PAMAM-calix-dendrimers: Synthesis and Thiacalixarene Conformation Effect on DNA Binding
Abstract
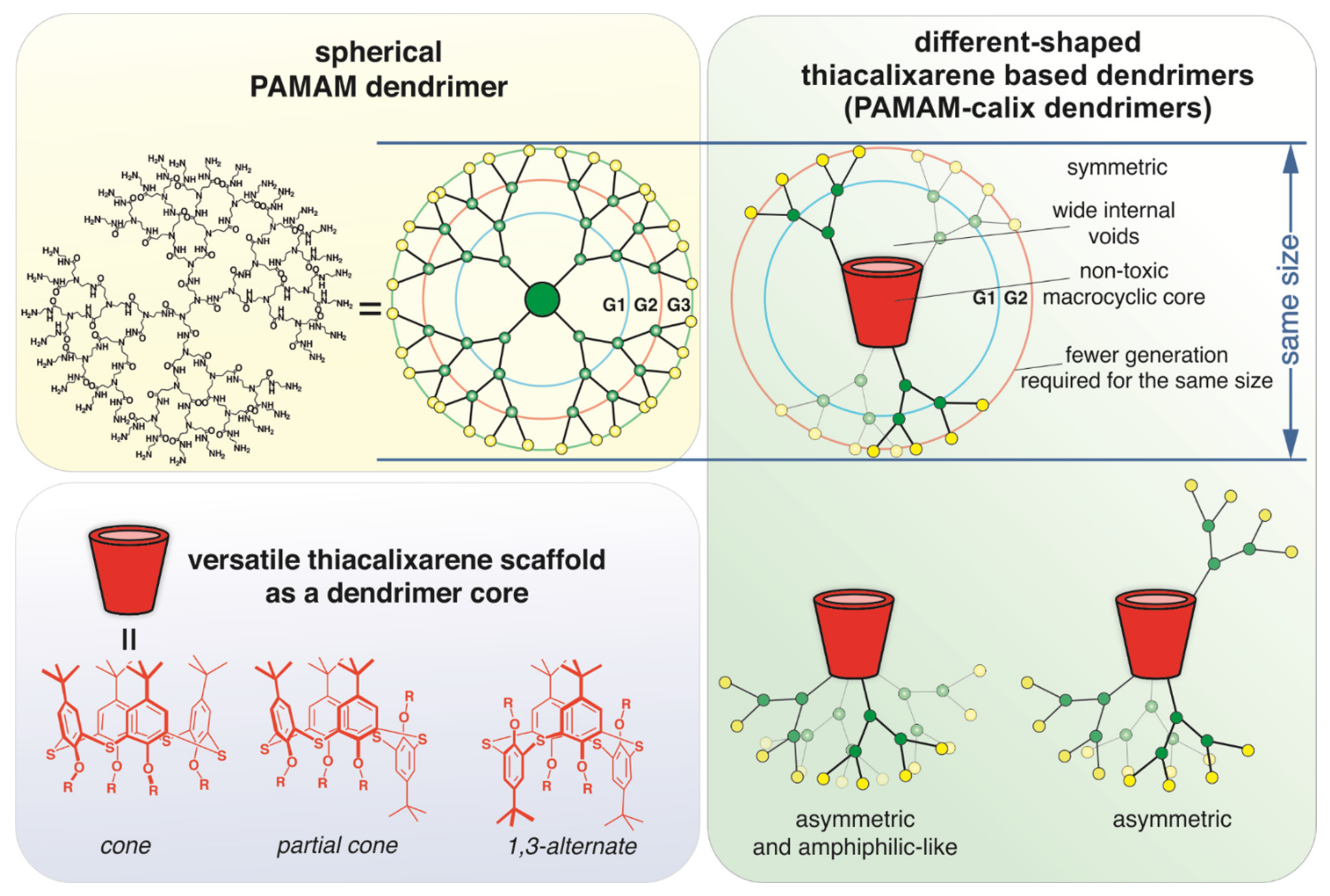
1. Introduction
2. Results and Discussion
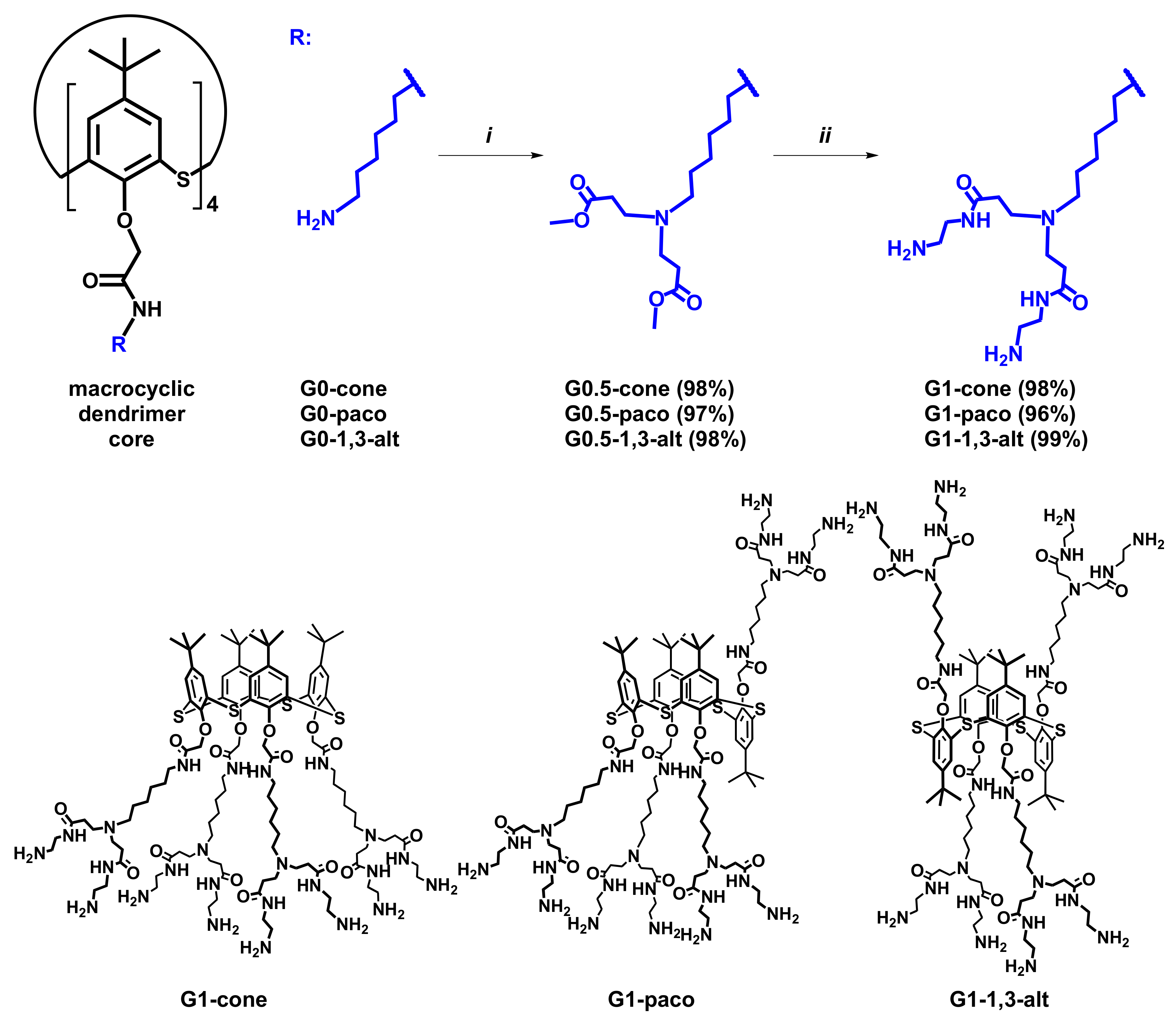
2.1. Synthesis of (Poly)Amidoamine Dendrimers Based on the p-tert-Buthylthiacalix[4]arene (PAMAM-Calix-Dendrimer) in Different Conformations
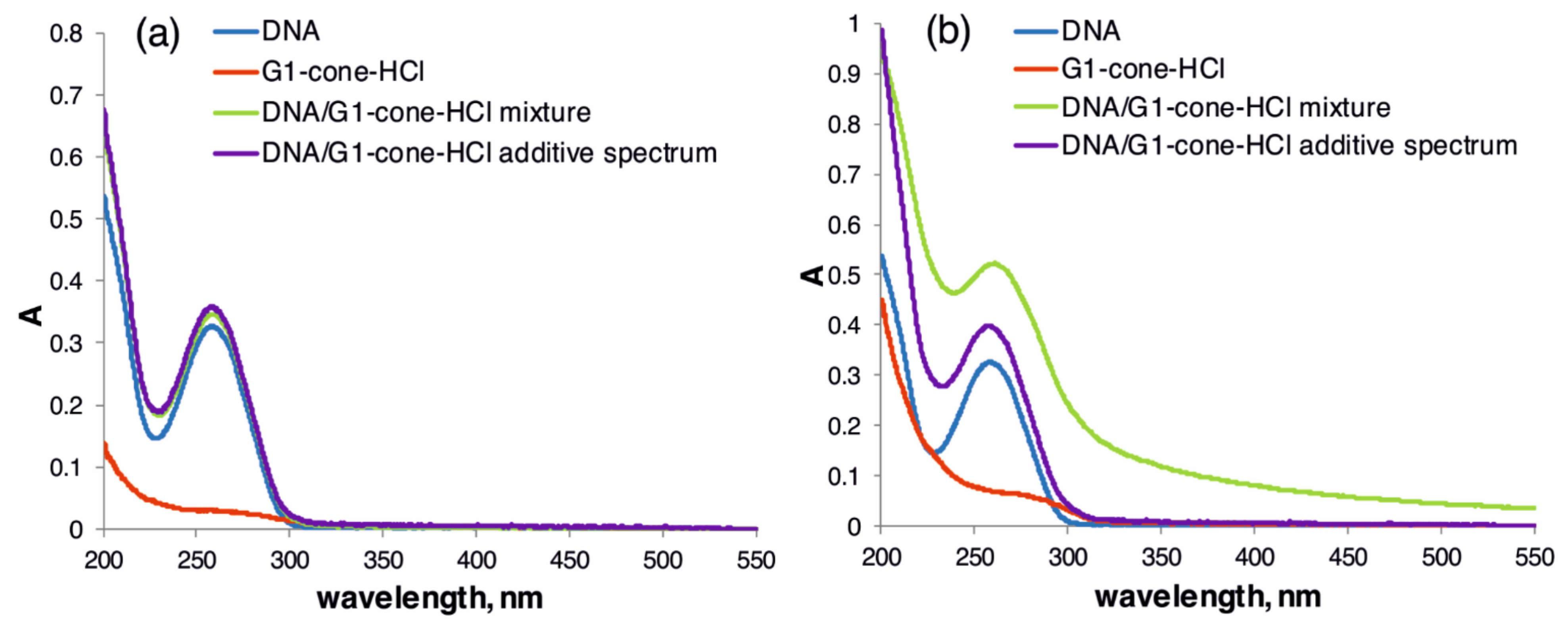
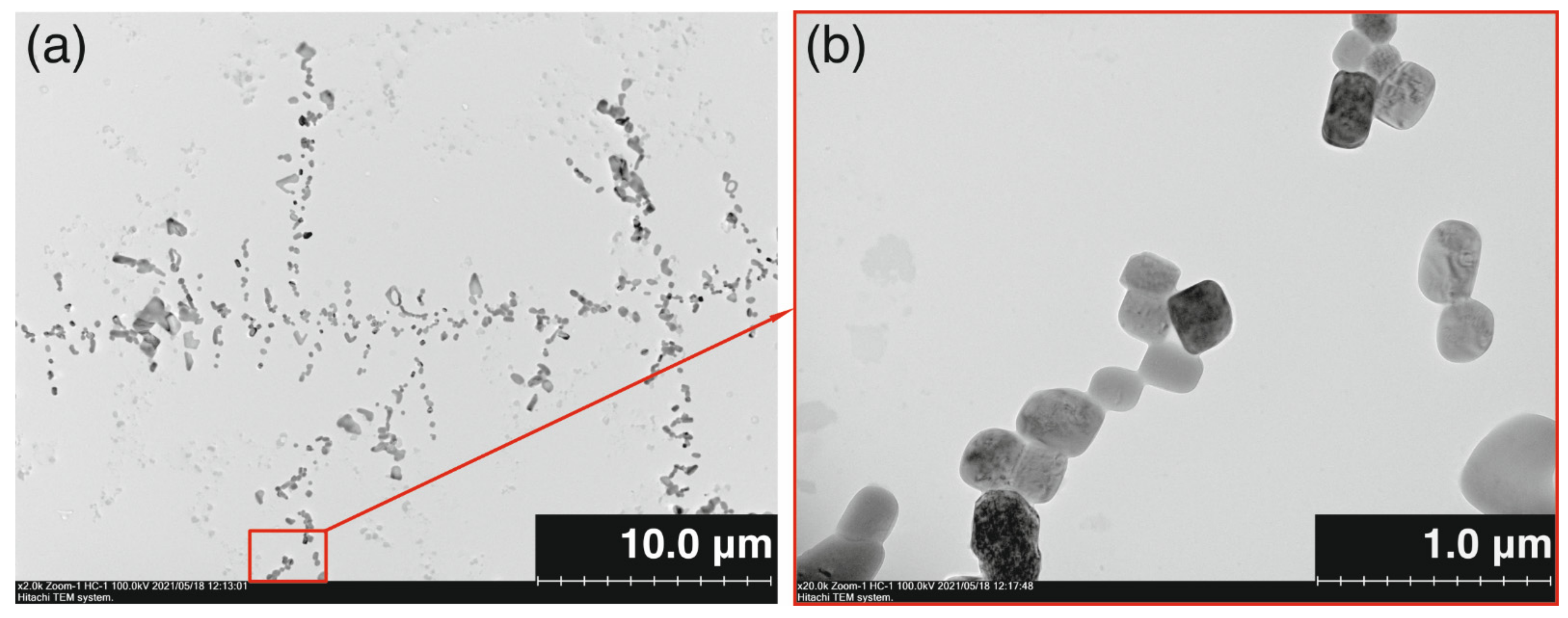
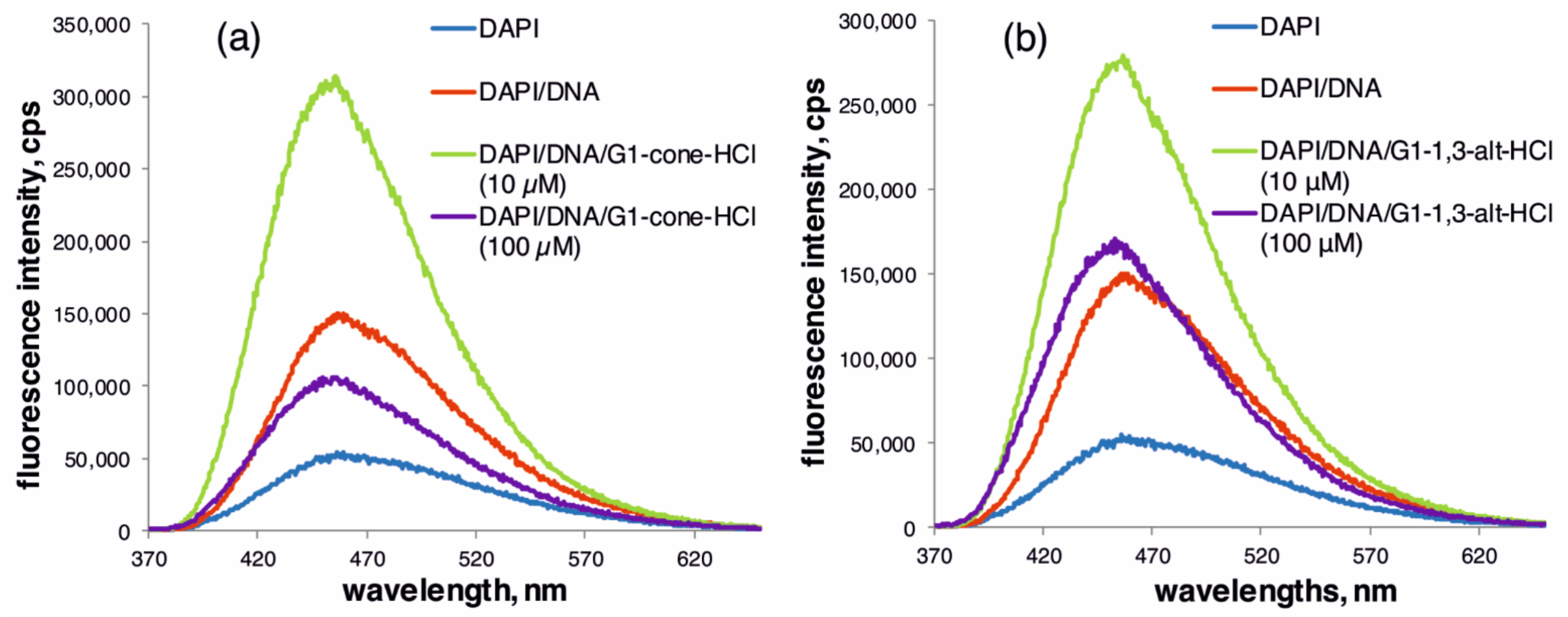
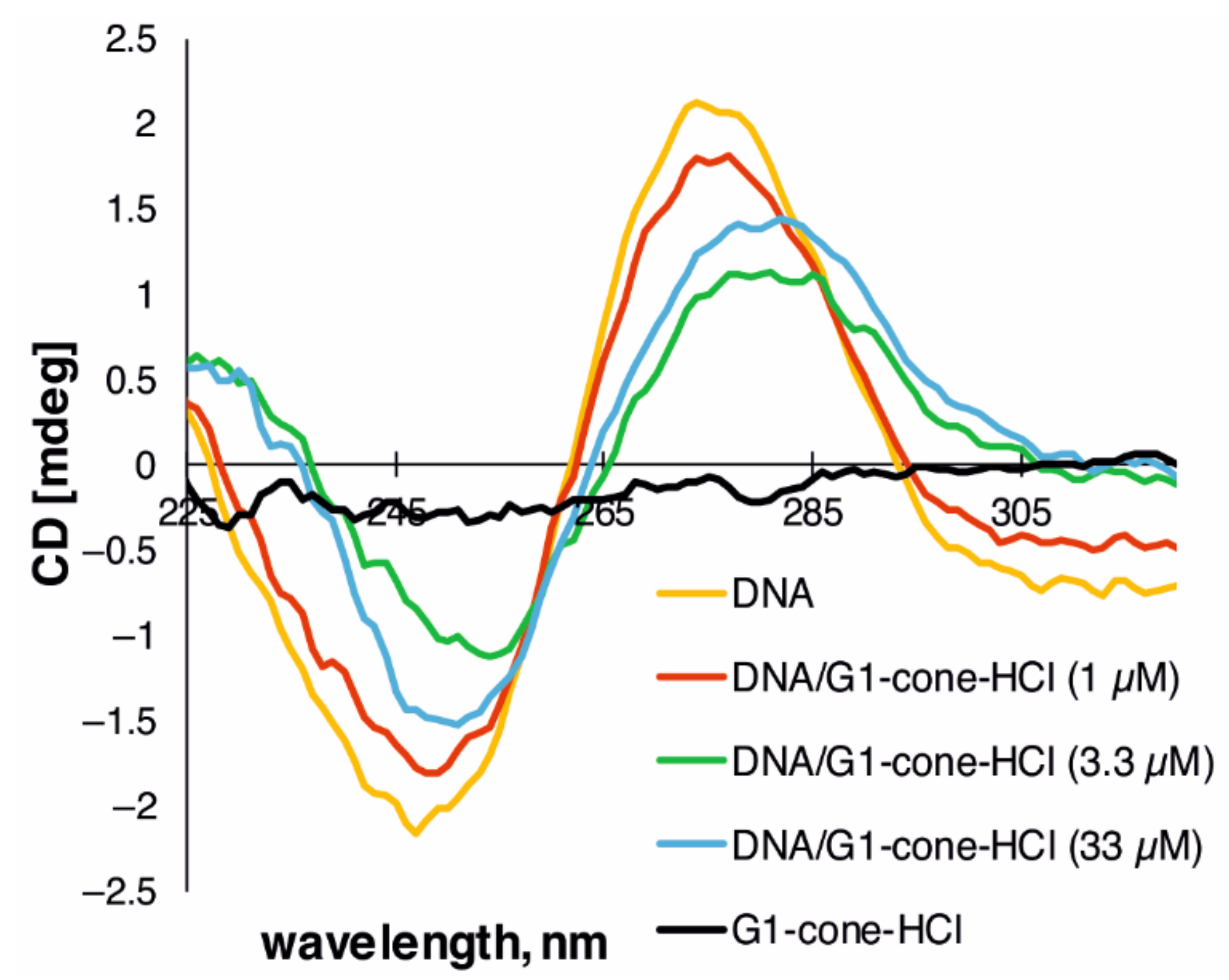
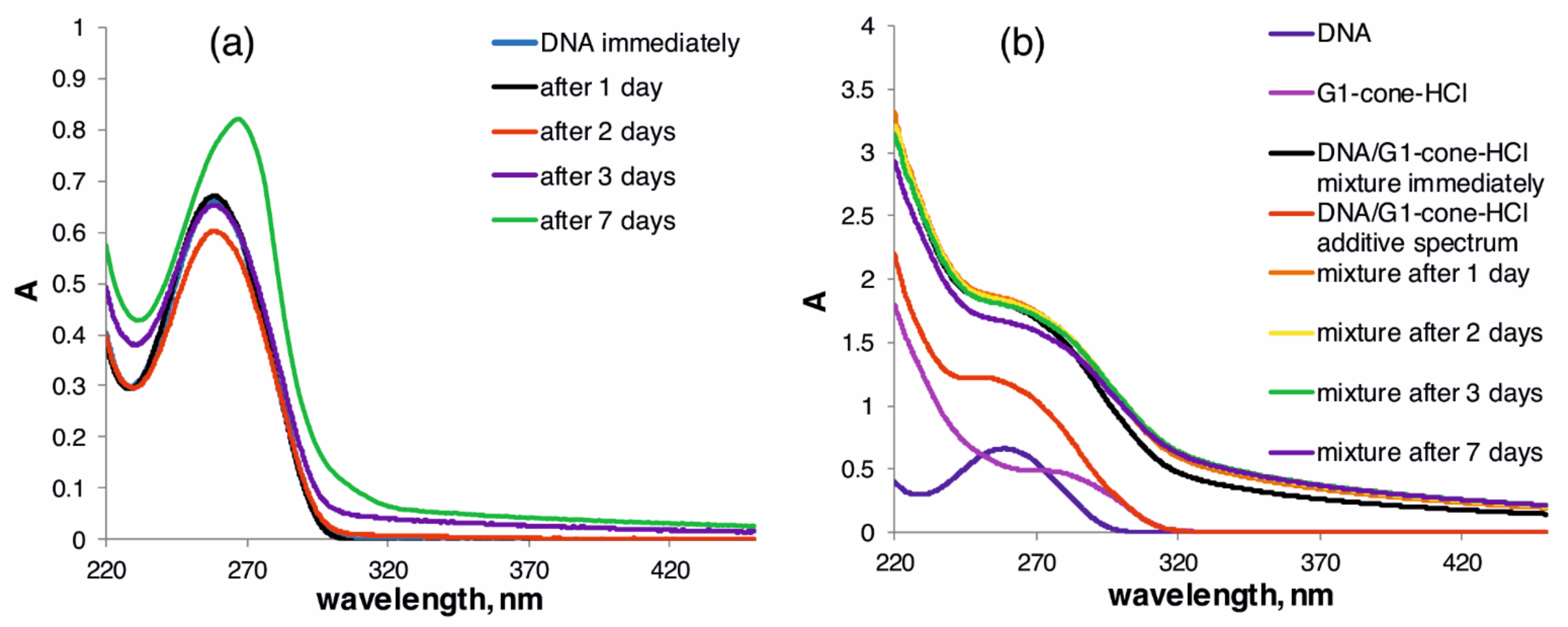
2.2. The Study of Binding of the Obtained PAMAM-Calix-Dendrimers G1 with Salmon Sperm DNA and the Effect on DNA Temporal Stability
3. Materials and Methods
3.1. General Experimental Information
3.2. General Procedure for the Synthesis of the Compounds G0.5 (Generation 0.5 Dendrimers with Thiacalix[4]arene Core in Cone (G0.5-cone), Partial Cone (G0.5-paco) and 1,3-Alternate (G0.5-1,3-Alt) Conformation)
3.2.1. G0.5-cone, 5,11,17,23-Tetra-tert-butyl-25,26,27,28-tetrakis[N-(6-(N,N-di(methoxycarbonylethyl)amino)hexyl)carbamoylmethoxy]-2,8,14,20-tetrathiacalix[4]arene
3.2.2. G0.5-paco, 5,11,17,23-Tetra-tert-butyl-25,26,27,28-tetrakis[N-(6-(N,N-di(methoxycarbonylethyl)amino)hexyl)carbamoylmethoxy]-2,8,14,20-tetrathiacalix[4]arene
3.2.3. G0.5-1,3-alt, 5,11,17,23-Tetra-tert-butyl-25,26,27,28-tetrakis[N-(6-(N,N-di(methoxycarbonylethyl)amino)hexyl)carbamoylmethoxy]-2,8,14,20-tetrathiacalix[4]arene
3.3. General Procedure for the Synthesis of the Compounds G1 (Generation 1 Dendrimers with Thiacalix[4]arene Core in Cone (G1-Cone), Partial Cone (G1-Paco), and 1,3-Alternate (G1-1,3-alt) Conformation)
3.3.1. G1-cone, 5,11,17,23-Tetra-tert-butyl-25,26,27,28-tetrakis[N-(6-(N,N-di(N-(2-aminoethyl)carbamoylethyl)amino)hexyl)carbamoylmethoxy]-2,8,14,20-tetrathiacalix[4]arene
3.3.2. G1-paco, 5,11,17,23-Tetra-tert-butyl-25,26,27,28-tetrakis[N-(6-(N,N-di(N-(2-aminoethyl)carbamoylethyl)amino)hexyl)carbamoylmethoxy]-2,8,14,20-tetrathiacalix[4]arene
3.3.3. G1-1,3-alt, 5,11,17,23-Tetra-tert-butyl-25,26,27,28-tetrakis[N-(6-(N,N-di(N-(2-aminoethyl)carbamoylethyl)amino)hexyl)carbamoylmethoxy]-2,8,14,20-tetrathiacalix[4]arene
3.4. Preparation of the Compounds G1-HCl
3.5. UV-Visible Spectroscopy
3.6. Study of DNA Stability in the Presence of G1-HCl Compounds by UV-Visible Spectroscopy
3.7. Fluorescence Spectroscopy
