Abstract
As the population ages, the incidence of neurodegenerative diseases is increasing. Due to intensive research, important steps in the elucidation of pathogenetic cascades have been made and significantly implicated mitochondrial dysfunction and oxidative stress. However, the available treatment in Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis is mainly symptomatic, providing minor benefits and, at most, slowing down the progression of the disease. Although in preclinical setting, drugs targeting mitochondrial dysfunction and oxidative stress yielded encouraging results, clinical trials failed or had inconclusive results. It is likely that by the time of clinical diagnosis, the pathogenetic cascades are full-blown and significant numbers of neurons have already degenerated, making it impossible for mitochondria-targeted or antioxidant molecules to stop or reverse the process. Until further research will provide more efficient molecules, a healthy lifestyle, with plenty of dietary antioxidants and avoidance of exogenous oxidants may postpone the onset of neurodegeneration, while familial cases may benefit from genetic testing and aggressive therapy started in the preclinical stage.
1. Introduction
Aging associates a series of physiologic deficits and a variable degree of cognitive impairment, being also a major risk factor for neurodegenerative diseases such as Alzheimer’s disease (AD), Parkinson’s disease (PD), or amyotrophic lateral sclerosis (ALS) [1]. From the enormous amount of research aimed at unraveling the mechanisms of aging and neurodegeneration several pieces of the puzzle emerged, but we have still a long way to go to grasp the whole picture. However, it appears that oxidative damage induced by free radicals and mitochondrial dysfunction play a major role in both processes.
2. Normal Aging
Aging of the brain occurs at molecular, cellular, and histological levels [2]. It associates lower levels of neuronal metabolic activity, subtle alterations in neuronal structure in several neuronal circuits, as well as synaptic atrophy, cytoskeletal abnormalities, accumulation of fluorescent pigments, and reactive astrocytes and microglia [3,4]. Research points toward the hypothalamus as initiating and controlling the gradual decline of energy metabolism of the entire body [5,6]. Through the secretion of neurohormones, the connections with the endocrine system, and projections of the orexinergic nucleus to the reticular activating system [5], the hypothalamus regulates the stress levels, metabolism, sleep, and influences the subjectively perceived quality of life and establishing of social relationships [7,8,9]. Degeneration of the suprachiasmatic nucleus may additionally contribute to circadian rhythm disorders and impaired sleep [10,11,12]. Limiting sleep may cause neuronal toxic waste products to accumulate and limit neurogenesis in the aging brain, igniting a vicious cycle which augments the neurodegenerative process [13].
Additional factors contribute to the declining metabolism of the brain cells. Cerebral metabolism relies mainly on a constant supply of glucose and oxygen through the blood flow, and, to a limited extent, on lactate [14]. Neuronal glucose uptake is mediated by glucose transporters (GLUTs), after which it is converted into glucose-6-phosphate (G6P) by hexokinase. As such, the availability of ATP, depending on oxygen supply and mitochondrial oxidative phosphorylation (OXPHOS), interferes with glucose uptake [5,15]. OXPHOS produces considerable higher amounts of ATP as compared to glycolysis [16,17]. In aging, reduced metabolism can be caused by mitochondrial dysfunction and reduced ATP synthesis, as well as by vascular changes which lead to limited oxygen supply.
The high cellular energy expenditure in the nervous system, used for synaptic transmission (around 80% of the energy consumption of the brain) [18,19], synaptogenesis, and synaptic pruning [18,20] associates a high rate of generation of reactive oxygen species mainly by electrons leaked from the mitochondrial electron transport chain (ETC). The excessive generation of free radicals cannot be neutralized by the antioxidant defenses and leads to oxidative stress, implicated in aging since the 1950s, when Harman suggested that free radical-induced damage of biomolecules, such as proteins, lipids, and DNA, causes a reduction of their biochemical and physiological function in aging [21]. Indeed, research has demonstrated altered composition of phospholipids in the brains of aged humans and animals together with increased malondialdehyde (a marker of lipid peroxidation) generation, which forms deposits connected with intraneuronal lipofuscin [22], and elevated carbonyl residues (a marker of protein oxidation) [23]. In addition, aging decreases the antioxidant defense systems, such as the astrocytic glutathione system [24], which further potentiates oxidative stress [2].
Mitochondria undergo a series of age-related changes such as fragmentation or enlargement [25], increased mitochondrial DNA (mtDNA) oxidative damage [26], exhibit dysfunctions of the respiratory chain [27] and of calcium homeostasis [28]. These changes associate a reduction of the intracellular NAD+ levels which impairs the function of NA dependent enzymes such as sirtuins (SIRT) and histone deacetylases [29,30]. Sirtuins are highly evolutionary conserved enzymes involved in the regulation of lifespan and aging in different organisms, from yeast to mammals [31]. In mammals, SIRT 3, 4, and 5 are located in the mitochondria, SIRT 2 in the cytosol, and SIRT 1, 6, and 7 are located in the nucleus [32]. Research has shown that SIRT 1, mainly in the hypothalamus, is a key player in controlling aging and longevity in mammalian organisms [33,34].
Shortening of the telomeres, which promote chromosomal stability during cell replication [35], was initially disregarded as having important influence in the brain since neurons are essentially postmitotic cells which no longer replicate. However, this view has been challenged by demonstrating cell cycle activity in 10–20% of neurons in the cortex of healthy aging brains and in AD [36,37], as well as by the presence of neural stem cells in the subventricular and subgranular zones, in the choroid plexuses and meninges [38]. In addition, glial cells (especially microglia) do replicate actively, with telomeres gradually shortening after each cellular replication [39].
Aging induces also an inflammatory phenotype during which, in response to mutations and DNA damage, nuclear factor-κB (NF-κB) initiates the transcription of tumor necrosis factor-α (TNF-α) and various inflammatory interleukins (IL-1β, IL-6, IL-8) [40]. ROS have a crucial role in this process, since they phosphorylate and degrade IκB, which binds to and inactivates NF-κB [41]. Low amounts of ROS initiate pro-survival signaling cascades, the activated NF-κB suppressing c-Jun N-terminal kinases (JNKs) and apoptosis and upregulating antioxidant and anti-apoptotic genes such as manganese superoxide dismutase (MnSOD) [42]. However, high ROS concentrations activate NF-κB through protein kinases and initiate cellular stress signaling pathways. Damaged neurons alter the ionic balance in the interstitial space and lead to cytokine release, thereby activating microglia [43,44]. Although TNF-α has important roles in learning and synaptic plasticity [45], excessive microglial activation will cause degeneration of synapses and functional impairments, characteristic of aging and neurodegeneration [46]. In addition, the inflammatory responses induce other transcription factors, such as signal transducer and activator of transcription (STAT-1) and peroxisome proliferator-activated receptor-gamma (PPARγ), the latter playing an important role in mitochondrial biogenesis [47]. As such, the secretory phenotype associated with senescence (SASP), particularly in astrocytes, can trigger several age-related neurodegenerative diseases [48].
3. Mitochondria in the Brain
The high cerebral metabolic activity relies mainly on oxidative phosphorylation for ATP production. However, mitochondria exert other important functions in the brain as well. The constant signaling leads to continuous variations in the cytosolic calcium concentrations and mitochondria in collaboration with the endoplasmic reticulum have a crucial role in regulating neurotransmission by buffering calcium in presynaptic terminals and regulating the somato-dendritic calcium levels [49,50]. Moreover, mitochondria regulate cell cycle and control cell death [51]. In order to accomplish these diverse functions, maintenance of mitochondrial fitness is crucial, requiring efficient quality control mechanisms [52], achieved through mitochondrial biogenesis and fission (separation of a single organelle into two or more daughter organelles, essential for populating dividing or growing cells with an adequate number of mitochondria), fusion (a process through which mitochondria share essential components), and mitophagy (elimination of damaged mitochondria before they lead to apoptosis of the whole cell) [53,54]. In addition, mitochondria must be trafficked along axons to provide energy all across the axon [2].
3.1. Mitochondrial Respiratory Chain and ROS Production
A mitochondrion has a spongy outer mitochondrial membrane (OMM), which allows free movement of small ions and uncharged molecules, and an impermeable inner mitochondrial membrane (IMM), which envelops the mitochondrial matrix. Between the IMM and OMM lies the intermembrane space [55]. The mitochondrial electron transport chain (ETC) consists of several of protein complexes situated in the IMM which use the electrons removed by reduced nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FADH2) from the Krebs cycle to pump protons from the matrix into the intermembrane space, thereby generating a potential gradient across the IMM, which will be used in the final step of OXPHOS to synthesize ATP [56]. In order to function properly, these complexes must be assembled by the folded IMM (mitochondrial cristae) into specifically configured structures [57]. Even under normal conditions, 1–2% of the total oxygen consumed leaks and generates ROS [58]. At least eight mitochondrial sites are able to generate ROS, with complexes I, II, and III being the main contributors [55,59]. Through lipid peroxidation, protein oxidation, and DNA damage, the generated ROS can cause alter mitochondrial function and increase the rate of ROS production, culminating in degeneration of neurons [60,61].
3.2. Mitochondria and Cellular Calcium Homeostasis
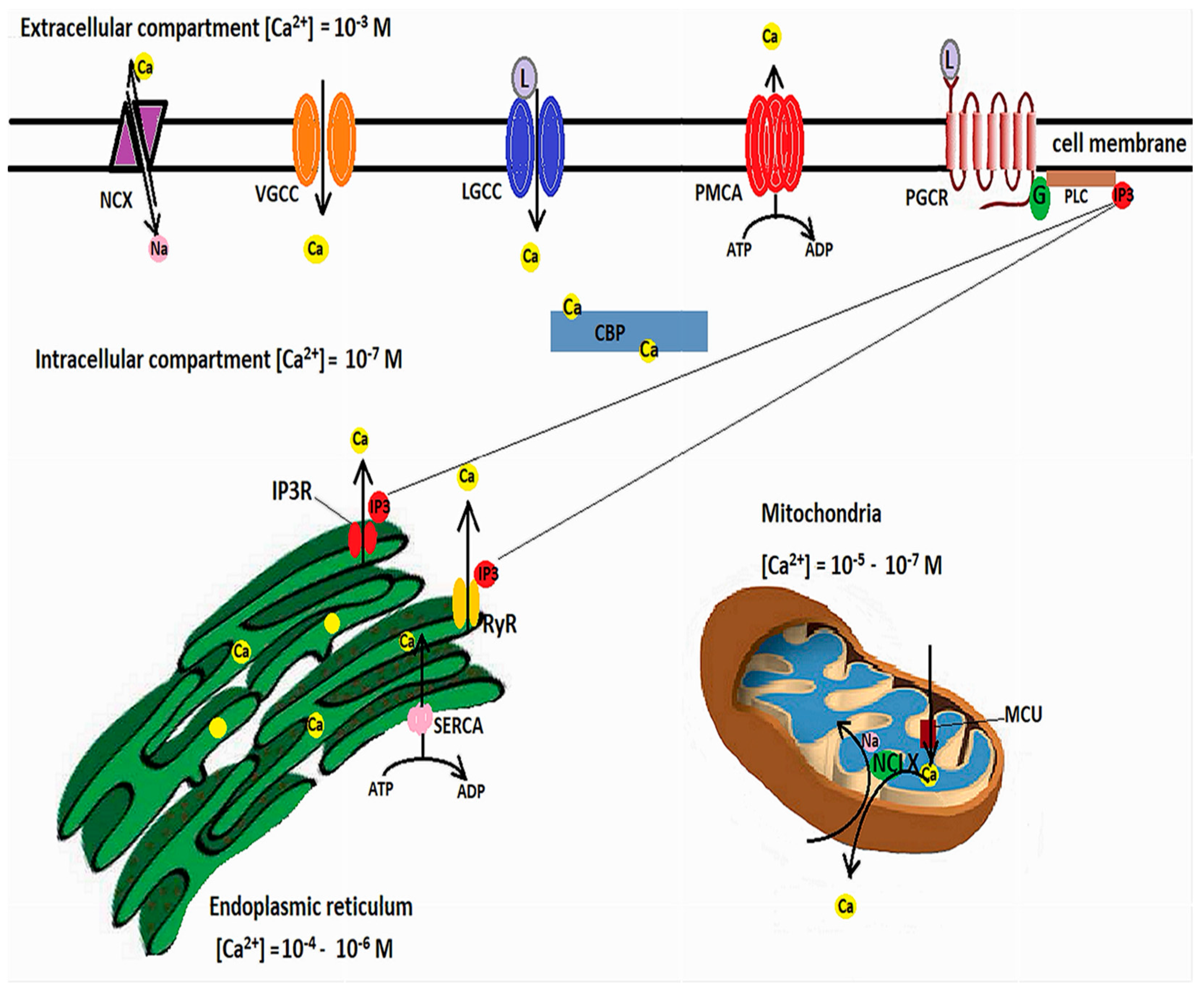
Calcium is involved in many neuronal functions, such as differentiation, vesicle release and synaptic transmission, or cell death and survival [62,63]. Transient fluctuations in the cytosolic Ca2+ act as second messengers. Free cytosolic Ca2+ levels are in nanomolar ranges, while extracellular levels are millimolar. Calcium influx occurs through ligand-operated or voltage-gated calcium channels (VGCCs), but Ca2+ can also be released from intracellular Ca2+ stores, among which the endoplasmic reticulum (ER) has a pivotal role [64]. Agonist binding to inositol 1,4,5-triphosphate (IP3) receptors or to ryanodine receptors (RyRs) causes a release of Ca2+ from the ER [65]. However, in order for the increases of cytosolic Ca2+ to be brief, calcium must be rapidly cleared through calcium efflux, binding to Ca2+-buffering proteins [66], or uptake into the ER or mitochondria [62].
Ca2+ efflux is achieved by the plasma membrane Ca2+-ATPase, which pumps Ca2+ against the concentration gradient while hydrolyzing ATP, and the Na+/Ca2+ exchanger (NCX), which relies the sodium gradient to extrude Ca2+ [67]. ER Ca2+ uptake is performed by the ATP-dependent sarco-endoplasmic reticulum Ca2+-ATPase (SERCA) [68], while mitochondrial Ca2+ uptake is mediated by voltage-dependent anion-selective channel proteins (VDCAs), which mediate Ca2+ transfer into the intermembrane space, from where the IMM-located mitochondrial Ca2+ uniporter (MCU) further transfers the calcium into the mitochondrial matrix. Increases in mitochondrial calcium activate the ETC dehydrogenases and ATP production [69]. However, calcium overload can alter the mitochondrial membrane potential, open the mitochondrial permeability transition pore (MPTP), and lead to cytochrome c release [70]. As such, mitochondrial Ca2+ concentrations must be finely tuned. A mitochondrial Na+/Ca2+ exchanger located in the IMM, and termed NCLX because it can also able to exchange Li+ for Ca2+, extrudes Ca2+ from the mitochondrial matrix by using the electrochemical gradient of Na+ [71], while from the intermembrane space Ca2+ is extruded by the Na+/Ca2+ exchanger 3 and VDACs [72]. Figure 1 illustrates the mechanisms involved in cellular calcium homeostasis.
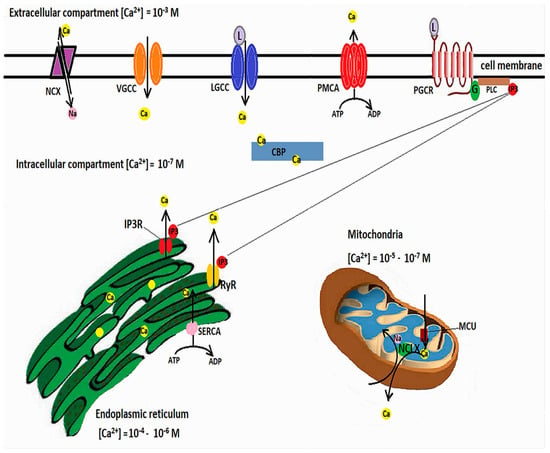
Figure 1.
Intracellular calcium homeostasis. Cellular Ca2+ influx is mediated by voltage-gated calcium channels (VGCC), ligand-gated calcium channels (LGCC), and, in exceptional circumstances, by reverse functioning of the sodium/calcium exchanger (NCX). In addition, Ca2+ can be released from the ER following inositol-1,4,5-triphosphate (IP3) binding to specific receptors (IP3R) or the ryanodine receptors (RyR). IP3 is generated by binding of ligands to plasmalemmal G-protein-coupled receptors, which activates phospholipase C to cleave phosphatidylinositol 4,5-biphosphate, resulting in the second messenger IP3. Excess cytosolic calcium is removed through efflux through the NCX and plasma membrane Ca2+ ATPase (PMCA) and uptake into ER by the sarcoendoplasmic reticulum Ca2+-ATPase (SERCA). Mitochondria buffers cytosolic calcium through the mitochondrial calcium uniporter (MCU) and extrudes excess Ca2+ through the Na+/Ca2+ exchanger (NCLX). In addition, cytosolic Ca2+ binding proteins (CBP) act as signal transducers.
In controlling the intracellular Ca2+ concentrations, mitochondria interact with the ER through mitochondria-associated ER membranes (MAMs) [73], microdomains where the OMM is just 10–100 nanometers apart from the ER [74,75]. These areas are enriched in inositol 1,4,5-triphosphate receptors (IP3Rs) [76] which form functional complexes with VDACs through a chaperone, Grp75 (glucose-regulated protein 75), belonging to the heat shock protein 70 family [77]. IP3R-Grp75-VDAC complexes regulate Ca2+ transfer from the ER to mitochondria [74]. The apposition of ER to mitochondria is controlled by phosphofurin acidic cluster sorting protein 2 (PACS2) [78]. Decreased contact sites between ER and mitochondria caused by PACS2 deletion result in mitochondrial fragmentation and apoptosis [79]. PACS2 is functionally linked to phosphatidylserine synthase-1 (PSS1), an enzyme located in MAMs which mediates the transfer of lipids between ER and mitochondria [80]. PACS2 and PSS1 were found upregulated in AD transgenic mice as well as human patients with late-onset AD [74,81]. Several other components of MAMs involved in calcium homeostasis and signaling cascades have been described, such as:
- -
- Bap31 (B cell receptor-associated protein 31), which interacts with the OMM protein Fis1 [82];
- -
- VAPB (Vesicle-associated membrane protein-associated protein B), which interacts with the OMM protein tyrosine phosphatase-interacting protein 51 [83];
- -
- Sig-1R (Sigma non-opioid intracellular 1-receptor 1), a chaperone which binds Grp78; under ER stress conditions, Grp78 dissociates from the ER lipid rafts and activates the unfolded protein response (UPR) [84,85];
- -
- Protein kinase-like endoplasmic reticulum kinase (PERK), which, when activated reduces protein synthesis until the accumulated unfolded protein is cleared [86].
3.3. Mitochondrial Dynamics
Mitochondria are dynamic organelles, being able to modulate their number, shape, size, and position in the cytoplasm through a careful balancing of two opposite processes: mitochondrial fusion and fission [87].
Fission is regulated by two proteins: Drp1 (dynamin-related/-like protein 1) and Dnm2 (dynamin 2) [88]. The initial step is wrapping of the endoplasmic reticulum around the mitochondria and reducing the diameter of the latter from 300–500 nm to about 150 nm [89]. The spatial association of replicating mtDNA with the ER-mitochondria contact sites explains the mtDNA distribution in the replicating organelles [90]. Following this step, the cytosolic protein Drp1 is recruited to the already marked constriction site on the OMM and bound to the phospholipid membrane by adaptor proteins such as MFF (mitochondrial fission factor) and mitochondrial dynamics proteins 49 and 51 (MiD49 and MiD51) [91]. Following Drp1 recruitment, a ring-like structure is formed around the mitochondria [92], after which GTP hydrolysis potentiates the constriction of the mitochondrial membrane [87]. The final step is recruitment of Dnm2, a GTPase which assembles at the marked site and completes the fission process [93]. The constriction and division of the IMM is calcium-dependent and occurs at ER–mitochondria contact sites, possibly even before Drp1 recruitment [94].
Mitochondrial fusion is the opposite process, by which the membranes of two mitochondria fuse, giving rise to one single mitochondrion and allowing for sharing of essential components between the two organelles. The process is regulated by two other proteins with GTPase activity, Mfn1 and Mfn2 (mitofusins 1 and 2) for fusion of the OMM, while fusion of the IMM is under control of another GTPase, OPA1 (optic atrophy 1) [95]. After tethering of two mitochondria (mediated through the GTP domains), the two adjacent OMMs increase their contact surface area followed by their fusion due to GTP hydrolysis, subsequent conformational changes, and oligomerization of Mfns [96]. Following OMM fusion, IMM fusion is mediated by IMM-inserted OPA1, which can be cleaved by two membrane-bound metalloproteases, OMA1 and YME1L, resulting in two high molecular weight fragments (L-OPA1) and three shorter fragments (S-OPA1) [97]. The interaction of L-OPA1 with cardiolipin, inserted in the IMM, is crucial for driving membrane fusion [98]. The balance between OMA1 or YME1L cleavage of OPA1 regulates mitochondrial fission [99]. Stimulation of OXPHOS induces YME1L cleavage of OPA1 and mitochondrial fusion, while OPA1 cleavage by OMA1 is a stress response, and may induce mitochondrial fragmentation as well [87].
Several post-translational modifications also regulate mitochondrial dynamics. Drp1 phosphorylation can stimulate either fission or fusion depending on the phosphorylation site [100,101]. Phosphorylation of MFF increases Drp1 recruitment, and subsequent mitochondrial fission [102], while ubiquitination of acetylated Mfn1 promotes its proteasomal degradation [103]; phosphorylation of Mfn1 by ERK inhibits mitochondrial fusion, promoting apoptosis [104]. Specific nutrient states also indirectly regulate the balance between these two processes. While starvation leads to fused and elongated mitochondria, a nutrient-rich environment is accompanied by fragmented mitochondria [105,106].
3.4. Autophagy
Autophagy is another essential step in maintaining the balance between protein synthesis and clearance, organelle biogenesis, and degradation, thereby promoting cellular health [107]. Based on the way in which the targeted cargo is conveyed for degradation to lysosomes, autophagy can be classified into [107,108]:
- -
- Macroautophagy, in which the autophagosome, a double-membraned vesicle, forms and fuses with lysosomes after which their content is degraded by the acidic hydrolases of the lysosomes;
- -
- Microautophagy, a process during which lysosomes wrap around various cytosolic compounds which are degraded after the involution of the membrane [109];
- -
- Chaperone-mediated autophagy, a process during which chaperones bind to damaged proteins and to receptors on the lysosomal membrane, followed by translocation of the protein into the lysosome for degradation [110].
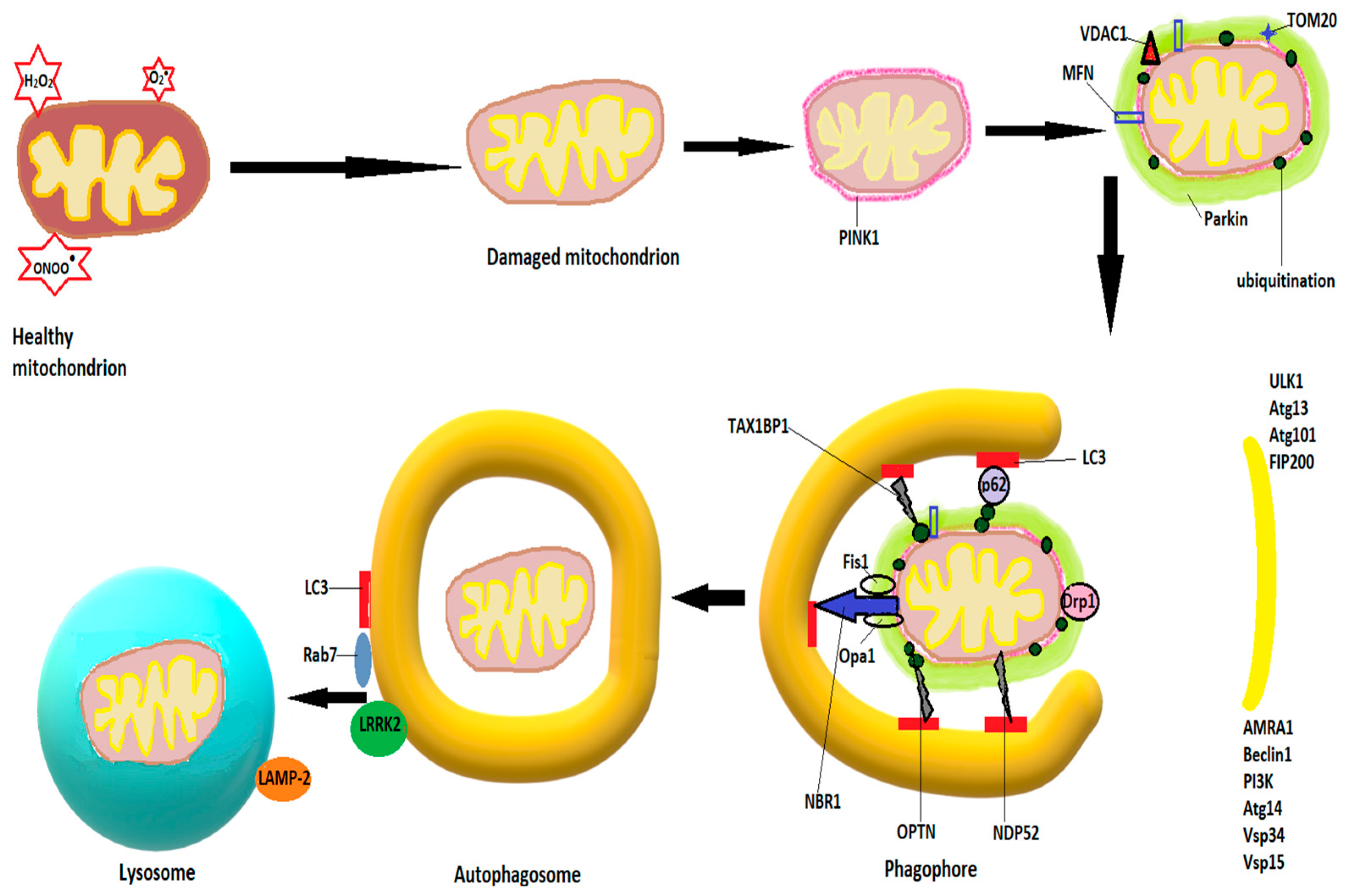
Autophagy for damaged mitochondria is also known as mitophagy. The first step in mitophagy initiation is the formation of an isolation membrane, the autophagosome, believed to derive from MAMs, membranes of the ER, or plasma membrane [111,112], followed by activation of the pre-initiation complex, containing ULK1 (Unc-51-like kinase 1, Atg 13 and 101 (autophagy-related proteins), and FIP200 (focal adhesion kinase family interacting partner 200) [113]. The pre-initiation complex recruits class III phosphatidylinositide 3-kinase (PI3K), beclin1, Atg 14, autophagy, and beclin 1 regulator (AMBRA1), as well as vascular protein sorting 34 and 15 (Vps 34 and 15) to produce phosphatidylinositol 3-phosphate (PI3P). Pi3P is also known as the initiation complex [114]. After activation, both complexes translocate to the nucleation site of the phagophore [108]. PI3P is recognized and interacts with other IM-located proteins, such as WD repeat protein interacting with phosphoinositide (WIPI) and FYVE domain containing proteins [115] and leads to a series of conjugations of Atgs on the phagophore, culminating in cleavage of pro-LC3 (light chain 3) by Atg4 to LC3-I, further transformed by phosphatidylethanolamine to LC3-II, and leading to elongation and closure of the isolation membrane [116,117]. Υ-aminobutiric acid type A-receptor-associated protein (GABARAP) and GABARAP-like 1 protein (GABARAPL1) are believed to play similar roles with LC3 in autophagosome expansion [118]. The signaling protein mTOR (mammalian target of rapamycin) strongly modulates autophagy. Inhibition of mTOR, as occurs in starvation, dephosphorylates and activates Atg13, igniting the mitophagy process [119]. When growth factors and cellular nutrients are abundant, mTOR phosphorylates Atg 13, preventing its binding to ULK1 and recruitment of FIP200 [107]. Fusion of autophagosomes to lysosomes is mediated by Rab7 and LAMP-2, a lysosomal transmembrane protein [120,121]. After fusion, lysosomal enzymes, mainly cathepsins, degrade the autophagosomal content [122].
