The Impact of NLRP3 Activation on Hematopoietic Stem Cell Transplantation
Abstract
1. Introduction
2. NLRP3 in Hematopoiesis and Leukemogenesis
3. Role of NLRP3 in TRM and Infection Complications after HSCT
4. Role of NLRP3 in the Development of GvHD
5. NLRP3 and Disease Relapse
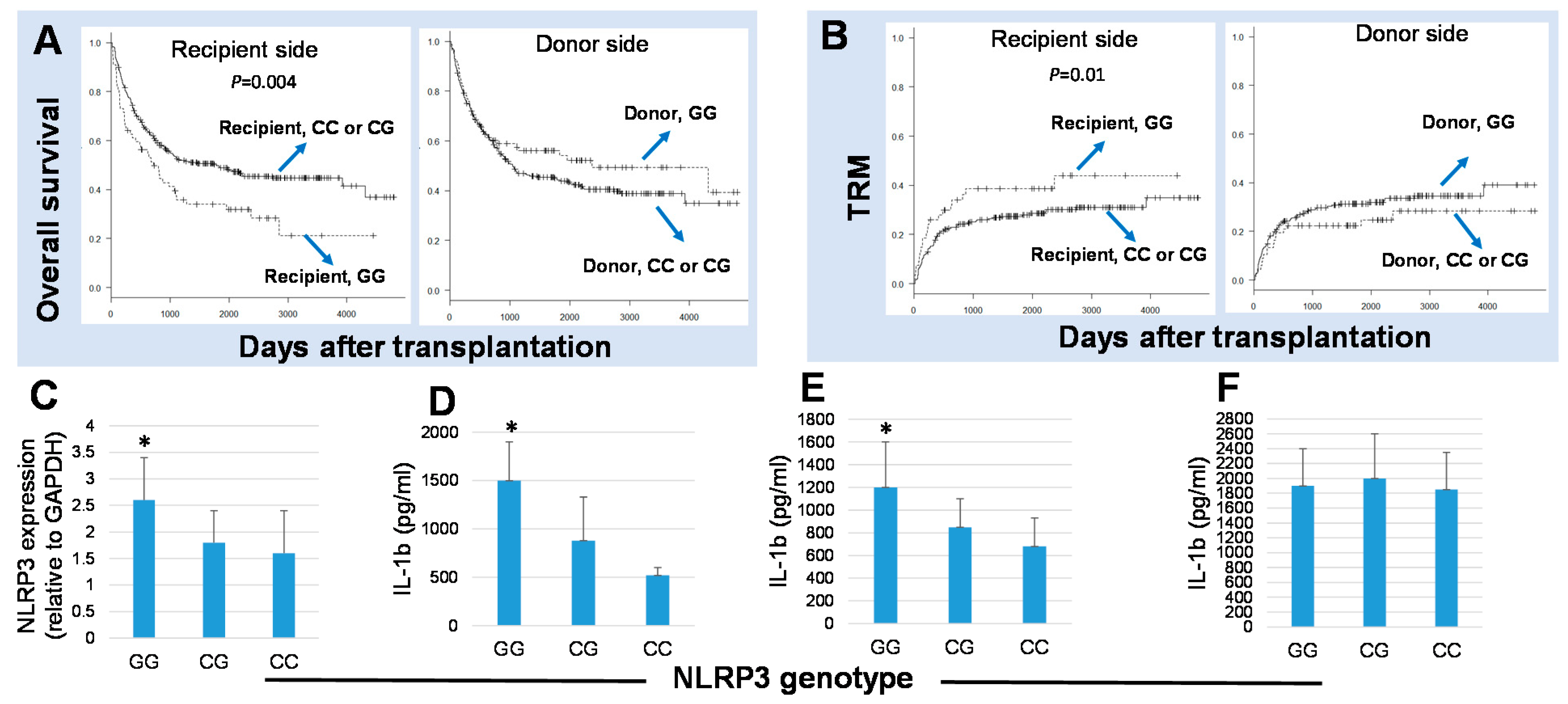
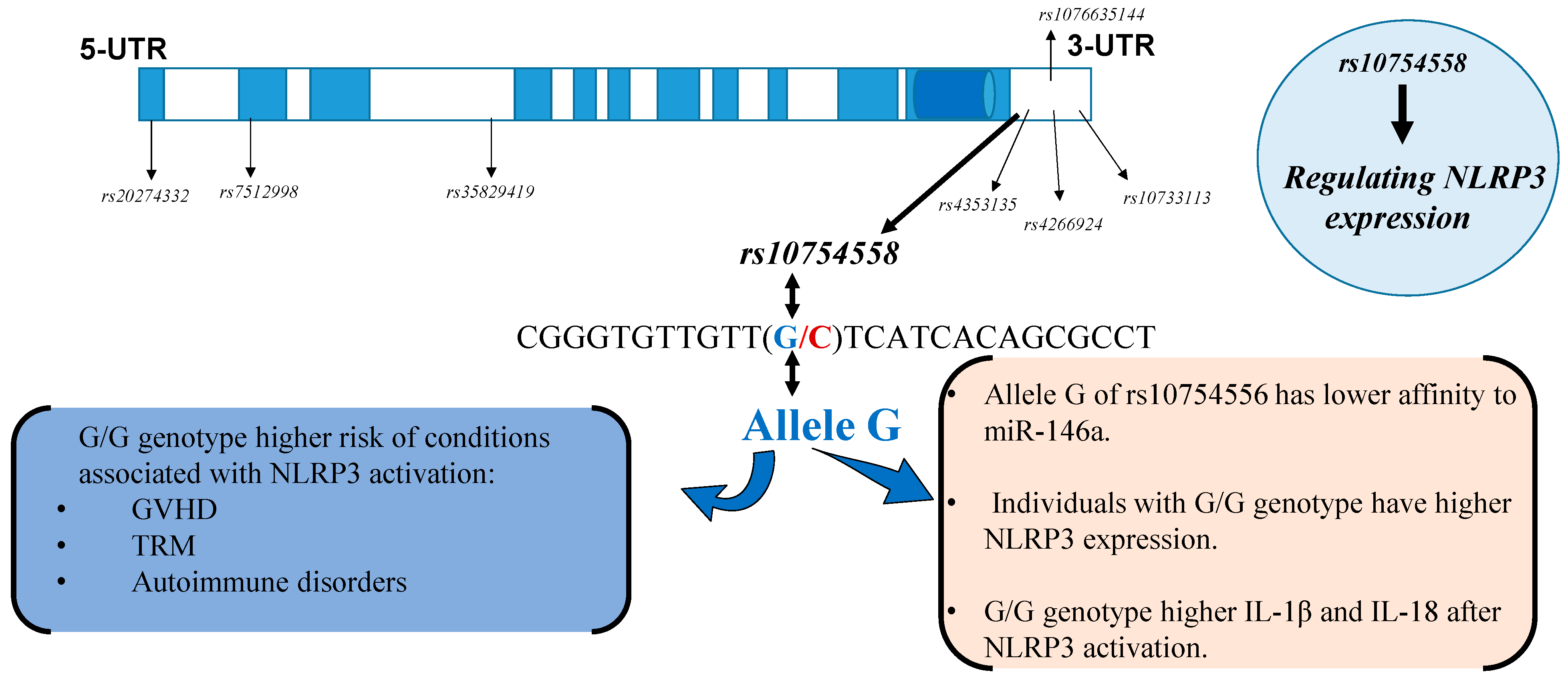
6. The Effects in Polymorphisms in NLRP3 Expression and Function
7. Concluding Remarks and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Espinoza, J.L.; Wadasaki, Y.; Takami, A. Infection Complications in Hematopoietic Stem Cells Transplant Recipients: Do Genetics Really Matter? Front. Microbiol 2018, 9, 2317. [Google Scholar] [CrossRef]
- Gratwohl, A.; Baldomero, H.; Aljurf, M.; Pasquini, M.C.; Bouzas, L.F.; Yoshimi, A.; Szer, J.; Lipton, J.; Schwendener, A.; Gratwohl, M.; et al. Hematopoietic stem cell transplantation: A global perspective. JAMA 2010, 303, 1617–1624. [Google Scholar] [CrossRef]
- Gratwohl, A.; Pasquini, M.C.; Aljurf, M.; Atsuta, Y.; Baldomero, H.; Foeken, L.; Gratwohl, M.; Bouzas, L.F.; Confer, D.; Frauendorfer, K.; et al. One million haemopoietic stem-cell transplants: A retrospective observational study. Lancet Haematol. 2015, 2, e91–e100. [Google Scholar] [CrossRef]
- Styczyński, J.; Tridello, G.; Koster, L.; Iacobelli, S.; van Biezen, A.; van der Werf, S.; Mikulska, M.; Gil, L.; Cordonnier, C.; Ljungman, P.; et al. Death after hematopoietic stem cell transplantation: Changes over calendar year time, infections and associated factors. Bone Marrow Transplant. 2020, 55, 126–136. [Google Scholar] [CrossRef]
- Niederwieser, D.; Baldomero, H.; Szer, J.; Gratwohl, M.; Aljurf, M.; Atsuta, Y.; Bouzas, L.F.; Confer, D.; Greinix, H.; Horowitz, M.; et al. Hematopoietic stem cell transplantation activity worldwide in 2012 and a SWOT analysis of the Worldwide Network for Blood and Marrow Transplantation Group including the global survey. Bone Marrow Transplant. 2016, 51, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, J.L. Machine learning for tackling microbiota data and infection complications in immunocompromised patients with cancer. J. Intern. Med. 2018, 284, 189–192. [Google Scholar] [CrossRef] [PubMed]
- Andermann, T.M.; Rezvani, A.; Bhatt, A.S. Microbiota Manipulation with Prebiotics and Probiotics in Patients Undergoing Stem Cell Transplantation. Curr. Hematol. Malig. Rep. 2016, 11, 19–28. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, W.; Zhou, R. NLRP3 inflammasome activation and cell death. Cell Mol. Immunol. 2021, 18, 2114–2127. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, M.Z.; Kucia, M. The Nlrp3 inflammasome—The evolving story of its positive and negative effects on hematopoiesis. Curr. Opin. Hematol. 2021, 28, 251–261. [Google Scholar] [CrossRef]
