Current Approaches in Supersecondary Structures Investigation
Abstract
1. Introduction
2. Structural Organization of Proteins
2.1. Levels of Structural Organization
- Hydrogen bonds between amino acid residues;
- Electrostatic interactions between the side groups of charged amino acids;
- Hydrophobic interactions between side groups of hydrophobic amino acids;
- Disulfide covalent bonds; and
- Interactions with cofactors.
2.2. Supersecondary Structure
2.3. Methods for Experimental Analysis of the Secondary Structure of a Protein
2.4. Protein Conformational Stability
3. Protein Families
- α proteins (46,456 protein structures and 289 folds);
- β proteins (48,724 protein structures and 178 folds);
- α/β proteins (51,349 protein structures and 148 folds);
- α + β proteins (53,931 protein structures and 388 folds);
- multidomain α and β proteins (56,572 protein structures and 71 folds); and
- membrane proteins, surface proteins, and peptides (56,835 protein structures and 60 folds).
- Hydrophobic amino acid residues should be at positions essential for the organization of hydrophobic clusters of A- and B-helices.
- There should be a 1-3-8 gap between clusters.
- The last position of the A-helix should be occupied by small or flexible residues, such as glycine, arginine, and lysine.
- The first position of the B-helix should be occupied by a polar uncharged amino acid residue (according to the Kyte–Doolittle scale [82]) or residue with small side group (glycine, alanine, and proline).
- In cytochrome C, a protein of the respiratory electron transport chain involved in electron transfer;
- In papain, an enzyme of unripe papaya fruit that is used for the development of enzyme-linked immunosorbent assays;
- In hexokinase, which catalyzes the transfer of a phosphoryl group from ATP to glucose;
- In the lambda repressor, a protein that affects transcription from the RM and R promoters.
4. Methods for Bioinformatical Analysis of Protein Structure
- Functional tasks → presentation, storage, and dissemination of experimental data.
- Analytical tasks → development of data analysis tools to generate new knowledge.
4.1. Protein Amino Acid Sequence Databases
- Swiss-Prot—protein database containing manually curated and verified records [87].
- TrEMBL—protein database of automatically annotated entries.
- Design of automated systems for data storing and analysis for molecular biology, biomedicine, and genetics tasks;
- Computer processing of biological data; and
- Popularization of databases and software for researchers.
4.2. Databases on Protein Structure
Alignment of Amino Acid Sequences
- Sequential pairwise alignment of sequences; and
- Multiple alignments.
- A ranking function to assess the effectiveness of the alignment; and
- An alignment strategy with the ability to identify amino acid substitutions, insertions, and deletions.
5. Methods for Predicting Protein Structure
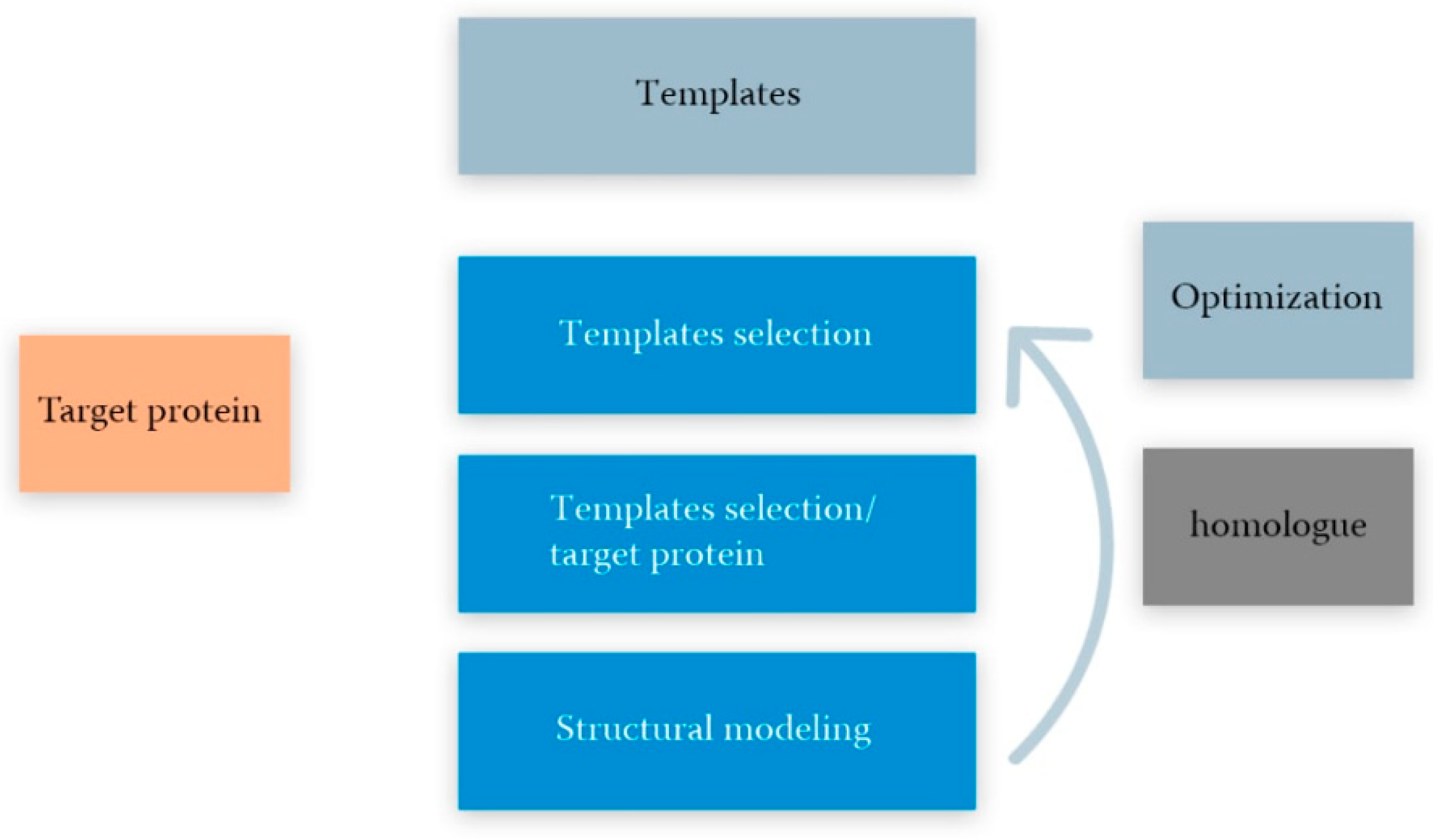
5.1. Template-Based Modeling
- The choice of a template(s) for the sequence of the modeled protein as a query and PDB as a database using a basic local alignment (BLAST, blast.ncbi.nlm.nih.gov, accessed on 20 September 2021);
- Initial alignment and correction of amino acid sequences of the modeling structure and the template(s). Usually performed using the blocks substitution matrix (e.g., BLOSUM80, BLOSUM62 and BLOSUM45) [99];
- Copying structurally variable regions of the template(s);
- Construction of structurally variable regions; for example, using CODA runs two programs for the prediction of the structurally variable regions of protein structures: FREAD, a knowledge-based method using a database of fragments taken from the PDB and PETRA, an ab initio method using a database of computer-generated conformers [109];
- Model optimization, including optimization of stereochemistry energy minimization, molecular dynamics, and estimation of prediction errors for homologous proteins using the support vector machine (SVM) regression method [112];
- Validation (experiment) is the final step of the theoretical model. Experimental data ranging from ligand binding to spectroscopy or X-ray crystallography can be used for the evaluation. The method for validating a three-dimensional structure of homology according to its experimental analog is the root-mean-square deviation (RMSD), which gives the average value of the distances between all atoms for two three-dimensional structures [107].