In non-receptor mediated mitophagy, the mitophagy induction process activates PTEN-induced kinase 1 (PINK1), which accumulates on the OMM and recruits and phosphorylates Parkin [123,124]. Parkin accumulates on the OMM and ubiquitinates OMM proteins, which leads to increased activity of PINK1 and more Parkin recruitment [113]. Among the proteins ubiquitinated by Parkin are voltage-dependent anion channels 1 (VDAC1), Mfn1 and Mfn2, as well as TOM20 (translocase of the outer mitochondrial membrane 20), which govern mitochondrial fusion. Ubiquitination of Mfn1/2 will block the fusion process and allow the isolation of the small and damaged mitochondria [125]. The ubiquitinated proteins recruit autophagy adaptor proteins, such as OPTN (optineurin), NBR1 (neighbor BRCA1), TAX1BP1 (Tax-1 binding protein), NDP52 (nuclear dot protein 52), or sequestosome-1, which interact with autophagosome proteins like GABARAP or LC3 through LC3 interacting regions (LIR) to mediate autophagosome formation and fusion with lysosomes [126,127]. Figure 2 shows schematically the mitophagy process.
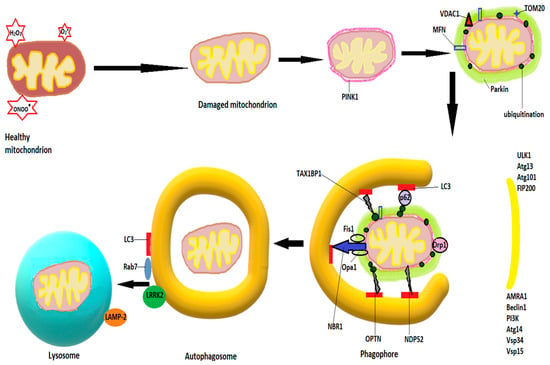
Figure 2.
Schematic diagram of mitophagy. The mitophagy induction process activates PINK1, which accumulates on the OMM and recruits and phosphorylates Parkin. The latter ubiquitinates OMM proteins, such as mitofusins 1 and 2 (Mfn), voltage-dependent anion channels 1 (VDAC1), and TOM20 (translocase of the outer mitochondrial membrane 20). Autophagosome formation starts with activation of the pre-initiation complex, containing ULK1 (Unc-51-like kinase 1), Atg 13 and 101 (autophagy-related proteins), and FIP200 (focal adhesion kinase family interacting partner 200), and continues with recruitment of class III phosphatidylinositide 3-kinase (PI3K), beclin1, Atg 14, AMBA1 (autophagy and beclin 1 regulator), and Vps 34 and 15 (vascular protein sorting), resulting in the production of phosphatidylinositol 3-phosphate (PI3P). The ubiquitinated proteins recruit autophagy adaptor proteins, such as neighbor BRCA1 gene (NBR1), optineurin (OPTN), Tax-1 binding protein (TAX1BP1), nuclear dot protein 52 (NDP52), or sequestosome-1, which interact with autophagosome proteins like GABARAP or LC3 to mediate autophagosome formation. Fusion of the autophagosome with lysosomes is mediated by LC3, Rab7, LRRK2, and LAMP-2 (see text).
However, Parkin-independent pathways of mitophagy exist as well [113], such as receptor-mediated mitophagy. The most studied proteins involved in receptor-mediated mitophagy are AMBRA1, FUNDC1 (FUN14 domain-containing protein 1), NIX (Nip3-like protein), and BNIP3, located on the OMM, as well as cardiolipin and prohibitin 2 (PHB2) on the IMM [113]. These receptors can bind to LC3 in a Parkin-independent manner [128] and induce mitophagy. Their transcription can be activated under various conditions. For example, transcription of BNIP3 and NIX are activated by hypoxia via hypoxia inducible factor 1 alpha (HIF1α) [129], which, after phosphorylation have a high binding affinity for LC3 [130]. Dephosphorylation of FUNDC1 by hypoxia facilitates its binding to LC3 [131].
In recent years, researchers have shown that mitochondria can be extruded from cells and taken up by endocytosis or phagocytosis by neighboring cells, where they ultimately undergo mitophagy, a phenomenon termed transcellular mitophagy [113,132]. It is reasonable to assume that transferring mitochondria back to the cell soma from dendrites or axons would be energetically unfavorable, which is why neurons release mitochondria at synapses to be degraded by glial cells [133]. In turn, glial cells can transfer mitochondria to neurons and protect the latter from hypoxia and energetic failure [134]. The precise pathways for mitochondrial transfer are still investigated, but some studies suggested an important role for a protein connecting mitochondria to cytoskeletal motor proteins, namely MIRO1 [135] while others showed involvement of astrocytic GFAP (glial acidic fibrillary protein) and neuronal UCP2 (uncoupling protein 2) [136].
Another way of disposing of damaged mitochondria has been identified in 1992 [137] and named mitoptosis. It enables the cells to degrade mitochondria without opening of the MPTP and igniting apoptosis [113]. Mitoptosis is likely activated by mitochondrial membrane depolarization, damage of the mitochondrial DNA (mtDNA), and ROS [113]. The exact mechanisms of mitoptosis require further study, but several situations have been described, such as swelling and fragmentation of cristae followed by cytoplasmic extrusion of cristae fragments through bursting of the OMM [138], or deterioration of cristae through coalescence of the IMM with preservation of intact OMM [138].
ROS play a key role in regulation of autophagy and mitophagy. One pathway, mentioned above, is the mTOR pathway. An amino acid-rich environment leads to translocation of mTOR complex 1 (mTORC1) to the lysosomal surface, where it interacts with Rheb and activates mTOR [139], while in starvation mTOR colocalizes with LC3 and initiates autophagy [140]. Considering that mTOR oxidation inhibits its activity, it is very likely that ROS regulate this step [141]. Further, S-nitrosation of IκB kinase β and JNK1 by nitric oxide inhibits their activity, which, in turn, prevents mTOR inactivation and release of Beclin from the Beclin-Bcl-2 complex [93,142].
Another pathway is the beclin-1-class III PI3K complex, with a series of cofactors such as AMBRA1, Bax-interacting factor 1 (Bif-1), or Rubicon (RUN domain- and cysteine-rich domain-containing beclin-1-interacting protein) [143]. Other regulators of autophagy include IP3 receptor, AMPK (5′-AMP-activated protein kinase) and DAPK (death-associated protein kinase) [144,145].
ROS and reactive nitrogen species induce post-translational protein modifications which also regulate the activity of transcription factors. For example, in the Nrf2 (nuclear factor-erythroid 2-related factor 2)/Keap1 (Kelch-like enoyl-CoA hydratase-associated protein 1) pathway, modifications of Keap1 lead to release of Nrf2, which binds to ARE (antioxidant-response element) and translocates to the nucleus, where it activates the transcription of antioxidant enzyme genes and proteins, such as p62 or p53. While p62 activates autophagy [146,147], p5 is linked to both autophagy-inhibiting and -promoting genes through TIGAR (tumor protein 53-induced glycolysis and apoptosis regulator) and DRAM (damage-regulated autophagy modulator), respectively [148,149]. In turn, impaired regulation of autophagy leads to increased oxidative stress and accumulation of ubiquitinated proteins, the latter causing mitochondrial dysfunction and further augmenting ROS generation in a feed forward loop [93].
4. The Brain and Oxidative Stress
Oxygen is crucial for proper cellular functioning, being involved in the generation of ATP [61]. Unfortunately, due to the univalent metabolic reduction status of oxygen, with the two lone electrons of oxygen spinning in parallel, it can accept only one electron at a time [150], leading to the generation of species having one unpaired electron which can exist on their own, defined by Halliwell as free radicals [151]. The oxygen derivatives are either free radicals, such as the superoxide anion (•O2−), hydroxyl radical (HO•), hydroperoxyl radical (HO2•), and peroxyl radicals (ROO•), or non-radicals which can be transformed into radicals, such as hydrogen peroxide (H2O2) [61,152]. Redox signaling is extensively used in the brain [153], being involved in signal transduction and gene transcription. For example, NADPH oxidases (NOXs) regulate hippocampal long-term potentiation [154] and NOX2-derived superoxide and hydrogen peroxide regulate hippocampal progenitor cell growth in adult brain via the phosphatidyl inositol 3 kinase (PI3K)/Akt signaling pathway [155]. Similarly, NOX-derived H2O2 have beneficial roles in axonal regeneration and axonal pathfinding during wiring of the developing brain [156,157]. As a consequence of hypoxia, mitochondria-derived superoxide-induced signaling leads to adaptive responses [158]. To counterbalance the possible deleterious effects of excessive free radicals, the biological systems have a series of antioxidant defenses, which can be divided into enzymatic ones (superoxide dismutases, catalase, glutathione peroxidases, glutathione transferases, thioredoxins, and peroxiredoxin) and non-enzymatic defenses, such as vitamins A, C, E, beta-carotene, or glutathione [159]. Whenever the rate of free radical production exceeds the biological system’s ability to neutralize them, oxidative stress ensues.
4.1. Vulnerability of the Brain to Oxidative Stress
The nervous system is very sensitive to oxidative stress, due to a series of reasons [153,160,161,162]:
- -
- Action potentials cause calcium influx and raise the intracellular calcium concentration from approximately 0.001 μm to roughly 100 μm [163]. High intracellular Ca2+ activates nNOS (neuronal nitric oxide synthase) and leads to NO (nitric oxide) formation [164], which binds to cytochrome c oxidase and inhibits mitochondrial respiration [165]. Mitochondria attempt to buffer intracellular calcium, but the subsequent calcium overload causes prolonged opening of the MPTP and inhibits ATP generation, inducing apoptosis [166].
- -
- The brain has very high energy demands to maintain the ionic gradients and support synaptic transmission [19] and relies mainly on synaptic mitochondria for the generation of required energy [167]. For example, neurotransmitter vesicle release requires 1.64 × 105 ATP/s/vesicle [19].
- -
- The brain has low antioxidant defenses. Neuronal cells have 50 times less catalase than hepatocytes [168], while cytosolic glutathione is about 50% lower in neurons compared with other cells [153], and this might diminish peroxiredoxin activity [169].
- -
- Microglia, the immune cells of the brain, are activated by H2O2 [170] and produce superoxide via NADPH oxidase isoforms, needed for bacterial killing [171].
- -
- The metabolism of neurotransmitters, such as dopamine metabolism via monoamine oxidases, generates ROS [153,172].
- -
- Neurotransmitters, such as dopamine, serotonin, or adrenaline, can auto-oxidize and generate superoxide [173,174].
- -
- The brain is enriched in redox active transition metals, such as Cu+ or Fe2+ [175]. Iron is a catalyzer in the hydroxyl radical-generating Fenton reaction, and also catalyzes peroxyl and alkoxyl radical generation, thereby contributing to ferroptosis, a form of cell death dependent on lipid peroxidation and Fe2+ [176]. Cu+ is a co-factor for Cu/ZnSOD and is important for cell signaling [177,178] but enhances copper-catalyzed Fenton reaction [175].
- -
- The brain is particularly rich in cholesterol, which may undergo auto-oxidation [179] and brain cells have a higher membrane surface/cytoplasmic volume ratio, cellular membranes being rich in polyunsaturated fatty acids (PUFA), which are highly susceptible to peroxidation through free radical attack [153].
- -
- Brain development and plasticity relies on non-coding RNAs (long non-coding RNAs and microRNAs) [180], but these molecules lack protective histones and are easily oxidized [181]. Oxidized messenger RNA results in truncated and mutated proteins, prone to misfolding [182].
4.2. Sources of Free Radicals
Excess free radicals can be generated from many sources.
4.2.1. Mitochondria and Oxidative Stress
Mitochondria are traditionally regarded as main sources of ROS. At least 10 potential sources of ROS production have been identified [183] but complexes I (NADH dehydrogenase) and III (ubiquinone cytochrome c reductase) of the ETC [184] are the most important ones. Transferring electrons to coenzyme Q or ubiquinone by complexes I and II results in ubiquinol (reduced ubiquinone, QH2), which will regenerate coenzyme Q via semiquinone anion (•Q−), an unstable intermediate which can transfer electrons to molecular oxygen, resulting in superoxide formation [60]. Being a non-enzymatic reaction, higher metabolic rates lead to increased superoxide production [185]. Superoxide is highly unstable and is transformed by the mitochondrial superoxide dismutase 2 (SOD2, manganese SOD) and the cytosolic SOD1 (copper zinc SOD) into the more stable hydrogen peroxide (H2O2). The latter can exit the IMM through aquaporin channels and diffuse through the OMM into the cytoplasm, where it serves for redox signaling, or is further reduced to water by catalase, glutathione peroxidases, and peroxiredoxins [186,187,188,189]. Other mitochondrial components contributing to ROS formation include monoamine oxidase, glycerol phosphate dehydrogenase, α-ketoglutarate dehydrogenase, and p66shc [60,183,190].
Mitochondrial ROS production is subject to variations induced by metabolic factors. For example, the NADH/NAD+ ratio affects the rate of ROS generation, which increases almost linearly with NADH reduction [191]. Succinate levels can fluctuate even in normal conditions between 0.3 and 1 mM [192], with increased succinate concentrations strongly increasing mitochondrial ROS generation [183,193]. The rate of ROS production is also influenced by the mitochondrial membrane potential [183]. Active mitochondrial phosphorylation of ADP or mitochondrial calcium uptake decreases the membrane potential, which influences the redox potential of the ETC and decreases ROS production [194]. Oxygen deprivation or ischemia significantly increase mitochondrial ROS generation [195], although this effect may not be attributed to mitochondria per se, but rather to signaling pathways triggered by hypoxia [196].
4.2.2. NADPH Oxidase as a Source of ROS
NADPH oxidase (NOX) was first described in phagocytes [197], after which seven NOX genes have been identified: NOX 1–5 and DUOX 1 and 2 [198]. The brain expresses mainly NOX2, as well as NOX4, both being described in the cortex and CA1 hippocampal areas [199]. The NOX2 enzyme complex has a membrane-bound cytochrome b558, several cytosolic proteins, and the Rac G-protein. Following phosphorylation of the cytosolic proteins and activation of Rac, the enzyme translocates to the membrane and forms active NOX2 with cytochrome b558 [200]. Further, NOX2 transfers protons across the membrane and leads to superoxide generation [198]. NOX4 produces mainly H2O2, used as a second messenger for cell proliferation and differentiation [201]. NOX has been described in neurons, astrocytes, and microglia [202], while at the cellular level, NOX isoforms localize to the endoplasmic reticulum, nucleus, plasma membrane, and mitochondria [203,204]. The ROS generated by activated NOX can depolarize the mitochondrial membrane and, together with calcium, can lead to opening of the MPTP [205] as well as activate phospholipase C with subsequent changes in membrane structure [198].
4.2.3. Monoamine Oxidase as a Source of ROS
The monoamine oxidases (MAO A and B) are flavoenzymes located on the OMM which catabolize amine neurotransmitters, such as serotonin, epinephrine, and dopamine [206]. MAO A is expressed in neurons, while MAO A and B can be found in glial cells [198]. They use FAD to break down monoamines, a process during which aldehydes are produced, while H2O2 results from the FAD-FADH2 cycle [198].
4.2.4. Peroxisomes and ROS Production
Although the major metabolic peroxisomal process leading to H2O2 generation is β-oxidation of free fatty acids [60], several other peroxisomal enzymes, such as xanthine oxidase, D-aspartate oxidase, acyl CoA oxidases, D-amino acid oxidase, urate oxidase, or L-α-hydroxy oxidase, can produce a variety of ROS, such as superoxide, hydrogen peroxide, nitric oxide, or hydroxyl radicals [207].
4.2.5. Exogenous Sources of ROS
In addition to the multiple sources of endogenous ROS, exogenous ROS can augment oxidative stress. The most common sources are water and air pollution, ultraviolet light exposure, alcohol and tobacco smoke, pesticides, industrial solvents, unhealthy diets (with smoked meat, high-fat diet), exposure to heavy metals or transition metals (Fe, Cr, Co, Cu, Hg, Pb, As), as well as certain drugs, such as Doxorubicin, Bleomycin, Metronidazole, or even Paracetamol [60].
4.3. Targets of ROS
The highly reactive free radicals damage proteins, lipids, and nucleic acids [152].
4.3.1. Proteins and ROS
Proteins can be oxidized by radicals, such as superoxide, hydroxyl radical, peroxyl, hydroperoxyl, or alkoxyl radicals, as well as by non-radical species or singlet oxygen [208]. Oxidation of amino acids such as lysine, arginine, proline, or threonine leads to carbonyl derivatives, used as markers of oxidative stress [209]. Methionine and cysteine, as sulphur-containing amino acids, are very susceptible to oxidation, leading to disulphides and methionine sulphoxide [210]. Protein oxidation leads to protein–protein cross linkages, altering of function, loss of enzymatic activity, and functional modifications of receptor and transport proteins [211]. Moreover, hydrogen peroxide and hydroxyl radicals inhibit glutamate uptake by astrocytes, augmenting excitotoxicity [212]. As a consequence, these altered proteins must be cleared, either by the autophagy-lysosome pathway or by the ubiquitin-proteasome system [213,214]. The ubiquitin-proteasomal system (UPS), the main degradation pathway of ubiquitinated misfolded proteins and short-lived signaling molecules, contains a 19S subunit and a catalytically active 20S core [215]. The regulatory cap with chaperone proteins unfolds the target protein, removing the ubiquitin tag in an ATP-dependent process, after which the target protein is fed into the catalytic core and is degraded by the proteasomal enzymes. Under severe stress, the UPS is overwhelmed, and the autophagy-lysosome pathway compensates for the increased protein damage [107]. A series of autophagy receptors, such as p62, NDP52, or NBR1 recognize the ubiquitin moiety and target tagged proteins to the autophagosome by binding to Atg8/LC3 [216,217]. FOXO3 (forkhead box O3) is a transcription factor activated by oxidative stress, which regulates the transcription of genes involved in proteasomal as well as autophagic protein degradation [218]. In addition, Parkin, ubiquitin ligases, and CHIP (C-terminus of Hsc70-interacting protein) contribute to both proteasomal and autophagosomal degradation of proteins [107,219]. The relative levels between BAG (Bcl-2 associated atanogene) 1 and BAG3, co-chaperone proteins, direct the cellular protein degradation pathway towards the proteasomal or phagosomal one [220].
4.3.2. Lipids and ROS
Free radicals or non-radical oxidative species attack the C-C double bonds of lipids, which is why polyunsaturated fatty acids are very vulnerable to oxidative attack [221]. In the initial phase, a free radical interacts with a methylene group in the fatty acid and generates a lipid radical by dissociating a hydrogen atom [222]. Further, the lipid radicals react with molecular O2 to form peroxyl radicals (ROO•) [223], which initiate a chain of self-sustained reactions amplifying the process. This results in cyclic peroxides and hydroperoxides, which can further be degraded to aldehydes, the final products being malondialdehyde (MDA), hydroxynonenal (HNE), and acrolein [224]. The process is terminated either through interaction of lipid radicals with lipid peroxides, resulting in non-reactive stable species [221], or through intervention of endogenous or exogenous antioxidants (vitamins C and E) [222,225].
In low concentrations, 4-HNE plays important homeostatic roles by acting as a signaling molecule and modulating gene expression by inducing post-translational protein modifications. The most common targets are thiol residues [225]. By interacting with cysteine thiols in Keap1, 4-HNE leads to release of Nrf2, which after translocating to the nucleus activates the expression of ARE genes such as glutathione-S-transferase, NADPH-dependent quinone reductase, or heme oxygenase-1 [226]. Another target of 4-HNE is NF-κB, a transcription factor for pro-inflammatory cytokines, normally maintained quiescent by binding to IκBα (inhibitor kappa B). Under cellular stress, IκB kinase phosphorylates IκBα facilitating the release of NF-κB, a transcription factor which mediates the transcription of antiapoptotic Bcl-2 proteins and of inflammatory cytokines, such as interleukin-6 (IL-6). 4-HNE inhibits IκB kinase, thus preventing IκBα phosphorylation and NF-κB nuclear translocation [227].
However, as a consequence of membrane lipid peroxidation, the membrane alters its permeability, increases its rigidity, and may even loose integrity [228]. In addition, lipid peroxidation products are involved in complex signaling pathways. Intracellular accumulation of 4-HNE can lead to apoptosis through both the intrinsic and extrinsic pathways [225]. 4-HNE increases the expression of p53, followed by activation of p21, JNK, Bax, and caspase 3 [229], leading to caspase-mediated apoptosis. In addition, 4-HNE initiates the binding of the death-associated protein Daxx to the intracellular surface of Fas [230], thereby being involved in modulation of the extrinsic pathway of apoptosis through the down-stream signaling proteins ASK1 and JNK [225]. Moreover, high levels of HNE can form conjugates with JNK, responsible for histone modification and facilitating of nuclear translocation [231] or can activate JNK through SPKK1 (stress-activated protein kinase kinase-1) activation [232]. Similarly, HNE can activate ERK via activation of MEK1/2 and p38MAPK (mitogen activated protein kinase) [233,234].
4.3.3. DNA
ROS can induce many types of DNA damage, such as base modification, deoxyribose modification, single strand breaks (SSBs) and double strand breaks (DSBs), DNA cross-links, or abasic sites [235]. Oxidized DNA is probably the most common DNA lesion in neurons [236]. As under normal conditions ROS can cause up to 50,000 DNA lesions/cell/day [237], delays in repair of DNA damage can cause genomic instability and induce signaling cascades leading to cell death [238]. The cells have various repair mechanisms, such as base excision repair (BER), mismatch repair (MMR), nucleotide excision repair (NER), and single- (SSBR) and double-strand break repair (DSBR) mechanisms [239].
Oxidized DNA bases are removed mainly by the BER/SSBR mechanism and involve the removal of the damaged base by a specific DNA glycosylase (DG), incision of the abasic site by an AP-endonuclease (APE1), filling of the resulting gap by a DNA polymerase, and sealing of the damaged DNA strand by a DNA ligase [240]. Several DGs have been identified, such as uracil-DNA glycosylases (UDGs), thymine DNA glycosylase (TDG), or oxidized base-specific DGs, such as 8-oxoGuanine (8-oxoG) DNA glycosylase (OGG1) [241,242]. OGG1-1a is involved in nuclear DNA repair, while OGG1-2a contributes to mitochondrial DNA repair [243]. APE1 cleaves the sugar-phosphate backbone of DNA at the abasic site. Phosphorylation of the enzyme, as happens after 1-methyl-4-phenylpyridinium (MPP+) exposure, reduces its enzyme activity and results in accumulation of damaged DNA in neurons [244].
Mitochondrial DNA is even more prone to oxidative damage because the mitochondrial genome is situated close to the IMM, the site of mitochondrial ROS generation, and because it lacks protective histones [245]. As mitochondrial DNA quality control is crucial for the communication with the nucleus, mitochondria contain antioxidant and DNA repair enzymes, such as OGG1 and MUTYH (mutY DNA glycosylase) [246]. Mitochondrial dysfunction, as happens after mtDNA deletions, can lead to activation of NF- κB through a calcineurin-dependent signaling pathway [247] and can, in turn, be induced by various signaling molecules. For example, PPAR-γ coactivator 1 and SIRT1 regulate mitochondrial function by activating the expression of mitochondrial transcription factor A (TFAM) [248].