- Church, L.D.; Cook, G.P.; McDermott, M.F. Primer: Inflammasomes and interleukin 1beta in inflammatory disorders. Nat. Clin. Pract. Rheumatol. 2008, 4, 34–42. [Google Scholar] [CrossRef]
- Guo, H.; Callaway, J.B.; Ting, J.P. Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat. Med. 2015, 21, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, M.Z.; Bujko, K.; Cymer, M.; Thapa, A.; Adamiak, M.; Ratajczak, J.; Abdel-Latif, A.K.; Kucia, M. The Nlrp3 inflammasome as a “rising star” in studies of normal and malignant hematopoiesis. Leukemia 2020, 34, 1512–1523. [Google Scholar] [CrossRef]
- Bai, B.; Yang, Y.; Wang, Q.; Li, M.; Tian, C.; Liu, Y.; Aung, L.H.H.; Li, P.F.; Yu, T.; Chu, X.M. NLRP3 inflammasome in endothelial dysfunction. Cell Death Dis. 2020, 11, 776. [Google Scholar] [CrossRef] [PubMed]
- Hatscher, L.; Amon, L.; Heger, L.; Dudziak, D. Inflammasomes in dendritic cells: Friend or foe? Immunol. Lett. 2021, 234, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Guarda, G.; Zenger, M.; Yazdi, A.S.; Schroder, K.; Ferrero, I.; Menu, P.; Tardivel, A.; Mattmann, C.; Tschopp, J. Differential expression of NLRP3 among hematopoietic cells. J. Immunol. 2011, 186, 2529–2534. [Google Scholar] [CrossRef] [PubMed]
- Adamiak, M.; Abdel-Latif, A.; Bujko, K.; Thapa, A.; Anusz, K.; Tracz, M.; Brzezniakiewicz-Janus, K.; Ratajczak, J.; Kucia, M.; Ratajczak, M.Z. Nlrp3 Inflammasome Signaling Regulates the Homing and Engraftment of Hematopoietic Stem Cells (HSPCs) by Enhancing Incorporation of CXCR4 Receptor into Membrane Lipid Rafts. Stem Cell Rev. Rep. 2020, 16, 954–967. [Google Scholar] [CrossRef]
- Yang, L.; Hu, M.; Lu, Y.; Han, S.; Wang, J. Inflammasomes and the Maintenance of Hematopoietic Homeostasis: New Perspectives and Opportunities. Molecules 2021, 26, 309. [Google Scholar] [CrossRef]
- Price, E.A. Aging and erythropoiesis: Current state of knowledge. Blood Cells Mol. Dis. 2008, 41, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Wang, R.; Hua, M.; Zhang, C.; Han, F.; Xu, M.; Yang, X.; Li, G.; Hu, X.; Sun, T.; et al. NLRP3 Inflammasome Promotes the Progression of Acute Myeloid Leukemia. Front. Immunol. 2021, 12, 661939. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Wu, Y.; Wen, X.; Li, P.; Lu, F.; Shang, H. Chronic stress promotes acute myeloid leukemia progression through HMGB1/NLRP3/IL-1β signaling pathway. J. Mol. Med. 2021, 99, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Hua, M.; Zhang, C.; Wang, R.; Liu, J.; Yang, X.; Han, F.; Hou, M.; Ma, D. NLRP3-activated bone marrow dendritic cells play antileukemic roles via IL-1β/Th1/IFN-γ in acute myeloid leukemia. Cancer Lett. 2021, 520, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Toprak, S.K.; Atilla, P.A.; Atilla, E.; Demirer, T. An overview of infectious complications after allogeneic hematopoietic stem cell transplantation. J. Infect. Chemother. 