3.8. Dynamic Light Scattering (DLS) Determination of the Hydrodynamic Size of the Particles
3.9. Electrokinetic Potentials
3.10. Circular Dichroism (CD) Studies
3.11. Transmission Electron Microscopy (TEM)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodríguez-Gascón, A.; del Pozo-Rodríguez, A.; Solinis, M.A. Development of nucleic acid vaccines: Use of self-amplifying RNA in lipid nanoparticles. Int. J. Nanomed. 2014, 9, 1833–1843. [Google Scholar] [CrossRef] [PubMed]
- Zygmuntowicz, A.; Burmańczuk, A.; Markiewicz, W. Selected Biological Medicinal Products and Their Veterinary Use. Animals 2020, 10, 2343. [Google Scholar] [CrossRef]
- Sharpe, H.R.; Gilbride, C.; Allen, E.; Belij-Rammerstorfer, S.; Bissett, C.; Ewer, K.; Lambe, T. The early landscape of coronavirus disease 2019 vaccine development in the UK and rest of the world. Immunology 2020, 160, 223–232. [Google Scholar] [CrossRef]
- Yamakawa, K.; Nakano-Narusawa, Y.; Hashimoto, N.; Yokohira, M.; Matsuda, Y. Development and Clinical Trials of Nucleic Acid Medicines for Pancreatic Cancer Treatment. Int. J. Mol. Sci. 2019, 20, 4224. [Google Scholar] [CrossRef] [PubMed]
- Pescador, R.; Capuzzi, L.; Mantovani, M.; Fulgenzi, A.; Ferrero, M. Defibrotide: Properties and clinical use of an old/new drug. Vasc. Pharmacol. 2013, 59, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Squadrito, F.; Bitto, A.; Irrera, N.; Pizzino, G.; Pallio, G.; Minutoli, L.; Altavilla, D. Pharmacological Activity and Clinical Use of PDRN. Front. Pharmacol. 2017, 8, 224. [Google Scholar] [CrossRef]
- Lee, D.-W.; Hyun, H.; Lee, S.; Kim, S.Y.; Kim, G.-T.; Um, S.; Hong, S.O.; Chun, H.J.; Yang, D.H. The Effect of Polydeoxyribonucleotide Extracted from Salmon Sperm on the Restoration of Bisphosphonate-Related Osteonecrosis of the Jaw. Mar. Drugs 2019, 17, 51. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-H.; Heo, S.-Y.; Oh, G.-W.; Heo, S.-J.; Jung, W.-K. Applications of Marine Organism-Derived Polydeoxyribonucleotide: Its Potential in Biomedical Engineering. Mar. Drugs 2021, 19, 296. [Google Scholar] [CrossRef]
- Matange, K.; Tuck, J.M.; Keung, A.J. DNA stability: A central design consideration for DNA data storage systems. Nat. Commun. 2021, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mendes, L.P.; Pan, J.; Torchilin, V.P. Dendrimers as Nanocarriers for Nucleic Acid and Drug Delivery in Cancer Therapy. Molecules 2017, 22, 1401. [Google Scholar] [CrossRef] [PubMed]