In addition to increased oxidative damage caused by oxidative stress, defective DNA repair mechanisms have been described in neurodegenerative diseases, such as AD or PD [235]. Dopaminergic neurons of PD patients showed upregulation of mitochondrial OGG1 and higher levels of phosphorylated APE1 [249]. Neurons from AD patients had higher levels of oxidized DNA bases in nuclear as well as mitochondrial DNA [250], exhibited a decrease of OGG1 activity [251] and lower levels of UDG [252] in comparison with neurons from healthy individuals.
4.3.4. RNA and Oxidative Damage
RNA is more abundant, accounting for 80–90% of the nucleic acid in cells [253] and is more vulnerable to oxidative damage due to its single stranded structure, proximity to the mitochondria, lack of oxidized RNA repair mechanisms, and less protection from proteins as compared to DNA [60,254]. The functions of RNA molecules in the cell are diverse, comprising the ribosomal RNA (rRNAs), which together with the transfer RNA (tRNA) and messenger RNA (mRNA) is responsible for protein synthesis, microRNAs (miRNAs), which are post-translational regulators of gene expression, and small nuclear and nucleolar RNAs. Traditionally, mRNA was regarded as the coding RNA [255]. The non-coding RNAs play important roles in mRNA splicing regulation and mRNA translation [256].
The most aggressive radical is the hydroxyl radical, produced in the Fenton and Haber–Weiss reactions [257]. It reacts with guanine to form 8-hydroxyguanosine (8-OHG), one of the most commonly used biomarkers of RNA oxidation [253]. The oxidative attack-induced damage to RNA leads to modified bases and nucleosides, cleavage and fragmentation of tRNA, aminoacylation, or defects in codon–anticodon pairing [258] and cause direct RNA strand breaks [259], base mismatches on tRNAs, translation errors [182], and disordered protein synthesis [260] with significant impact on cell viability. Although cells have several mechanisms to degrade altered transcripts, such as the nonsense-mediated mRNA decay (NMD) [261], these mechanisms tend to be overwhelmed with aging. As such, altered misfolded proteins accumulate [262].
5. Selective Neuron Vulnerability in Neurodegenerative Diseases
Although the many pathways leading to increased ROS generation and their consequences apply to all cells, each neurodegenerative disease leads to degeneration of particular groups of neurons, which led researchers to look for explanations for this selective vulnerability of neuronal populations.
5.1. Selective Neuronal Vulnerability in Alzheimer’s Disease
The memory loss and cognitive decline characteristic of AD are caused by atrophy of the entorhinal cortex (EC), mainly of neurons in layer II (ECII), and hippocampus, particularly the CA1 region [72]. Research has shown that neurons in these areas have high energetic demands and are very sensitive to decreased oxygen and glucose supply [263]. In addition, CA1 and EC II pyramidal neurons are glutamatergic and thereby more vulnerable to NMDA excitotoxicity and the damaging effects of increased intracellular calcium concentrations [264], as opposed to neocortical inhibitory interneurons, which have high levels of Ca2+-binding proteins [265]. A series of molecular hallmarks of the vulnerable neurons have been recently described. For example, ECII pyramidal neurons have an impaired activity of a regulator of tau splicing, likely linked to disturbed microtubule dynamics [266], which may facilitate tau spreading to other brain regions via CA1 neurons [267]. Selectively vulnerable excitatory neuron subpopulations were shown to express RORB (RAR-related Orphan Receptor B), while also exhibiting differences in the expression of genes encoding synapse- versus axon-localized proteins, subunits of the potassium channels, G-protein signaling molecules and neurotransmitter receptor signaling molecules [268]. However, the connections between RORB expression, accumulation of phosphorylated tau, and neural degeneration remain to be further characterized.
5.2. Selective Neuronal Vulnerability in Parkinson’s Disease
The motor symptoms of PD are caused by the loss of nigral dopaminergic neurons leading to dopamine depletion in the dorsal striatum [269]. Structurally, these neurons have very long and branched axons (up to 4.5 m), being connected to a large number of neurons (up to 2.4 million synapses) [270,271], which requires a high density of axonal mitochondria and challenges mitochondrial bioenergetics [272].
At the molecular level, nigral dopaminergic neurons have low Ca2+-buffering capacity despite high activity-dependent Ca2+ loads, which increase OXPHOS and ROS generation [273], and may promote mtDNA damage [274]. In addition, the metabolism of dopamine itself leads to ROS generation and causes accumulation of mtDNA deletions [275].
5.3. Motor Neuron Vulnerability in Amyotrophic Lateral Sclerosis
The pathological hallmark of ALS is degeneration of upper and lower motor neurons, which both have very long axons and, thus, depend on proper mitochondrial function and trafficking [72]. The large motor unit size imposes high energetic demands on spinal motor neurons to maintain neurotransmission and muscle contraction [276]. The relative preservation of motor neurons in the oculomotor, trochlear and abducens nuclei may be linked to the small number of innervated muscle fibers (up to five muscle fibers as opposed to at least 300 fibers innervated by spinal motor neurons) [277], the particular grape-like structure of the neuromuscular junction [278], as well as the high Ca2+ buffering capacity of these neurons [279].
6. Oxidative Stress in Neurodegenerative Diseases
Research has increasingly shown the presence of oxidative stress markers in the nervous system of patients having neurodegenerative diseases, but the molecular pathways are just beginning to be elucidated. The description of genetic defects leading to AD, PD, HD, or ALS helped in identifying downstream effects of the mutant proteins, similar pathways being subsequently demonstrated in idiopathic forms of the diseases. However, the pathophysiology is still incompletely elucidated, despite accumulated knowledge having already led to therapeutic attempts.
6.1. Oxidative Stress in Alzheimer’s Disease
Familial cases of AD with mutations of Presenilin 1 (PS1) and 2 (PS2) have linked AD pathogenesis with disturbed calcium homeostasis [280]. PSs interact with the ryanodine receptors [281] and influence ER–mitochondria coupling [282]. Indeed, high MAM numbers have been described in animal models of AD and in fibroblasts or brain tissue of AD patients [283]. In addition, Aβ aggregates can mediate Ca2+ transfer from ER to the mitochondria through the MCU [284], while tau inhibits mitochondrial calcium efflux [285]. Moreover, Aβ can form calcium-permeable channels in membranes [286,287], while tau can form non-selective ion channels in lipid bilayers [288]. The deleterious effects of increased mitochondrial calcium in inducing mitochondrial dysfunction and oxidative stress have been described in the previous sections. In astrocytes, Aβ interacting with the calcium sensing receptors (CaSRs) induces their downregulation, leading neighboring neurons to secrete newly synthesized Aβ, nitric oxide, and peroxynitrite [289].
In the mitochondria, one of the earliest alterations linked to AD is lipoxidation of ATP synthase with consequent reduction of its function, described in EC neurons as early as Braak stages I-II [290,291]. Located inside the IMM, inserted in a PUFA-rich lipid bilayer and close to the mitochondrial matrix, the enzyme is an easy target for free radicals generated by complexes I and III [292]. In addition to energetic failure, ATP synthase modification further increases ROS production with subsequent potentiation of oxidative modifications of biological molecules [293]. In this cascade, most modified lipoxidation products are involved in energy metabolism, increasing energy failure, followed by proteins involved in neurotransmission, antioxidant defenses, and ion channels, as shown in Table 1.

Table 1.
The modified mitochondrial proteins described in AD and their involvement in cellular processes (adapted from Jové et al. [291]).
The communication between mitochondria and nucleus, and the import of nuclear-coded mitochondrial subunits are impaired by the bioenergetic defects and dysfunctional mitochondrial proteins, leading to altered expression of regulatory and structural mitochondrial complexes and enzymes [299]. The lipid metabolism is also altered, with changes in lipidomic profiles described in AD, especially in the EC [300]. The oxidative damage to PUFAs leads to the reduction of these fatty acids in the lipid rafts in EC from early stages of AD [301], with consequences on membrane thickness, fluidity, curvature, as well as activity of membrane-bound proteins, which favor amyloidogenic processing of amyloid precursor protein. It appears that mitochondrial oxidative stress, altered lipid metabolism, lipid peroxidation, and bioenergetic defects are all part of a self-sustained loop which contributes to augmentation of the altered mitochondrial dynamics, mitochondrial trafficking, impaired mitophagy, and impaired ER–mitochondrial interaction [290].
The intracellular build-up of amyloid beta (Aβ) and phosphorylated tau induces much of the altered mitochondrial dynamics, expressed as excessive mitochondrial fragmentation, with increased number and decreased size of mitochondria [302]. Both Aβ and phosphorylated tau increased the GTPase activity of Drp1, which leads to excessive mitochondrial fragmentation [303,304]. In vitro, mutant APP cellular lines exhibited high concentrations of mRNA and proteins of mitochondrial fission and diminished levels of mRNA and proteins of mitochondrial fusion [305]. Phosphorylated tau has also been shown to downregulate Opa1 and upregulate Mfn1 and Mfn2 [306]. In addition, cytoplasmic accumulation of Aβ leads to depletion of Parkin and PINK1 levels, thereby interfering with the mitophagy pathway. Defective mitophagy results in accumulation of autophagic vacuoles in the neuronal soma and dysfunctional neurites [307], a process potentiated by the particularities of mitophagy in neurons, where mature lysosomes are concentrated in the cell body whereas mitochondria extend along the axons and dendrites of neurons, making neuronal mitophagy a slower process [308]. In addition, Aβ oligomers interact with autophagic vacuoles in the distal parts of axons, leading to inhibition of mitochondrial axonal transport [309]. In addition, in laboratory models, mutations in the PS1 gene altered lysosomal acidification [310] and led to diminished expression of autophagy-related genes through the ERK/CREB signaling pathway [311].
Protein tau has also important contributions to the protection of the cellular genome in physiological conditions, by binding chromatin [236]. The hyperphosphorylated state of tau interferes with this function, creating the premises for infliction of more oxidative DNA damage and longer time needed to repair these DNA lesions [236].
One must not overlook the role of microglial activation and chronic inflammation in the pathogenesis of AD. Microglia, the innate immune macrophage-like cells of the nervous system, account for about 10% of the cellular population in healthy adult brain [312]. In AD, microglia have increased expression of complement receptors leading to upregulation of the NF-κB signaling pathway, Aβ-activated Fc receptors (which induces the expression of MIP-macrophage inflammatory protein-1α), and increased expression of scavenger receptors A-1 and B (SCARA and SCARB) [313,314]. Binding of Aβ to SCARB-2 activates microglia, to produce proinflammatory cytokines, chemokines, and ROS [315]. Increased levels of tau, by inducing the expression of toll-like receptors, also lead to the release of pro-inflammatory cytokines IL-1β, IL-6, IL-8 through the NF-κB signaling pathway [316]. Although inflammation activates autophagy in cells [108], since impairment of autophagy affects microglial cells as well, the chronic inflammatory microglial phenotype supplementary contributes to neuronal damage in AD [317].
6.2. Oxidative Stress in Parkinson’s Disease
PD is the second most common neurodegenerative disease, exhibiting both cognitive and neuromuscular impairments. Although most cases are sporadic, several mutations have been described in familial and early-onset forms of PD, such as mutations in PARK1 (encoding PINK1), PARK2 (encoding for Parkin), PARK1/4 (α-synuclein), PARK7 (DJ1), PARK8 (LRRK2), PARK9 (ATP13A2) PARK17 (Vsp35), FBX07, GIGYF2, or HTRA2, highlighting the involvement of the ubiquitin protein degradation pathway, oxidative stress, cell survival pathway, mitochondrial function, and apoptosis in PD pathogenesis [113,318]. The pathological hallmark of PD is the accumulation of insoluble inclusions consisting predominantly of synuclein, (Lewy bodies) mainly in the nigral dopaminergic neurons [61].
The study of toxin-induced parkinsonian syndromes has helped in elucidating PD pathogenesis. In the 20th century, exposure to 1-methyl-phenyl 4-phenyl-1,2,3,6-tetrahydropyridine was shown to cause symptoms resembling severe parkinsonism [319] by interfering with complex I of the ETC. Further, in experimental setting, rotenone, a complex I inhibitor, led to apoptosis in human neuroblastoma cells, and this finding was further expanded to discuss the involvement of pesticide exposure in the etiology of PD [320]. Subsequently, several researchers consistently reported a defect of complex I of the mitochondrial ETC leading to a 30–40% decline in its activity [48,321], caused by a diminished rate of production of complex I subunits, destruction of its structure, and oxidative damage [322]. α-synuclein targets to mitochondria and leads to a decrease in complex I activity [323]. LLRK2 (leucine-rich repeat kinase 2) mutations can increase α-synuclein levels [324]. PINK1 and Parkin regulate mitophagy, as described earlier. DJ1 is important for mitochondrial integrity and dynamics; overexpression of DJ1 has been shown to reduce mitochondrial fragmentation induced by rotenone, independently of PINK1 [325].
Convincing evidence has shown accumulation of increased lipid, protein, and DNA oxidation products in the degenerating neurons in PD [326,327,328] together with reduction in the antioxidant GSH [329], thereby implicating oxidative stress in the pathogenesis of PD. Initially, ROS probably originate from the ETC, external factors, and dopamine auto-oxidation [330], potentiated later by monoamine oxidase B metabolism of dopamine, inflammatory responses, or the contribution of heavy metals [331]. ROS have been shown to induce unorthodox activation of Iron Regulatory protein 1, thereby contributing to iron accumulation in dopaminergic cells [332]. Hydrogen peroxide can easily diffuse to adjacent neurons, where it can generate hydroxyl radicals through its interaction with iron [55]. PINK1 mutations in the kinase domain of the mitochondrially located molecule also increase the cells’ susceptibility to oxidative stress [55,333].
Another commonly described aspect in PD is impaired mitochondrial dynamics and altered mitochondrial morphology, with either elongated or fragmented mitochondria [334,335]. Impaired mitochondrial fusion, possibly related to the diminished levels of the short form of Opa1 described in brain samples of patients with idiopathic PD [336], contributes to the degeneration of nigral dopaminergic neurons, as does interaction of mutant LRRK2 with Drp1, which promotes mitochondrial fission [337]. α-synuclein can also bind to the OMM and decrease the mitochondrial fusion rate [338], which, together with increasing the Mfn1 and Mfn2 levels, promotes fragmentation and shortening of mitochondria [339]. In addition, mutant LRRK2 affects mitochondrial trafficking by interfering with MIRO1 removal, resulting in accumulation of MIRO1, which bind dynein and kinesin (microtubule motors) to mitochondria [340]. An interesting finding is the need to acidify the synaptic vesicles for proper loading of dopamine into these vesicles [341], a task performed by vacuolar ATPases and which require proper levels of ATP [330]. As such, in energy-deficient conditions, dopamine is improperly packed into synaptic vesicles and its cytoplasmic levels increase, creating conditions for its auto-oxidation [342].
6.3. Oxidative Stress in Amyotrophic Lateral Sclerosis
ALS is the third most common neurodegenerative disease, after AD and PD [16], occurring both sporadically (90–95% of cases) and as inherited disease caused by genetic mutations. Many mutations leading to familial ALS cases have been described, the most common ones being mutations in SOD1, FUS (fused in sarcoma/translocated in liposarcoma or heterogenous nuclear ribonucleoprotein P2), C9orf72 (chromosome 9 open reading frame 72), and TARDP (transactive response DNA binding protein 43) [343,344], the study of which can help identify the pathophysiologic mechanisms of the disease.
Decreased glucose metabolism with reduced ATP generation was reported in the cerebral cortex of SOD1G93A mice before the clinical picture of ALS emerged, while the spinal cord exhibited these abnormalities only in later stages [345]. Similar abnormalities were observed in human patients as well [346], possibly related to downregulation of two key enzymes in glycolysis, PGK (phosphoglycerate kinase) and PGM2L1 (phosphoglucomutase-2-like 1) [347]. It appears that this metabolic disturbance does not affect neighboring astrocytes [348]. In response, neurons upregulate glycolysis [349] at the expense of increasing oxidative stress or may turn toward alternative sources of energy such as ketone bodies [350]. Beta-hydroxybutyrate facilitates oxidation of NADH and increases the NAD+/NADH ratio, thereby inhibiting mitochondrial ROS production [351] and activating SIRT1 and SIRT3 [31]. SIRT1, through deacetylation, alters the activity of the PGC-1α (peroxisome proliferator-activated receptor gamma coactivator 1-α)/ERR-α (estrogen-related receptor α) complex, with important roles in mitochondrial biogenesis regulation [350,352].
Disturbed cellular calcium handling is another feature described in motor neurons of patients with ALS as well as in vitro and in vivo models expressing mutant SOD1 [353]. After AMPA activation, the recovery of physiological calcium concentrations is delayed in motor neurons of ALS models [354], rendering them susceptible to Ca2+-induced excitotoxicity [355]. In addition, analysis of motor neurons from patients with TDP-43 mutations showed an upregulation of AMPA and NMDA receptors and an imbalance between MICU1 and MICU2 leading to reduced mitochondrial calcium uptake [356].
Mitochondrial quality control was also found deficient, with abnormally swollen and vacuolated organelles distributed mainly in the cell body and proximal axon of motor neurons [357]. Normally, mitochondrial mitophagic receptors OPTN and NDP52 target to mitochondria through their ubiquitin-binding domain and recruit autophagosomes [358]. Mutations of OPTN and TBK1, encoding an OPTN-containing kinase, were found in patients with ALS [359], leading to impairments of mitophagy and accumulation of damaged mitochondria which further decrease glucose metabolism and ATP generation. Excessive ROS activate Drp1, with consequent increased mitochondrial fission, while reduced ATP levels cause impaired autophagy and decrease proteasomal protein degradation with subsequent accumulation of protein aggregates which can trigger ER stress [360].
Although oxidative stress may not be the triggering factor, it is likely that it exacerbates ALS progression, and, again, mitochondria significantly contribute to the generation of ROS. The SOD1 mutation diminishes the affinity of Cu/ZnSOD for Zn, altering its antioxidant properties [361], and to a higher load of DNA damage, possibly due to a loss of nuclear protection [350]. The oxidants/antioxidants ratio is supplementally altered by the reduced levels of GSH found in the motor cortex of ALS patients in comparison with healthy volunteers [362]. The altered calcium homeostasis leads to mitochondrial dysfunction and to the activation of intrinsic (through mitochondrial release of death signals) or extrinsic (through specific ligands binding to death receptors like Fas or DR6—death receptor 6) pathways of apoptosis [363]. Motor neurons in affected regions of ALS patients show increased levels of p53 [364], possibly caused by reduced proteasomal degradation of p53 [365], which leads to a decrease in Bcl-2 and increase in Bax, Fas, and caspases 8 and 3 [366]. In addition, astrocytes and microglia are activated, leading to chronic inflammation and supplemental generation of ROS. Cultures of rat astroglia exposed to CSF from ALS patients showed increased release of inflammatory cytokines IL-6 and TNFα, reduced anti-inflammatory cytokine IL10, increased generation of cyclooxygenase-2 and prostaglandin E2, and downregulation of trophic factors such as GDNF (glial cell-derived neurotrophic factor) or VEGF (vascular endothelial growth factor) [367]. Moreover, astrocytes from ALS patients release neurotoxic factors able to damage motor neurons through a Bax-dependent mechanism [368] and proliferate to surround degenerating motor neurons and produce molecules which inhibit axonal regrowth [369]. Activated microglia are also toxic for motor neurons. In studying the effects of TDP43 mutation, microglial activation led to a NF-κB and NLRP3 (NLR family pyrin domain containing 3) inflammasome-dependent cascade which damaged motor neurons, while in the absence of microglia, motor neurons were able to survive [370]. Further, blocking the NF-κB signaling pathway in microglia rescued motor neurons in an animal model of ALS [362].
Table 2 summarizes the abnormal proteins associated with the three neurodegenerative diseases discussed and their deleterious actions on mitochondrial function.

Table 2.
Abnormal proteins associated with impaired mitochondrial function in the three most frequent neurodegenerative diseases (adapted form Paß et al. [72]). ↑ = increases; ↓ = decreases.
7. Translating Theoretical Knowledge into Therapy
7.1. Targeting Oxidative Stress and Mitochondrial Dysfunction in Alzheimer’s Disease
Clinical trials on disease-modifying therapies have mostly failed, AD treatment proving one of the most challenging areas in modern medicine despite the increasing incidence and prevalence of the disease [387]. Currently, there are six FDA-approved drugs used for treatment of AD, four of which (tacrine, donepezil, rivastigmine, and galantamine) are acetyl cholinesterase inhibitors, memantine is a NMDA antagonist, while the very controversially recently approved aducanumab is an amyloid beta-directed monoclonal antibody [388]. Except for the last molecule, none of them are disease-modifying.
In a long list of trials marked by inconclusive results or failures, several antioxidant molecules have been repeatedly evaluated.
Curcumin is able to scavenge free radicals (ROS or reactive nitrogen species) [389], inhibit the activity of enzymes involved in free radical generation such as cyclooxygenases and xanthine oxidase [390], to modulate the activity of endogenous antioxidants such as glutathione peroxidase, catalase, and superoxide dismutases [391], to diminish lipid peroxidation, and to reduce the expression of NF-κB, ERK, inducible nitric oxide synthase, and inflammatory cytokines IL-1β and IL-6 [392]. The efficacy of 24-week curcumin supplementation on AD progression has been evaluated with inconclusive results in a phase 2 randomized, double-blind, placebo-controlled trial (NCT 00099710) [393], followed by a second pilot study of curcumin and Gingko extracts administered over 6 months (NCT00164749), the results of which were not released to date [394]. Another active trial, NCT01811381, aims at evaluating the efficacy of curcumin in association with yoga in patients with mild cognitive impairment [394].
Resveratrol has been shown to enhance the PI3K/Akt pathway and the nuclear translocation of Nrf2 [395], to suppress NF-κB and MAPK pathway activation, and to attenuate the microglial release of TNF-α and pro-inflammatory IL-1β [396]. A phase 2, double-blind, placebo-controlled study (NCT01504854) of the effect of resveratrol supplements in patients with probable AD has not yet published the results [394]. NCT00678431 is another completed study evaluating whether dietary supplementation with Resveratrol, glucose, and malate is able to slow the progression of AD. According to the published results, the differences between the active and placebo arms were not statistically significant [397]. A currently recruiting phase 1 study (NCT02502253) will assess BBB penetration and bioavailability of a grape seed extract as well as its efficacy in mild cognitive impairment, prediabetes, and type 2 diabetes mellitus [394].
As an antioxidant, quercetin scavenges free radicals [398], upregulates antioxidant enzymes such as glutathione transferase, glutathione peroxidase, SOD, catalase and thioredoxin [398], and induces the Nrf2-ARE pathway [399]. The SToMP-AD trial (Senolytic Therapy to Modulate Progression of Alzheimer’s Disease, NCT04063124) is an ongoing phase I/II trial assessing bioavailability and safety of dasatinib + quercetin in old adults with early-stage AD, while ALSENLITE, NCT04785300 is currently recruiting by invitation participants in a phase I/II trial to assess safety and tolerance of the combination [394].
Sulforaphane was able in animal models of AD to modulate the Nrf2/ARE pathway, to inhibit NFκB and upregulate neurotrophin expression [400]. An ongoing randomized, double-blind, placebo-controlled trial (NCT04213391) aims at assessing safety and efficacy of sulforaphane in mild to moderate Alzheimer’s disease patients [394].
Modest cognitive beneficial effects in AD patients were obtained with another dietary antioxidant supplement, soy isoflavone, in a placebo-controlled, double-blind pilot study (NCT00205179) which randomized 73 patients [401]. 8-hydroxydaidzein, a compound from fermented soy, exhibits antioxidant properties by quenching ROS, inhibiting microglial TNF-α and IL-6 release, and by upregulating the Nrf2 antioxidant and Akt/NF-κB anti-inflammatory pathways [402].
Chlorogenic acid, the main compound of coffee, was able in vitro to reduce intracellular ROS accumulation, stop the activation of α-secretases, BACE-1, or MAPK, and attenuate GSH depletion [403]. In animal models of AD, polyphenols from coffee reduced hippocampal amyloid plaque burden, thereby attenuating memory impairments and cognitive dysfunction [404]. CAFCA (NCT04570085) is a multicenter, randomized, double-blind, placebo-controlled trial planned by the University of Lille, aiming at evaluating the effect of a 30-week caffeine treatment on cognitive function in early and moderate stages of Alzheimer’s disease [394].
Lipoic acid is a molecule with anti-inflammatory functions, able to reduce NF-κB activity in vitro in cells stimulated with TNF-α [405], and able to recycle vitamins C, E, and glutathione [406]. A small phase I/II trial randomized 39 AD patients to receive lipoic acid, fish oil, or placebo and evaluated the rate of cognitive impairment over 12 months, showing promising results [407] and has been followed by another phase I/II study, NCT01058941, with 67 participants followed over 18 months, the results of which have not yet been published.
Other trials with antioxidants have also been performed or are under way. For example, NCT00117403 was a phase 1 trial with the active arm taking a combination of vitamins E, C, lipoic acid, and coenzyme Q10 for 4 months. Although CSF analysis indicated a reduction of markers of oxidative stress, the investigators cautioned against a more rapid cognitive decline [408]. A prospective cohort study with 4246 participants (PREADVISE, NCT00040378) followed for 7–12 years concluded that neither vitamin E nor selenium could influence the rate of cognitive decline [409]. Moreover, a phase 3 trial comparing the added effect of tocopherol (TEAM-AD, NCT00235716) revealed that vitamin E showed a modest benefit over placebo in slowing cognitive decline but did not add to the effect of memantine [410]. A currently recruiting phase 1 study, NCT04430517, will assess the effect of oral 1000 mg of nicotinamide riboside intake for 12 weeks on redox status, GSH levels, and mitochondrial function as well as on cognition in participants with mild cognitive impairment od AD [394], while a phase 1 trial of supplementation of glycine, alanine, and N-acetylcysteine (Glutathione in Alzheimer’s disease, NCT04430517) for 24 weeks is registered but not yet recruiting [394].
