: How viruses orchestrate ER functions to infect cells. Nat. Rev. Microbiol. 2016, 14, 407–420. [Google Scholar] [CrossRef]
- Vermeire, K.; Schols, D. Specific CD4 down-modulating compounds with potent anti-HIV activity. J. Leukoc. Biol. 2003, 74, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Heaton, N.S.; Moshkina, N.; Fenouil, R.; Gardner, T.J.; Aguirre, S.; Shah, P.S.; Zhao, N.; Manganaro, L.; Hultquist, J.F.; Noel, J.; et al. Targeting Viral Proteostasis Limits Influenza Virus, HIV, and Dengue Virus Infection. Immunity 2016, 44, 46–58. [Google Scholar] [CrossRef] [Green Version]
- Gillece, P.; Pilon, M.; Römisch, K. The protein translocation channel mediates glycopeptide export across the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA 2000, 97, 4609–4614. [Google Scholar] [CrossRef] [Green Version]
- Vermeire, K.; Bell, T.W.; Choi, H.-J.; Jin, Q.; Samala, M.F.; Sodoma, A.; De Clercq, E.; Schols, D. The Anti-HIV Potency of Cyclotriazadisulfonamide Analogs Is Directly Correlated with Their Ability to Down-Modulate the CD4 Receptor. Mol. Pharmacol. 2003, 63, 203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vermeire, K.; Zhang, Y.; Princen, K.; Hatse, S.; Samala, M.F.; Dey, K.; Choi, H.J.; Ahn, Y.; Sodoma, A.; Snoeck, R.; et al. CADA inhibits human immunodeficiency virus and human herpesvirus 7 replication by down-modulation of the cellular CD4 receptor. Virology 2002, 302, 342–353. [Google Scholar] [CrossRef] [Green Version]
- Foster, C.A.; Dreyfuss, M.; Mandak, B.; Meingassner, J.G.; Naegeli, H.U.; Nussbaumer, A.; Oberer, L.; Scheel, G.; Swoboda, E.-M. Pharmacological Modulation of Endothelial Cell-associated Adhesion Molecule Expression: Implications for Future Treatment of Dermatological Diseases. J. Dermatol. 1994, 21, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Hommel, U.; Weber, H.P.; Oberer, L.; Naegeli, H.U.; Oberhauser, B.; Foster, C.A. The 3D-structure of a natural inhibitor of cell adhesion molecule expression. FEBS Lett. 1996, 379, 69–73. [Google Scholar] [CrossRef] [Green Version]
- Boger, D.L.; Chen, Y.; Foster, C.A. Synthesis and evaluation of aza HUN-7293. Bioorg. Med. Chem. Lett. 2000, 10, 1741–1744. [Google Scholar] [CrossRef]
- Chen, Y.; Bilban, M.; Foster, C.A.; Boger, D.L. Solution-phase parallel synthesis of a pharmacophore library of HUN-7293 analogues: A general chemical mutagenesis approach to defining structure-function properties of naturally occurring cyclic (depsi)peptides. J. Am. Chem. Soc. 2002, 124, 5431–5440. [Google Scholar] [CrossRef]
- Harant, H.; Lettner, N.; Hofer, L.; Oberhauser, B.; de Vries, J.E.; Lindley, I.J. The translocation inhibitor CAM741 interferes with vascular cell adhesion molecule 1 signal peptide insertion at the translocon. J. Biol. Chem. 2006, 281, 30492–30502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrison, J.L.; Kunkel, E.J.; Hegde, R.S.; Taunton, J. A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. Nature 2005, 436, 285–289. [Google Scholar] [CrossRef]
- Klein, W.; Westendorf, C.; Schmidt, A.; Conill-Cortes, M.; Rutz, C.; Blohs, M.; Beyermann, M.; Protze, J.; Krause, G.; Krause, E.; et al. Defining a conformational consensus motif in cotransin-sensitive signal sequences: A proteomic and site-directed mutagenesis study. PLoS ONE 2015, 10, e0120886. [Google Scholar] [CrossRef]