- Ceze, L.; Nivala, J.; Strauss, K. Molecular digital data storage using DNA. Nat. Rev. Genet. 2019, 20, 456–466. [Google Scholar] [CrossRef]
- Tan, X.; Ge, L.; Zhang, T.; Lu, Z. Preservation of DNA for data storage. Russ. Chem. Rev. 2021, 90, 280–291. [Google Scholar] [CrossRef]
- Gibson, D.S. Drug–DNA interactions and novel drug design. Pharmacogenomics J. 2002, 2, 275–276. [Google Scholar] [CrossRef] [PubMed]
- Portugal, J.; Barceló, F. Noncovalent Binding to DNA: Still a Target in Developing Anticancer Agents. Curr. Med. Chem. 2016, 23, 4108–4134. [Google Scholar] [CrossRef] [PubMed]
- Khadieva, A.; Mostovaya, O.; Padnya, P.; Kalinin, V.; Grishaev, D.; Tumakov, D.; Stoikov, I. Arylamine Analogs of Methylene Blue: Substituent Effect on Aggregation Behavior and DNA Binding. Int. J. Mol. Sci. 2021, 22, 5847. [Google Scholar] [CrossRef]
- Pedziwiatr-Werbicka, E.; Milowska, K.; Dzmitruk, V.; Ionov, M.; Shcharbin, D.; Bryszewska, M. Dendrimers and hyperbranched structures for biomedical applications. Eur. Polym. J. 2019, 119, 61–73. [Google Scholar] [CrossRef]
- Padnya, P.L.; Andreyko, E.A.; Mostovaya, O.A.; Rizvanov, I.K.; Stoikov, I.I. The synthesis of new amphiphilic p-tert-butylthiacalix[4]arenes containing peptide fragments and their interaction with DNA. Org. Biomol. Chem. 2015, 13, 5894–5904. [Google Scholar] [CrossRef]
- Yakimova, L.S.; Nugmanova, A.R.; Mostovaya, O.A.; Vavilova, A.A.; Shurpik, D.N.; Mukhametzyanov, T.A.; Stoikov, I.I. Nanostructured Polyelectrolyte Complexes Based on Water-Soluble Thiacalix[4]Arene and Pillar[5]Arene: Self-Assembly in Micelleplexes and Polyplexes at Packaging DNA. Nanomaterials 2020, 10, 777. [Google Scholar] [CrossRef]
- Gallego-Yerga, L.; Lomazzi, M.; Franceschi, V.; Sansone, F.; Mellet, C.O.; Donofrio, G.; Casnati, A.; Fernández, J.M.G. Cyclodextrin- and calixarene-based polycationic amphiphiles as gene delivery systems: A structure–activity relationship study. Org. Biomol. Chem. 2014, 13, 1708–1723. [Google Scholar] [CrossRef]
- Basilotta, R.; Mannino, D.; Filippone, A.; Casili, G.; Prestifilippo, A.; Colarossi, L.; Raciti, G.; Esposito, E.; Campolo, M. Role of Calixarene in Chemotherapy Delivery Strategies. Molecules 2021, 26, 3963. [Google Scholar] [CrossRef] [PubMed]
- Shurpik, D.N.; Padnya, P.L.; Stoikov, I.I.; Cragg, P.J. Antimicrobial Activity of Calixarenes and Related Macrocycles. Molecules 2020, 25, 5145. [Google Scholar] [CrossRef] [PubMed]