- Level of similarity between the target sequence and the template,
- The presence of an experimentally solved structure with high resolution, and
- The presence of ligands or cofactors.
5.2. Template-Free Modeling
5.3. Algorithms for Predicting Some Supersecondary Structures
6. Study of the Geometric Parameters of Supersecondary Structures in Proteins
6.1. Revelation of Supersecondary Structures
6.2. Analysis of the Geometries of SSS and Tertiary Protein Structures
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Efimov, A.V. Standard structures in proteins. Prog. Biophys. Mol. Biol. 1993, 60, 201–239. [Google Scholar] [CrossRef]
- Efimov, A.V. New structural motifs in alpha-helical proteins. Bioorg. Khim. 1997, 23, 243–250. [Google Scholar] [PubMed]
- Flot, M.; Mishra, A.; Kuchi, A.S.; Hoque, M.T. StackSSSPred: A Stacking-Based Prediction of Supersecondary Structure from Sequence. Methods Mol. Biol. 2019, 1958, 101–122. [Google Scholar] [CrossRef]
- Tsai, F.C.; Sherman, J.C. Circular dichroism analysis of a synthetic peptide corresponding to the alpha, alpha-corner motif of hemoglobin. Biochem. Biophys. Res. Commun. 1993, 196, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Rudnev, V.R. Recognition and Stability Analysis of Structural Motifs of α-α-corner Type in Globular Proteins. Mat. Biolog. Bioinform. 2013, 8, 398–406. [Google Scholar] [CrossRef]
- Rudnev, V.R. Conformational Analysis of Structural Motifs of α-α-Corner in the Computational Experiment of Molecular Dynamics. Mat. Biolog. Bioinform. 2014, 9, 575–584. [Google Scholar] [CrossRef]
- Liang, H.; Chen, H.; Fan, K.; Wei, P.; Guo, X.; Jin, C.; Zeng, C.; Tang, C.; Lai, L. De novo design of a beta alpha beta motif. Angew. Chem. Int. Ed. Engl. 2009, 48, 3301–3303. [Google Scholar] [CrossRef]
- Kier, B.L.; Shu, I.; Eidenschink, L.A.; Andersen, N.H. Stabilizing capping motif for β-hairpins and sheets. Proc. Natl. Acad. Sci. USA 2010, 107, 10466–10471. [Google Scholar] [CrossRef]
- Alberts, B.; Bray, D.; Hopkin, K.; Johnson, A.D.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Essential Cell Biology; Garland Science: New York, NY, USA, 2015. [Google Scholar]
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed]
- Kargatov, A.M.; Efimov, A.V. Unique Combinations of βαβ-Units and Π-Like Modules in Proteins and Specific Features of Their Amino Acid Sequences. Mol. Biol. 2018, 52, 43–50. [Google Scholar] [CrossRef]
- Berndt, K. Lectures. School of Biomedical Sciences. Curtin University. 2004. Available online: http://www.cryst.bbk.ac.uk/ (accessed on 10 July 2021).