- Schreiner, E.P.; Kern, M.; Steck, A.; Foster, C.A. Synthesis of ether analogues derived from HUN-7293 and evaluation as inhibitors of VCAM-1 expression. Bioorg. Med. Chem. Lett. 2004, 14, 5003–5006. [Google Scholar] [CrossRef]
- Besemer, J.; Harant, H.; Wang, S.; Oberhauser, B.; Marquardt, K.; Foster, C.A.; Schreiner, E.P.; de Vries, J.E.; Dascher-Nadel, C.; Lindley, I.J. Selective inhibition of cotranslational translocation of vascular cell adhesion molecule 1. Nature 2005, 436, 290–293. [Google Scholar] [CrossRef]
- Harant, H.; Wolff, B.; Schreiner, E.P.; Oberhauser, B.; Hofer, L.; Lettner, N.; Maier, S.; de Vries, J.E.; Lindley, I.J. Inhibition of vascular endothelial growth factor cotranslational translocation by the cyclopeptolide CAM741. Mol. Pharm. 2007, 71, 1657–1665. [Google Scholar] [CrossRef] [PubMed]
- Westendorf, C.; Schmidt, A.; Coin, I.; Furkert, J.; Ridelis, I.; Zampatis, D.; Rutz, C.; Wiesner, B.; Rosenthal, W.; Beyermann, M.; et al. Inhibition of biosynthesis of human endothelin B receptor by the cyclodepsipeptide cotransin. J. Biol. Chem. 2011, 286, 35588–35600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maifeld, S.V.; MacKinnon, A.L.; Garrison, J.L.; Sharma, A.; Kunkel, E.J.; Hegde, R.S.; Taunton, J. Secretory protein profiling reveals TNF-alpha inactivation by selective and promiscuous Sec61 modulators. Chem. Biol. 2011, 18, 1082–1088. [Google Scholar] [CrossRef] [Green Version]
- MacKinnon, A.L.; Paavilainen, V.O.; Sharma, A.; Hegde, R.S.; Taunton, J. An allosteric Sec61 inhibitor traps nascent transmembrane helices at the lateral gate. eLife 2014, 3, e01483. [Google Scholar] [CrossRef] [Green Version]
- MacKinnon, A.L.; Garrison, J.L.; Hegde, R.S.; Taunton, J. Photo-leucine incorporation reveals the target of a cyclodepsipeptide inhibitor of cotranslational translocation. J. Am. Chem. Soc. 2007, 129, 14560–14561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palladino, M.A.; Bahjat, F.R.; Theodorakis, E.A.; Moldawer, L.L. Anti-TNF-alpha therapies: The next generation. Nat. Rev. Drug Discov. 2003, 2, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Saenz, A.; Sandhu, M.; Carrasco, Y.; Maglathlin, R.L.; Taunton, J.; Moasser, M.M. Targeting HER3 by interfering with its Sec61-mediated cotranslational insertion into the endoplasmic reticulum. Oncogene 2015, 34, 5288–5294. [Google Scholar] [CrossRef] [Green Version]
- Junne, T.; Wong, J.; Studer, C.; Aust, T.; Bauer, B.W.; Beibel, M.; Bhullar, B.; Bruccoleri, R.; Eichenberger, J.; Estoppey, D.; et al. Decatransin, a new natural product inhibiting protein translocation at the Sec61/SecYEG translocon. J. Cell Sci. 2015, 128, 1217–1229. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.C.; Chen, Z.; Jiang, Y.; Akare, S.; Kolber-Simonds, D.; Condon, K.; Agoulnik, S.; Tendyke, K.; Shen, Y.; Wu, K.M.; et al. Apratoxin A Shows Novel Pancreas-Targeting Activity through the Binding of Sec 61. Mol. Cancer Ther. 2016, 15, 1208–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paatero, A.O.; Kellosalo, J.; Dunyak, B.M.; Almaliti, J.; Gestwicki, J.E.; Gerwick, W.H.; Taunton, J.; Paavilainen, V.O. Apratoxin Kills Cells by Direct Blockade of the Sec61 Protein Translocation Channel. Cell Chem. Biol. 2016, 23, 561–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hau, A.M.; Greenwood, J.A.; Lohr, C.V.; Serrill, J.D.; Proteau, P.J.; Ganley, I.G.; McPhail, K.L.; Ishmael, J.E. Coibamide A induces mTOR-independent autophagy and cell death in human glioblastoma cells. PLoS ONE 2013, 8, e65250. [Google Scholar] [CrossRef] [Green Version]