- Morohashi, N.; Narumi, F.; Iki, N.; Hattori, A.T.; Miyano, S. Thiacalixarenes. Chem. Rev. 2006, 106, 5291–5316. [Google Scholar] [CrossRef] [PubMed]
- Mostovaya, O.A.; Gorbachuk, V.V.; Padnya, P.; Vavilova, A.A.; Evtugyn, G.A.; Stoikov, I.I. Modification of Oligo- and Polylactides with Macrocyclic Fragments: Synthesis and Properties. Front. Chem. 2019, 7, 554. [Google Scholar] [CrossRef]
- Fasting, C.; Schalley, C.A.; Weber, M.; Seitz, O.; Hecht, S.; Koksch, B.; Dernedde, J.; Graf, C.; Knapp, E.-W.; Haag, R. Multivalency as a Chemical Organization and Action Principle. Angew. Chem. Int. Ed. 2012, 51, 10472–10498. [Google Scholar] [CrossRef]
- Antipin, I.S.; Alfimov, M.V.; Arslanov, V.V.; Burilov, V.A.; Vatsadze, S.Z.; Voloshin, Y.Z.; Volcho, K.P.; Gorbatchuk, V.V.; Gorbunova, Y.G.; Gromov, S.P.; et al. Functional supramolecular systems: Design and applications. Russ. Chem. Rev. 2021, 90, 895–1107. [Google Scholar] [CrossRef]
- Mostovaya, O.; Padnya, P.; Shurpik, D.; Shiabiev, I.; Stoikov, I. Novel lactide derivatives of p-tert-butylthiacalix[4]arene: Directed synthesis and molecular recognition of catecholamines. J. Mol. Liq. 2020, 327, 114806. [Google Scholar] [CrossRef]
- Ahmed, M. Peptides, polypeptides and peptide–polymer hybrids as nucleic acid carriers. Biomater. Sci. 2017, 5, 2188–2211. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Z.; Ding, L.; Huang, A.-T.; Kao, C.-L.; Peng, L. Poly(amidoamine) dendrimers: Covalent and supramolecular synthesis. Mater. Today Chem. 2019, 13, 34–48. [Google Scholar] [CrossRef]
- Tomalia, D.A.; Frechet, J. Discovery of dendrimers and dendritic polymers: A brief historical perspective*. J. Polym. Sci. Part A: Polym. Chem. 2002, 40, 2719–2728. [Google Scholar] [CrossRef]
- Kallos, G.J.; Tomalia, D.A.; Hedstrand, D.M.; Lewis, S.; Zhou, J. Molecular weight determination of a polyamidoamine Starburst polymer by electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 1991, 5, 383–386. [Google Scholar] [CrossRef]
- Allikmaa, V.; Lopp, M.; Pehk, T.; Peterson, J. Fragmentation of pamam dendrimers in methanol. Proc. Estonian Acad. Sci. Chem. 2001, 50, 167–172. [Google Scholar] [CrossRef]
- Fischer, D.; Li, Y.; Ahlemeyer, B.; Krieglstein, J.; Kissel, T. In vitro cytotoxicity testing of polycations: Influence of polymer structure on cell viability and hemolysis. Biomaterials 2002, 24, 1121–1131. [Google Scholar] [CrossRef]