2016, 22, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Mikulska, M.; Raiola, A.M.; Galaverna, F.; Balletto, E.; Borghesi, M.L.; Varaldo, R.; Gualandi, F.; Giannoni, L.; Pastori, G.; Giacobbe, D.R.; et al. Pre-Engraftment Bloodstream Infections after Allogeneic Hematopoietic Cell Transplantation: Impact of T Cell-Replete Transplantation from a Haploidentical Donor. Biol. Blood Marrow Transplant. 2018, 24, 109–118. [Google Scholar] [CrossRef]
- Cho, S.Y.; Lee, H.J.; Lee, D.G. Infectious complications after hematopoietic stem cell transplantation: Current status and future perspectives in Korea. Korean J. Intern. Med. 2018, 33, 256–276. [Google Scholar] [CrossRef]
- Gudiol, C.; Garcia-Vidal, C.; Arnan, M.; Sánchez-Ortega, I.; Patiño, B.; Duarte, R.; Carratalà, J. Etiology, clinical features and outcomes of pre-engraftment and post-engraftment bloodstream infection in hematopoietic SCT recipients. Bone Marrow Transplant. 2014, 49, 824–830. [Google Scholar] [CrossRef]
- da Costa, L.S.; Outlioua, A.; Anginot, A.; Akarid, K.; Arnoult, D. RNA viruses promote activation of the NLRP3 inflammasome through cytopathogenic effect-induced potassium efflux. Cell Death Dis. 2019, 10, 346. [Google Scholar] [CrossRef]
- He, W.T.; Wan, H.; Hu, L.; Chen, P.; Wang, X.; Huang, Z.; Yang, Z.H.; Zhong, C.Q.; Han, J. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 2015, 25, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhao, W. NLRP3 Inflammasome-A Key Player in Antiviral Responses. Front. Immunol. 2020, 11, 211. [Google Scholar] [CrossRef]
- Tartey, S.; Kanneganti, T.D. Differential role of the NLRP3 inflammasome in infection and tumorigenesis. Immunology 2019, 156, 329–338. [Google Scholar] [CrossRef]
- Ng, J.; Hirota, S.A.; Gross, O.; Li, Y.; Ulke-Lemee, A.; Potentier, M.S.; Schenck, L.P.; Vilaysane, A.; Seamone, M.E.; Feng, H.; et al. Clostridium difficile toxin-induced inflammation and intestinal injury are mediated by the inflammasome. Gastroenterology 2010, 139, 542–552; discussion 552.e1–552.e3. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Fernandes-Alnemri, T.; Alnemri, E.S. Involvement of the AIM2, NLRC4, and NLRP3 inflammasomes in caspase-1 activation by Listeria monocytogenes. J. Clin. Immunol 2010, 30, 693–702. [Google Scholar] [CrossRef]