- Luesch, H.; Chanda, S.K.; Raya, R.M.; DeJesus, P.D.; Orth, A.P.; Walker, J.R.; Izpisúa Belmonte, J.C.; Schultz, P.G. A functional genomics approach to the mode of action of apratoxin A. Nat. Chem. Biol. 2006, 2, 158–167. [Google Scholar] [CrossRef]
- Liu, Y.; Law, B.K.; Luesch, H. Apratoxin a reversibly inhibits the secretory pathway by preventing cotranslational translocation. Mol. Pharm. 2009, 76, 91–104. [Google Scholar] [CrossRef] [Green Version]
- Wan, X.; Serrill, J.D.; Humphreys, I.R.; Tan, M.; McPhail, K.L.; Ganley, I.G.; Ishmael, J.E. ATG5 Promotes Death Signaling in Response to the Cyclic Depsipeptides Coibamide A and Apratoxin A. Mar. Drugs 2018, 16, 77. [Google Scholar] [CrossRef] [Green Version]
- Medina, R.A.; Goeger, D.E.; Hills, P.; Mooberry, S.L.; Huang, N.; Romero, L.I.; Ortega-Barria, E.; Gerwick, W.H.; McPhail, K.L. Coibamide A, a potent antiproliferative cyclic depsipeptide from the Panamanian marine cyanobacterium Leptolyngbya sp. J. Am. Chem. Soc. 2008, 130, 6324–6325. [Google Scholar] [CrossRef] [Green Version]
- Serrill, J.D.; Wan, X.; Hau, A.M.; Jang, H.S.; Coleman, D.J.; Indra, A.K.; Alani, A.W.; McPhail, K.L.; Ishmael, J.E. Coibamide A, a natural lariat depsipeptide, inhibits VEGFA/VEGFR2 expression and suppresses tumor growth in glioblastoma xenografts. Investig. New Drugs 2016, 34, 24–40. [Google Scholar] [CrossRef]
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug development from marine natural products. Nat. Rev. Drug Discov. 2009, 8, 69–85. [Google Scholar] [CrossRef]
- Gerwick, W.H.; Tan, L.T.; Sitachitta, N. Nitrogen-containing metabolites from marine cyanobacteria. Alkaloids. Chem. Biol. 2001, 57, 75–184. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.T. Bioactive natural products from marine cyanobacteria for drug discovery. Phytochemistry 2007, 68, 954–979. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Puddick, J.; Prinsep, M.R.; Rottmann, M.; Tan, L.T. Lagunamides A and B: Cytotoxic and antimalarial cyclodepsipeptides from the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2010, 73, 1810–1814. [Google Scholar] [CrossRef]
- Luesch, H.; Yoshida, W.Y.; Moore, R.E.; Paul, V.J.; Corbett, T.H. Total structure determination of apratoxin A, a potent novel cytotoxin from the marine cyanobacterium Lyngbya majuscula. J. Am. Chem. Soc. 2001, 123, 5418–5423. [Google Scholar] [CrossRef] [PubMed]
- Naidoo, D.; Roy, A.; Kar, P.; Mutanda, T.; Anandraj, A. Cyanobacterial metabolites as promising drug leads against the M(pro) and PL(pro) of SARS-CoV-2: An in silico analysis. J. Biomol. Struct. Dyn. 2021, 39, 6218–6230. [Google Scholar] [CrossRef] [PubMed]
- Tranter, D.; Paatero, A.O.; Kawaguchi, S.; Kazemi, S.; Serrill, J.D.; Kellosalo, J.; Vogel, W.K.; Richter, U.; Mattos, D.R.; Wan, X.; et al. Coibamide A Targets Sec61 to Prevent Biogenesis of Secretory and Membrane Proteins. ACS Chem. Biol. 2020, 15, 2125–2136. [Google Scholar] [CrossRef]