Latrepirdine has been shown in preclinical studies to prevent lipid peroxidation and inhibit opening of the MPTP as well as voltage-gated calcium ion (Ca2+) channels in neurons, protecting against Aβ-induced neurotoxicity [411], while having very low acetylcholinesterase-inhibitory action, as opposed to other antihistamine drugs. In an 8-week open-label pilot study on 14 AD patients, latrepirdine showed clinical benefits [411], which prompted a phase 2 randomized, double-blind, placebo-controlled trial of 60 mg latrepirdine orally for 26 weeks in 183 AD patients (NCT00377715), which confirmed the positive effect on cognition, behavior, and global function [412]. However, the CONNECTION trial (NCT00675623), a phase 3 clinical trial of 60 mg latrepirdine for 6 months in 598 patients could not demonstrate any significant effect of the drug compared to placebo [413], and neither could CONCERT (NCT00829374), a phase 3 clinical trial comparing two doses of latrepirdine to placebo in 1003 patients. A meta-analysis of the several studies with latrepirdine in mild-to-moderate AD patients concluded that despite some modest beneficial effects on behavior, cognition and functional status are not influenced [414].
7.2. Targeting Oxidative Stress and Mitochondrial Dysfunction in Parkinson’s Disease
Currently prescribed treatments for PD are levodopa with 1-amino acid decarboxylase inhibitors, dopamine agonists (pramipexole, ropinirole, rotigotine), MAO-B inhibitors (selegiline, rasagiline), catechol-O-methyltransferase inhibitors (entacapone), and anticholinergic drugs [415], while refractory cases may undergo surgical deep brain stimulation procedures. Although helpful in relieving symptoms, none of these approaches interfere with the neurodegenerative process, and some concerns regarding the accelerated neurodegeneration caused by levodopa metabolism and subsequent increase in oxidative stress have been raised [416]. Recent developments in the treatment of PD focus on [417] immunotherapies to restrict the propagation of α-synuclein, neurotrophic factors, such as GDNF (glial cell-derived neurotrophic factor), regenerative therapies using cell-based and genetic approaches to replace the function of the lost dopaminergic neurons, re-establishing the balance between neurotransmitters by targeting non-dopaminergic neurotransmission, interfering with the neuroinflammatory response, or improving the stereotactical surgical treatments. As for supporting/improving mitochondrial function and targeting oxidative stress, the clinical trials performed so far and their conclusions are described below.
Creatine acts as an antioxidant and improves mitochondrial function [418]. Although a pilot study reported in 2006 could not detect any improvement in the UPDRS scores of PD patients after 2–4 g creatine/day for 2 years [419], a subsequent phase 2 multi-center, double-blind, pilot study of minocycline and creatine was conducted in 195 patients with early untreated PD to test efficacy in slowing disease progression (National Institute for Neurological Disorders and Stroke Parkinson’s Disease Neuroprotection Trial, NCT00063193) and showed promising results [420], followed by a larger phase 3 double-blind, parallel-group, placebo controlled study (NET-PD LS-1, NCT00449865), which enrolled 1741 participants who were administered 10 creatine/day for 5 years, which failed to show improvements in clinical outcome [421].
Vitamin E has also been evaluated in several studies, with mixed results. After a population-based study showed a negative association between vitamin E intake and incident PD [422], similar subsequent studies failed to confirm the decreased PD risk associated with dietary antioxidants [423]. A pilot open label study evaluated the efficacy of tocopherol in slowing down the disease progression and claimed that vitamin E can postpone the need for levodopa therapy by 2.5 years [424], a claim contradicted by the findings of a subsequent multicenter, randomized, placebo-controlled study (DATATOP) carried out on 800 patients to evaluate the effect of selegiline and/or tocopherol versus placebo on the onset of disability prompting the need for levodopa therapy [425] and which found beneficial effects for selegiline but not for tocopherol. At present, a phase 2 pilot, randomized, double blind, placebo-controlled trial (NCT04491383) is recruiting 100 participants in Singapore to assess efficacy of tocotrienols delaying motor disability in PD [394].
Since complex I activity was found reduced in PD, coenzyme Q10, an antioxidant and electron acceptor for complexes I and II seemed a reasonable approach. A randomized, double-blind, placebo-controlled trial which enrolled 80 early-stage PD patients showed that 300–1200 mg coenzyme Q10/day could significantly reduce disability in a dose-dependent manner [426]. However, a large phase 3 multi-center, randomized, double-blind, placebo-controlled clinical trial which included 600 participants (QE3, NCT00740714) failed to show clinical benefits for 1200 or 2400 mg of coenzyme Q10 daily [427]. To overcome the reduced BBB permeability of coenzyme Q10, it was delivered in a nanodispersed solution in doses of 300 mg (equivalent to 1200 mg coenzyme Q10)/day to 132 participants in a phase 3 clinical trial (NCT00180037), which also showed null results [428]. A mitochondria-targeted synthetic coenzyme Q10 analog, MitoQ, has positive charges and lipophilic properties, enabling it to easily cross the BBB and accumulate within mitochondria. However, it also failed to alter the disease course in a phase 2 clinical trial which enrolled 128 participants (NCT00329056) [429].
Glutathione is an endogenous antioxidant molecule which has been tested in a number of clinical trials. While the intravenous administration of 1200 mg/day of reduced glutathione was shown to significantly improve disability [430], a subsequent trial with 700 mg glutathione/day given also IV failed to confirm these results [431]. After a phase 1 safety trial (NCT01398748), intranasally administered glutathione was tested in a phase 2b study on 45 participants over 12 weeks for efficacy on disease progression ((in) GSH, NCT02424708) but showed null results [432]. Studies with other dietary antioxidants are in preclinical phases, but, considering the poor BBB penetration of these compounds [433], it is unlikely that they will successfully translate in clinical settings.
Melatonin, being amphiphilic, can cross the BBB and exhibit antioxidant activity in the central nervous system. After promising results in animal models, the effect of 3 mg melatonin/day for 4 weeks on motor performances and quality of sleep was assessed in a small study involving 18 PD patients. Sleep disturbances are common complaints in PD and may herald a more aggressive course and a more rapid progression toward cognitive decline [434]. The trial showed that melatonin improved the subjective quality of sleep but had no effect on motor performances [435].
Given the high levels of iron in nigral neurons and the involvement of iron in ROS production, the efficacy of iron chelators was assessed first in a pilot study on 22 participants (DeferipronPD, NCT01539837) and showed non-significant motor improvement [436] followed by a phase 2/3 trial enrolling 37 patients in the active arm and 40 in the placebo arm (FAIR PARK-I, NCT00943748) and which showed reduction of iron in the substantia nigra as well as motor improvement [437]. Currently, an extended clinical trial, enrolling 372 participants (FAIR PARK-II, NCT02655315), is active but not recruiting [394].
Other agents, such as PPARγ coactivator-1α (PGC-1α) agonists, Nrf2 enhancers, or natural antioxidant compounds are only in preclinical stages [415] except for pioglitazone, a PPARγ coactivator-1α agonist, which, administered in doses of 15–45 mg/day in 210 PD patients (NCT01280123) could not show significant motor benefits [438].
7.3. Targeting Oxidative Stress and Mitochondria in ALS
For years the only approved therapy in ALS was riluzole, a glutamate antagonist which interferes with excitotoxic neuronal death [439].
Based on the results of several studies evaluating the effect of edaravone (3-methyl-1-phenyl-2-pyrazoline-5-one, or MCI-186), the FDA approved the molecule in 2017 for use in ALS [440]. Edaravone acts as a free radical scavenger of hydroxyl and peroxyl radicals, hydrogen peroxide and peroxynitrite [441], and activates the Nrf2/HO-1 signaling pathway, protecting cells against apoptosis [440], thereby offering a modest clinical benefit.
Melatonin, an endogenous molecule involved in regulation of the sleep–wake cycle, exhibits also antioxidant properties [442]. In ALS patients it slowed the progression of the motor impairment [443]. However, no clinical trial with melatonin in ALS is currently ongoing [394].
Alpha-lipoic acid is a hydroxyl radical scavenger and induces the ERK/PI3K/Akt pathway, thereby regulating the expression of antioxidant genes. Its safety and efficacy in ALS are currently being explored and compared to riluzole in the Explore Neuroprotective Effect of Lipoic Acid in Amyotrophic Lateral Sclerosis (NCT04518540) trial, conducted by the Zhejiang University School of Medicine [394].
Dopaminergic drugs, such as pramipexole, reduce oxidative stress [444] and glutamate excitotoxicity [445]. The safety and efficacy of dexpramipexole was evaluated in several phase 1, 2, and 3 clinical studies. So far, results published from a phase 3 randomized, double-blind, placebo-controlled, multi-center study which enrolled 942 participants (EMPOWER, NCT 01281189) revealed a good safety profile but non-significant efficacy of the treatment arm compared to placebo [446] despite promising results of a phase 2 randomized, double blind safety and tolerability study (NCT00647296) [447]. The results of the extension phase, NCT01622088 are still awaited [394]. Another drug currently used in the treatment of PD, rasagiline, a monoamine oxidase B inhibitor, exhibits also antioxidative and anti-apoptotic activities, making it a potential therapeutic option in ALS. Its safety and efficacy were evaluated in two phase 2 trials, an open-label one enrolling 36 participants (NCT01232738), which showed no improved clinical course but suggested reduced apoptosis [448] and a second phase 2, double-blind, placebo-controlled trial (NCT017866030) evaluating efficacy, which showed null results after 12 months of treatment [449].
Dietary antioxidants and antioxidant food supplements have also been tested in several trials. A phase 2 trial with high doses of coenzyme Q10 in ALS (NCT00243932) concluded that there is insufficient evidence to justify a phase 3 trial [450], while NCT02588807, a phase 1 trial with food supplements for the treatment of patients with ALS is suspended [394]. However, two further studies, planned but not yet recruiting, will evaluate safety and efficacy of coenzyme Q10 with vitamin E, N-acetyl cysteine, and L-cysteine (MICABO-ALS, NCT04244630) and of liposomed polyphenols resveratrol and curcumin (NCT04654689) in patients with ALS.
8. Concluding Remarks
From the repeated failures of drugs targeting mitochondrial dysfunction and oxidative stress in neurodegenerative diseases, it appears that starting these interventions by the time of clinical diagnosis is probably too late. Due to the resilience of the brain to insults, the described pathogenetic cascades and loops are already full-blown and significant neuronal loss has already occurred when clinical symptoms enable diagnosis. Genetic testing in familial neurodegenerative diseases could allow starting these therapies in the preclinical stage. Since the present healthcare systems cannot afford extensive and invasive evaluations of the population at large, promoting a healthy lifestyle, with plenty of dietary antioxidant intake and avoidance of exogenous oxidants could postpone the onset of neurodegenerative diseases. Hopefully, ongoing research will provide more efficient, multimodal molecules to interfere with the pathogenesis of these diseases, which, due to aging of the population, are a global threat.
Funding
This research received no external funding.
Acknowledgments
The author wishes to thank José L. Quiles for editing this interesting topical collection, as well as the two anonymous reviewers for appreciating this review.
Conflicts of Interest
The author declares no conflict of interest.
References
- Albers, D.S.; Flint Beal, M. Mitochondrial Dysfunction and Oxidative Stress in Aging and Neurodegenerative Disease. In Advances in Dementia Research; Jellinger, K., Schmidt, R., Windisch, M., Eds.; Springer: Vienna, Austria, 2000. [Google Scholar]
- Zia, A.; Pourbagher-Shahri, A.M.; Farkhondeh, T.; Samarghandian, S. Molecular and cellular pathways contributing to brain aging. Behav. Brain Funct. 2021, 17, 6. [Google Scholar] [CrossRef]
- Huffman, K.J. The developing, aging neocortex: How genetics and epigenetics influence early developmental patterning and age-related change. Front. Genet. 2012, 3, 212. [Google Scholar] [CrossRef] [PubMed]
- Wanagat, J.; Allison, D.B.; Weindruch, R. Caloric intake and aging: Mechanisms in rodents and a study in nonhuman primates. Toxicol. Sci. 1999, 52, 35–40. [Google Scholar] [CrossRef]
- Błaszczyk, J.W. Energy metabolism decline in the aging brain–pathogenesis of neurodegenerative disorders. Metabolites 2020, 10, 450. [Google Scholar] [CrossRef]
- Camandola, S.; Mattson, M.P. Brain metabolism in health, aging, and neurodegeneration. EMBO J. 2017, 36, 1474–1492. [Google Scholar] [CrossRef]
- Kmiec, Z. Central regulation of food intake in ageing. J. Physiol. Pharmacol. 2006, 57, 7–16. [Google Scholar] [PubMed]
- Zhang, X.Y.; Yu, L.; Zhuang, Q.X.; Zhu, J.N.; Wang, J.J. Central functions of the orexinergic system. Neurosci. Bull. 2013, 29, 355–365. [Google Scholar] [CrossRef]
- Waterson, M.J.; Horvath, T.L. Neuronal regulation of energy homeostasis: Beyond the hypothalamus and feeding. Cell Metab. 2015, 22, 962–970. [Google Scholar] [CrossRef] [PubMed]
- Rolls, A. Hypothalamic control of sleep in aging. Neuromol. Med. 2012, 14, 139–153. [Google Scholar] [CrossRef]
- Cahine, L.M.; Amara, A.W.; Videnovic, A. A systematic review of the literature on disorder of sleep and wakefulness in Parkinson’s disease from 2005 to 2015. Sleep Med. Rev. 2016, 35, 33–50. [Google Scholar] [CrossRef] [PubMed]
- Brzecka, A.; Leszek, J.; Ashraf, G.M.; Ejma, M.; Ávila-Rodriguez, M.F.; Yarla, N.S.; Tarasov, V.V.; Chubarev, V.N.; Samsonova, A.N.; Barreto, G.E.; et al. Sleep disorders associated with Alzheimer’s disease: A perspective. Front. Neurosci. 2018, 12, 330. [Google Scholar] [CrossRef]
- Aalling, N.N.; Nedergaard, M.; DiNuzzo, M. Cerebral metabolic changes during sleep. Curr. Neurol. Neurosci. Rep. 2018, 18, 57. [Google Scholar] [CrossRef] [PubMed]
- Dombrowski, G.J.; Swiatek, K.R.; Chao, K.L. Lactate, 3-hydroxybutyrate, and glucose as substrates for the early postnatal rat brain. Neurochem. Res. 1989, 14, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Leithner, C.; Royl, G. The oxygen paradox of neurovascular coupling. J. Cerebr. Blood Flow Metab. 2014, 34, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Liang, J.; Zhou, B. Glucose metabolic dysfunction in neurodegenerative diseases–new mechanistic insights and the potential of hypoxia as a prospective therapy targeting metabolic reprogramming. Int. J. Mol. Sci. 2021, 22, 5887. [Google Scholar] [CrossRef]
- Romano, A.H.; Conway, T. Evolution of carbohydrate metabolic pathways. Res. Microbiol. 1996, 147, 448–455. [Google Scholar] [CrossRef]
- Attwell, D.; Laughlin, S.B. An energy budget for signaling in the grey matter of the brain. Br. J. Pharmacol. 2001, 21, 1133–1145. [Google Scholar] [CrossRef]
- Harris, J.J.; Jolivet, R.; Attwell, D. Synaptic energy use and supply. Neuron 2012, 75, 762–777. [Google Scholar] [CrossRef]
- Rangaraju, V.; Lauterbach, M.; Schuman, E.M. Spatially stable mitochondrial compartments fuel local translation during plasticity. Cell 2019, 176, 73–84. [Google Scholar] [CrossRef]
- Harman, D. Aging: A theory based on free radical and radiation chemistry. J. Gerontol. 1956, 11, 298–300. [Google Scholar] [CrossRef]
- Cini, M.; Moretti, A. Studies on lipid peroxidation and protein oxidation in the aging brain. Neurobiol. Aging 1995, 16, 53–57. [Google Scholar] [CrossRef]
- Smith, C.D.; Carney, J.M.; Starke-Reed, P.E.; Oliver, C.N.; Stadtman, E.R.; Floyd, R.A.; Markesbery, W.R. Excess brain protein oxidation and enzyme dysfunction in nortmal aging and Alzheimer disease. Proc. Natl. Acad. Sci. USA 1991, 88, 10540–10543. [Google Scholar] [CrossRef] [PubMed]
- Dringen, R.; Hirrlinger, J. Glutathione pathways in the brain. Biol. Chem. 2003, 384, 505–516. [Google Scholar] [CrossRef]
- Stahon, K.E.; Bastian, C.; Griffith, S.; KJidd, G.J.; Brunet, S.; Baltan, S. Age-related changes in axonal and mitochondrial ultrastructure and function in white matter. J. Neurosci. 2016, 36, 9990–10001. [Google Scholar] [CrossRef]
- Santos, R.X.; Correia, S.C.; Zhu, X.; Smith, M.A.; Moreira, P.I.; Castellani, R.J.; Nunomura, A.; Perry, G. Mitochondrial DNA oxidative damage and repair in aging and Alzheimer’s disease. Antioxid. Redox Signal. 2013, 18, 2444–2457. [Google Scholar] [CrossRef]
- Pollard, A.K.; Craig, E.L.; Chakrabarti, L. Mitochondrial complex I activity measured by spectrophotometry is reduced across all brain regions in ageing and more specifically in neurodegeneration. PLoS ONE 2016, 11, e0157405. [Google Scholar] [CrossRef]
- Leslie, S.W.; Chandler, L.J.; Barr, E.M.; Farrar, R.P. Reduced calcium uptake by rat brain mitochondria and synaptosomes in response to aging. Brain Res. 1985, 329, 177–183. [Google Scholar] [CrossRef]
- Fang, E.F.; Lautrup, S.; Hou, Y.; Demarest, T.G.; Croteau, D.L.; Mattson, M.P.; Bohr, V.A. NAD+ in aging: Molecular mechanisms and translational implications. Trends Mol. Med. 2017, 23, 899–916. [Google Scholar] [CrossRef] [PubMed]
- Feldman, J.L.; Dittenhafer-Reed, K.E.; Kudo, N.; Thelen, J.N.; Ito, A.; Yoshida, M.; Denu, J.M. Kinetic and structural basis for acyl-group selectivity and NAD+ dependence in sirtuin-catalyzed deacylation. Biochemistry 2015, 54, 3037–3050. [Google Scholar] [CrossRef] [PubMed]
- Imai, S.-I.; Guarente, L. It takes two to tango: NAD+ and sirtuins in aging/longevity control. NPJ Aging Mech. Dis. 2016, 2, 16017. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S. Emerging therapeutic roles for NAD+ metabolism in mitochondrial and age-related disorders. Clin. Transl. Med. 2016, 16, 1–11. [Google Scholar] [CrossRef]
- Satoh, A.; Brace, C.S.; Rensing, N.; Cliften, P.; Wozniak, D.F.; Herzog, E.D.; Yamada, K.A.; Imai, S.-I. Sirt 1 extends life span and delays aging in mice through the regulation of Nk2 homebox 1 in the DMH and LH. Cell Metab. 2013, 18, 416–430. [Google Scholar] [CrossRef]
- Satoh, A.; Imai, S.-I. Systemic regulation of mammalian ageing and longevity by brain sirtuins. Nat. Commun. 2014, 5, 4211. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, E.H.; Epel, E.S.; Lin, J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science 2015, 350, 1193–1198. [Google Scholar] [CrossRef]
- Fischer, H.G.; Morawski, M.; Bruckner, M.K.; Mittag, A.; Tarnok, A.; Arendt, T. Changes in neuronal DNA content variation in the human brain during aging. Aging Cell 2012, 11, 628–633. [Google Scholar] [CrossRef]
- Mosch, B.; Morawski, M.; Mittag, A.; Lenz, D.; Tarnok, A.; Arendt, T. Aneuploidy and DNA replication in the normal human brain and Alzheimer’s disease. J. Neurosci. 2007, 27, 6859–6967. [Google Scholar] [CrossRef]
- Ceafalan, L.C.; Popescu, B.O. Juxtacerebral tissue regeneration potential: Telocytes contribution. Adv. Exp. Med. Biol. 2016, 913, 397–402. [Google Scholar]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres shorten during ageing of human fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef]
- Salminen, A.; Kauppinen, A.; Kaarniranta, A. Emerging role of NF-κB signalling in the induction of senescence-associated secretory phenotype (SASP). Cell Signal 2012, 24, 835–845. [Google Scholar] [CrossRef]
- Baldwin, A.S., Jr. The NF-κB and IκB proteins: New discoveries and insights. Annu. Rev. Immunol. 1996, 14, 649–681. [Google Scholar] [CrossRef]
- Patten, D.A.; Germain, M.; Kelly, M.A.; Slack, R.S. Reactive oxygen species: Stuck in the middle of neurodegeneration. J. Alzheime. Dis. 2010, 20, S357–S367. [Google Scholar] [CrossRef] [PubMed]
- Von Bernhardi, R.; Eugenin-von Bernhardi, L.; Eugenin, J. Microglial cell dysregulation in brain aging and neurodegeneration. Front. Aging Neurosci. 2015, 7, 124. [Google Scholar] [CrossRef]
- Mosher, K.I.; Wyss-Coray, T. Microglial dysfunction in brain aging and Alzheimer’s disease. Biochem. Pharmacol. 2014, 88, 594–604. [Google Scholar] [CrossRef]
- Snow, W.M.; Stoesz, B.M.; Kelly, D.M.; Albensi, B.C. Roles for NF-κB and gene targets of NF-κB in synaptic plasticity, memory, and navigation. Mol. Neurobiol. 2014, 49, 757–770. [Google Scholar] [CrossRef]
- Okun, E.; Griffioen, K.J.; Mattson, M.P. Toll-like receptor signaling in neural plasticity and disease. Trends Neurosci. 2011, 34, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Sastie, M.; Klockgether, T.; Heneka, M.T. Contribution of inflammatory processes to Alzheimer’s disease. Int. J. Dev. Neurosci. 2006, 24, 167–176. [Google Scholar] [CrossRef]
- Sazanova, M.A.; Sinyov, V.V.; Ryzhkova, A.I.; Sazanova, M.D.; Kirichenko, T.V.; Khotina, V.A.; Khasanova, Z.B.; Doroschuk, N.A.; Karagodin, V.P.; Orekhov, A.N.; et al. Some molecular and cellular stress mechanisms associated with neurodegenerative diseases and atherosclerosis. Int. J. Mol. Sci. 2021, 22, 699. [Google Scholar] [CrossRef]
- Hirabayashi, Y.; Kwon, S.-K.; Paek, H.; Pernice, W.M.; Paul, M.A.; Lee, J.; Erfani, P.; Raczkowski, A.; Petrey, D.S.; Pon, L.A.; et al. ER-mitochondria tethering by PDZD8 regulates Ca2+ dynamics in mammalian neurons. Science 2017, 358, 623–630. [Google Scholar] [CrossRef]