- Kasper, L.; König, A.; Koenig, P.A.; Gresnigt, M.S.; Westman, J.; Drummond, R.A.; Lionakis, M.S.; Groß, O.; Ruland, J.; Naglik, J.R.; et al. The fungal peptide toxin Candidalysin activates the NLRP3 inflammasome and causes cytolysis in mononuclear phagocytes. Nat. Commun. 2018, 9, 4260. [Google Scholar] [CrossRef]
- Saïd-Sadier, N.; Padilla, E.; Langsley, G.; Ojcius, D.M. Aspergillus fumigatus stimulates the NLRP3 inflammasome through a pathway requiring ROS production and the Syk tyrosine kinase. PLoS ONE 2010, 5, e10008. [Google Scholar] [CrossRef] [PubMed]
- Vonk, A.G.; Netea, M.G.; van Krieken, J.H.; Iwakura, Y.; van der Meer, J.W.; Kullberg, B.J. Endogenous interleukin (IL)-1 alpha and IL-1 beta are crucial for host defense against disseminated candidiasis. J. Infect. Dis. 2006, 193, 1419–1426. [Google Scholar] [CrossRef]
- Shen, H.; Yu, Y.; Chen, S.M.; Sun, J.J.; Fang, W.; Guo, S.Y.; Hou, W.T.; Qiu, X.R.; Zhang, Y.; Chen, Y.L.; et al. Dectin-1 Facilitates IL-18 Production for the Generation of Protective Antibodies against. Front. Microbiol. 2020, 11, 1648. [Google Scholar] [CrossRef]
- Alves, A.B.R.M.; David, M.A.; de Castro, L.F.; da Silva, R.M.; Longhi, L.N.A.; Blotta, M.H.S.L.; Mamoni, R.L. Differential production of interleukin-1 family cytokines (IL-1β, IL-18, IL-33 and IL-37) in patients with paracoccidioidomycosis: Correlation with clinical form and antifungal therapy. Med. Mycol. 2018, 56, 332–343. [Google Scholar] [CrossRef]
- Griffiths, J.S.; Camilli, G.; Kotowicz, N.K.; Ho, J.; Richardson, J.P.; Naglik, J.R. Role for IL-1 Family Cytokines in Fungal Infections. Front. Microbiol. 2021, 12, 633047. [Google Scholar] [CrossRef]
- McAuley, J.L.; Tate, M.D.; MacKenzie-Kludas, C.J.; Pinar, A.; Zeng, W.; Stutz, A.; Latz, E.; Brown, L.E.; Mansell, A. Activation of the NLRP3 inflammasome by IAV virulence protein PB1-F2 contributes to severe pathophysiology and disease. PLoS Pathog. 2013, 9, e1003392. [Google Scholar] [CrossRef]
- Jankovic, D.; Ganesan, J.; Bscheider, M.; Stickel, N.; Weber, F.C.; Guarda, G.; Follo, M.; Pfeifer, D.; Tardivel, A.; Ludigs, K.; et al. The Nlrp3 inflammasome regulates acute graft-versus-host disease. J. Exp. Med. 2013, 210, 1899–1910. [Google Scholar] [CrossRef] [PubMed]
- Penack, O.; Peczynski, C.; van der Werf, S.; Finke, J.; Ganser, A.; Schoemans, H.; Pavlu, J.; Niittyvuopio, R.; Schroyens, W.; Kaynar, L.; et al. Association of uric acid levels before start of conditioning with mortality after allogeneic hematopoietic stem cell transplantation—A prospective, non-interventional study of the EBMT Transplant Complication Working Party. Haematologica 2020, 105, 1977–1983. [Google Scholar] [CrossRef] [PubMed]