- Shi, W.; Lu, D.; Wu, C.; Li, M.; Ding, Z.; Li, Y.; Chen, B.; Lin, X.; Su, W.; Shao, X.; et al. Coibamide A kills cancer cells through inhibiting autophagy. Biochem. Biophys. Res. Commun. 2021, 547, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.; Simmonds, R. Pleiotropic molecular effects of the Mycobacterium ulcerans virulence factor mycolactone underlying the cell death and immunosuppression seen in Buruli ulcer. Biochem. Soc. Trans. 2014, 42, 177–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morel, J.D.; Paatero, A.O.; Wei, J.; Yewdell, J.W.; Guenin-Mace, L.; Van Haver, D.; Impens, F.; Pietrosemoli, N.; Paavilainen, V.O.; Demangel, C. Proteomics Reveals Scope of Mycolactone-mediated Sec61 Blockade and Distinctive Stress Signature. Mol. Cell. Proteom. MCP 2018, 17, 1750–1765. [Google Scholar] [CrossRef] [Green Version]
- McKenna, M.; Simmonds, R.E.; High, S. Mechanistic insights into the inhibition of Sec61-dependent co- and post-translational translocation by mycolactone. J. Cell Sci. 2016, 129, 1404–1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demangel, C.; High, S. Sec61 blockade by mycolactone: A central mechanism in Buruli ulcer disease. Biol. Cell 2018, 110, 237–248. [Google Scholar] [CrossRef] [Green Version]
- Mve-Obiang, A.; Lee, R.E.; Umstot, E.S.; Trott, K.A.; Grammer, T.C.; Parker, J.M.; Ranger, B.S.; Grainger, R.; Mahrous, E.A.; Small, P.L.C. A newly discovered mycobacterial pathogen isolated from laboratory colonies of Xenopus species with lethal infections produces a novel form of mycolactone, the Mycobacterium ulcerans macrolide toxin. Infect. Immun. 2005, 73, 3307–3312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guenin-Macé, L.; Ruf, M.-T.; Pluschke, G.; Demangel, C. Mycolactone: More than Just a Cytotoxin. In Buruli Ulcer: Mycobacterium Ulcerans Disease; Pluschke, G., Röltgen, K., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 117–134. [Google Scholar] [CrossRef] [Green Version]
- George, K.M.; Chatterjee, D.; Gunawardana, G.; Welty, D.; Hayman, J.; Lee, R.; Small, P.L. Mycolactone: A polyketide toxin from Mycobacterium ulcerans required for virulence. Science 1999, 283, 854–857. [Google Scholar] [CrossRef] [Green Version]
- Guenin-Macé, L.; Baron, L.; Chany, A.C.; Tresse, C.; Saint-Auret, S.; Jönsson, F.; Le Chevalier, F.; Bruhns, P.; Bismuth, G.; Hidalgo-Lucas, S.; et al. Shaping mycolactone for therapeutic use against inflammatory disorders. Sci. Transl. Med. 2015, 7, 289ra285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baron, L.; Paatero, A.O.; Morel, J.D.; Impens, F.; Guenin-Mace, L.; Saint-Auret, S.; Blanchard, N.; Dillmann, R.; Niang, F.; Pellegrini, S.; et al. Mycolactone subverts immunity by selectively blocking the Sec61 translocon. J. Exp. Med. 2016, 213, 2885–2896. [Google Scholar] [CrossRef] [PubMed]
- Ogbechi, J.; Hall, B.S.; Sbarrato, T.; Taunton, J.; Willis, A.E.; Wek, R.C.; Simmonds, R.E. Inhibition of Sec61-dependent translocation by mycolactone uncouples the integrated stress response from ER stress, driving cytotoxicity via translational activation of ATF4. Cell Death Dis. 2018, 9, 397. [Google Scholar] [CrossRef] [Green Version]
- McKenna, M.; Simmonds, R.E.; High, S. Mycolactone reveals the substrate-driven complexity of Sec61-dependent transmembrane protein biogenesis. J. Cell Sci. 2017, 130, 1307–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerard, S.F.; Hall, B.S.; Zaki, A.M.; Corfield, K.A.; Mayerhofer, P.U.; Costa, C.; Whelligan, D.K.; Biggin, P.C.; Simmonds, R.E.; Higgins, M.K. Structure of the Inhibited State of the Sec Translocon. Mol. Cell 2020, 79, 406–415.e7. [Google Scholar] [CrossRef] [PubMed]