- Kwon, S.K.; Sando, R., 3rd; Lewis, T.L.; Hirabayashi, Y.; Maximov, A.; Polleux, F. LKB1 regulates mitochondria-dependent presynaptic calcium clearance and neurotransmitter release properties at excitatory synapses along cortical axons. PLoS Biol. 2016, 14, e1002516. [Google Scholar] [CrossRef]
- Lin, M.T.; Beal, M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 2006, 443, 787–795. [Google Scholar] [CrossRef]
- Rugarli, E.I.; Langer, T. Mitochondrial quality control: A matter of life and death for neurons. EMBO J. 2012, 31, 1336–1349. [Google Scholar] [CrossRef]
- Youle, R.J.; van der Bliek, A.M. Mitochondrial fission, fusion, and stress. Science 2012, 337, 1062–1065. [Google Scholar] [CrossRef]
- Benard, G.; Karbowski, M. Mitochondrial fusion and division: Regulation and role in cell viability. Semin. Cell Dev. Biol. 2009, 20, 365–374. [Google Scholar] [CrossRef]
- Kausar, S.; Wang, F.; Cui, H. The role of mitochondria in reactive oxygen species generation and its implications for neurodegenerative diseases. Cells 2018, 7, 274. [Google Scholar] [CrossRef] [PubMed]
- Osellame, L.D.; Blacker, T.S.; Duchen, M.R. Cellular and molecular mechanisms of mitochondrial function. Best Pract. Res. Clin. Endocrinol. Metab. 2012, 26, 711–723. [Google Scholar] [CrossRef]
- Zhao, R.-Z.; Jiang, S.; Zhang, L.; Yu, Z.-B. Mitochondrial electron transport chain, ROS generation, and uncoupling. Int. J. Mol. Med. 2019, 44, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Cadenas, E.; Davies, K.J.A. Mitochondrial free radical generation, oxidative stress, and aging. Free Rad. Biol. Med. 2000, 29, 222–230. [Google Scholar] [CrossRef]
- Brand, M.D. The sites and topology of mitochondrial superoxide production. Exp. Gerontol. 2010, 45, 466–472. [Google Scholar] [CrossRef]
- Phaniendra, A.; Jestadi, D.B.; Periyasamy, L. Free radicals: Properties, sources, targets, and their implication in various diseases. Indian J. Clin. Biochem. 2015, 30, 11–26. [Google Scholar] [CrossRef]
- Singh, A.; Kukreti, R.; Saso, L.; Kukreti, S. Oxidative stress: A key modulator in neurodegenerative diseases. Molecules 2019, 24, 1583. [Google Scholar] [CrossRef]
- Esteras, N.; Abramov, A.Y. Mitochondrial calcium deregulation in the mechanism of beta-amyloid and tau pathology. Cells 2020, 9, 2135. [Google Scholar] [CrossRef]
- Horigane, S.-I.; Ozawa, Y.; Yamada, H.; Takemoto-Kimura, S. Calcium signalling: A key regulator of neuronal migration. J. Biochem. 2019, 165, 401–409. [Google Scholar] [CrossRef]
- Verkhratsky, A. Endoplasmic reticulum calcium signaling in nerve cells. Biol. Res. 2004, 37, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Karagas, N.E.; Venkatachalam, K. Roles for the endoplasmic reticulum in regulation of neuronal calcium homeostasis. Cells 2019, 8, 1232. [Google Scholar] [CrossRef] [PubMed]
- Schwaller, B. Cytosolic Ca2+ buffers. Cold Spring Harb. Perspect. Biol. 2010, 2, a004051. [Google Scholar] [CrossRef]
- Brini, M.; Carafoli, E. The plasma membrane Ca2+ ATPase and the plasma membrane sodium calcium exchanger cooperate in the regulation of cell calcium. Cold Spring Harb. Perspect. Biol. 2010, 3, a004168. [Google Scholar] [CrossRef] [PubMed]
- Primeau, J.O.; Armanious, G.P.; Fisher, M.E.; Young, H.S. The SarcoEndoplasmic Reticulum Calcium ATPase. Subcell. Biochem. 2018, 87, 229–258. [Google Scholar]
- McCormack, J.G.; Denton, R.M. The role of intramitochondrial Ca2+ in the regulation of oxidative phosphorylation in mammalian tissues. Biochem. Soc. Trans. 1993, 21, 793–799. [Google Scholar] [CrossRef]
- Briston, T.; Selwood, D.; Szabadkai, G.; Duchen, M.R. Mitochondrial permeability transition: A molecular lesion with multiple drug targets. Trends Pharm. Sci. 2019, 40, 50–70. [Google Scholar] [CrossRef]
- Palty, R.; Silverman, W.F.; Hershfinkel, M.; Caporale, T.; Sensi, S.L.; Parnis, J.; Nolte, C.; Fishman, D.; Shoshan-Barmatz, V.; Herrmann, S.; et al. NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc. Natl. Acad. Sci. USA 2009, 107, 436–441. [Google Scholar] [CrossRef]
- Paß, T.; Wiesner, R.J.; Pla-Martin, D. Selective neuron vulnerability in common and rare diseases–mitochondria in the focus. Front. Mol. Biosci. 2021, 8, 676187. [Google Scholar] [CrossRef] [PubMed]
- Csordás, G.; Renken, C.; Várnai, P.; Walter, L.; Weaver, D.; Buttle, K.F.; Balla, T.; Manella, C.A.; Hajnóczky, G. Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell. Biol. 2006, 174, 915–921. [Google Scholar] [CrossRef] [PubMed]
- Eysert, F.; Kinoshita, P.F.; Mary, A.; Vaillant-Beuchot, L.; Checler, F.; Chami, M. Molecular dysfunctions of mitochondria-associated membranes (MAMs) in Alzheimer’s disease. Int. J. Mol. Sci. 2020, 21, 9521. [Google Scholar] [CrossRef]
- Rizzuto, P.; Pinton, P.; Carrington, W.; Fay, F.S.; Fogarty, K.E.; Lifshitz, L.M.; Tuft, R.A.; Pozzan, T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 1998, 280, 1763–1766. [Google Scholar] [CrossRef] [PubMed]
- Mikoshiba, K. IP3 receptor/Ca2+ channel: From discovery to new signaling concepts. J. Neurochem. 2007, 102, 1426–1446. [Google Scholar] [CrossRef]
- Szabadkai, G.; Bianchi, K.; Várnai, P.; De Stefani, D.; Wieckowski, M.R.; Cavagna, D.; Nagy, A.I.; Balla, T.; Rizzuto, R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 2006, 175, 901–911. [Google Scholar] [CrossRef]
- Simmen, T.; Aslan, J.E.; Blagoveshchenskaya, A.D.; Thomas, L.; Wan, L.; Xiang, Y.; Feliciangeli, S.F.; Hing, C.-H.; Crump, C.M.; Thomas, G. PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 2005, 24, 717–729. [Google Scholar] [CrossRef]
- Youker, R.T.; Shinde, U.; Day, R.; Thomas, G. At the crossroads of homeostasis and disease: Roles of the PACS proteins in membrane traficking and apoptosis. Biochem. J. 2009, 421, 1–15. [Google Scholar] [CrossRef]
- Stone, S.J.; Vance, J.E. Phosphatidylserine synthase-1 and -2 are localized to mitochondria-associated membranes. J. Biol. Chem. 2000, 275, 34534–34540. [Google Scholar] [CrossRef]
- Hedskog, L.; Pinho, C.M.; Filadi, R.; Rönnbäck, A.; Hertwig, L.; Wiehager, B.; Larssen, P.; Gellhaar, S.; Sandebring, A.; Westerlund, M.; et al. Modulation of the endoplasmic reticulum-mitochondria interface in Alzheimer’s disease and related models. Proc. Natl. Acad. Sci. USA 2013, 110, 7916–7921. [Google Scholar] [CrossRef]
- Iwasawa, R.; Mahul-Mellier, A.-L.; Datler, C.; Pazarentzos, E.; Grimm, S. Fis1 and Bap31 bridge the mitochondria-ER interface to establish a platform for apoptosis induction. EMBO J. 2011, 30, 556–568. [Google Scholar] [CrossRef]
- De Vos, K.J.; Mórotz, G.M.; Stoica, R.; Tudor, E.L.; Lau, K.-F.; Ackerley, S.; Warley, A.; Shaw, C.E.; Miller, C.C.J. VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum. Mol. Genet. 2012, 21, 1299–1311. [Google Scholar] [CrossRef]
- Hayashi, T.; Su, T.P. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca2+ signaling and cell survival. Cell 2007, 131, 569–610. [Google Scholar] [CrossRef] [PubMed]
- Prasad, M.; Pawlak, K.J.; Burak, W.E.; Perry, E.E.; Marshall, B.; Whittal, R.M.; Bose, H.S. Mitochondrial metabolic regulation by GRP78. Sci. Adv. 2017, 3, e1602038. [Google Scholar] [CrossRef]
- Hetz, C.; Papa, F.R. The unfolded protein response and cell fate control. Mol. Cell 2018, 69, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Tilokani, L.; Nagashima, S.; Paupe, V.; Minczuk, M. Mitochondrial dynamics: Overview of molecular mechanisms. Essays Biochem. 2018, 62, 341–360. [Google Scholar]
- Kraus, F.; Ryan, M.T. The constriction and scission machineries involved in mitochondrial fission. J. Cell Sci. 2017, 30, 2953–2960. [Google Scholar] [CrossRef]
- Friedman, J.R.; Lackner, L.L.; West, M.; DiBenedetto, J.R.; Nunnari, J.; Voeltz, G.K. ER tubules mark sites of mitochondrial division. Science 2011, 334, 358–362. [Google Scholar] [CrossRef]
- Lewis, S.C.; Uchiyama, L.F.; Nunnari, J. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 2016, 353, aaf5549. [Google Scholar] [CrossRef] [PubMed]
- Losón, O.C.; Song, Z.; Chen, H.; Chan, D.C. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell 2013, 24, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, C.; Grabiger, S.; Schwefel, D.; Faelber, K.; Rosenbaum, E.; Mears, J.; Rocks, O.; Daumke, O. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J. 2013, 32, 1280–1292. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Westrate, L.M.; Wu, H.; Page, C.; Voeltz, G.K. Multiple dynamin family members collaborate to drive mitochondrial division. Nature 2016, 540, 139–143. [Google Scholar] [CrossRef]
- Cho, B.; Cho, H.M.; Jo, Y.; Kim, H.D.; Song, M.; Moon, C.; Kim, H.; Kim, K.; Sesaki, H.; Rhyu, I.J.; et al. Constriction of the mitochondrial inner compartment is a priming event for mitochondrial division. Nat. Commun. 2017, 8, 15754. [Google Scholar] [CrossRef] [PubMed]
- Eura, Y.; Ishihara, N.; Yokota, S.; Mihara, K. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J. Biochem. 2003, 134, 333–344. [Google Scholar] [CrossRef]
- Ishihara, N.; Eura, Y.; Mihara, K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J. Cell Sci. 2004, 117, 6535–6546. [Google Scholar] [CrossRef] [PubMed]
- Griparic, L.; Kanazawa, T.; Van der Bliek, A.M. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J. Cell Biol. 2007, 178, 757–764. [Google Scholar] [CrossRef]
- Ban, T.; Ishihara, T.; Kohno, H.; Saita, S.; Ichimura, A.; Maenaka, K.; Oka, T.; Mihara, K.; Ishihara, N. Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat. Cell Biol. 2017, 19, 856–863. [Google Scholar] [CrossRef]
- MacVicar, T.; Langer, T. OPA1 processing in cell death and disease-the long and short of it. J. Cell Sci. 2016, 129, 2297–2306. [Google Scholar] [CrossRef]
- Taguchi, N.; Ishihara, N.; Jofuku, A.; Oka, T.; Mihara, K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 2007, 282, 11521–11529. [Google Scholar] [CrossRef]
- Gomes, L.C.; Di Benedetto, G.; Scorrano, L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011, 13, 589–598. [Google Scholar] [CrossRef]
- Toyama, E.Q.; Herzig, S.; Courchet, J.; Lewis, T.L., Jr.; Loson, O.C.; Hellberg, K.; Young, N.P.; Chen, H.; Polleux, F.; Chan, D.C.; et al. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 2016, 351, 275–281. [Google Scholar] [CrossRef]
- Park, Y.Y.; Nguyen, O.T.; Kang, H.; Cho, H. MARCH5-mediated quality control on acetylated Mfn1 facilitates mitochondrial homeostasis and cell survival. Cell Death Dis. 2014, 5, e1172. [Google Scholar] [CrossRef]
- Pyakurel, A.; Savoia, C.; Hess, D.; Scorrano, L. Extracellular regulated kinase phosphorylates mitofusin 1 to control mitochondrial morphology and apoptosis. Mol. Cell 2015, 58, 244–254. [Google Scholar] [CrossRef]
- Liesa, M.; Shirihai, O.S. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013, 17, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, R.; Gilkerson, R.; Aggeler, R.; Yamagata, K.; Remington, S.J.; Capaldi, R.A. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004, 64, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Giordano, S.; Zhang, J. Autophagy, mitochondria and oxidative stress: Cross-talk and redox signalling. Biochem. J. 2012, 441, 523–540. [Google Scholar] [CrossRef]
- Eshraghi, M.; Adlimoghaddam, A.; Mahmoodzadeh, A.; Sharifzad, F.; Yasavoli-Sharahi, H.; Lorzadeh, S.; Albensi, B.C.; Ghavami, S. Alzheimer’s disease pathogenesis: Role of autophagy and mitophagy focusing in microglia. Int. J. Mol. Sci. 2021, 22, 3330. [Google Scholar] [CrossRef]
- Mijaljica, D.; Prescott, M.; Devenish, R.J. Microautophagy in mammalian cells: Revisiting a 40-year-old conundrum. Autophagy 2011, 7, 673–682. [Google Scholar] [CrossRef]
- Bejarano, E.; Cuervo, A.M. Chaperone-mediated autophagy. Proc. Am. Thorac. Soc. 2010, 7, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, B.; Moreau, K.; Jahreiss, L.; Puri, C.; Rubinsztein, D.C. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 2010, 12, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Zhang, M.; Kenny, S.J.; Liu, D.; Maeda, M.; Saito, K.; Mathur, A.; Xu, K.; Schekman, R. Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep. 2017, 18, 1586–1603. [Google Scholar] [CrossRef]
- Swerdlow, N.S.; Wilkins, H.M. Mitophagy and the brain. Int. J. Mol. Sci. 2020, 21, 9661. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.H.; Young, L.N. Mechanisms of autophagy initiation. Annu. Rev. Biochem. 2017, 86, 225–244. [Google Scholar] [CrossRef] [PubMed]
- Itakura, E.; Mizushima, N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 2010, 6, 764–776. [Google Scholar] [CrossRef]
- Otomo, C.; Metlagel, Z.; Takaesu, G.; Otomo, T. Structure of human Atg12-Atg5 conjugate required for LC3 lipidation in autophagy. Nat. Struct. Mol. Biol. 2013, 20, 59. [Google Scholar] [CrossRef] [PubMed]
- Romanov, J.; Walczak, M.; Ibiricu, I.; Schuchner, S.; Ogris, E.; Kraft, C.; Martens, S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012, 31, 4304–4317. [Google Scholar] [CrossRef]
- Le Grand, J.N.; Chakrama, F.Z.; Seguin-Py, S.; Fraichard, A.; Delage-Mourroux, R.; Jouvenot, M.; Boyer-Guittaut, M. GABARAPL1 (GEC1): Original or copycat? Autophagy 2011, 7, 1098–1107. [Google Scholar] [CrossRef] [PubMed]
- Kundu, M. ULK1, mammalian target of rapamycin, and mitochondria: Linking nutrient availability and autophagy. Antioxid. Redox Signal. 2011, 14, 1953–1958. [Google Scholar] [CrossRef] [PubMed]
- Saftig, P.; Beertsen, W.; Eskelinen, E.L. LAMP-2: A control step for phagosome and autophagosome maturation. Autophagy 2008, 4, 510–512. [Google Scholar] [CrossRef]
- Gutierrez, M.G.; Munafo, D.B.; Beron, W.; Colombo, M.I. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell Sci. 2004, 117, 2687–2697. [Google Scholar] [CrossRef]
- Qiao, L.; Hamamichi, S.; Caldwell, K.A.; Caldwell, G.A.; Yacoubian, T.A.; Wilson, S.; Xie, Z.L.; Speake, L.D.; Parks, R.; Crabtree, D.; et al. Lysosomal enzyme cathepsin D protects against α-synuclein aggregation and toxicity. Mol. Brain 2008, 1, 17. [Google Scholar] [CrossRef]
- Vives-Bauza, C.; Zhou, C.; Huang, Y.; Cui, M.; De Vries, R.L.; Kim, J.; May, J.; Tocilescu, M.A.; Liu, W.; Ko, H.S.; et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA 2012, 107, 378–383. [Google Scholar] [CrossRef]
- Jin, M.S.; Youle, R.J. PINK1- and Parkin-mediated mitophagy at a glance. J. Cell Sci. 2012, 125, 795–799. [Google Scholar] [CrossRef]
- Gegg, M.E.; Schapira, A.H. PINK1-parkin-dependent mitophagy involves ubiquitination of mitofusins 1 and 2: Implications for Parkinson’s disease pathogenesis. Autophagy 2011, 7, 243–245. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Fung, G.; Deng, H.; Zhang, J.; Fiesel, F.C.; Springer, W.; Li, X.; Luo, H. NBR1 is dispensable for PARK2-mediated mitophagy regardless of the presence or absence of SQSTM1. Cell Death Dis. 2015, 6, e1943. [Google Scholar] [CrossRef]
- Wong, Y.C.; Holzbaur, E.L. Temporal dynamics of PARK2/parkin and OPTN/optineurin recruitment during the mitophagy of damaged mitochondria. Autophagy 2015, 11, 422–424. [Google Scholar] [CrossRef] [PubMed]
- Strappazzon, F.; Nazio, F.; Corrado, M.; Cianfanelli, V.; Romagnoli, A.; Fimia, G.M.; Campello, S.; Nardacci, R.; Piacentini, M.; Campanella, M.; et al. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015, 22, 517. [Google Scholar] [CrossRef] [PubMed]
- Sowter, H.M.; Ratcliffe, P.J.; Watson, P.; Greenberg, A.H.; Harris, A.L. HIF-1-dependent regulation of hypoxic induction of cell death factors BNIP3 and NIX in human tumors. Cancer Res. 2001, 61, 6669–6673. [Google Scholar] [PubMed]
- Rogov, V.V.; Suzuki, H.; Marinkovic, M.; Lang, V.; Kato, R.; Kawasaki, M.; Buljubasic, M.; Sprung, M.; Rogova, N.; Wakatsuki, S.; et al. Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins. Sci. Rep. 2017, 7, 1131. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Feng, D.; Chen, G.; Chen, M.; Zheng, Q.; Song, P.; Ma, Q.; Zhu, C.; Wang, R.; Qi, W.; et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 2012, 14, 177–185. [Google Scholar] [CrossRef]
- Davis, C.H.; Marsh-Armstrong, N. Discovery and implications of transcellular mitophagy. Autophagy 2014, 10, 2383–2384. [Google Scholar] [CrossRef]
- Davis, C.H.; Kim, K.Y.; Bushong, E.A.; Mills, E.A.; Boassa, D.; Shih, T.; Kinebuchi, M.; Phan, S.; Zhou, Y.; Bihlmeyer, N.A.; et al. Transcellular degradation of axonal mitochondria. Proc. Natl. Acad. Sci. USA 2014, 111, 9633–9638. [Google Scholar] [CrossRef]
- Hayakawa, K.; Esposito, E.; Wang, X.; Terasaki, Y.; Liu, Y.; Xing, C.; Ji, X.; Lo, E.H. Transfer of mitochondria from astrocytes to neurons after stroke. Nature 2016, 535, 551–555. [Google Scholar] [CrossRef]
- Ahmad, T.; Mukherjee, S.; Pattnaik, B.; Kumar, M.; Singh, S.; Kumar, M.; Rehman, R.; Tiwari, B.K.; Jha, K.A.; Barhanpurkar, A.P.; et al. Miro1 regulates intercellular mitochondrial transport & enhances mesenchymal stem cell rescue efficacy. EMBO J. 2014, 33, 994–1010. [Google Scholar] [PubMed]
- Hass, D.T.; Barnstable, C.J. Mitochondrial uncoupling protein 2 knock-out promotes mitophagy to decrease retinal ganglion cell death in a mouse model of glaucoma. J. Neurosci. 2019, 39, 3582–3596. [Google Scholar] [PubMed]
- Zorov, D.B.; Kinally, K.W.; Tedeschi, H. Voltage activation of heart inner mitochondrial membrane channels. J. Bioenerg. Biomembr. 1992, 24, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Tinari, A.; Garofalo, T.; Sorice, M.; Degli Esposti, M.; Malorni, W. Mitoptosis: Different pathways for mitochondrial execution. Autophagy 2007, 3, 282–284. [Google Scholar] [CrossRef]
- Sancak, Y.; Bar-Peled, L.; Zoncu, R.; Markhard, A.L.; Nada, S.; Sabatini, D.M. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010, 141, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; McPhee, C.K.; Zheng, L.; Mardones, C.A.; Rong, Y.; Peng, J.; Mi, N.; Zhao, Y.; Liu, Z.; Wan, F.; et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 2010, 465, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Dames, S.A.; Mulet, J.M.; Rathgeb-Szabo, K.; Hall, M.N.; Grzesiek, S. The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability. J. Biol. Chem. 2005, 280, 20558–20564. [Google Scholar] [CrossRef]
- Sarkar, S.; Korolchuk, V.I.; Renna, M.; Imarisio, S.D.; Fleming, A.; Williams, A.; Garcia-Arencibia, M.; Rose, C.; Luo, S.; Underwood, B.R.; et al. Complex inhibitory effects of nitric oxide on autophagy. Mol. Cell. 2011, 43, 19–32. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Q.J.; Li, X.; Yan, Y.; Backer, J.M.; Chait, B.T.; Heintz, N.; Yus, Z. Distinct regulation opf autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009, 11, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Rubinsztein, S.C. Inositol and IP3 levels regulate autophagy: Biology and therapeutic speculations. Autophagy 2006, 2, 132–134. [Google Scholar] [CrossRef] [PubMed]
- Harrison, B.; Kraus, M.; Burch, L.; Stevens, C.; Craig, A.; Gordon-Weeks, P.; Hupp, T.R. DAPK-1 binding to a linear peptide motif in MAP18 stimulates autophagy and membrane blebbing. J.Biol. Chem. 2008, 283, 9999–10014. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Itoh, K.; Yamamoto, M. Roles of Nrf2 in activation of antioxidant enzyme genes via antioxidant responsive elements. Methods Enzymol. 2002, 348, 182–190. [Google Scholar] [PubMed]
- Fujita, K.; Maeda, D.; Xiao, Q.; Srinivasuta, S.M. Nrf2-mediated induction of p62 controls Toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc. Natl. Acad. Sci. USA 2011, 108, 1427–1432. [Google Scholar] [CrossRef]
- Li, P.F.; Dietz, R.; Von Harsdorf, R. p53 regulates mitochondrial membrane potential through reactive oxygen species and induces cytochrome c-independent apoptosis blocked by Bcl-2. EMBO J. 1999, 18, 6027–6036. [Google Scholar] [CrossRef]
- Crighton, D.; Wilkinson, S.; O’Prey, J.; Syed, N.; Smith, P.; Harrison, P.R.; Gasco, M.; Garrone, O.; Crook, T.; Ryan, K.M. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006, 126, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Fukuto, J.M.; Carrington, S.J.; Tantillo, D.J.; Harrison, J.G.; Ignarro, L.J.; Freeman, B.A.; Chen, A.; Wink, D.A. Small molecule signaling agents: The integrated chemistry and biochemistry of nitrogen oxides, oxides of carbon, dioxygen, hydrogen sulfide, and their derived species. Chem. Res. Toxicol. 2012, 25, 769–793. [Google Scholar] [CrossRef]
- Halliwell, B. Reactive species and antioxidants. Redox biology is a fundamenta theme of aerobic life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef]
- Dröge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [CrossRef]
- Cobley, N.J.; Fiorello, M.L.; Bailey, D.M. 13 reasons why the brain is susceptible to oxidative stress. Redox Biol. 2018, 15, 490–513. [Google Scholar] [CrossRef]
- Massad, C.; Klann, E. Reactive oxygen species in the regulation of synaptic plasticity and memory. Antioxid. Redox Signal. 2011, 14, 2013–2054. [Google Scholar] [CrossRef]
- Dickinson, B.C.; Peltier, J.; Store, D.; Schaffer, D.V.; Chang, C.J. Nox2 redox signaling maintains essential cell populations in the brain. Nat. Chem. Biol. 2011, 7, 106–112. [Google Scholar] [CrossRef]
- Gauron, C.; Meda, F.; Dupont, E.; Albadri, S.; Quenech’Du, N.; Ipendey, E.; Volovitch, M.; Del Bene, F.; Joliot, A.; Rampon, C.; et al. Hydrogen peroxide (H2O2) controls axon pathfinding during zebrafish development. Dev. Biol. 2016, 414, 133–141. [Google Scholar] [CrossRef]
- Pasterkamp, R.J. Getting neural circuits into shape with semaphorins. Nat. Rev. Neurosci. 2012, 13, 605–618. [Google Scholar] [CrossRef]
- Bailey, D.M.; Bärtsch, P.; Knauth, M.; Baumgartner, R.W. Emerging concepts in acute mountain sickness and high-altitude cerebral edema: From the molecular to the morphological. Cell. Moll. Life Sci. 2009, 66, 3583–3594. [Google Scholar] [CrossRef] [PubMed]
- Birben, E.; Sahiner, U.M.; Sackesen, C.; Erzurum, S.; Kalayci, O. Oxidative stress and antioxidant defense. World Allergy Organ. J. 2012, 5, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Kalogeris, T.; Baines, C.P.; Krenz, M.; Korthuis, R.J. Cell biology of ischemia/reperfusion injury. Int. Rev. Cell. Mol. Biol. 2012, 298, 229–317. [Google Scholar] [PubMed]