- Hall, V.; Nash, A.; Hines, E.; Rodger, A. Elucidating protein secondary structure with circular dichroism and a neural network. J. Comput. Chem. 2013, 34, 2774–2786. [Google Scholar] [CrossRef]
- Doherty, A.J.; Serpell, L.C.; Pontig, C.P. The helix-hairpin-helix DNA-binding motif: A structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996, 24, 2488–2497. [Google Scholar] [CrossRef]
- Tikhonov, D.; Kulikova, L.; Kopylov, A.; Malsagova, K.; Stepanov, A.; Rudnev, V.; Kaysheva, A. Super Secondary Structures of Proteins with Post-Translational Modifications in Colon Cancer. Molecules 2020, 25, 3144. [Google Scholar] [CrossRef]
- Tikhonov, D.; Kulikova, L.; Kopylov, A.T.; Rudnev, V.; Stepanov, A.; Malsagova, K.; Izotov, A.; Kulikov, D.; Zulkarnaev, A.; Enikeev, D.; et al. Proteomic and molecular dynamic investigations of PTM-induced structural fluctuations in breast and ovarian cancer. Sci. Rep. 2021, 11, 193. [Google Scholar] [CrossRef]
- Rudnev, V.; Kulikova, L.; Kaysheva, A.; Efimov, A.; Tikhonov, D. Use of the Molecular Dynamics Method to Investigate the Stability of α-α-Corner Structural Motifs in Proteins. Symmetry 2021, 13, 1193. [Google Scholar] [CrossRef]
- Gunasekaran, K.; Ramakrishnan, C.; Balaram, P. Beta-hairpins in proteins revisited: Lessons for de novo design. Protein Eng. 1997, 10, 1131–1141. [Google Scholar] [CrossRef]
- Kumar, M.; Bhasin, M.; Natt, N.K.; Raghava, G.P.S. BhairPred: Prediction of β-hairpins in a protein from multiple alignment information using ANN and SVM techniques. Nucleic Acids Res. 2005, 33, W154–W159. [Google Scholar] [CrossRef]
- Blanco, F.J.; Rivas, G.; Serrano, L. A short linear peptide that folds into a native stable β-hairpin in aqueous solution. Nat. Struct. Mol. Biol. 1994, 1, 584–590. [Google Scholar] [CrossRef]
- DeGrado, W.F.; Summa, C.M.; Pavone, V.; Nastri, F.; Lombardi, A. De Novo Design and Structural Characterization of Proteins and Metalloproteins. Annu. Rev. Biochem. 1999, 68, 779–819. [Google Scholar] [CrossRef]
- Batalha, I.L.; Lychko, I.; Branco, R.; Iranzo, O.; Roque, A.C.A. β-Hairpins as peptidomimetics of human phosphoprotein-binding domains. Org. Biomol. Chem. 2019, 17, 3996–4004. [Google Scholar] [CrossRef]
- Efimov, A. Structure of coiled β-β-hairpins and β-β-corners. FEBS Lett. 1991, 284, 288–292. [Google Scholar] [CrossRef]
- Boshkova, E.A.; Efimov, A.V. Structures closed into cycles in proteins containing 3β-corners. Biochemistry 2010, 75, 1258–1263. [Google Scholar] [CrossRef]
- Kadamuri, R.V.; Irukuvajjula, S.S.; Vadrevu, R. βαβ Super-Secondary Motifs: Sequence, Structural Overview, and Pursuit of Potential Autonomously Folding βαβ Sequences from (β/α)8/TIM Barrels. Breast Cancer 2019, 1958, 221–236. [Google Scholar]
- Sun, L.; Hu, X.; Li, S.; Jiang, Z.; Li, K. Prediction of complex super-secondary structure βαβ motifs based on combined features. Saudi J. Biol. Sci. 2016, 23, 66–71. [Google Scholar] [CrossRef]
- Tomii, K.; Kanehisa, M. Systematic Detection of Protein Structural Motifs. In Systematic Detection of Protein Structural Motifs; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Richardson, J.S.; Richardson, D.C. Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl. Acad. Sci. USA 2002, 99, 2754–2759. [Google Scholar] [CrossRef]
- Thompson, M.C.; Yeates, T.O.; Rodriguez, J.A. Advances in methods for atomic resolution macromolecular structure determination. F1000Research 2020, 9, 667. [Google Scholar] [CrossRef]
- Knapman, K. High-throughput protein crystallography—Large amounts of data, analyzed simultaneously, provide new insights into protein-ligand interactions, accelerating the pace of drug discovery. Chem. Innov. 2001, 31, 22–27. [Google Scholar]
- Chen, C.-J.; Lin, Y.-H.; Huang, Y.-C.; Liu, M.-Y. Crystal structure of rubredoxin from Desulfovibrio gigas to ultra-high 0.68Å resolution. Biochem. Biophys. Res. Commun. 2006, 349, 79–90. [Google Scholar] [CrossRef]
- Howard, E.; Sanishvili, R.; Cachau, R.; Mitschler, A.; Chevrier, B.; Barth, P.; Lamour, V.; Van Zandt, M.; Sibley, E.; Bon, C.; et al. Ultrahigh resolution drug design I: Details of interactions in human aldose reductase-inhibitor complex at 0.66 Å. Proteins 2004, 55, 792–804. [Google Scholar] [CrossRef]
- Takeda, K.; Kusumoto, K.; Hirano, Y.; Miki, K. Detailed assessment of X-ray induced structural perturbation in a crystalline state protein. J. Struct. Biol. 2010, 169, 135–144. [Google Scholar] [CrossRef]
- Schmidt, A.; Teeter, M.; Weckert, E.; Lamzin, V.S. Crystal structure of small protein crambin at 0.48 Å resolution. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 424–428. [Google Scholar] [CrossRef]
- Wang, J.; Dauter, M.; Alkire, R.; Joachimiak, A.; Dauter, Z. Triclinic lysozyme at 0.65 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 2007, 63, 1254–1268. [Google Scholar] [CrossRef]
- Camilloni, C.; De Simone, A.; Vranken, W.; Vendruscolo, M. Determination of Secondary Structure Populations in Disordered States of Proteins Using Nuclear Magnetic Resonance Chemical Shifts. Biochemistry 2012, 51, 2224–2231. [Google Scholar] [CrossRef] [PubMed]
- Sborgi, L.; Verma, A.; Sadqi, M.; De Alba, E.; Muñoz, V. Protein Folding at Atomic Resolution: Analysis of Autonomously Folding Supersecondary Structure Motifs by Nuclear Magnetic Resonance. Methods Mol. Biol. 2013, 932, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Opella, S.J. Structure Determination of Membrane Proteins by Nuclear Magnetic Resonance Spectroscopy. Annu. Rev. Anal. Chem. 2013, 6, 305–328. [Google Scholar] [CrossRef]
- Frueh, D.P.; Goodrich, A.C.; Mishra, S.H.; Nichols, S.R. NMR methods for structural studies of large monomeric and multimeric proteins. Curr. Opin. Struct. Biol. 2013, 23, 734–739. [Google Scholar] [CrossRef]
- Hendrickx, P.; Gutmanas, A.; Kleywegt, G.J. Vivaldi: Visualization and validation of biomacromolecular NMR structures from the PDB. Proteins 2013, 81, 583–591. [Google Scholar] [CrossRef]
- Greenfield, N.J.; Kotlyanskaya, L.; Hitchcock-DeGregori, S.E. Structure of the N Terminus of a Nonmuscle α-Tropomyosin in Complex with the C Terminus: Implications for Actin Binding. Biochemistry 2009, 48, 1272–1283. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef]
- Cressey, D.; Callaway, E. Cryo-electron microscopy wins chemistry Nobel. Nature 2017, 550, 167. [Google Scholar] [CrossRef]
- Glaeser, R.; Chiu, W.; Frank, J.; DeRosier, D.; Downing, K. Electron. Crystallography of Biological Macromolecules; Oxford University Press: Oxford, UK, 2007. [Google Scholar]
- Yip, K.M.; Fischer, N.; Paknia, E.; Chari, A.; Stark, H. Atomic-resolution protein structure determination by cryo-EM. Nature 2020, 587, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Scheffen, M.; Marchal, D.G.; Beneyton, T.; Schuller, S.K.; Klose, M.; Diehl, C.; Lehmann, J.; Pfister, P.; Carrillo, M.; He, H.; et al. A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation. Nat. Catal. 2021, 4, 105–115. [Google Scholar] [CrossRef]
- Merk, A.; Bartesaghi, A.; Banerjee, S.; Falconieri, V.; Rao, P.; Davis, M.I.; Pragani, R.; Boxer, M.B.; Earl, L.A.; Milne, J.L.; et al. Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery. Cell 2016, 165, 1698–1707. [Google Scholar] [CrossRef] [PubMed]
- Bartesaghi, A.; Aguerrebere, C.; Falconieri, V.; Banerjee, S.; Earl, L.A.; Zhu, X.; Grigorieff, N.; Milne, J.L.; Sapiro, G.; Wu, X.; et al. Atomic Resolution Cryo-EM Structure of β-Galactosidase. Structure 2018, 26, 848–856.e3. [Google Scholar] [CrossRef] [PubMed]
- Bank, R.P.D. RCSB PDB-7KFR: Adeno-Associated Virus (AAV-DJ)-cryo-EM Structure at 1.56 Angstrom Resolution. Available online: https://www.rcsb.org/structure/7KFR (accessed on 8 October 2021).