- Zong, G.; Barber, E.; Aljewari, H.; Zhou, J.; Hu, Z.; Du, Y.; Shi, W.Q. Total Synthesis and Biological Evaluation of Ipomoeassin F and Its Unnatural 11R-Epimer. J. Org. Chem. 2015, 80, 9279–9291. [Google Scholar] [CrossRef] [Green Version]
- Cao, S.; Norris, A.; Wisse, J.H.; Miller, J.S.; Evans, R.; Kingston, D.G. Ipomoeassin F, a new cytotoxic macrocyclic glycoresin from the leaves of Ipomoea squamosa from the Suriname rainforest. Nat. Prod. Res. 2007, 21, 872–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zong, G.; Hu, Z.; O’Keefe, S.; Tranter, D.; Iannotti, M.J.; Baron, L.; Hall, B.; Corfield, K.; Paatero, A.O.; Henderson, M.J.; et al. Ipomoeassin F Binds Sec61alpha to Inhibit Protein Translocation. J. Am. Chem. Soc. 2019, 141, 8450–8461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Keefe, S.; Roboti, P.; Duah, K.B.; Zong, G.; Schneider, H.; Shi, W.Q.; High, S. Ipomoeassin-F inhibits the in vitro biogenesis of the SARS-CoV-2 spike protein and its host cell membrane receptor. J. Cell Sci. 2021, 134, jcs257758. [Google Scholar] [CrossRef]
- Roboti, P.; O’Keefe, S.; Duah, K.B.; Shi, W.Q.; High, S. Ipomoeassin-F disrupts multiple aspects of secretory protein biogenesis. Sci. Rep. 2021, 11, 11562. [Google Scholar] [CrossRef]
- Zong, G.; Hu, Z.; Duah, K.B.; Andrews, L.E.; Zhou, J.; O’Keefe, S.; Whisenhunt, L.; Shim, J.S.; Du, Y.; High, S.; et al. Ring Expansion Leads to a More Potent Analogue of Ipomoeassin F. J. Org. Chem. 2020, 85, 16226–16235. [Google Scholar] [CrossRef] [PubMed]
- Kazemi, S.; Kawaguchi, S.; Badr, C.E.; Mattos, D.R.; Ruiz-Saenz, A.; Serrill, J.D.; Moasser, M.M.; Dolan, B.P.; Paavilainen, V.O.; Oishi, S.; et al. Targeting of HER/ErbB family proteins using broad spectrum Sec61 inhibitors coibamide A and apratoxin A. Biochem. Pharm. 2021, 183, 114317. [Google Scholar] [CrossRef]
- Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef]
- Pauwels, E.; Rutz, C.; Provinciael, B.; Stroobants, J.; Schols, D.; Hartmann, E.; Krause, E.; Stephanowitz, H.; Schulein, R.; Vermeire, K. A Proteomic Study on the Membrane Protein Fraction of T Cells Confirms High Substrate Selectivity for the ER Translocation Inhibitor Cyclotriazadisulfonamide. Mol. Cell. Proteom. MCP 2021, 20, 100144. [Google Scholar] [CrossRef]
- Claeys, E.; Pauwels, E.; Humblet-Baron, S.; Provinciael, B.; Schols, D.; Waer, M.; Sprangers, B.; Vermeire, K. Small Molecule Cyclotriazadisulfonamide Abrogates the Upregulation of the Human Receptors CD4 and 4-1BB and Suppresses In Vitro Activation and Proliferation of T Lymphocytes. Front. Immunol. 2021, 12, 1340. [Google Scholar] [CrossRef] [PubMed]
- Van Puyenbroeck, V.; Claeys, E.; Schols, D.; Bell, T.W.; Vermeire, K. A Proteomic Survey Indicates Sortilin as a Secondary Substrate of the ER Translocation Inhibitor Cyclotriazadisulfonamide (CADA). Mol. Cell. Proteom. MCP 2017, 16, 157–167. [Google Scholar] [CrossRef] [Green Version]
- Cross, B.C.; McKibbin, C.; Callan, A.C.; Roboti, P.; Piacenti, M.; Rabu, C.; Wilson, C.M.; Whitehead, R.; Flitsch, S.L.; Pool, M.R.; et al. Eeyarestatin I inhibits Sec61-mediated protein translocation at the endoplasmic reticulum. J. Cell Sci. 2009, 122, 4393–4400. [Google Scholar] [CrossRef] [Green Version]