- Duncan, R.; Izzo, L. Dendrimer biocompatibility and toxicity. Adv. Drug Deliv. Rev. 2005, 57, 2215–2237. [Google Scholar] [CrossRef] [PubMed]
- Labieniec-Watala, M.; Watala, C. PAMAM Dendrimers: Destined for Success or Doomed to Fail? Plain and Modified PAMAM Dendrimers in the Context of Biomedical Applications. J. Pharm. Sci. 2015, 104, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Baklouti, L.; Cheriaa, N.; Mahouachi, M.; Abidi, R.; Kim, J.S.; Kim, Y.; Vicens, J. Calixarene-Based Dendrimers. A Timely Review. J. Incl. Phenom. Macrocycl. Chem. 2006, 54, 1–7. [Google Scholar] [CrossRef]
- Rahimi, M.; Karimian, R.; Mostafidi, E.; Noruzi, E.B.; Taghizadeh, S.; Shokouhi, B.; Kafil, H.S. Highly branched amine-functionalized p-sulfonatocalix[4]arene decorated with human plasma proteins as a smart, targeted, and stealthy nano-vehicle for the combination chemotherapy of MCF7 cells. New J. Chem. 2018, 42, 13010–13024. [Google Scholar] [CrossRef]
- Ficker, M.; Paolucci, V.; Christensen, J.B. Improved large-scale synthesis and characterization of small and medium generation PAMAM dendrimers. Can. J. Chem. 2017, 95, 954–964. [Google Scholar] [CrossRef]
- Tomalia, D.A.; Naylor, A.M.; Iii, W.A.G. Starburst Dendrimers: Molecular-Level Control of Size, Shape, Surface Chemistry, Topology, and Flexibility from Atoms to Macroscopic Matter. Angew. Chem. Int. Ed. 1990, 29, 138–175. [Google Scholar] [CrossRef]
- Mostovaya, O.A.; Gorbachuk, V.V.; Bazanova, O.B.; Gerasimov, A.V.; Evtugyn, V.G.; Osin, Y.N.; Myakushev, V.D.; Rizvanov, I.K.; Stoikov, I.I. Thiacalixarene “knot” effect on protein binding by oligolactic acid particles. Mater. Chem. Front. 2018, 3, 292–300. [Google Scholar] [CrossRef]
- Mostovaya, O.A.; Padnya, P.; Shurpik, D.; Vavilova, A.; Evtugyn, V.G.; Osin, Y.; Stoikov, I.I. Iminodiacetic Derivatives of p-tert-Butylthiacalix[4]arene: Synthesis and Influence of Conformation on the Aggregation with Bismarck Brown Y. Macroheterocycles 2017, 10, 154–163. [Google Scholar] [CrossRef]
- Padnya, P.; Shibaeva, K.; Arsenyev, M.; Baryshnikova, S.; Terenteva, O.; Shiabiev, I.; Khannanov, A.; Boldyrev, A.; Gerasimov, A.; Grishaev, D.; et al. Catechol-Containing Schiff Bases on Thiacalixarene: Synthesis, Copper (II) Recognition, and Formation of Organic-Inorganic Copper-Based Materials. Molecules 2021, 26, 2334. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Tajmir-Riahi, H.A. Polyamine–DNA interactions and development of gene delivery vehicles. Amino Acids 2016, 48, 2423–2431. [Google Scholar] [CrossRef]