- Nakane, T.; Kotecha, A.; Sente, A.; McMullan, G.; Masiulis, S.; Brown, P.M.G.E.; Grigoras, I.T.; Malinauskaite, L.; Malinauskas, T.; Miehling, J.; et al. Single-particle cryo-EM at atomic resolution. Nature 2020, 587, 152–156. [Google Scholar] [CrossRef]
- Degréve, L.; Fuzo, C.; Caliri, A. Extended secondary structures in proteins. Biochim. Biophys. Acta 2014, 1844, 384–388. [Google Scholar] [CrossRef] [PubMed]
- DuPai, C.D.; Davies, B.W.; Wilke, C.O. A systematic analysis of the beta hairpin motif in the Protein Data Bank. Protein Sci. 2021, 30, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Mahalakshmi, R. Aromatic interactions in β-hairpin scaffold stability: A historical perspective. Arch. Biochem. Biophys. 2019, 661, 39–49. [Google Scholar] [CrossRef]
- Popp, A.; Wu, L.; Keiderling, T.A.; Hauser, K. Effect of Hydrophobic Interactions on the Folding Mechanism of β-Hairpins. J. Phys. Chem. B 2014, 118, 14234–14242. [Google Scholar] [CrossRef]
- Jiménez, M.A. Design of Monomeric Water-Soluble β-Hairpin and β-Sheet Peptides. Metabolomics 2014, 1216, 15–52. [Google Scholar]
- Nelson, D.L.; Nelson, D.L.; Lehninger, A.L.; Cox, M.M. Lehninger Principles of Biochemistry; W.H. Freeman: New York, NY, USA, 2008. [Google Scholar]
- Hollingsworth, S.A.; Karplus, P.A. A fresh look at the Ramachandran plot and the occurrence of standard structures in proteins. Biomol. Concepts 2010, 1, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, G.; Sasisekharan, V. Conformation of Polypeptides and Proteins. Adv. Protein Chem. 1968, 23, 283–437. [Google Scholar] [CrossRef] [PubMed]
- Read, R.J.; Adams, P.; Arendall, W.B.; Brunger, A.; Emsley, P.; Joosten, R.; Kleywegt, G.J.; Krissinel, E.B.; Lütteke, T.; Otwinowski, Z.; et al. A New Generation of Crystallographic Validation Tools for the Protein Data Bank. Structure 2011, 19, 1395–1412. [Google Scholar] [CrossRef] [PubMed]
- Ting, D.; Wang, G.; Shapovalov, M.; Mitra, R.; Jordan, M.; Dunbrack, R.L. Neighbor-Dependent Ramachandran Probability Distributions of Amino Acids Developed from a Hierarchical Dirichlet Process Model. PLoS Comput. Biol. 2010, 6, e1000763. [Google Scholar] [CrossRef] [PubMed]
- Berkholz, D.S.; Krenesky, P.B.; Davidson, J.R.; Karplus, P.A. Protein Geometry Database: A flexible engine to explore backbone conformations and their relationships to covalent geometry. Nucleic Acids Res. 2009, 38, D320–D325. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Porter, L.L.; Rose, G.D. Redrawing the Ramachandran plot after inclusion of hydrogen-bonding constraints. Proc. Natl. Acad. Sci. USA 2011, 108, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Kleywegt, G.J.; Jones, T.A. Phi/Psi-chology: Ramachandran revisited. Structure 1996, 4, 1395–1400. [Google Scholar] [CrossRef]
- Davis, I.W.; Murray, L.W.; Richardson, J.; Richardson, D.C. MOLPROBITY: Structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004, 32, W615–W619. [Google Scholar] [CrossRef]
- Garbuzynskiy, S.; Kondratova, M. Structural features of protein folding nuclei. FEBS Lett. 2008, 582, 768–772. [Google Scholar] [CrossRef] [PubMed]
- Boshkova, E.A.; Gordeev, A.B.; Efimov, A.V. A novel structural tree for wrap-proteins, a subclass of (α + β)-proteins. J. Biomol. Struct. Dyn. 2014, 32, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Efimov, A.V. Super-secondary Structures and Modeling of Protein Folds. Methods Mol. Biol. 2012, 932, 177–189. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Gordeev, A.B.; Kargatov, A.M.; Efimov, A.V. PCBOST: Protein classification based on structural trees. Biochem. Biophys. Res. Commun. 2010, 397, 470–471. [Google Scholar] [CrossRef] [PubMed]
- Gordeev, A.B.; Efimov, A.V. Modeling of folds and folding pathways for some protein families of (α + β)- and (α/β)-classes. J. Biomol. Struct. Dyn. 2013, 31, 4–16. [Google Scholar] [CrossRef]