- Yeh, A.C.; Brunner, A.M.; Spitzer, T.R.; Chen, Y.B.; Coughlin, E.; McAfee, S.; Ballen, K.; Attar, E.; Caron, M.; Preffer, F.I.; et al. Phase I study of urate oxidase in the reduction of acute graft-versus-host disease after myeloablative allogeneic stem cell transplantation. Biol. Blood Marrow Transplant. 2014, 20, 730–734. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Anasetti, C. What are the most important donor and recipient factors affecting the outcome of related and unrelated allogeneic transplantation? Best Pract. Res. Clin. Haematol. 2008, 21, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Crux, N.B.; Elahi, S. Human Leukocyte Antigen (HLA) and Immune Regulation: How Do Classical and Non-Classical HLA Alleles Modulate Immune Response to Human Immunodeficiency Virus and Hepatitis C Virus Infections? Front. Immunol. 2017, 8, 832. [Google Scholar] [CrossRef]
- Uchino, K.; Vu Quang, L.; Mizuno, S.; Horio, T.; Yamamoto, H.; Hanamura, I.; Kodera, Y.; Luis Espinoza, J.; Onizuka, M.; Kashiwase, K.; et al. Donor UNC-93 Homolog B1 genetic polymorphism predicts survival outcomes after unrelated bone marrow transplantation. Genes Immun. 2021, 22, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Horio, T.; Morishita, E.; Mizuno, S.; Uchino, K.; Hanamura, I.; Espinoza, J.L.; Morishima, Y.; Kodera, Y.; Onizuka, M.; Kashiwase, K.; et al. Donor Heme Oxygenase-1 Promoter Gene Polymorphism Predicts Survival after Unrelated Bone Marrow Transplantation for High-Risk Patients. Cancers 2020, 12, 424. [Google Scholar] [CrossRef]
- Espinoza, J.L.; Takami, A.; Onizuka, M.; Sao, H.; Akiyama, H.; Miyamura, K.; Okamoto, S.; Inoue, M.; Kanda, Y.; Ohtake, S.; et al. NKG2D gene polymorphism has a significant impact on transplant outcomes after HLA-fully-matched unrelated bone marrow transplantation for standard risk hematologic malignancies. Haematologica 2009, 94, 1427–1434. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, J.L.; Takami, A.; Nakata, K.; Onizuka, M.; Kawase, T.; Akiyama, H.; Miyamura, K.; Morishima, Y.; Fukuda, T.; Kodera, Y.; et al. A genetic variant in the IL-17 promoter is functionally associated with acute graft-versus-host disease after unrelated bone marrow transplantation. PLoS ONE 2011, 6, e26229. [Google Scholar] [CrossRef]
- Gam, R.; Shah, P.; Crossland, R.E.; Norden, J.; Dickinson, A.M.; Dressel, R. Genetic Association of Hematopoietic Stem Cell Transplantation Outcome beyond Histocompatibility Genes. Front. Immunol. 2017, 8, 380. [Google Scholar] [CrossRef][Green Version]
- Schoemans, H.M.; Lee, S.J.; Ferrara, J.L.; Wolff, D.; Levine, J.E.; Schultz, K.R.; Shaw, B.E.; Flowers, M.E.; Ruutu, T.; Greinix, H.; et al. EBMT-NIH-CIBMTR Task Force position statement on standardized terminology & guidance for graft-versus-host disease assessment. Bone Marrow Transplant. 2018, 53, 1401–1415. [Google Scholar] [CrossRef]