- Lang, K.; Prošková, P.; Kroupa, J.; Morávek, J.; Stibor, I.; Pojarová, M.; Lhoták, P. The synthesis and complexation of novel azosubstituted calix[4]arenes and thiacalix[4]arenes. Dye. Pigment. 2008, 77, 646–652. [Google Scholar] [CrossRef]
- Luo, J.; Xie, Z.; Lam, J.W.Y.; Cheng, L.; Tang, B.Z.; Chen, H.; Qiu, C.; Kwok, H.S.; Zhan, X.; Liu, Y.; et al. Aggregation-induced emission of 1-methyl-1,2,3,4,5-pentaphenylsilole. Chem. Commun. 2001, 1740–1741. [Google Scholar] [CrossRef]
- Puplampu, J.B.; Yakimova, L.; Vavilova, A.; Rizvanov, I.K.; Stoikov, I.I. p-tert-Butyl Thiacalix[4]arene Derivatives Functionalized in the Lower Rim with Bis(3-aminopropyl)amine: Synthesis and Interaction with DNA. Macroheterocycles 2015, 8, 75–80. [Google Scholar] [CrossRef]
- Yakimova, L.; Padnya, P.; Tereshina, D.; Kunafina, A.; Nugmanova, A.; Osin, Y.; Evtugyn, V.; Stoikov, I. Interpolyelectrolyte mixed nanoparticles from anionic and cationic thiacalix[4]arenes for selective recognition of model biopolymers. J. Mol. Liq. 2019, 279, 9–17. [Google Scholar] [CrossRef]
- Freifelder, D.M. Physical Biochemistry: Applications to Biochemistry and Molecular Biology (Life Sciences/Biochemistry), 2nd ed.; W. H. Freeman: San Francisco, CA, USA, 1982; 761p. [Google Scholar]
- Kamaly, N.; He, J.C.; Ausiello, D.A.; Farokhzad, N.K.O.C. Nanomedicines for renal disease: Current status and future applications. Nat. Rev. Nephrol. 2016, 12, 738–753. [Google Scholar] [CrossRef]
- Hao, M.; Chen, B.; Zhao, X.; Zhao, N.; Xu, F.-J. Organic/inorganic nanocomposites for cancer immunotherapy. Mater. Chem. Front. 2020, 4, 2571–2609. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer: Boston, MA, USA, 2006; p. 954. [Google Scholar]
- Khan, G.S.; Shah, A.; Rehman, Z.-U.; Barker, D. Chemistry of DNA minor groove binding agents. J. Photochem. Photobiol. B Biol. 2012, 115, 105–118. [Google Scholar] [CrossRef]
- Kapuściński, J.; Skoczylas, B. Fluorescent complexes of DNA with DAPI 4′,6-diamidine-2-phenyl indole 2HCl or DCI 4′,6-dicarboxyamide-2-pnenyl indole. Nucleic Acids Res. 1978, 5, 3775–3800. [Google Scholar] [CrossRef][Green Version]