- Sigrist, C.J.A.; Cerutti, L.; de Castro, E.; Langendijk-Genevaux, P.; Bulliard, V.; Bairoch, A.; Hulo, N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2009, 38, D161–D166. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Sillitoe, I.; Bordin, N.; Dawson, N.; Waman, V.P.; Ashford, P.; Scholes, H.M.; Pang, C.S.M.; Woodridge, L.; Rauer, C.; Sen, N.; et al. CATH: Increased structural coverage of functional space. Nucleic Acids Res. 2021, 49, D266–D273. [Google Scholar] [CrossRef]
- Chothia, C.; Levitt, M.; Richardson, D. Structure of proteins: Packing of alpha-helices and pleated sheets. Proc. Natl. Acad. Sci. USA 1977, 74, 4130–4134. [Google Scholar] [CrossRef]
- Richardson, J.S. Handedness of crossover connections in beta sheets. Proc. Natl. Acad. Sci. USA 1976, 73, 2619–2623. [Google Scholar] [CrossRef]
- Richardson, J.S. β-Sheet topology and the relatedness of proteins. Nature 1977, 268, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, M.; Thornton, J. On the conformation of proteins: The handedness of the β-strand-α-helix-β-strand unit. J. Mol. Biol. 1976, 105, 367–382. [Google Scholar] [CrossRef]
- Finkelstein, A.V.; Ptitsyn, O.B. Why do globular proteins fit the limited set of folding patterns? Prog. Biophys. Mol. Biol. 1987, 50, 171–190. [Google Scholar] [CrossRef]
- Fox, N.K.; Brenner, S.E.; Chandonia, J.-M. SCOPe: Structural Classification of Proteins—Extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res. 2014, 42, D304–D309. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, A.; Howorth, D.; Chothia, C.; Kulesha, E.; Murzin, A.G. SCOP2 prototype: A new approach to protein structure mining. Nucleic Acids Res. 2014, 42, D310–D314. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Efimov, A.V. Favoured structural motifs in globular proteins. Structure 1994, 2, 999–1002. [Google Scholar] [CrossRef]
- Efimov, A.V. Structural trees for protein superfamilies. Proteins Struct. Funct. Bioinform. 1997, 28, 241–260. [Google Scholar] [CrossRef]
- Gibas, C.; Jambeck, P. Developing Bioinformatics Computer Skills, 1st ed.; O’Reilly: Sebastopol, CA, USA, 2001. [Google Scholar]
- Veuthey, A.-L.; Bridge, A.; Gobeill, J.; Ruch, P.; McEntyre, J.R.; Bougueleret, L.; Xenarios, I. Application of text-mining for updating protein post-translational modification annotation in UniProtKB. BMC Bioinform. 2013, 14, 104. [Google Scholar] [CrossRef]
- Jungo, F.; Bougueleret, L.; Xenarios, I.; Poux, S. The UniProtKB/Swiss-Prot Tox-Prot program: A central hub of integrated venom protein data. Toxicon 2012, 60, 551–557. [Google Scholar] [CrossRef]
- Poux, S.; Magrane, M.; Arighi, C.; Bridge, A.; O’Donovan, C.; Laiho, K. Expert curation in UniProtKB: A case study on dealing with conflicting and erroneous data. Database 2014, 2014, bau016. [Google Scholar] [CrossRef] [PubMed]
- Acland, A.; Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bryant, S.H.; Canese, K.; Church, D.M.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2014, 42, D7–D17. [Google Scholar] [CrossRef]
- Benson, D.A.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2014, 42, D32–D37. [Google Scholar] [CrossRef] [PubMed]
- London, S.; Gürdal, O.; Gall, C. Automatic Export of PubMed® Citations to EndNote®. Med. Ref. Serv. Q. 2010, 29, 146–153. [Google Scholar] [CrossRef]
- Sayle, R.A. RASMOL: Biomolecular graphics for all. Trends Biochem. Sci. 1995, 20, 374–376. [Google Scholar] [CrossRef]
- Dwivedi, V.D.; Arora, S.; Pandey, A. Computational analysis of physico-chemical properties and homology modeling of carbonic anhydrase from Cordyceps militaris. Netw. Model. Anal. Health Inform. Bioinform. 2013, 2, 209–212. [Google Scholar] [CrossRef][Green Version]