- Hill, G.R.; Betts, B.C.; Tkachev, V.; Kean, L.S.; Blazar, B.R. Current Concepts and Advances in Graft-Versus-Host Disease Immunology. Annu. Rev. Immunol. 2021, 39, 19–49. [Google Scholar] [CrossRef]
- Okoev, G.; Weisdorf, D.J.; Wagner, J.E.; Blazar, B.R.; MacMillan, M.L.; DeFor, T.; Lazaryan, A.; El Jurdi, N.; Holtan, S.G.; Brunstein, C.G.; et al. Outcomes of chronic graft-versus-host disease following matched sibling donor versus umbilical cord blood transplant. Bone Marrow Transplant. 2021, 56, 1373–1380. [Google Scholar] [CrossRef]
- Koehn, B.H.; Apostolova, P.; Haverkamp, J.M.; Miller, J.S.; McCullar, V.; Tolar, J.; Munn, D.H.; Murphy, W.J.; Brickey, W.J.; Serody, J.S.; et al. GVHD-associated, inflammasome-mediated loss of function in adoptively transferred myeloid-derived suppressor cells. Blood 2015, 126, 1621–1628. [Google Scholar] [CrossRef]
- Qiao, J.; Huang, Y.; Xia, Y.; Chu, P.; Yao, H.; Xu, L.; Qi, K.; Liu, Y.; Xu, K.; Zeng, L. Busulfan and cyclosphamide induce liver inflammation through NLRP3 activation in mice after hematopoietic stem cell transplantation. Sci. Rep. 2015, 5, 17828. [Google Scholar] [CrossRef] [PubMed]
- Luft, T.; Dreger, P.; Radujkovic, A. Endothelial cell dysfunction: A key determinant for the outcome of allogeneic stem cell transplantation. Bone Marrow Transplant. 2021, 56, 2326–2335. [Google Scholar] [CrossRef]
- Horowitz, M.; Schreiber, H.; Elder, A.; Heidenreich, O.; Vormoor, J.; Toffalori, C.; Vago, L.; Kröger, N. Epidemiology and biology of relapse after stem cell transplantation. Bone Marrow Transplant. 2018, 53, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, J.L.; Nguyen, V.H.; Ichimura, H.; Pham, T.T.; Nguyen, C.H.; Pham, T.V.; Elbadry, M.I.; Yoshioka, K.; Tanaka, J.; Trung, L.Q.; et al. A functional polymorphism in the NKG2D gene modulates NK-cell cytotoxicity and is associated with susceptibility to Human Papilloma Virus-related cancers. Sci. Rep. 2016, 6, 39231. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Blobe, G.C. Role of transforming growth factor-beta in hematologic malignancies. Blood 2006, 107, 4589–4596. [Google Scholar] [CrossRef] [PubMed]
- Gökbuget, N.; Canaani, J.; Nagler, A.; Bishop, M.; Kröger, N.; Avigan, D. Prevention and treatment of relapse after stem cell transplantation with immunotherapy. Bone Marrow Transplant. 2018, 53, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Zhao, Y.; Pang, Y.; Ji, M.; Sun, Y.; Wang, H.; Zou, J.; Wang, Y.; Li, G.; Sun, T.; et al. NLRP3 inflammasome upregulates PD-L1 expression and contributes to immune suppression in lymphoma. Cancer Lett. 2021, 497, 178–189. [Google Scholar] [CrossRef]