- Unksov, I.N.; Slita, A.V.; Petrova, A.V.; Pereviazko, I.; Bakulev, V.; Rolich, V.I.; Bondarenko, A.; Kasyanenko, N.A. Interaction between cationic agents and small interfering RNA and DNA molecules. J. Physics Conf. Ser. 2016, 769, 012023. [Google Scholar] [CrossRef]
- Kasyanenko, N.A.; Tikhomirov, R.A.; Bakulev, V.M.; Demidov, V.N.; Chikhirzhina, E.V.; Moroshkina, E. DNA Complexes with Cobalt(II) Phthalocyanine Disodium Disulfonate. ACS Omega 2019, 4, 16935–16942. [Google Scholar] [CrossRef]
- Alves, J.E.F.; de Oliveira, J.F.; Souza, T.R.C.D.L.; de Moura, R.O.; Júnior, L.B.D.C.; de Lima, M.D.C.A.; de Almeida, S.M.V. Novel indole-thiazole and indole-thiazolidinone derivatives as DNA groove binders. Int. J. Biol. Macromol. 2021, 170, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Kharwade, R.; More, S.; Warokar, A.; Agrawal, P.; Mahajan, N. Starburst pamam dendrimers: Synthetic approaches, surface modifications, and biomedical applications. Arab. J. Chem. 2020, 13, 6009–6039. [Google Scholar] [CrossRef]
- Nordén, B.; Kurucsev, T. Analysing DNA complexes by circular and linear dichroism. J. Mol. Recognit. 1994, 7, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Lohani, N.; Rajeswari, M.R. Preferential binding of anticancer drugs to triplex DNA compared to duplex DNA: A spectroscopic and calorimetric study. RSC Adv. 2016, 6, 39903–39917. [Google Scholar] [CrossRef]
- Gubendran, A.; Kesavan, M.P.; Ayyanaar, S.; Mitu, L.; Athappan, P.; Rajesh, J. Non-enolisable Knoevenagel condensate appended Schiff bases-metal (II) complexes: Spectral characteristics, DNA-binding and nuclease activities. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 181, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Haque, L.; Bhuiya, S.; Giri, I.; Chowdhury, S.; Das, S. Structural alteration of low pH, low temperature induced protonated form of DNA to the canonical form by the benzophenanthridine alkaloid nitidine: Spectroscopic exploration. Int. J. Biol. Macromol. 2018, 119, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- Rey-Rico, A.; Cucchiarini, M. Supramolecular Cyclodextrin-Based Hydrogels for Controlled Gene Delivery. Polymers 2019, 11, 514. [Google Scholar] [CrossRef] [PubMed]
- De Laporte, L.; Rea, J.; Shea, L.D. Design of modular non-viral gene therapy vectors. Biomaterials 2006, 27, 947–954. [Google Scholar] [CrossRef]