- Lowe, E.; Fan, R.; Jiang, J.; Johnson, H.; Kirk, C.; McMinn, D.; Millare, B.; Muchamuel, T.; Qian, Y.; Tuch, B.; et al. Preclinical evaluation of KZR-261, a novel small molecule inhibitor of Sec61. J. Clin. Oncol. 2020, 38, 3582. [Google Scholar] [CrossRef]
- Vermeire, K.; Princen, K.; Hatse, S.; De Clercq, E.; Dey, K.; Bell, T.W.; Schols, D. CADA, a novel CD4-targeted HIV inhibitor, is synergistic with various anti-HIV drugs in vitro. AIDS 2004, 18, 2115–2125. [Google Scholar] [CrossRef]
- Berger, K.; Pauwels, E.; Parkinson, G.; Landberg, G.; Le, T.; Demillo, V.G.; Lumangtad, L.A.; Jones, D.E.; Islam, M.A.; Olsen, R.; et al. Reduction of Progranulin-Induced Breast Cancer Stem Cell Propagation by Sortilin-Targeting Cyclotriazadisulfonamide (CADA) Compounds. J. Med. Chem. 2021, 64, 12865–12876. [Google Scholar] [CrossRef] [PubMed]
- Bell, T.W.; Anugu, S.; Bailey, P.; Catalano, V.J.; Dey, K.; Drew, M.G.; Duffy, N.H.; Jin, Q.; Samala, M.F.; Sodoma, A.; et al. Synthesis and structure-activity relationship studies of CD4 down-modulating cyclotriazadisulfonamide (CADA) analogues. J. Med. Chem. 2006, 49, 1291–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chawla, R.; Van Puyenbroeck, V.; Pflug, N.C.; Sama, A.; Ali, R.; Schols, D.; Vermeire, K.; Bell, T.W. Tuning Side Arm Electronics in Unsymmetrical Cyclotriazadisulfonamide (CADA) Endoplasmic Reticulum (ER) Translocation Inhibitors to Improve their Human Cluster of Differentiation 4 (CD4) Receptor Down-Modulating Potencies. J. Med. Chem. 2016, 59, 2633–2647. [Google Scholar] [CrossRef] [PubMed]
- Vermeire, K.; Lisco, A.; Grivel, J.C.; Scarbrough, E.; Dey, K.; Duffy, N.; Margolis, L.; Bell, T.W.; Schols, D. Design and cellular kinetics of dansyl-labeled CADA derivatives with anti-HIV and CD4 receptor down-modulating activity. Biochem. Pharm. 2007, 74, 566–578. [Google Scholar] [CrossRef] [PubMed]
- Lumangtad, L.A.; Claeys, E.; Hamal, S.; Intasiri, A.; Basrai, C.; Yen-Pon, E.; Beenfeldt, D.; Vermeire, K.; Bell, T.W. Syntheses and anti-HIV and human cluster of differentiation 4 (CD4) down-modulating potencies of pyridine-fused cyclotriazadisulfonamide (CADA) compounds. Bioorg. Med. Chem. 2020, 28, 115816. [Google Scholar] [CrossRef] [PubMed]
- Demillo, V.G.; Goulinet-Mateo, F.; Kim, J.; Schols, D.; Vermeire, K.; Bell, T.W. Unsymmetrical cyclotriazadisulfonamide (CADA) compounds as human CD4 receptor down-modulating agents. J. Med. Chem. 2011, 54, 5712–5721. [Google Scholar] [CrossRef] [PubMed]
- Bell, T.W.; Demillo, V.G.; Schols, D.; Vermeire, K. Improving potencies and properties of CD4 down-modulating CADA analogs. Expert Opin. Drug Discov. 2012, 7, 39–48. [Google Scholar] [CrossRef]
- Vermeire, K.; Bell, T.W.; Van Puyenbroeck, V.; Giraut, A.; Noppen, S.; Liekens, S.; Schols, D.; Hartmann, E.; Kalies, K.U.; Marsh, M. Signal peptide-binding drug as a selective inhibitor of co-translational protein translocation. PLoS Biol. 2014, 12, e1002011. [Google Scholar] [CrossRef] [Green Version]
- Van Puyenbroeck, V.; Pauwels, E.; Provinciael, B.; Bell, T.W.; Schols, D.; Kalies, K.U.; Hartmann, E.; Vermeire, K. Preprotein signature for full susceptibility to the co-translational translocation inhibitor cyclotriazadisulfonamide. Traffic 2020, 21, 250–264. [Google Scholar] [CrossRef]