- Seeliger, D.; De Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aid. Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef]
- Kuhlman, B.; Bradley, P. Advances in protein structure prediction and design. Nat. Rev. Mol. Cell Biol. 2019, 20, 681–697. [Google Scholar] [CrossRef]
- Silva, D.-A.; Yu, S.; Ulge, U.Y.; Spangler, J.B.; Jude, K.; Labão-Almeida, C.; Ali, L.; Quijano-Rubio, A.; Ruterbusch, M.; Leung, I.; et al. De novo design of potent and selective mimics of IL-2 and IL-15. Nature 2019, 565, 186–191. [Google Scholar] [CrossRef]
- Wu, F.; Xu, J. Deep template-based protein structure prediction. PLoS Comput. Biol. 2021, 17, e1008954. [Google Scholar] [CrossRef]
- Fiser, A. Template-Based Protein Structure Modeling. Methods Mol. Biol. 2010, 673, 73–94. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Shen, Y. Template-based prediction of protein structure with deep learning. BMC Genom. 2020, 21, 878. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Wuyun, Q.; Li, Y.; Mortuza, S.M.; Zhang, C.; Pearce, R.; Ruan, J.; Zhang, Y. Detecting distant-homology protein structures by aligning deep neural-network based contact maps. PLoS Comput. Biol. 2019, 15, e1007411. [Google Scholar] [CrossRef] [PubMed]
- Chothia, C.; Lesk, A. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986, 5, 823–826. [Google Scholar] [CrossRef]
- Rost, B. Twilight zone of protein sequence alignments. Protein Eng. 1999, 12, 85–94. [Google Scholar] [CrossRef]
- Croll, T.I.; Sammito, M.D.; Kryshtafovych, A.; Read, R.J. Evaluation of template-based modeling in CASP13. Proteins Struct. Funct. Bioinform. 2019, 87, 1113–1127. [Google Scholar] [CrossRef]
- Pieper, U.; Webb, B.M.; Dong, G.Q.; Schneidman-Duhovny, D.; Fan, H.; Kim, S.J.; Khuri, N.; Spill, Y.; Weinkam, P.; Hammel, M.; et al. ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2014, 42, D336–D346. [Google Scholar] [CrossRef]
- Yang, J.; Anishchenko, I.; Park, H.; Peng, Z.; Ovchinnikov, S.; Baker, D. Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci. USA 2020, 117, 1496–1503. [Google Scholar] [CrossRef]
- Xu, J. Distance-based protein folding powered by deep learning. Proc. Natl. Acad. Sci. USA 2019, 116, 16856–16865. [Google Scholar] [CrossRef]
- Haddad, Y.; Adam, V.; Heger, Z. Ten quick tips for homology modeling of high-resolution protein 3D structures. PLoS Comput. Biol. 2020, 16, e1007449. [Google Scholar] [CrossRef]
- Krogh, A.; Brown, M.; Mian, I.; Sjölander, K.; Haussler, D. Hidden Markov Models in Computational Biology: Applications to Protein Modeling. J. Mol. Biol. 1994, 235, 1501–1531. [Google Scholar] [CrossRef]
- Deane, C.M.; Blundell, T.L. CODA: A combined algorithm for predicting the structurally variable regions of protein models. Protein Sci. 2001, 10, 599–612. [Google Scholar] [CrossRef]
- Wang, Q.; Canutescu, A.A.; Dunbrack, R.L., Jr. SCWRL and MolIDE: Computer programs for side-chain conformation prediction and homology modeling. Nat. Protoc. 2008, 3, 1832–1847. [Google Scholar] [CrossRef]
- Huang, X.; Pearce, R.; Zhang, Y. FASPR: An open-source tool for fast and accurate protein side-chain packing. Bioinformatics 2020, 36, 3758–3765. [Google Scholar] [CrossRef]
- Eramian, D.; Shen, M.-Y.; Devos, D.; Melo, F.; Sali, A.; Marti-Renom, M.A. A composite score for predicting errors in protein structure models. Protein Sci. 2006, 15, 1653–1666. [Google Scholar] [CrossRef] [PubMed]
- Oda, T.; Lim, K.; Tomii, K. Simple adjustment of the sequence weight algorithm remarkably enhances PSI-BLAST performance. BMC Bioinform. 2017, 18, 288. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2017, 27, 293–315. [Google Scholar] [CrossRef]