- Simion, A.; Jurcau, A. The role of antioxidant treatment in acute ischemic stroke: Past, present and future. Neurol. Res. Surg. 2019, 2, 1–7. [Google Scholar] [CrossRef]
- Jurcau, A.; Simion, A. Oxidative stress in the pathogenesis of Alzheimer’s disease and cerebrovascular disease with therapeutic implications. CNS Neurol. Disord. Drug Targets 2020, 19, 24–108. [Google Scholar] [CrossRef]
- Zucker, R.S. Calcium- and activity-dependent synaptic plasticity. Curr. Opin. Neurobiol. 1999, 9, 305–313. [Google Scholar] [CrossRef]
- Lipton, S.A.; Choi, Y.-P.; Pan, Z.-H.; Lei, S.Z.; Chen, H.-S.V.; Sucher, N.J.; Loscalzo, J.; Singel, D.J.; Stamler, J.S. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature 1993, 364, 626–632. [Google Scholar] [CrossRef]
- Brown, G.C. Nitric oxide and mitochondrial respiration. Biochim. Biophys Acta-Bioenerg. 1999, 1411, 351–369. [Google Scholar] [CrossRef]
- Rizzuto, R.; De Stefani, D.; Raffaello, A.; Mammucari, C. Mitochondria as sensors and regulators of calcium signalling. Nat. Rev. Mol. Cell Biol. 2012, 13, 566–578. [Google Scholar] [CrossRef]
- Vos, M.; Lauwers, E.; Verstreken, P. Synaptic mitochondria in synaptic transmission and organization of vesicle pools in health and disease. Front. Synaptic Neurosci. 2010, 2, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Zou, L.; Zhang, X.; Branco, V.; Wang, J.; Carvalho, C.; Holmgren, A.; Lu, J. Redox signaling mediated by thioredoxin and glutathione systems in the central nervous system. Antioxid. Redox Signal. 2017, 27, 989–1010. [Google Scholar] [CrossRef]
- Fisher, A.B. Peroxiredoxin 6 in the repair of peroxidized cell membranes and cell signaling. Arch. Biochem. Biophys. 2017, 617, 68–83. [Google Scholar] [CrossRef]
- Mander, P.K.; Jekabsone, A.; Brown, G.C. Microglia proliferation is regulated by hydrogen peroxide from NADPH oxidase. J. Immunol. 2006, 176, 1046–1052. [Google Scholar] [CrossRef]
- Colton, C.A.; Gilbert, D.L. Production of superoxide anion by a CNS macrophage, the microglia. FEBS Lett. 1987, 223, 284–288. [Google Scholar] [CrossRef]
- Edmondson, D. Hydrogen peroxide produced by mitochondrial monoamine oxidase catalysis: Biological implications. Curr. Pharm. Des. 2014, 20, 155–160. [Google Scholar] [CrossRef]
- Heikkila, R.E.; Cohen, G. 6-Hydroxydopamine: Evidence for superoxide radical as an oxidative intermediate. Science 1973, 181, 456–457. [Google Scholar] [CrossRef]
- Misra, H.P.; Fridovich, I. The role of superoxide anion in the autoxidation of epinephrine and a simple assay for superoxide dismutase. J. Biol. Chem. 1972, 247, 3170–3175. [Google Scholar] [CrossRef]
- Que, E.L.; Domaille, D.W.; Chang, C.J. Metals in neurobiology: Probing their chemistry and biology with molecular imaging. Chem. Rev. 2008, 39, 1517–1549. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of non-apoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Abreu, I.A.; Cabelli, D.E. Superoxide dismutases––A review of the metal-associated mechanistic variations. Biochim. Biophys. Acta-Proteins Proteom. 2010, 1804, 263–274. [Google Scholar] [CrossRef]
- Chang, C.J. Searching for harmony in transition-metal signaling. Nat. Chem. Biol. 2015, 11, 744–747. [Google Scholar] [CrossRef]
- Zerbinati, C.; Iuliano, L. Cholesterol and related sterols autoxidation. Free Radic. Biol. Med. 2017, 111, 151–155. [Google Scholar] [CrossRef]
- Briggs, J.A.; Wolvetang, J.E.; Mattick, J.S.; Rinn, J.L.; Barry, G. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron 2015, 88, 861–877. [Google Scholar] [CrossRef]
- Simms, C.L.; Zaher, H.S. Quality control of chemically damaged RNA. Cell. Mol. Life Sci. 2016, 73, 3639–3653. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Chock, P.B.; Stadtman, E.R. Oxidized messenger RNA induces translation errors. Proc. Natl. Acad. Sci. USA 2006, 104, 66–71. [Google Scholar] [CrossRef]
- Starkov, A.A. The role of mitochondria in reactive oxygen species metabolism and signaling. Ann. N. Y. Acad. Sci. 2006, 1147, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Andreyev, A.Y.; Kushnareva, Y.E.; Starkov, A.A. Mitochondrial metabolism of reactive oxygen species. Biochemistry 2005, 70, 200–214. [Google Scholar] [CrossRef]
- Finkel, T.; Holbrook, N.J. Oxidants, oxidative stress and the biology of ageing. Nature 2000, 408, 239–247. [Google Scholar] [CrossRef]
- Fukai, T.; Ushio-Fukai, M. Superoxide dismutases: Role in redox signaling, vascular function, and disease. Antioxid. Redox Signal. 2011, 15, 1583–1606. [Google Scholar] [CrossRef] [PubMed]
- Handy, D.E.; Loscalzo, J. Redox regulation of mitochondrial function. Antioxid. Redox Signal. 2012, 16, 1323–1367. [Google Scholar] [CrossRef] [PubMed]
- Bienert, G.P.; Chaumont, F. Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; He, W.; Liou, Y.-C. The redox language in neurodegenerative diseases: Oxidative post-translational modifications by hydrogen peroxide. Cell Death Dis. 2021, 12, 58. [Google Scholar] [CrossRef] [PubMed]
- Starkov, A.A. Mitochondrial alpha-ketoglutarate dehydrogenase complex generates reactive oxygen species. J. Neurosci. 2004, 24, 7779–7788. [Google Scholar] [CrossRef]
- Tretter, L.; Adam-Vizi, V. Generation of reactive oxygen species in the reaction catalyzed by alpha-ketoglutarate dehydrogenase. J. Neurosci. 2004, 24, 7771–7778. [Google Scholar] [CrossRef]
- Sato, K. Insulin, ketone bodies, and mitochondrial energy transduction. FASEB J. 1995, 9, 651–658. [Google Scholar] [CrossRef]
- Zoccarato, F. Succinate modulation of H2O2 release at NADH: Ubiquinone oxidoreductase (Complex I) in brain mitochondria. Biochem. J. 2007, 406, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Adam-Vizi, V.; Chinopoulos, C. Bioenergetics and the formation of mitochondrial reactive oxygen species. Trends Pharmacol. Sci. 2006, 27, 639–645. [Google Scholar] [CrossRef]
- Abramov, A.Y.; Scorziello, A.; Duchen, M.R. Three distinct mechanisms generate oxygen free radicals in neurons and contribute to cell death during anoxia and reoxygenation. J. Neurosci. 2007, 27, 1129–1138. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, D.L.; Salter, J.D.; Brookes, P.S. Response of mitochondrial reactive oxygen species generation to steady-state oxygen tension: Implications for hypoxic cell signaling. Am. J. Physiol. Heart. Circ. Physiol. 2007, 292, H101–H108. [Google Scholar] [CrossRef] [PubMed]
- Babior, B.M.; Kipnes, R.S.; Curnutte, J.T. Biological defense mechanisms. The production by leukocytes of superoxide, a potential bactericidal agent. J. Clin. Investig. 1973, 52, 741–744. [Google Scholar] [CrossRef]
- Ghandi, S.; Abramov, A.Y. Mechanism of oxidative stress in neurodegeneration. Oxid. Med. Cell. Longev. 2012, 2012, 428010. [Google Scholar]
- Pradeep, H.; Diya, J.B.; Shashikumar, S.; Rajanikant, G.K. Oxidative stress- assassin behind the ischemic stroke. Folia Neuropathol. 2012, 50, 219–230. [Google Scholar] [CrossRef]
- Babior, B.M. NADPH oxidase. Curr. Opin. Immunol. 2004, 16, 42–47. [Google Scholar] [CrossRef]
- Guo, S.; Chen, X. The human Nox4: Gene, structure, physiological function and pathological significance. J. Drug Target 2015, 23, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Abramov, A.Y.; Jacobson, J.; Wientjes, F.; Hothersall, J.; Canevari, L.; Duchen, M.R. Expression and modulation of an NADPH oxidase in mammalian astrocytes. J. Neurosci. 2005, 25, 9176–9184. [Google Scholar] [CrossRef]
- Kuroda, J.; Nakagawa, K.; Yamasaki, T.; Nakamura, K.-I.; Takeya, R.; Kuribayashi, F.; Imajoh-Ohmi, S.; Igarashi, K.; Shibata, Y.; Sueishi, K.; et al. The superoxide-producing NAD(P)H oxidase Nox4 in the nucleus of human vascular endothelial cells. Genes Cells 2005, 10, 1139–1151. [Google Scholar] [CrossRef] [PubMed]
- Graham, K.A.; Kulawiec, M.; Owens, K.M.; Li, X.; Desouki, M.M.; Chandra, D.; Singh, K.K. NADPH oxidase 4 is an oncoprotein localized to mitochondria. Cancer Biol. Ther. 2010, 10, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Abramov, A.Y.; Canevari, L.; Duchen, M.R. Beta-amyloid peptides induce mitochondrial dysfunction and oxidative stress in astrocytes and death of neurons through activation of NADPH oxidase. J. Neurosci. 2004, 24, 565–575. [Google Scholar] [CrossRef]
- Edmondson, D.E.; Binda, C.; Wang, J.; Upadhyay, A.K.; Mattevi, A. Molecular and mechanistic properties of the membrane-bound mitochondrial monoamine oxidases. Biochemistry 2009, 48, 4220–4230. [Google Scholar] [CrossRef]
- Schrader, M.; Fahimi, H.D. Peroxisomes and oxidative stress. Biochim. Biophys. Acta 2006, 1763, 1755–1766. [Google Scholar] [CrossRef]
- Dean, R.T.; Fu, S.; Stocker, R.; Davies, M.J. Biochemistry and pathology of radical-mediated protein oxidation. Biochem J. 1997, 324, 1–18. [Google Scholar] [CrossRef]
- Chevion, M.; Berenshtein, E.; Stadtman, E.R. Human studies related to protein oxidation: Protein carbonyl content as a marker of damage. Free Radic. Res. 2000, 33, S99–S108. [Google Scholar] [PubMed]
- Pryor, W.A.; Jin, X.; Squadrito, G.L. One- and two-electron oxidations of methionine by peroxynitrite. Proc. Natl. Acad. Sci. USA 1994, 91, 11173–11177. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Koppal, T.; Howard, B.; Subramaniam, R.; Hall, N.; Hensley, K.; Yatin, S.; Allen, K.; Aksenov, M.; Aksenova, M.; et al. Structural and functional changes in proteins induced by free radical-mediated oxidative stress and protective action of the antioxidants N-tert-butyl-alpha-phenylnitrone and vitamin E. Ann. N. Y. Acad. Sci. 1998, 854, 448–462. [Google Scholar] [CrossRef] [PubMed]
- Sorg, O.; Horn, T.F.; Yu, N.; Gruol, D.; Bloom, F.E. Inhibition of astrocyte glutamate uptake by reactive oxygen species: Role of antioxidant enzymes. Mol. Med. 1997, 3, 431–440. [Google Scholar] [CrossRef]
- Grune, T.; Reinheckel, T.; Davies, K.J. Degradation of oxidized proteins in mammalian cells. FASEB J. 1997, 11, 526–534. [Google Scholar] [CrossRef]
- Grune, D.; Merker, K.; Sandig, G.; Davies, K.J. Selective degradation of oxidatively modified protein substrates by proteasome. Biochem. Biophys. Res. Commun. 2003, 305, 709–718. [Google Scholar] [CrossRef]
- Varshavsky, A. The ubiquitin system. Trends Biochem. Sci. 1997, 22, 383–387. [Google Scholar] [CrossRef]
- Kirkin, V.; Lamark, T.; Sou, Y.S.; Bjørkøy, G.; Nunn, J.L.; Bruun, J.A.; Shvets, E.; McEwan, D.G.; Clausen, T.H.; Wild, P.; et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 2009, 33, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Kirkin, V.; Lamark, T. Johansen, T.; Dikic. I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy 2009, 5, 732–733. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Brault, J.J.; Schild, A.; Cao, P.; Sandri, M.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007, 6, 472–483. [Google Scholar] [CrossRef] [PubMed]
- Olzmann, J.A.; Li, L.; Chudaev, M.V.; Chen, J.; Perez, F.A.; Palmiter, R.D.; Chin, L.S. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell Biol. 2007, 178, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Gamerdinger, M.; Hajieva, P.; Kaya, M.; Wolfrum, U.; Hartl, F.U.; Behl, C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009, 28, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Ayala, A.; Munoz, M.F.; Arguelles, S. Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef]
- Perluigi, M.; Coccia, R.; Butterfield, D.A. 4-Hydroxy-2-nonenal, a reactive product of lipid peroxidation, and neurodegenerative diseases: A toxic combination illuminated by redox proteomics studies. Antioxid. Redox Signal. 2012, 17, 1590–1609. [Google Scholar] [CrossRef]
- Erejuwa, O.O.; Sulaiman, S.A.; Ab Wahab, M.S. Evidence in support of potential applications of lipid peroxidation products in cancer treatment. Oxid. Med. Cell. Longev. 2013, 2013, 931251. [Google Scholar] [CrossRef]
- Bradley-Whitman, M.A.; Lovell, M.A. Biomarkers of lipid peroxidation in Alzheimer disease (AD): An update. Arch. Toxicol. 2015, 89, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Fritz, K.S.; Petersen, D.R. An overview of the chemistry and biology of reactive aldehydes. Free Radic. Biol. Med. 2013, 59, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Schopfer, F.J.; Cipollina, C.; Freeman, B.A. Formation and signaling actions of electrophilic lipids. Chem Rev. 2011, 111, 5997–6021. [Google Scholar] [CrossRef]
- Kansanen, E.; Jyrkkanen, H.K.; Levonen, A.L. Activation of stress signaling pathways by electrophilic oxidized and nitrated lipids. Free Radic. Biol. Med. 2012, 52, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Kozak, K.R.; Marnett, L.J. IkappaB kinase, a molecular target for inhibition by 4-hydroxy-2-nonenal. J. Biol. Chem. 2001, 276, 18223–18228. [Google Scholar] [CrossRef] [PubMed]
- Sultana, R.; Perluigi, M.; Allan Butterfield, D. Lipid peroxidation triggers neurodegeneration: A redox proteomics view into the Alzheimer disease brain. Free Radic. Biol. Med. 2013, 62, 157–169. [Google Scholar] [CrossRef]
- Laurora, S.; Tamagno, E.; Briatore, F.; Bardini, P.; Pizzimenti, S.; Toaldo, C.; Reffo, P.; Costelli, P.; Dianzani, M.U.; Danni, O.; et al. 4-Hydroxynonenal modulation of p53 family gene expression in the SK-N-BE neuroblastoma cell line. Free Radic. Biol. Med. 2005, 38, 215–225. [Google Scholar] [CrossRef]
- Li, J.; Sharma, R.; Patrick, B.; Sharma, A.; Jeyabal, P.V.; Reddy, P.M.; Saini, M.K.; Dwivedi, S.; Dhanani, S.; Ansari, N.-H.; et al. Regulation of CD95 (Fas) expression and Fas-mediated apoptotic signaling in HLE B-3 cells by 4-hydroxynonenal. Biochemistry 2006, 45, 12253–12264. [Google Scholar] [CrossRef]
- Parola, M.; Robino, G.; Marra, F.; Pinzani, M.; Bellomo, G.; Leonarduzzi, G.; Chiarugi, P.; Camandola, S.; Poli, G.; Waeg, G.; et al. HNE interacts directly with JNK isoforms in human hepatic stellate cells. J. Clin. Investig. 1998, 102, 1942–1950. [Google Scholar] [CrossRef]
- Song, B.J.; Soh, Y.; Bae, M.; Pie, J.; Wan, J.; Jeong, K. Apoptosis of PC12 cells by 4-hydroxy-2-nonenal is mediated through selective activation of the c-Jun N-Terminal protein kinase pathway. Chem. Biol. Interact. 2001, 130–132, 943–954. [Google Scholar] [CrossRef]
- Shibata, N.; Kato, Y.; Inose, Y.; Hiroi, A.; Yamamoto, T.; Morikawa, S.; Sawada, M.; Kobayashi, M. 4-hydroxy-2-nonenal upregulates and phosphorylates cytosolic phospholipase A2 in cultured Ra2 microglial cells via MAPK pathways. Neuropathology 2011, 31, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Kim, C.E.; Yun, M.R.; Seo, K.W.; Park, H.M.; Yun, J.W.; Shin, H.K.; Bae, S.S.; Kim, C.D. 4-hydroxynonenal enhances MMP-9 production in murine macrophages via 5-lipoxygenase-mediated activation of ERK and p38 MAPK. Toxicol. Appl. Pharmacol. 2010, 242, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Narciso, L.; Parlanti, E.; Racaniello, M.; Simonelli, V.; Cardinale, A.; Merlo, D.; Dogliotti, E. The response to oxidative DNA damage in neurons: Mechanisms and disease. Neural Plast. 2016, 2016, 3619274. [Google Scholar] [CrossRef] [PubMed]
- Colnaghi, L.; Rondelli, D.; Muzi-Falconi, M.; Sertic, S. Tau and DNA damage in neurodegeneration. Brain Sci. 2020, 10, 946. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef]
- Rodier, F.; Coppe, J.P.; Patil, C.K.; Hoeijmakers, W.A.; Munoz, D.P.; Raza, S.R.; Freund, A.; Campeau, E.; Davalos, A.R.; Campisi, J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell. Biol. 2009, 11, 973–979. [Google Scholar] [CrossRef]
- Maynard, S.; Fang, E.F.; Scheibye-Knudsen, M.; Croteau, D.L.; Bohr, V.A. DNA damage, DNA repair, aging, and neurodegeneration. Cold Spring Harb. Perspect. Med. 2015, 5, a025130. [Google Scholar] [CrossRef]
- Hegde, M.L.; Mantha, A.K.; Hazra, T.K.; Bhakat, K.K.; Mitra, S.; Szczesny, B. Oxidative genome damage and its repair: Implications in aging and neurodegenerative diseases. Mech. Ageing Dev. 2012, 133, 157–168. [Google Scholar] [CrossRef]
- Jacobs, A.L.; Schär, P. DNA glycosylases: In DNA repair and beyond. Chromosoma 2012, 121, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Hegde, M.L.; Hazra, T.K.; Mitra, S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008, 18, 27–47. [Google Scholar] [CrossRef]
- Nishioka, K.; Ohtsubo, T.; Oda, H.; Fujiwara, T.; Kang, D.; Sugimachi, K.; Nakabeppu, Y. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol. Biol. Cell 1999, 10, 1637–1652. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.; Qu, D.; Zhang, Y.; Venderova, K.; Haque, M.E.; Rousseaux, M.W.C.; Slack, R.S.; Woulfe, J.M.; Park, D.S. The role of Cdk5-mediated apurinic/apyrimidinic endonuclease 1 phosphorylation in neuronal death. Nat. Cell Biol. 2010, 12, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Cha, M.-Y.; Kim, D.K.; Mook-Yung, I. The role of mitochondrial DNA mutation on neurodegenerative diseases. Exp. Mol. Med. 2015, 47, e150. [Google Scholar] [CrossRef] [PubMed]
- Reeve, A.K.; Krishnan, K.J.; Turnbull, D. Mitochondrial DNA mutations in disease, aging, and neurodegeneration. Ann. N. Y. Acad. Sci. 2008, 1147, 21–29. [Google Scholar] [CrossRef]
- Biswas, G.; Anandatheerthavarada, H.K.; Zaidi, M.; Avadhani, N.G. Mitochondria to nucleus stress signaling: A distinctive mechanism of NFkappaB/Rel activation through calcineurin-mediated inactivation of IkappaBbeta. J. Cell Biol. 2003, 161, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Finley, L.W.S.; Haigis, M.C. The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Ageing Res. Rev. 2009, 8, 173–188. [Google Scholar] [CrossRef]
- Fukae, J.; Takanashi, M.; Kubo, S.-I.; Nishioka, K.-I.; Nakabeppu, Y.; Mori, H.; Mizuno, Y.; Hattori, N. Expression of 8-oxoguanine DNA glycosylase (OGG1) in Parkinson’s disease and related neurodegenerative disorders. Acta Neuropathol. 2005, 109, 256–262. [Google Scholar] [CrossRef]
- Coppedè, F.; Migliore, L. DNA damage in neurodegenerative diseases. Mutat. Res. 2015, 776, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Lovell, M.A.; Xie, C.; Markesbery, W.R. Decreased base excision repair and increased helicase activity in Alzheimer’s disease brain. Brain Res. 2000, 855, 116–123. [Google Scholar] [CrossRef]
- Weissman, L.; Jo, D.-G.; Sørensen, M.M.; De Souza-Pinto, M.C.; Markesbery, W.R.; Mattson, M.P.; Bohr, V.A. Defective DNA base excision repair in brain from individuals with Alzheimer’s disease and amnestic mild cognitive impairment. Nucleic Acids Res. 2007, 35, 5545–5555. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chenm, X.; Lim, Z.; Yem, W.; Dingm, H.; Lim, P.; Aung, L.H.H. Role of RNA oxidation in neurodegenerative diseases. Int. J. Mol. Sci. 2020, 21, 5022. [Google Scholar] [CrossRef]
- Hofer, T.; Badouard, C.; Bajak, E.; Ravanat, J.L.; Mattsson, A.; Cotgreave, I.A. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol Chem. 2005, 386, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Mattick, J.S. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput. Biol. 2008, 4, e1000176. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Yeo, G.; Muotri, A.R.; Kuwabara, T.; Gage, F.H. Noncoding RNAs in the mammalian central nervous system. Annu. Rev. Neurosci. 2006, 29, 77–103. [Google Scholar] [CrossRef]
- Barciszewski, J.; Barciszewska, M.Z.; Siboska, G.; Rattan, S.; Clark, B.F. Some unusual nucleic acid bases are products of hydroxyl radical oxidation of DNA and RNA. Mol. Biol. Rep. 1999, 26, 231–238. [Google Scholar] [CrossRef]
- Nunomura, A.; Lee, H.-G.; Zhu, X.; Perry, G. Consequences of RNA oxidation on protein synthesis rate and fidelity: Implications for the pathophysiology of neuropsychiatric disorders. Biochem. Soc. Trans. 2017, 45, 1053–1066. [Google Scholar] [CrossRef]
- Jacobs, A.C.; Resendiz, M.J.E.; Greenberg, M.M. Direct strand scission from a nucleobase radical in RNA. J. Am. Chem. Soc. 2010, 132, 3668–3669. [Google Scholar] [CrossRef][Green Version]
- Ding, Q.; Dimayuga, E.; Bruce-Keller, A.J. Oxidative stress alters neuronal RNA- and protein-synthesis: Implications for neural viability. Free. Radic. Res. 2007, 41, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.L.; Zaher, H.S. How do cells cope with RNA damage and its consequences? J. Biol. Chem. 2019, 294, 15158–15171. [Google Scholar] [CrossRef]
- Khanam, H.; Ali, A.; Asif, M.; Shamsuzzaman. Neurodegenerative diseases linked to misfolded proteins and their therapeutic approaches: A review. Eur. J. Med. Chem. 2016, 124, 1121–1141. [Google Scholar] [CrossRef] [PubMed]
- Hevner, R.F.; Wong-Riley, M.T. Entorhinal cortex of the human, monkey, and rat: Metabolic map as revealed by cytochrome oxidase. J. Comp. Neurol. 1992, 326, 451–469. [Google Scholar] [CrossRef]
- Wang, R.; Reddy, P.H. Role of glutamate and NMDA receptors in Alzheimer’s disease. J. Alzheimer’s Dis. 2017, 57, 1041–1048. [Google Scholar] [CrossRef]
- Hof, P.R.; Nimchinsky, E.A.; Celio, M.R.; Bouras, C.; Morrison, J.H. Calretinin-immunoreactive neocortical interneurons are unaffected in Alzheimer’s diseaase. Neurosci. Lett. 1993, 152, 145–148. [Google Scholar] [CrossRef]
- Roussarie, J.-P.; Yao, V.; Rodriguez-Rodriguez, P.; Oughtred, R.; Rust, J.; Plautz, Z.; Kasturia, S.; Albornoz, C.; Wang, W.; Schmidt, E.F.; et al. Selective neuronal vulnerability in Alzheimer’s disease: A network-based analysis. Neuron 2020, 107, 821–835. [Google Scholar] [CrossRef]
- Kaufman, S.K.; Del Tredici, K.; Thomas, T.L.; Braak, H.; Diamond, M.I. Tau seeding activity begins in the transentorhinal/entorhinal regions and anticipates phospho-tau pathology in Alzheimer’s disease and PART. Acta Neuropathol. 2018, 136, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Leng, K.; Li, E.; Eser, R.; Piergies, A.; Sit, R.; Tan, M.; Neff, N.; Li, S.H.; Diehl Rodriguez, R.; Suemoto, C.K.; et al. Molecular characterization of selectively vulnerable neurons in Alzheimer’s disease. Nat. Neurosci. 2021, 24, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Michel, P.P.; Hirsch, E.C.; Hunot, S. Understanding dopaminergic cell death pathways in Parkinson disease. Neuron 2016, 90, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, W.; Furuta, T.; Nakamura, K.C.; Hioki, H.; Fujiyama, F.; Arai, R.; Kaneko, T. Single nigrostriatal dopaminergic neurons form widely spread and highly dense axonal arborizations in the neostriatum. J. Neurosci. 2009, 29, 444–453. [Google Scholar] [CrossRef]