- Fiebiger, E.; Hirsch, C.; Vyas, J.M.; Gordon, E.; Ploegh, H.L.; Tortorella, D. Dissection of the dislocation pathway for type I membrane proteins with a new small molecule inhibitor, eeyarestatin. Mol. Biol. Cell 2004, 15, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, L.; Ye, Y. Inhibition of p97-dependent protein degradation by Eeyarestatin I. J. Biol. Chem. 2008, 283, 7445–7454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, D.M. Could a Common Mechanism of Protein Degradation Impairment Underlie Many Neurodegenerative Diseases? J. Exp. Neurosci. 2018, 12, 1179069518794675. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Shinkre, B.A.; Lee, J.G.; Weniger, M.A.; Liu, Y.; Chen, W.; Wiestner, A.; Trenkle, W.C.; Ye, Y. The ERAD inhibitor Eeyarestatin I is a bifunctional compound with a membrane-binding domain and a p97/VCP inhibitory group. PLoS ONE 2010, 5, e15479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Mora-Jensen, H.; Weniger, M.A.; Perez-Galan, P.; Wolford, C.; Hai, T.; Ron, D.; Chen, W.; Trenkle, W.; Wiestner, A.; et al. ERAD inhibitors integrate ER stress with an epigenetic mechanism to activate BH3-only protein NOXA in cancer cells. Proc. Natl. Acad. Sci. USA 2009, 106, 2200–2205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, S.; Erdmann, F.; Jung, M.; Wagner, R.; Cavalie, A.; Zimmermann, R. Sec61 complexes form ubiquitous ER Ca2+ leak channels. Channels 2011, 5, 228–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamayun, I.; O’Keefe, S.; Pick, T.; Klein, M.C.; Nguyen, D.; McKibbin, C.; Piacenti, M.; Williams, H.M.; Flitsch, S.L.; Whitehead, R.C.; et al. Eeyarestatin Compounds Selectively Enhance Sec61-Mediated Ca(2+) Leakage from the Endoplasmic Reticulum. Cell Chem. Biol. 2019, 26, 571–583.e6. [Google Scholar] [CrossRef] [Green Version]
- Ding, R.; Zhang, T.; Xie, J.; Williams, J.; Ye, Y.; Chen, L. Eeyarestatin I derivatives with improved aqueous solubility. Bioorg. Med. Chem. Lett. 2016, 26, 5177–5181. [Google Scholar] [CrossRef]
- Du, R.; Sullivan, D.K.; Azizian, N.G.; Liu, Y.; Li, Y. Inhibition of ERAD synergizes with FTS to eradicate pancreatic cancer cells. BMC Cancer 2021, 21, 237. [Google Scholar] [CrossRef] [PubMed]
- Aletrari, M.O.; McKibbin, C.; Williams, H.; Pawar, V.; Pietroni, P.; Lord, J.M.; Flitsch, S.L.; Whitehead, R.; Swanton, E.; High, S.; et al. Eeyarestatin 1 interferes with both retrograde and anterograde intracellular trafficking pathways. PLoS ONE 2011, 6, e22713. [Google Scholar] [CrossRef] [Green Version]
- McKibbin, C.; Mares, A.; Piacenti, M.; Williams, H.; Roboti, P.; Puumalainen, M.; Callan, A.C.; Lesiak-Mieczkowska, K.; Linder, S.; Harant, H.; et al. Inhibition of protein translocation at the endoplasmic reticulum promotes activation of the unfolded protein response. Biochem. J. 2012, 442, 639–648. [Google Scholar] [CrossRef]
- Auner, H.W.; Moody, A.M.; Ward, T.H.; Kraus, M.; Milan, E.; May, P.; Chaidos, A.; Driessen, C.; Cenci, S.; Dazzi, F.; et al. Combined inhibition of p97 and the proteasome causes lethal disruption of the secretory apparatus in multiple myeloma cells. PLoS ONE 2013, 8, e74415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steenhuis, M.; Koningstein, G.M.; Oswald, J.; Pick, T.; O’Keefe, S.; Koch, H.G.; Cavalie, A.; Whitehead, R.C.; Swanton, E.; High, S.; et al. Eeyarestatin 24 impairs SecYEG-dependent protein trafficking and inhibits growth of clinically relevant pathogens. Mol. Microbiol. 2021, 115, 28–40. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).