- Jensen, L.B.; Pavan, G.M.; Kasimova, M.R.; Rutherford, S.; Danani, A.; Nielsen, H.M.; Foged, C. Elucidating the molecular mechanism of PAMAM–siRNA dendriplex self-assembly: Effect of dendrimer charge density. Int. J. Pharm. 2011, 416, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Turro, N.J.; Tomalia, D.A. Using Ethidium Bromide To Probe the Interactions between DNA and Dendrimers. Langmuir 1999, 16, 15–19. [Google Scholar] [CrossRef]
| Compound | G1 Concentration, M | d, nm (PDI) | ζ-Potential, mV |
|---|---|---|---|
| G1-1,3-alt-HCl | 5 × 10−4 | 115.6 ± 1.4 (0.167 ± 0.011) | 44.9 ± 2.6 |
| 1 × 10−4 | 169.4 ± 1.1 (0.099 ± 0.009) | 30.8 ± 1.1 | |
| 5 × 10−5 | 265.2 ± 5.2 (0.162 ± 0.013) | 21.1 ± 0.7 | |
| 1 × 10−5 | 204.3 ± 2.5 (0.250 ± 0.009) | −33.0 ± 1.2 | |
| 5 × 10−6 | 149.4 ± 1.4 (0.197 ± 0.010) | −44.6 ± 2.9 | |
| G1-cone-HCl | 5 × 10−4 | 122.9 ± 0.9 (0.181 ± 0.007) | 49.0 ± 1.7 |
| 1 × 10−4 | 162.4 ± 1.2 (0.176 ± 0.014) | 44.8 ± 2.9 | |
| 5 × 10−5 | 170.4 ± 0.6 (0.188 ± 0.011) | 33.5 ± 1.1 | |
| 1 × 10−5 | 196.3 ± 3.2 (0.254 ± 0.007) | −28.0 ± 2.4 | |
| 5 × 10−6 | 129.8 ± 1.1 (0.178 ± 0.012) | −37.7 ± 4.3 | |
| G1-paco-HCl | 5 × 10−4 | 112.2 ± 0.4 (0.155 ± 0.007) | 45.2 ± 2.2 |
| 1 × 10−4 | 159.8 ± 0.8 (0.118 ± 0.016) | 36.9 ± 1.6 | |
| 5 × 10−5 | 196.1 ± 1.9 (0.099 ± 0.014) | 25.7 ± 0.7 | |
| 1 × 10−5 | 172.7 ± 1.0 (0.107 ± 0.020) | −30.7 ± 2.3 | |
| 5 × 10−6 | 152.6 ± 1.8 (0.174 ± 0.011) | −34.4 ± 1.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mostovaya, O.; Padnya, P.; Shiabiev, I.; Mukhametzyanov, T.; Stoikov, I. PAMAM-calix-dendrimers: Synthesis and Thiacalixarene Conformation Effect on DNA Binding. Int. J. Mol. Sci. 2021, 22, 11901. https://doi.org/10.3390/ijms222111901
Mostovaya O, Padnya P, Shiabiev I, Mukhametzyanov T, Stoikov I. PAMAM-calix-dendrimers: Synthesis and Thiacalixarene Conformation Effect on DNA Binding. International Journal of Molecular Sciences. 2021; 22(21):11901. https://doi.org/10.3390/ijms222111901
Chicago/Turabian StyleMostovaya, Olga, Pavel Padnya, Igor Shiabiev, Timur Mukhametzyanov, and Ivan Stoikov. 2021. "PAMAM-calix-dendrimers: Synthesis and Thiacalixarene Conformation Effect on DNA Binding" International Journal of Molecular Sciences 22, no. 21: 11901. https://doi.org/10.3390/ijms222111901
APA StyleMostovaya, O., Padnya, P., Shiabiev, I., Mukhametzyanov, T., & Stoikov, I. (2021). PAMAM-calix-dendrimers: Synthesis and Thiacalixarene Conformation Effect on DNA Binding. International Journal of Molecular Sciences, 22(21), 11901. https://doi.org/10.3390/ijms222111901