- Bolam, J.P.; Pissadaki, E.K. Living on the edge with too many mouths to feed: Why dopamine neurons die. Mov. Disord. 2012, 27, 1478–1483. [Google Scholar] [CrossRef] [PubMed]
- Pacelli, C.; Giguère, N.; Bourque, M.-J.; Lévesque, M.; Slack, R.S.; Trudeau, L.-É. Elevated mitochondrial bioenergetics and axonal arborization size are key contributors to the vulnerability of dopamine neurons. Curr. Biol. 2015, 25, 2349–2360. [Google Scholar] [CrossRef]
- Guzman, J.N.; Sanchez-Padilla, J.; Wokosin, D.; Kondapalli, J.; Ilijic, E.; Schumacker, P.T.; Surmeier, D.J. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature 2010, 468, 696–700. [Google Scholar] [CrossRef]
- Lawless, C.; Greaves, L.; Reeve, A.K.; Turnbull, D.M.; Vincent, A.E. The rise and rise of mitochondrial DNA mutations. Open Biol. 2020, 10, 200061. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, J.F.; Baris, O.R.; Hess, S.; Moser, N.; Schroder, H.; Chinta, S.J.; Andersen, J.K.; Kloppenburg, P.; Wiesner, R.J. Catecholamine metabolism drives generation of mitochondrial DNA deletions in dopaminergic neurons. Brain 2014, 137, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Burke, R.E.; Tsairis, P. Anatomy and innervation ratios in motor units of cat gastrocnemius. J. Physiol. 1973, 234, 749–765. [Google Scholar] [CrossRef]
- Enoka, R.M. Morphological features and activation patterns of motor units. J. Clin. Neurophysiol. 1995, 12, 538–559. [Google Scholar] [CrossRef]
- Zimmermann, L.; Morado-Díaz, C.J.; Davis-López de Carrizosa, M.A.; De la Cruz, R.R.; May, P.J.; Streicher, J.; Pastor, A.M.; Blumer, R. Axons giving rise to the palisade endings of feline extraocular muscle display motor features. J. Neurosci. 2013, 33, 2784–2793. [Google Scholar] [CrossRef] [PubMed]
- Alexianu, M.E.; Ho, B.-K.; Mohamed, A.H.; La Bella, V.; Smith, R.G.; Appel, S.H. The role of calcium-binding proteins in selective motoneuron vulnerability in amyotrophic lateral sclerosis. Ann. Neurol. 1994, 36, 846–858. [Google Scholar] [CrossRef]
- LaFerla, F.M. Calcium dyshomeostasis in intracellular signalling in Alzheimer’s disease. Nat. Rev. Neurosci. 2002, 3, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.L.; Mayne, M.; Holden, C.P.; Geiger, J.D.; Mattson, M.P. Presenilin-1 mutations increase levels of ryanodine receptors and calcium release in PC12 cells and cortical neurons. J.Biol. Chem. 2000, 275, 18195–18200. [Google Scholar] [CrossRef]
- Filadi, R.; Greotti, E.; Turacchio, G.; Luini, A.; Pozzan, T.; Pizzo, P. Presenilin 2 modulates endoplasmic reticulum-mitochondria coupling by tuning the antagonistic effect of mitofusin 2. Cell. Rep. 2016, 15, 2226–2238. [Google Scholar] [CrossRef]
- Area-Gomez, E.; Schon, E.A. On the pathogenesis of Alzheimer’s disease: The MAM hypothesis. FASEB J. 2017, 31, 864–867. [Google Scholar] [CrossRef] [PubMed]
- Calvo-Rodriguez, M.; Hou, S.S.; Snyder, A.C.; Kharitonova, E.K.; Russ, A.N.; Das, S.; Fan, Z.; Muzikansky, A.; Garcia-Alloza, M.; Serrano-Pozo, A.; et al. Increased mitochondrial calcium levels associated with neuronal death in a mouse model of Alzheimer’s disease. Nat. Commun. 2020, 11, 2146. [Google Scholar] [CrossRef]
- Britti, E.; Ros, J.; Esteras, N.; Abramov, A.Y. Tau inhibits mitochondrial calcium efflux and makes neurons vulnerable to calcium-induced cell death. Cell. Calcium 2020, 86, 102150. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Morales, J.-C.; Arranz-Tagarro, J.-A.; Calvo-Gallardo, E.; Maroto, M.; Padín, J.-F.; García, A.G. Stabilizers of neuronal and mitochondrial calcium cycling as a strategy for developing a medicine for Alzheimer’s disease. ACS Chem. Neurosci. 2012, 3, 873–883. [Google Scholar] [CrossRef]
- Magi, S.; Castaldo, P.; Macrí, M.L.; Maiolino, M.; Matteucci, A.; Bastioli, G.; Gratteri, S.; Amoroso, S.; Lariccia, V. Intracellular calcium dysregulation: Implications for Alzheimer’s disease. Biomed. Res. Int. 2016, 2016, 6701324. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; Ramachandran, S.; Azimov, R.; Kagan, B.L.; Lal, R. Ion channel formation bt Tau protein: Implications for Alzheimer’s disease and tauopathies. Biochemistry 2015, 50, 7320–7325. [Google Scholar] [CrossRef]
- Armato, U.; Chiarini, A.; Chakravarthy, B.; Chioffi, F.; Pacchiana, R.; Colarusso, E.; Whitfield, J.F.; Dal Prá, I. Calcium-sensing receptor antagonist (calcilytic) NPS 2143 specifically blocks the increased secretion of endogenous Aβ42 prompted by exogenous fibrillary or soluble Aβ25-35 in human cortical astrocytes and neurons–therapeutic relevance to Alzheimer’s disease. Biochim. Biophys. Acta 2013, 10, 1634–1652. [Google Scholar]
- Jové, M.; Mota-Martorell, N.; Torres, P.; Ayala, V.; Portero-Otin, M.; Ferrer, I.; Pamplona, R. The causal role of lipoxidative damage in mitochondrial bioenergetic dysfunction linked to Alzheimer’s disease pathology. Life 2021, 11, 388. [Google Scholar] [CrossRef]
- Terni, B.; Boada, J.; Portero-Otin, M.; Pamplona, R.; Ferrer, I. Mitochondrial ATP-synthase in the entorhinal cortex is a target of oxidative stress at stages I/II of Alzheimer’s disease pathology. Brain Pathol. 2010, 20, 222–233. [Google Scholar] [CrossRef]
- Jové, M.; Pradas, I.; Dominguez-Gonzalez, M.; Ferrer, I.; Pamplona, R. Lipids and lipoxidation in human brain aging. Mitochondrial ATP-synthase as a key lipoxidation target. Redox Biol. 2019, 23, 101083. [Google Scholar] [CrossRef] [PubMed]
- Tobore, T.O. On the central role of mitochondria dysfunction and oxidative stress in Alzheimer’s disease. Neurol. Sci. 2019, 40, 1527–1540. [Google Scholar] [CrossRef]
- Chang, R.Y.K.; Etheridge, N.; Dodd, P.R.; Nouwens, A.S. Targeted quantitative analysis of synaptic proteins in Alzheimer’s disease brain. Neurochem. Int. 2014, 75, 66–75. [Google Scholar] [CrossRef]
- Reed, T.T.; Pierce, W.M.; Markesbery, W.R.; Butterfield, D.A. Proteomic identification of HNE-bound proteins in early Alzheimer disease: Insights into the role of lipid peroxidation in the progression of AD. Brain Res. 2009, 1274, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Perluigi, M.; Sultana, R.; Cenini, G.; Di Domenico, F.; Memo, M.; Pierce, W.M.; Coccia, R.; Butterfield, D.A. Redox proteomics identification of 4-hydroxynonenal-modified brain proteins in Alzheimer’s disease: Role of lipid peroxidation in Alzheimer’s disease pathogenesis. Proteom. Clin. Appl. 2009, 3, 682–693. [Google Scholar] [CrossRef]
- Di Domenico, F.; Tramutola, A.; Butterfield, D.A. Role of 4-hydroxynonenal (HNE) in the pathogenesis of Alzheimer disease and other selected age-related neurodegenerative disorders. Free Radic. Biol. Med. 2017, 111, 253–261. [Google Scholar] [CrossRef]
- Pamplona, R.; Dalfó, E.; Ayala, V.; Bellmunt, M.J.; Prat, J.; Ferrer, I.; Portero-Otín, M. Proteins in human brain cortex are modified by oxidation, glycoxidation, and lipoxidation. Effects of Alzheimer disease and identification of lipoxidation targets. J. Biol. Chem. 2005, 280, 21522–21530. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Xi, Y.; Gao, M.; Li, Z.; Xu, C.; Fan, S.; He, W. Gene expression profiles of entorhinal cortex in Alzheimer’s disease. Am. J. Alzheimer’s Dis. Other Demen. 2014, 29, 526–532. [Google Scholar] [CrossRef]
- Chan, R.B.; Oliveira, T.G.; Cortes, E.P.; Honig, L.S.; Duff, K.E.; Small, S.A.; Wenk, M.R.; Shui, G.; Di Paulo, G. Comparative lipidomic analysis of mouse and human brain with Alzheimer disease. J.Biol. Chem. 2012, 287, 2678–2688. [Google Scholar] [CrossRef]
- Fabelo, N.; Martín, V.; Marín, R.; Moreno, D.; Ferrer, I.; Díaz, M. Altered lipid composition in cortical lipid rafts occurs at early stages of sporadic Alzheimer’s disease and facilitates APP/BACE1 interactions. Neurobiol. Aging 2014, 35, 1801–1812. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.H.; Oliver, D.M. Amyloid beta and phosphorylated tau-induced defective autophagy and mitophagy in Alzheimer’s disease. Cells 2019, 8, 488. [Google Scholar] [CrossRef] [PubMed]
- Manczak, M.; Calkins, M.J.; Reddy, P.H. Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer’s disease: Implications for neuronal damage. Hum. Mol. Genet. 2011, 20, 2495–2509. [Google Scholar] [CrossRef]
- Manczak, M.; Reddy, P.H. Abnormal interaction between the mitochondrial fission protein Drp1 and hyperphosphorylated tau in Alzheimer’s disease neurons: Implications for mitochondrial dysfunction and neuronal damage. Hum. Mol. Genet. 2012, 21, 2538–2547. [Google Scholar] [CrossRef]
- Reddy, P.H.; Williams, J.; Smith, F.; Bhatti, J.S.; Kumar, S.; Vijayan, M.; Kandimalla, R.; Kuruva, C.S.; Wang, R.; Manczak, M.; et al. MicroRNAs, aging, cellular senescence, and Alzheimer’s disease. Prog. Mol. Biol. Transl. Sci. 2017, 146, 127–171. [Google Scholar]
- Crespo-Biel, N.; Theunis, C.; Van Leuven, F. Protein tau: Prime cause of synaptic and neuronal degeneration in Alzheimer’s disease. Int. J. Alzheimer’s Dis. 2012, 2012, 251426. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Tammineni, P. Alterations in mitochondrial quality control in Alzheimer’s disease. Front. Cell. Neurosci. 2016, 10, 24. [Google Scholar] [CrossRef]
- Cai, Q.; Zakaria, H.M.; Simone, A.; Sheng, Z.-H. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr. Biol. 2012, 22, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Tammineni, P.; Ye, X.; Feng, T.; Aikal, D.; Cai, Q. Impaired retrograde transport of axonal autophagosomes contributes to autophagic stress in Alzheimer’s disease neurons. eLife 2017, e21776. [Google Scholar] [CrossRef]
- Nixon, R.A.; Yang, D.S. Autophagy failure in Alzheimer’s disease–locating the primary defect. Neurobiol. Dis. 2011, 43, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Chong, C.M.; Ke, M.; Tan, Y.; Huang, Z.; Zhang, K.; Ai, N.; Ge, W.; Qin, D.; Lu, J.H.; Su, H. Presenilin 1 deficiency suppresses autophagy in human neural stem cells through reducing γ-secretase-independent ERK/CREB signaling. Cell Death Dis. 2018, 9, 879. [Google Scholar] [CrossRef] [PubMed]
- Augusto-Oliveira, M.; Arrifano, G.P.; Lopes-Araújo, A.; Santos-Sacramento, L.; Takeda, P.Y.; Anthony, D.C.; Malva, J.O.; Crespo-Lopez, M.E. What do microglia really do in healthy adult brain? Cells 2019, 8, 1293. [Google Scholar] [CrossRef] [PubMed]
- Loughlin, A.J.; Woodroofe, M.N.; Cuzner, M.L. Regulation of Fc receptor and major histocompatibility complex antigen expression on isolated rat microglia by tumour necrosis factor, interleukin-1 and lipopolysaccharide: Effects on interferon-gamma induced activation. Immunology 1992, 75, 170–175. [Google Scholar] [PubMed]
- Song, X.; Shapiro, S.; Goldman, D.L.; Casadevall, A.; Scharff, M.; Lee, S.C. Fcgamma receptor I- and III-mediated macrophage inflammatory protein 1alpha induction in primary human and murine microglia. Infect. Immun. 2002, 70, 5177–5184. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.J.; El Khoury, J.; Medeiros, L.A.; Terada, K.; Geula, C.; Luster, A.D.; Freeman, M.W. A CD36-initiated signaling cascade mediates inflammatory effects of beta-amyloid. J. Biol. Chem. 2002, 277, 47373–47379. [Google Scholar] [CrossRef]
- Anderson, K.V. Toll signaling pathways in the innate immune response. Curr. Opin. Immunol. 2000, 12, 13–19. [Google Scholar] [CrossRef]
- Calvo-Rodriguez, M.; García-Rodríguez, C.; Villalobos, C.; Núñez, L. Role of Toll Like Receptor 4 in Alzheimer’s Disease. Front. Immunol. 2020, 11, 1588. [Google Scholar] [CrossRef]
- Blauwendraat, C.; Nalls, M.A.; Singleton, A.B. The genetic architecture of Parkinson’s disease. Lancet Neurol. 2020, 19, 170–178. [Google Scholar] [CrossRef]
- Langston, J.W.; Ballard, P.; Tetrud, J.W.; Irwin, I. Chronic parkinsonism in humans due to a product of meperidine-analog synthesis. Science 1983, 219, 979–980. [Google Scholar] [CrossRef] [PubMed]
- Betarbet, R.; Sherer, T.B.; MacKenzie, G.; Garcia-Osuna, M.; Panov, A.V.; Greenmayre, J.T. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat. Neurosci. 2000, 3, 1301–1306. [Google Scholar] [CrossRef] [PubMed]
- Mann, V.M.; Cooper, J.M.; Daniel, S.E.; Srai, K.; Jenner, P.; Marsden, C.D.; Schapira, A.H. Complex I, iron, and ferritin in Parkinson’s disease substantia nigra. Ann. Neurol. 1994, 36, 876–881. [Google Scholar] [CrossRef]
- Dawson, T.M.; Dawson, V.L. Molecular pathways of neurodegeneration in Parkinson’s disease. Science 2003, 302, 819–822. [Google Scholar] [CrossRef]
- Chinta, S.J.; Mallajosyula, J.K.; Rane, A.; Andersen, J.K. Mitochondrial α-synuclein accumulation impairs complex I function in dopaminergic neurons and results in increased mitophagy in vivo. Neurosci. Lett. 2010, 486, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Quing, H.; Zhang, Y.; Deng, Y.; McGeer, E.G.; McGeer, P.L. Lrrk2 intercation with α-synucleinin diffuse Lewy body disease. Biochem. Biophys. Res. Commun. 2009, 390, 1229–1234. [Google Scholar] [CrossRef]
- Thomas, K.J.; McCoy, M.K.; Blackinton, J.; Beilina, A.; Van der Brug, M.; Sandebring, A.; Miller, D.; Maric, D.; Cedazo-Minguez, A.; Cookson, M.R. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum. Mol. Genet. 2011, 20, 40–50. [Google Scholar] [CrossRef]
- Dalfo, E.; Ferrer, I. Early α-synuclein lipoxidation in neocortex in Lewy body diseases. Neurobiol. Aging 2008, 29, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Yoritaka, A.; Hattori, N.; Uchida, K.; Tanaka, M.; Stadtman, E.R.; Mizuno, Y. Immunohistochemical detection of 4-hydroxynonenal protein adducts in Parkinson disease. Proc. Natl. Acad. Sci. USA 1996, 93, 2696–2701. [Google Scholar] [CrossRef]
- Floor, E.; Wetzel, M.G. Increased protein oxidation in human substantia nigra pars compacta in comparison with basal ganglia and prefrontal coretx measured with an improved dinitrophenylhydrazine assay. J. Neurochem. 1998, 70, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Sofic, E.; Lange, K.W.; Jellinger, K.; Riederer, P. Reduced and oxidized glutathione in the substantia nigra of patients with Parkinson’s disease. Neurosci. Lett. 1992, 142, 128–130. [Google Scholar] [CrossRef]
- Muddapu, V.R.; Chakravarthy, V.S. Influence of energy deficiency on the subcellular processes of substantia nigra pars compacta cell for understanding parkinsonian neurodegeneration. Sci. Rep. 2021, 11, 1754. [Google Scholar] [CrossRef]
- Pavlin, M.; Repič, M.; Vianello, R.; Mavri, J. The chemistry of neurodegeneration: Kinetic data and their implications. Mol. Neurobiol. 2016, 53, 3400–3415. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, H.; Yasuda, T. Iron accumulation in Parkinson’s disease. J. Neural Transm. 2012, 119, 1511–1514. [Google Scholar] [CrossRef]
- Valente, E.M.; Abou-Sleiman, P.M.; Caputo, V.; Muqit, M.M.; Harvey, K.; Gispert, S.; Ali, Z.; Del Turco, D.; Bentivoglio, A.R.; Healy, D.G.; et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 2004, 304, 1158–1160. [Google Scholar] [CrossRef] [PubMed]
- Mortiboys, H.; Johansen, K.K.; Aasly, J.O.; Bandmann, O. Mitochondrial impairment in patients with Parkinson disease with the G2019S mutation in LRRK2. Neurology 2010, 75, 2017–2020. [Google Scholar] [CrossRef]
- Stafa, K.; Tsika, E.; Moser, R.; Musso, A.; Glauser, L.; Jones, A.; Biskup, S.; Xiong, Y.; Bandopadhyay, R.; Dawson, V.L.; et al. Functional interaction of Parkinson’s disease-associated LRRK2 with membres of the dynamin GTPase superfamily. Hum. Mol. Genet. 2014, 23, 2055–2077. [Google Scholar] [CrossRef]
- Zilocchi, M.; Finzi, G.; Lualdi, M.; Sessa, F.; Fasano, M.; Alberio, T. Mitochondrial alterations in Parkinson’s disease human samples and cellular models. Neurochem. Int. 2018, 118, 61–72. [Google Scholar] [CrossRef]
- Perez-Carrion, M.; Pischedda, F.; Biosa, A.; Russo, I.; Straniero, L.; Civiero, L.; Guida, M.; Gloeckner, C.J.; Ticozzi, N.; Tiloca, C.; et al. The LRRK2 variant E193K prevents mitochondrial fission upon MPP+ treatment by altering LRRK2 binding to DRP1. Front. Mol. Neurosci. 2018, 11, 64. [Google Scholar] [CrossRef]
- Devoto, P.V.M.; Dimopoulos, N.; Alloatti, M.; Pardi, M.B.; Saez, T.M.; Otero, M.G.; Cromberg, L.E.; Marín-Burgin, A.; Scassa, M.E.; Stokin, G.B.; et al. αSynuclein control of mitochondrial homeostasis in human-derived neurons is disrupted by mutations associated with Parkinson’s disease. Sci. Rep. 2017, 7, 5042. [Google Scholar] [CrossRef]
- Xie, W.; Chung, K.K.K. Alpha-synuclein impairs normal dynamics of mitochondria in cell and animal models of Parkinson’s disease. J. Neurochem. 2012, 122, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Melkov, A.; Abdu, U. Regulation of long-distance transport of mitochondria along microtubules. Cell. Mol. Life Sci. 2018, 75, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Herman, M.A.; Trimbuch, T.; Rosenmund, C. Differential pH dynamics in synaptic vesicles from intact glutamatergic and GABAergic synapses. Front. Synaptic Neurosci. 2018, 10, 44. [Google Scholar] [CrossRef]
- Umek, N.; Gerŝak, B.; Vintar, N.; Ŝoŝtarič, M.; Mavri, J. Dopamine autoxidation is controlled by acidic pH. Front. Mol. Neurosci. 2018, 11, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Borchelt, D.R.; Lee, M.K.; Slunt, H.S.; Guarnieri, M.; Xu, Z.-S.; Wong, P.C.; Brown, R.H.; Price, D.L.; Sisodia, S.S.; Cleveland, D.W. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc. Nat. Acad. Sci. USA 1994, 91, 8292–8296. [Google Scholar] [CrossRef]
- Mejzini, R.; Flynn, L.L.; Pitout, I.L.; Fletcher, S.; Wilton, S.D.; Akkari, P.A. ALS genetics, mechanisms, and therapeutics: Where are we now? Front. Neurosci. 2019, 13, 1310. [Google Scholar] [CrossRef] [PubMed]
- Browne, S.E.; Yang, L.; DiMauro, J.P.; Fuller, S.W.; Licata, S.C.; Beal, M.F. Bioenergetic abnormalities in discrete cerebral motor pathways presage spinal cord pathology in the G93A SOD1 mouse model of ALS. Neurobiol. Dis. 2006, 22, 599–610. [Google Scholar] [CrossRef]
- Hatazawa, J.; Brooks, R.A.; Dalakas, M.C.; Mansi, L.; Di Chiro, G. Cortical motor-sensory hypometabolism in amyotrophic lateral sclerosis: A PET study. J. Comput. Assist. Tomogr. 1988, 12, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Raman, R.; Allen, S.P.; Goodall, E.F.; Kramer, S.; Ponger, L.L.; Heath, P.R.; Milo, M.; Hollinger, H.C.; Walsh, T.; Highley, J.R.; et al. Gene expression signatures in motor neurone disease fibroblasts reveal dysregulation of metabolism, hypoxia-response and RNA processing functions. Neuropathol. Appl. Neurobiol. 2015, 41, 201–226. [Google Scholar] [CrossRef]
- Tefera, T.W.; Borges, K. Neuronal glucose metabolism is impaired while astrocytic TCA cycling is unaffected at symptomatic stages in the HSOD1G93A mouse model of amyotrophic lateral sclerosis. J. Cereb. Blood Flow Metab. 2019, 39, 1710–1724. [Google Scholar] [CrossRef]
- Diaz-Garcia, C.M.; Mongeon, R.; Lahmann, C.; Koveal, D.; Zucker, H.; Yellen, G. Neuronal stimulation triggers neuronal glycolysis and not lactate uptake. Cell Metab. 2017, 26, 361–374. [Google Scholar] [CrossRef]
- Obrador, E.; Salvador-Palmer, R.; López-Blanch, R.; Jihad-Jebbar, A.; Vallés, S.; Estrela, J.M. The link between oxidative stress, redox status, bioenergetics and mitochondria in the pathophysiology of ALS. Int. J. Mol. Sci. 2021, 22, 6352. [Google Scholar] [CrossRef]
- Maalouf, M.; Sullivan, P.G.; Davis, L.; Kim, D.Y.; Rho, J.M. Ketones inhibit mitochondrial production of reactive oxygen species production following glutamate excitotoxicity by increasing NADH oxidation. Neuroscience 2007, 71, 1102–1110. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.L. Could sirtuin activities modify ALS onset and progression? Cell. Mol. Neurobiol. 2017, 37, 1147–1160. [Google Scholar] [CrossRef]
- Siklós, L.; Engelhardt, J.I.; Alexianu, M.E.; Gurney, M.E.; Siddique, T.; Appel, S.H. Intracellular calcium parallels motoneuron degeneration in SOD-1 mutant mice. J. Neuropathol. Exp. Neurol. 1998, 57, 571–587. [Google Scholar] [CrossRef]
- Guatteo, E.; Carunchio, I.; Pieri, M.; Albo, F.; Canu, N.; Mercuri, N.B.; Zona, C. Altered calcium homeostasis in motor neurons following AMPA receptor but not voltage-dependent calcium channels’ activation in a genetic model of amyotrophic lateral sclerosis. Neurobiol. Dis. 2007, 28, 90–100. [Google Scholar] [CrossRef]
- Van den Bosch, L.; Vandenberghe, W.; Klaassen, H.; Van Houtte, E.; Robberecht, W. Ca(2+)-permeable AMPA receptors and selective vulnerability of motor neurons. J. Neurol. Sci. 2000, 180, 29–34. [Google Scholar] [CrossRef]
- Dafinca, R.; Barbagallo, P.; Farrimond, L.; Candalija, A.; Scaber, J.; Ababneh, N.A.; Sathyaprakash, C.; Vowles, J.; Cowley, S.A.; Talbot, K. Impairment of mitochondrial calcium buffering links mutations in C9ORF72 and TARDBP in iPS-derived motor neurons from patients with ALS/FTD. Stem Cell Rep. 2020, 14, 892–908. [Google Scholar] [CrossRef]
- Sasaki, S.; Iwata, M. Mitochondrial alterations in the spinal cord of patients with sporadic amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 2007, 66, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Padman, B.S.; Nguyen, T.N.; Uoselis, L.; Skulsuppaisarn, M.; Nguyen, L.K.; Lazarou, M. LC3/GABARAPs drive ubiquitin-independent recruitment of optineurin and NDP52 to amplify mitophagy. Nat. Commun. 2019, 10, 408. [Google Scholar] [CrossRef]
- Feng, S.-M.; Che, C.-H.; Feng, S.-Y.; Liu, C.-Y.; Li, L.-Y.; Li, Y.-X.; Huang, H.-P.; Zou, Z.-Y. Novel mutation in optineurin causing aggressive ALS+/- frontotemporal dementia. Ann. Clin. Transl. Neurol. 2019, 6, 2377–2383. [Google Scholar] [CrossRef]
- Babbar, M.; Sheikh, M.S. Metabolic stress and disorders related to alterations in mitochondrial fission or fusion. Mol. Cell Pharmacol. 2013, 5, 109–133. [Google Scholar]
- Lyons, T.J.; Liu, H.; Goto, J.J.; Nersissian, A.; Roe, J.A.; Graden, J.A.; Café, C.; Ellerby, L.M.; Bredesen, D.E.; Gralla, E.B.; et al. Mutations in copper-zinc superoxide dismutase that cause amyotrophic lateral sclerosis alter the zinc binding site and the redox behavior of the protein. Proc. Natl. Acad. Sci. USA 1996, 93, 12240–12244. [Google Scholar] [CrossRef]