- Hooft, R.; Vriend, G.; Sander, C.; Abola, E.E. Errors in protein structures. Nature 1996, 381, 272. [Google Scholar] [CrossRef] [PubMed]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo—Distance constraints applied on model quality estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. Scoring function for automated assessment of protein structure template quality. Proteins 2004, 57, 702–710. [Google Scholar] [CrossRef]
- Olechnovič, K.; Venclovas, C. The CAD-score web server: Contact area-based comparison of structures and interfaces of proteins, nucleic acids and their complexes. Nucleic Acids Res. 2014, 42, W259–W263. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Buchan, D.W.A.; Jones, D.T. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 2019, 47, W402–W407. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Jia, Y.; Zhang, Y. Protein structure prediction. Int. J. Mod. Phys. B 2018, 32, 1840009. [Google Scholar] [CrossRef] [PubMed]
- Mortuza, S.M.; Zheng, W.; Zhang, C.; Li, Y.; Pearce, R.; Zhang, Y. Improving fragment-based ab initio protein structure assembly using low-accuracy contact-map predictions. Nat. Commun. 2021, 12, 5011. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Kryshtafovych, A.; Schwede, T.; Topf, M.; Fidelis, K.; Moult, J. Critical assessment of methods of protein structure prediction (CASP)—Round XIII. Proteins 2019, 87, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Fiser, A.; Šali, A. Modeller: Generation and Refinement of Homology-Based Protein Structure Models. Methods Enzymol. 2003, 374, 461–491. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Rohl, C.A.; Strauss, C.E.; Misura, K.M.; Baker, D. Protein Structure Prediction Using Rosetta. Methods Enzymol. 2004, 383, 66–93. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, J.; Roy, A.; Zhang, Y. Automated protein structure modeling in CASP9 by I-TASSER pipeline combined with QUARK-based ab initio folding and FG-MD-based structure refinement. Proteins 2011, 79 (Suppl. S10), 147–160. [Google Scholar] [CrossRef]
- MacCarthy, E.; Perry, D.; Kc, D.B. Advances in Protein Super-Secondary Structure Prediction and Application to Protein Structure Prediction. Methods Mol. Biol. 2019, 1958, 15–45. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Kurgan, L. Computational Prediction of Secondary and Supersecondary Structures. Methods Mol. Biol. 2012, 932, 63–86. [Google Scholar] [CrossRef]
- Yonge, F.; Gaoshan, K. Identify Beta-Hairpin Motifs with Quadratic Discriminant Algorithm Based on the Chemical Shifts. PLoS ONE 2015, 10, e0139280. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Costa, C.; Shanahan, H.P.; Jones, S.; Thornton, J.M. HTHquery: A method for detecting DNA-binding proteins with a helix-turn-helix structural motif. Bioinformatics 2005, 21, 3679–3680. [Google Scholar] [CrossRef] [PubMed]
- Matthes, D.; de Groot, B.L. Secondary Structure Propensities in Peptide Folding Simulations: A Systematic Comparison of Molecular Mechanics Interaction Schemes. Biophys. J. 2009, 97, 599–608. [Google Scholar] [CrossRef]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
- Feig, M.; MacKerell, A.D.; Brooks, C.L. Force Field Influence on the Observation of π-Helical Protein Structures in Molecular Dynamics Simulations. J. Phys. Chem. B 2003, 107, 2831–2836. [Google Scholar] [CrossRef]
- Oostenbrink, C.; Villa, A.; Mark, A.E.; Van Gunsteren, W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004, 25, 1656–1676. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, G.A.; Friesner, R.A.; Tirado-Rives, J.; Jorgensen, W.L. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides. J. Phys. Chem. B 2001, 105, 6474–6487. [Google Scholar] [CrossRef]
- Rueda, M.; Ferrer-Costa, C.; Meyer, T.; Pérez, A.; Camps, J.; Hospital, A.; Gelpí, J.L.; Orozco, M. A consensus view of protein dynamics. Proc. Natl. Acad. Sci. USA 2007, 104, 796–801. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Van Der Spoel, D.; Van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Carter, P.; Andersen, C.A.F.; Rost, B. DSSPcont: Continuous secondary structure assignments for proteins. Nucleic Acids Res. 2003, 31, 3293–3295. [Google Scholar] [CrossRef] [PubMed]
- Frishman, D.; Argos, P. Knowledge-based protein secondary structure assignment. Proteins 1995, 23, 566–579. [Google Scholar] [CrossRef]
- Labesse, G.; Colloc’H, N.; Pothier, J.; Mornon, J.-P. P-SEA: A new efficient assignment of secondary structure from Cα trace of proteins. Comput. Appl. Bioinform. 1997, 13, 291–295. [Google Scholar] [CrossRef]
- Kelleher, N.L. A Cell-Based Approach to the Human Proteome Project. J. Am. Soc. Mass Spectrom. 2012, 23, 1617–1624. [Google Scholar] [CrossRef] [PubMed]
- Indeykina, M.I.; Popov, I.A.; Kozin, S.A.; Kononikhin, A.S.; Kharybin, O.N.; Tsvetkov, P.O.; Makarov, A.A.; Nikolaev, E.N. Capabilities of MS for Analytical Quantitative Determination of the Ratio of α- and βAsp7 Isoforms of the Amyloid-β Peptide in Binary Mixtures. Anal. Chem. 2011, 83, 3205–3210. [Google Scholar] [CrossRef]
- Tilli, T.M.; Mello, K.D.; Ferreira, L.B.; Matos, A.R.; Accioly, M.T.S.; Faria, P.A.; Bellahcène, A.; Castronovo, V.; Gimba, E.R. Both osteopontin-c and osteopontin-b splicing isoforms exert pro-tumorigenic roles in prostate cancer cells. Prostate 2012, 72, 1688–1699. [Google Scholar] [CrossRef]
- Su, Z.-D.; Sun, L.; Yu, D.-X.; Li, R.-X.; Li, H.-X.; Yu, Z.-J.; Sheng, Q.-H.; Lin, X.; Zeng, R.; Wu, J.-R. Quantitative detection of single amino acid polymorphisms by targeted proteomics. J. Mol. Cell Biol. 2011, 3, 309–315. [Google Scholar] [CrossRef]
- Wang, Q.; Chaerkady, R.; Wu, J.; Hwang, H.J.; Papadopoulos, N.; Kopelovich, L.; Maitra, A.; Matthaei, H.; Eshleman, J.R.; Hruban, R.H.; et al. Mutant proteins as cancer-specific biomarkers. Proc. Natl. Acad. Sci. USA 2011, 108, 2444–2449. [Google Scholar] [CrossRef]
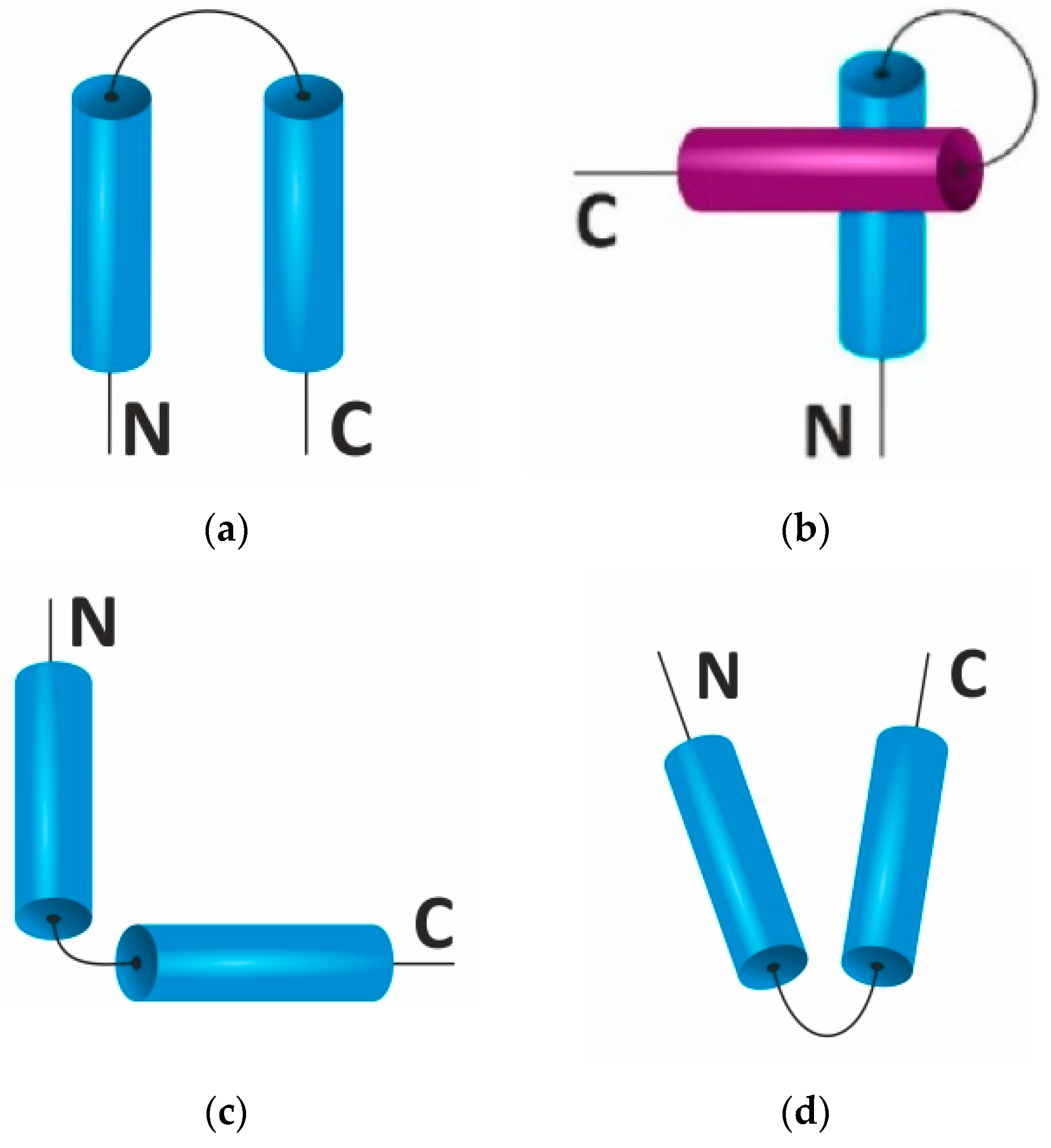
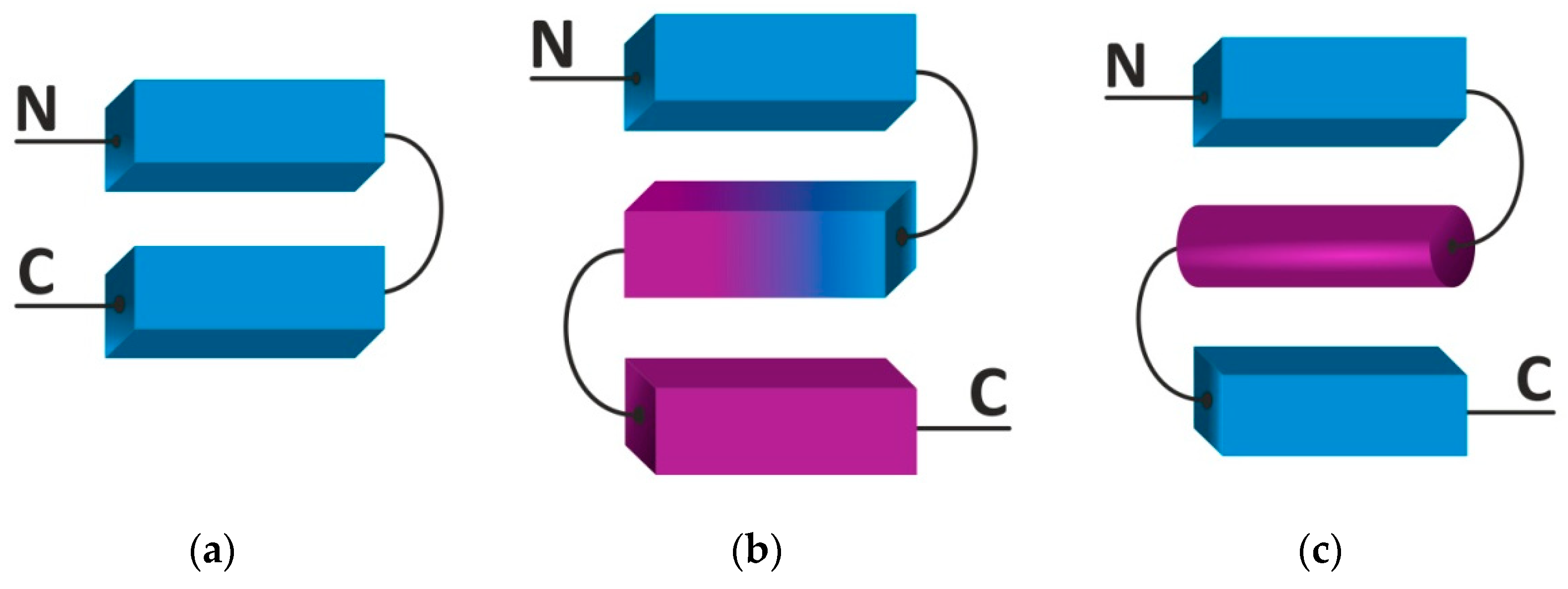



| Structure | Hydrogen Bond | Residue/Coil | Displacement/Residue (Å) | φ | ψ |
|---|---|---|---|---|---|
| Right α-helix | COo–HN+4 | +3.6 | 1.5 | −60° | −45° |
| Antiparallel β-sheet | between strands | −2.3 | 3.4 | −135° | +150° |
| Parallel β-sheet | between strands | −2.3 | 3.2 | −120° | +135° |
| Research Method | Total Number of Entries Available | Number of Protein Structures | The Number of Structures of Complexes of Proteins and Nucleic Acids |
|---|---|---|---|
| X-ray | 160,277 | 159,817 | 9715 |
| NMR | 13,500 | ||
| EM | 8870 | ||
| Mixed | 193 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rudnev, V.R.; Kulikova, L.I.; Nikolsky, K.S.; Malsagova, K.A.; Kopylov, A.T.; Kaysheva, A.L. Current Approaches in Supersecondary Structures Investigation. Int. J. Mol. Sci. 2021, 22, 11879. https://doi.org/10.3390/ijms222111879
Rudnev VR, Kulikova LI, Nikolsky KS, Malsagova KA, Kopylov AT, Kaysheva AL. Current Approaches in Supersecondary Structures Investigation. International Journal of Molecular Sciences. 2021; 22(21):11879. https://doi.org/10.3390/ijms222111879
Chicago/Turabian StyleRudnev, Vladimir R., Liudmila I. Kulikova, Kirill S. Nikolsky, Kristina A. Malsagova, Arthur T. Kopylov, and Anna L. Kaysheva. 2021. "Current Approaches in Supersecondary Structures Investigation" International Journal of Molecular Sciences 22, no. 21: 11879. https://doi.org/10.3390/ijms222111879
APA StyleRudnev, V. R., Kulikova, L. I., Nikolsky, K. S., Malsagova, K. A., Kopylov, A. T., & Kaysheva, A. L. (2021). Current Approaches in Supersecondary Structures Investigation. International Journal of Molecular Sciences, 22(21), 11879. https://doi.org/10.3390/ijms222111879