- Frakes, A.E.; Ferraiuolo, L.; Haidet-Phillips, A.M.; Schmelzer, L.; Braun, L.; Miranda, C.J.; Ladner, K.J.; Bevan, A.K.; Foust, K.D.; Godbout, J.P.; et al. Microglia induce motor neuron death via the classical NF-ΚB pathway in amyotrophic lateral sclerosis. Neuron 2014, 81, 1009–1023. [Google Scholar] [CrossRef] [PubMed]
- Putcha, G.V.; Harris, C.A.; Moulder, K.L.; Easton, R.M.; Thompson, C.B.; Johnson, E.M. Intrinsic and extrinsic pathway signaling during neuronal apoptosis: Lessons from the analysis of mutant mice. J. Cell Biol. 2002, 157, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.J. P53 is abnormally elevated and active in the CNS of patients with amyotrophic lateral sclerosis. Neurobiol. Dis. 2000, 7, 613–622. [Google Scholar] [CrossRef]
- Kabashi, E.; Agar, J.N.; Taylor, D.M.; Minotti, S.; Durham, H.D. Focal dysfunction of the proteasome: A pathogenic factor in a mouse model of amyotrophic lateral sclerosis. J. Neurochem. 2004, 89, 1325–1335. [Google Scholar] [CrossRef]
- Ranganathan, S.; Bowser, R. P53 and cell cycle proteins participate in spinal motor neuron cell death in ALS. Open Pathol. J. 2010, 4, 11–22. [Google Scholar] [CrossRef]
- Mishra, P.-S.; Dhull, D.K.; Nalini, A.; Vijayalakshmi, K.; Sathyaprabha, T.N.; Alladi, P.A.; Raju, T.R. Astroglia acquires a toxic neuroinflammatory role in response to the cerebrospinal fluid from amyotrophic lateral sclerosis patients. J. Neuroinflamm. 2016, 13, 212. [Google Scholar] [CrossRef] [PubMed]
- Re, D.B.; Le Verche, V.; Yu, C.; Amoroso, M.W.; Politi, K.A.; Phani, S.; Ikiz, B.; Hoffmann, L.; Koolen, M.; Nagata, T.; et al. Necroptosis drives motor neuron death in models of both sporadic and familial ALS. Neuron 2014, 81, 1001–1008. [Google Scholar] [CrossRef]
- Vargas, M.R.; Johnson, J.A. Astrogliosis in amyotrophic lateral sclerosis: Role and therapeutic potential of astrocytes. Neurotherapeutics 2010, 7, 471–481. [Google Scholar] [CrossRef]
- Zhao, W.; Beers, D.R.; Bell, S.; Wang, J.; Wen, S.; Baloh, R.H.; Appel, S.H. TDP-43 activates microglia through NF-ΚB and NLRP3 inflammasome. Exp. Neurol. 2015, 273, 24–35. [Google Scholar] [CrossRef]
- Cho, D.-H.; Nakamura, T.; Fang, J.; Cieplak, P.; Godzik, A.; Gu, Z.; Lipton, S.A. S-nitrosylation of Drp1 mediates β-amyloid-related mitochondrial fission and neuronal injury. Science 2009, 324, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-C.; Qi, X. Inhibition of excessive mitochondrial fission reduced aberrant autophagy and neuronal damage caused by LRRK2 G2019S mutation. Hum. Mol. Genet. 2013, 22, 4545–4561. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yamashita, T.; Tian, F.; Morimoto, N.; Ikeda, Y.; Deguchi, K.; Abe, K. Mitochondrial fusion and fission proteins expression dynamically change in a murine model of amyotrophic lateral sclerosis. Curr. Neurovasc. Res. 2013, 10, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, X.; Fujioka, H.; Hoppel, C.; Whone, A.L.; Caldwell, M.A.; Cullen, P.J.; Liu, J.; Zhu, X. Parkinson’s disease-associated mutant VPS35 causes mitochondrial dysfunction by recycling DLP1 complexes. Nat. Med. 2016, 22, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-F.; Gendron, T.F.; Zhang, Y.-J.; Lin, W.-L.; D’Alton, S.; Sheng, H.; Castanedes Casey, M.; Tong, J.; Knight, J.; Yu, X.; et al. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J. Neurosci. 2010, 30, 10851–10859. [Google Scholar] [CrossRef]
- Tang, F.-L.; Liu, W.; Hu, J.-X.; Erion, J.R.; Ye, J.; Mei, L.; Xiong, W.-C. VPS35 deficiency or mutation causes dopaminergic neuronal loss by impairing mitochondrial fusion and function. Cell Rep. 2015, 12, 1631–1643. [Google Scholar] [CrossRef]
- Hsieh, C.-H.; Shaltouki, A.; Gonzalez, A.E.; Bettencourt da Cruz, A.; Burbulla, L.F.; St. Lawrence, E.; Schüle, B.; Krainc, D.; Palmer, T.D.; Wang, X. Functional impairment in Miro degradation and mitophagy is a shared feature in familial and sporadic Parkinson’s disease. Cell Stem Cell. 2016, 19, 709–724. [Google Scholar] [CrossRef] [PubMed]
- Combs, B.; Mueller, R.L.; Morfini, G.; Brady, S.T.; Kanaan, N.M. Tau and axonal transport misregulation in tauopathies. Adv. Exp. Med. Biol. 2019, 1184, 81–95. [Google Scholar]
- Shaltouki, A.; Hsieh, C.-H.; Kim, M.J.; Wang, X. Alpha-synuclein delays mitophagy and targeting Miro rescues neuron loss in Parkinson’s models. Acta Neuropathol. 2018, 136, 607–620. [Google Scholar] [CrossRef]
- Wong, Y.C.; Holzbaur, E.L.F. Optineurin is an autophagy receptor for damaged mitochondria in Parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. USA 2014, 111, E4439–E4448. [Google Scholar] [CrossRef]
- Evans, C.S.; Holzbaur, E.L.F. Autophagy and mitophagy in ALS. Neurobiol. Dis. 2019, 122, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Goode, A.; Butler, K.; Long, J.; Cavey, J.; Scott, D.; Shaw, B.; Sollenberger, J.; Gell, C.; Johansen, T.; Oldham, N.J.; et al. Defective recognition of LC3 by mutant SQSTM1/p62 implicates impairment of autophagy as a pathogenic mechanism in ALS-FTLD. Autophagy 2016, 12, 1094–1104. [Google Scholar] [CrossRef] [PubMed]
- Grenier, K.; McLelland, G.L.; Fon, E.A. Parkin- and PINK1-dependent mitophagy in neurons: Will the real pathway please stand up? Front. Neurol. 2013, 4, 100. [Google Scholar] [CrossRef]
- Pickrell, A.M.; Huang, C.-H.; Kennedy, S.R.; Ordureau, A.; Sideris, D.P.; Hoekstra, J.G.; Harper, J.W.; Youle, R.J. Endogenous Parkin preserves dopaminergic substantia nigra neurons following mitochondrial DNA mutagenic stress. Neuron 2015, 87, 371–381. [Google Scholar] [CrossRef]
- Amo, T.; Saiki, S.; Sawayama, T.; Sato, S.; Hattori, N. Detailed analysis of mitochondrial respiratory chain defects caused by loss of PINK1. Neurosci. Lett. 2014, 580, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Hanss, Z.; Larsen, S.B.; Antony, P.; Mencke, P.; Massart, F.; Jarazo, J.; Schwamborn, J.C.; Barbuti, P.A.; Mellick, G.D.; Krüger, R. Mitochondrial and clearance impairment in p.D620N VPS35 patient-derived neurons. Mov. Disord. 2021, 36, 704–715. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Nguyen, T.T.D.; Nguyen, T.K.O.; Vo, T.K.; Vo, V.G. Advances in developing therapeutic strategies in Alzheimer’s disease. Biomed. Pharmacother. 2021, 139, 111623. [Google Scholar] [CrossRef]
- Dunn, B.; Stein, P.; Cavazzoni, P. Approval of Aducanumab for Alzheimer’s disease–the FDA’s perspective. JAMA Intern. Med. 2021, 181, 1276–1278. [Google Scholar] [CrossRef] [PubMed]
- Menon, V.P.; Sudheer, A.R. Antioxidant and anti-inflammatory properties of curcumin. Adv. Exp. Med. Biol. 2007, 595, 105–125. [Google Scholar]
- Lin, Y.G.; Kunnumakkara, A.B.; Nair, A.; Merritt, W.M.; Han, L.Y.; Armaiz-Pena, G.M.; Kamat, A.A.; Spannuth, W.A.; Gershenson, D.M.; Lutgendorf, S.K. Curcumin inhibits tumor growth and angiogenesis in ovarian carcinoma by targeting the nuclear factor-κB pathway. Clin. Cancer Res. 2007, 13, 3423–3430. [Google Scholar] [CrossRef] [PubMed]
- Marchiani, A.; Rozzo, C.; Fadda, A.; Delogu, G.; Ruzza, P. Curcumin and curcumin-like molecules: From spice to drugs. Curr. Med. Chem. 2014, 21, 204–222. [Google Scholar] [CrossRef]
- Nuzzo, D.; Amato, A.; Picone, P.; Terzo, S.; Galizzi, G.; Bonina, F.B.; Mulé, F.; Di Carlo, M. A natural dietary supplement with a combination of nutrients prevents neurodegeneration induced by a high fat diet in mice. Nutrients 2018, 10, 1130. [Google Scholar] [CrossRef]
- Ringman, J.M.; Frautschy, S.A.; Teng, E.; Begum, A.N.; Bardens, J.; Beige, M.; Gylys, K.H.; Badmaev, V.; Heath, D.D.; Apostolova, L.G.; et al. Oral curcumin for Alzheimer’s disease: Tolerability and efficacy in a 24-week randomized, double blind, placebo-controlled study. Alzheimer’s Res. Ther. 2012, 4, 43. [Google Scholar] [CrossRef]
- Available online: https://www.clinicaltrials.gov (accessed on 3 October 2021).
- Hannan, M.A.; Dash, R.; Sohag, A.M.; Haque, N. Neuroprotection against oxidative stress: Phytochemicals targeting TrkB signaling and the Nrf2-ARE antioxidant system. Front. Mol. Neurosci. 2020, 13, 116. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.L.; Yang, J.Y.; Dong, Y.X.; Wang, J.M.; Cui, Y.H.; Ikeshima, T.; Zhao, Y.Q.; Wu, C.F. Resveratrol inhibits nitric oxide and TNF-alpha production by lipopolysaccharide-activated microglia. Int. Immunopharmacol. 2005, 5, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.W.; Grossman, H.; Neugroschl, J.; Parker, S.; Burden, A.; Luo, X.; Sano, M. A randomized, double-blind, placebo-controlled trial of resveratrol with glucose and malate (RGM) to slow the progression of Alzheimer’s disease: A pilot study. Alzheimers Dement. 2018, 4, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.; Ullah, H.; Aschner, M.; Cheang, W.S.; Akkol, E.K. Neuroprotective effects of quercetin in Alzheimer’s disease. Biomolecules 2020, 10, 59. [Google Scholar] [CrossRef]
- Costa, L.G.; Garrick, J.M.; Roque, P.J.; Pellacani, G. Mechanisms of neuroprotection by quercetin: Counteracting oxidative stress and more. Oxid. Med. Cell Longev. 2016, 2016, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, R.; Zhan, Z.; Li, X.; Zhou, F.; Xing, A.; Jiang, C.; Chen, Y.; An, L. Beneficial effects of sulforaphane treatment in Alzheimer’s disease may be mediated through reduced HDAC1/3 and increased p75NTR expression. Front. Aging Neurosci. 2017, 9, 121. [Google Scholar] [CrossRef]
- Gleason, C.E.; Carlsson, C.M.; Barnet, J.H.; Meade, S.A.; Setchell, K.D.; Atwood, C.S.; Johnson, S.C.; Ries, M.L.; Asthana, S. A preliminary study of the safety, feasibility and cognitive efficacy of soy isoflavone supplements in older men and women. Age Ageing 2009, 38, 86–93. [Google Scholar] [CrossRef]
- Wu, P.-S.; Ding, H.-Y.; Yen, J.-H.; Chen, S.-F.; Lee, K.-H.; Wu, M.-J. Anti-inflammatory activity of 8-hydroxydaidzein in LPS-stimulated BV2 microglial cells via activation of the Nrf2-antioxidant and attenuation of Akt/NF-κB-inflammatory signaling pathways, as well as inhibition of COX-2 activity. J. Agric. Food Chem. 2018, 66, 5790–5801. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.S.; Jang, Y.J.; Hwang, M.K.; Kang, N.J.; Lee, K.W.; Lee, H.J. Attenuation of oxidative neuronal cell death by coffee phenolic phytochemicals. Mutat. Res. 2009, 661, 18–24. [Google Scholar] [CrossRef]
- Ishida, K.; Yamamoto, M.; Misawa, K.; Nishimura, H.; Misawa, K.; Ota, N.; Shimotoyodome, A. Coffee polyphenols prevent cognitive dysfunction and suppress amyloid β plaques in APP/PS2 transgenic mouse. Neurosci. Res. 2020, 154, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.A.; Hughes, D.A. Alpha-lipoic acid modulates NF-kappaB activity in human monocytic cells by direct interaction with DNA. Exp. Gerontol. 2002, 37, 401–410. [Google Scholar] [CrossRef]
- Packer, L.; Roy, S.; Sen, C.K. Alpha-lipoic acid: A metabolic antioxidant and potential redox modulator of transcription. Adv. Pharmacol. 1997, 38, 79–101. [Google Scholar]
- Shinto, L.; Quinn, J.; Montine, T.; Dodge, H.H.; Woodward, W.; Baldauf-Wagner, S.; Waichunas, D.; Bumgarner, L.; Bourdette, D.; Silbert, L.; et al. A randomized placebo-controlled pilot trial of omega-3 fatty acids and alpha lipoic acid in Alzheimer’s disease. J. Alzheimer’s Dis. 2014, 38, 111–210. [Google Scholar] [CrossRef]
- Galasko, D.R.; Peskind, E.; Clark, C.M.; Quinn, J.F.; Ringman, J.M.; Jicha, G.A.; Cotman, C.; Cottrell, B.; Montine, T.J.; Thomas, R.G.; et al. Alzheimer’s Disease Cooperative Study. Antioxidants for Alzheimer disease: A randomized clinical trial with cerebrospinal fluid biomarker measures. Arch. Neurol. 2012, 69, 836–841. [Google Scholar] [CrossRef]
- Kryscio, R.J.; Abner, E.L.; Caban-Holt, A.; Lovell, M.; Goodman, P.; Darke, A.K.; Yee, M.; Crowley, J.; Schmitt, F.A. Association of antioxidant supplement use and dementia in the Prevention of Alzheimer’s Disease by Vitamin E and Selenium Trial (PREADViSE). JAMA Neurol. 2017, 74, 567–573. [Google Scholar] [CrossRef]
- Dysken, M.W.; Sano, M.; Asthana, S.; Vertrees, J.E.; Pallaki, M.; Llorente, M.; Love, S.; Schellenberg, G.D.; McCarten, J.R.; Malphurs, J.; et al. Effect of vitamin E and memantine on functional decline in Alzheimer disease: The TEAM-AD VA cooperative randomized trial. J. Am. Med Assoc. 2014, 311, 33–44. [Google Scholar] [CrossRef]
- Bachurin, S.; Shevtsova, E.; Kireeva, E.; Oxenkrug, G.; Sablin, S. Mitochondria as a target for neurotoxins and neuroprotective agents. Ann. N. Y. Acad. Sci. 2003, 993, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Doody, R.S.; Gavrilova, S.I.; Sano, M.; Thomas, R.G.; Aisen, P.S.; Bachurin, S.O.; Seely, L.; Hung, D.; Dimebon Investigators. Effect of dimebon on cognition, activities of daily living, behaviour, and global function in patients with mild-to-moderate Alzheimer’s disease: A randomised, double-blind, placebo-controlled study. Lancet 2008, 372, 207–215. [Google Scholar] [CrossRef]
- Jones, R.W. Dimebon disappointment. Alzheime. Res Ther. 2010, 2, 25. [Google Scholar] [CrossRef][Green Version]
- Chau, S.; Herrmann, N.; Ruthirakuhan, M.T.; Chen, J.J.; Lanctôt, K.L. Latrepirdine for Alzheimer’s disease. Cochrane Database Syst. Rev. 2015, 4, CD009524. [Google Scholar] [CrossRef]
- Chang, K.-S.; Chen, C.-M. The role of oxidative stress in Parkinson’s disease. Antioxidants 2020, 9, 597. [Google Scholar] [CrossRef]
- Barzilai, A.; Melamed, E.; Shirvan, A. Is there a rationale for neuroprotection against dopamine toxicity in Parkinson’s disease? Cell. Mol. Neurobiol. 2001, 21, 215–235. [Google Scholar] [CrossRef]
- Stoker, T.B.; Barker, R.A. Recent developments in the treatment of Parkinson’s Disease. F1000Res. 2020, 9, F1000 Faculty Rev-862. [Google Scholar] [CrossRef] [PubMed]
- Rae, C.D.; Broer, S. Creatine as a booster for human brain function. How might it work? Neurochem. Int. 2015, 89, 249–259. [Google Scholar] [CrossRef]
- Bender, A.; Koch, W.; Elstner, M.; Schombacher, Y.; Bender, J.; Moeschl, M.; Gekeler, F.; Muller-Myhsok, B.; Gasser, T.; Tatsch, K.; et al. Creatine supplementation in Parkinson disease: A placebo-controlled randomized pilot trial. Neurology 2006, 67, 1262–1264. [Google Scholar] [CrossRef] [PubMed]
- The NINDS-NET PD Investigators. A randomized, double-blind, futility clinical trial of creatine and minocycline in early Parkinson’s disease. Neurology 2006, 67, 1262–1264. [Google Scholar]
- Kieburtz, K.; Tilley, B.C.; Elm, J.J.; Babcock, D.; Hauser, R.; Ross, G.W.; Augustine, A.H.; Augustine, E.U.; Aminoff, M.J.; Writing Group for the NINDS Exploratory Trials in Parkinson Disease (NET-PD) Investigators; et al. Effect of creatine monohydrate on clinical progression in patients with Parkinson disease: A randomized clinical trial. J. Am. Med Assoc. 2015, 313, 584–593. [Google Scholar] [CrossRef]
- De Rijk, M.C.; Breteler, M.M.; Den Breeijen, J.H.; Launer, L.J.; Grobbee, D.E.; Van der Meche, F.G.; Hofman, A. Dietary antioxidants and Parkinson disease. The Rotterdam Study. Arch. Neurol. 1997, 54, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.M.; Hernán, M.A.; Chen, H.; Spiegelman, D.; Willett, W.C.; Ascherio, A. Intakes of vitamins E and C, carotenoids, vitamin supplements, and PD risk. Neurology 2002, 59, 1161–1169. [Google Scholar] [CrossRef]
- Fahn, S. A pilot trial of high-dose alpha-tocopherol and ascorbate in early Parkinson’s disease. Ann. Neurol. 1992, 32, S128–S132. [Google Scholar] [CrossRef] [PubMed]
- The Parkinson Study Group. Effects of tocopherol and deprenyl on the progression of disability in early Parkinson’s disease. N. Engl. J. Med. 1993, 328, 176–183. [Google Scholar] [CrossRef]
- Shults, C.W.; Oakes, D.; Kieburtz, K.; Beal, M.F.; Haas, R.; Plumb, S.; Juncos, J.L.; Nutt, J.; Shoulson, I.; Carter, J.; et al. Effects of coenzyme Q10 in early Parkinson disease: Evidence of slowing of the functional decline. Arch. Neurol. 2002, 59, 1541–1550. [Google Scholar] [CrossRef] [PubMed]
- Beal, M.F.; Oakes, D.; Shoulson, I.; Henchcliffe, C.; Galpern, W.R.; Haas, R.; Juncos, J.L.; Nutt, J.G.; Voss, T.S.; Parkinson Study Group QE3 Investigators; et al. A randomized clinical trial of high-dosage coenzyme Q10 in early Parkinson disease: No evidence of benefit. JAMA Neurol. 2014, 71, 543–552. [Google Scholar] [PubMed]
- Storch, A.; Jost, W.H.; Vieregge, P.; Spiegel, J.; Greulich, W.; Durner, J.; Muller, T.; Kupsch, A.; Henningsen, H.; Oertel, W.H.; et al. Randomized, double-blind, placebo-controlled trial on symptomatic effects of coenzyme Q10 in Parkinson disease. Arch. Neurol. 2007, 64, 938–944. [Google Scholar] [CrossRef]
- Snow, B.J.; Rolfe, F.L.; Lockhart, M.M.; Frampton, C.M.; O’Sullivan, J.D.; Fung, V.; Smith, R.A.; Murphy, M.P.; Taylor, K.M.; Protect Study. A double-blind, placebo-controlled study to assess the mitochondria-targeted antioxidant MitoQ as a disease-modifying therapy in Parkinson’s disease. Mov. Disord. 2010, 25, 1670–1674. [Google Scholar] [CrossRef]
- Sechi, G.; Deledda, M.G.; Bua, G.; Satta, W.M.; Deiana, G.A.; Pes, G.M.; Rosati, G. Reduced intravenous glutathione in the treatment of early Parkinson’s disease. Prog. Neuropsychopharmacol. Biol. Psychiatry 1996, 20, 1159–1170. [Google Scholar] [CrossRef]
- Hauser, R.A.; Lyons, K.E.; McClain, T.; Carter, S.; Perlmutter, D. Randomized, double-blind, pilot evaluation of intravenous glutathione in Parkinson’s disease. Mov. Disord. 2009, 24, 979–983. [Google Scholar] [CrossRef]
- Mischley, L.K.; Lau, R.C.; Shankland, E.G.; Wilbur, T.K.; Padowski, J.M. Phase IIb study of intranasal glutathione in Parkinson’s disease. J. Parkinsons Dis. 2017, 7, 289–299. [Google Scholar] [CrossRef]
- Jurcau, A. The role of natural antioxidants in the prevention of dementia–where do we stand and future perspectives. Nutrients 2021, 13, 282. [Google Scholar] [CrossRef]
- Jurcau, A.; Nunkoo, V.S. Clinical markers may identify patients at risk for early Parkinson’s disease dementia: A prospective study. Am. J. Alzheime. Dis. Other Demen. 2021, 36, 15333175211021369. [Google Scholar] [CrossRef]
- Medeiros, C.A.; Carvalhedo de Bruin, P.F.; Lopes, L.A.; Magalhaes, M.C.; De Lourdes Seabra, M.; De Bruin, V.M. Effect of exogenous melatonin on sleep and motor dysfunction in Parkinson’s disease. A randomized, double blind, placebo-controlled study. J. Neurol. 2007, 254, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Martin-Bastida, A.; Ward, R.J.; Newbould, R.; Piccini, P.; Sharp, D.; Kabba, C.; Patel, M.C.; Spino, M.; Connelly, J.; Tricta, F.; et al. Brain iron chelation by deferiprone in a phase 2 randomised double-blinded placebo controlled clinical trial in Parkinson’s disease. Sci. Rep. 2017, 7, 1398. [Google Scholar] [CrossRef] [PubMed]
- Devos, D.; Moreau, C.; Devedjian, J.C.; Kluza, J.; Petrault, M.; Laloux, C.; Jonneaux, A.; Ryckewaert, G.; Garcon, G.; Rouaix, N.; et al. Targeting chelatable iron as a therapeutic modality in Parkinson’s disease. Antioxid. Redox Signal. 2014, 21, 195–210. [Google Scholar] [CrossRef] [PubMed]
- NINDS Exploratory Trials in Parkinson Disease (NET-PD) FS-ZONE Investigators. Pioglitazone in early Parkinson’s disease: A phase 2, multicentre, double-blind, randomised trial. Lancet Neurol. 2015, 14, 795–803. [Google Scholar] [CrossRef]
- Lacomblez, L.; Bensimon, G.; Leigh, P.N.; Guillet, P.; Meininger, V. Dose-ranging study of riluzole in amyotrophic lateral sclerosis. Amyotrophic Lateral Sclerosis/Riluzole Study Group II. Lancet 1996, 347, 1425–1431. [Google Scholar] [CrossRef]
- Cho, H.; Shukla, S. Role of edaravone as a treatment option for patients with amyotrophic lateral sclerosis. Pharmaceuticals 2020, 14, 29. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Tanaka, M.; Yuki, S.; Hirai, M.; Yamamoto, Y. How is edaravone effective against acute ischemic stroke and amyotrophic lateral sclerosis? J. Clin. Biochem. Nutr. 2018, 62, 20–38. [Google Scholar] [CrossRef]
- Ramis, M.R.; Esteban, S.; Miralles, A.; Tan, D.-X.; Reiter, R.J. Protective effects of melatonin and mitochondria-targeted antioxidants against oxidative stress: A review. Curr. Med. Chem. 2015, 22, 2690–2711. [Google Scholar] [CrossRef]
- Bald, E.M.; Nance, C.S.; Schultz, J.L. Melatonin may slow disease progression in amyotrophic lateral sclerosis: Findings from the pooled resource open-access ALS clinic trials database. Muscle Nerve 2021, 63, 572–576. [Google Scholar] [CrossRef]
- Pattee, G.L.; Post, G.R.; Gerber, R.E.; Bennett, J.P. Reduction of oxidative stress in amyotrophic lateral sclerosis following pramipexole treatment. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. 2003, 4, 90–95. [Google Scholar] [CrossRef]
- Izumi, Y.; Sawada, H.; Yamamoto, N.; Kume, T.; Katsuki, H.; Shimohama, S.; Akaike, A. Novel neuroprotective mechanisms of pramipexole, an anti-Parkinson drug, against endogenous dopamine-mediated excitotoxicity. Eur. J. Pharmacol. 2007, 557, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Cudkowicz, M.E.; Van den Berg, L.H.; Shefner, J.M.; Mitsumoto, H.; Mora, J.S.; Ludolph, A.; Hardiman, O.; Bozik, M.E.; Ingersoll, E.W.; Archibald, D.; et al. Dexpramipexole versus placebo for patients with amyotrophic lateral sclerosis (EMPOWER): A randomised, double-blind, phase 3 trial. Lancet Neurol. 2013, 12, 1059–1067. [Google Scholar] [CrossRef]
- Cudkowicz, M.; Bozik, M.E.; Ingersoll, E.W.; Miller, R.; Mitsumoto, H.; Shefner, J.; Moore, D.H.; Schoenfeld, D.; Mather, J.L.; Archibald, D.; et al. The effects of dexpramipexole (KNS-760704) in individuals with amyotrophic lateral sclerosis. Nat. Med. 2011, 17, 1652–1656. [Google Scholar] [CrossRef] [PubMed]
- Macchi, Z.; Wang, Y.; Moore, D.; Katz, J.; Saperstein, D.; Walk, D.; Simpson, E.; Genge, A.; Bertorini, T.; Fernandes, J.A.; et al. Western ALS (WALS) Rasagiline Study Group. A multi-center screening trial of rasagiline in patients with amyotrophic lateral sclerosis: Possible mitochondrial biomarker target engagement. Amyotroph. Lateral Scler. Front. Degener. 2015, 16, 345–352. [Google Scholar] [CrossRef]
- Statland, J.M.; Moore, D.; Wang, Y.; Walsh, M.; Mozaffar, T.; Elman, L.; Nations, S.P.; Mitsumoto, H.; Fernandes, J.A.; Saperstein, D.; et al. Rasagiline Investigators of the Muscle Study Group and Western ALS Consortium. Rasagiline for amyotrophic lateral sclerosis: A randomized, controlled trial. Muscle Nerve 2019, 59, 201–207. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).