- Theivanthiran, B.; Evans, K.S.; DeVito, N.C.; Plebanek, M.; Sturdivant, M.; Wachsmuth, L.P.; Salama, A.K.; Kang, Y.; Hsu, D.; Balko, J.M.; et al. A tumor-intrinsic PD-L1/NLRP3 inflammasome signaling pathway drives resistance to anti-PD-1 immunotherapy. J. Clin. Investig. 2020, 130, 2570–2586. [Google Scholar] [CrossRef]
- Paugh, S.W.; Bonten, E.J.; Savic, D.; Ramsey, L.B.; Thierfelder, W.E.; Gurung, P.; Malireddi, R.K.; Actis, M.; Mayasundari, A.; Min, J.; et al. NALP3 inflammasome upregulation and CASP1 cleavage of the glucocorticoid receptor cause glucocorticoid resistance in leukemia cells. Nat. Genet. 2015, 47, 607–614. [Google Scholar] [CrossRef]
- Singh, J.; Kumari, S.; Arora, M.; Verma, D.; Palanichamy, J.K.; Kumar, R.; Sharma, G.; Bakhshi, S.; Pushpam, D.; Ali, M.S.; et al. Prognostic Relevance of Expression of. Front. Oncol. 2021, 11, 606370. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Lerm, M.; Blomgran Julinder, R.; Eriksson, P.; Söderkvist, P.; Särndahl, E. Gene polymorphisms in the NALP3 inflammasome are associated with interleukin-1 production and severe inflammation: Relation to common inflammatory diseases? Arthritis Rheumatol. 2008, 58, 888–894. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.B.; Qing, Y.F.; He, Y.L.; Xie, W.G.; Zhou, J.G. Association of NLRP3 polymorphisms with susceptibility to primary gouty arthritis in a Chinese Han population. Clin. Rheumatol. 2018, 37, 235–244. [Google Scholar] [CrossRef]
- Zhang, Q.; Fan, H.W.; Zhang, J.Z.; Wang, Y.M.; Xing, H.J. NLRP3 rs35829419 polymorphism is associated with increased susceptibility to multiple diseases in humans. Genet. Mol. Res. 2015, 14, 13968–13980. [Google Scholar] [CrossRef]
- Hitomi, Y.; Ebisawa, M.; Tomikawa, M.; Imai, T.; Komata, T.; Hirota, T.; Harada, M.; Sakashita, M.; Suzuki, Y.; Shimojo, N.; et al. Associations of functional NLRP3 polymorphisms with susceptibility to food-induced anaphylaxis and aspirin-induced asthma. J. Allergy Clin. Immunol. 2009, 124, 779–785.e776. [Google Scholar] [CrossRef]
- Pontillo, A.; Brandão, L.A.; Guimarães, R.L.; Segat, L.; Athanasakis, E.; Crovella, S. A 3’UTR SNP in NLRP3 gene is associated with susceptibility to HIV-1 infection. J. Acquir. Immune Defic. Syndr. 2010, 54, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wu, S.; Liang, T. Association of NLRP3 rs35829419 and rs10754558 Polymorphisms with Risks of Autoimmune Diseases: A Systematic Review and Meta-Analysis. Front. Genet. 2021, 12, 690860. [Google Scholar] [CrossRef] [PubMed]
- Granell, M.; Urbano-Ispizua, A.; Pons, A.; Aróstegui, J.I.; Gel, B.; Navarro, A.; Jansa, S.; Artells, R.; Gaya, A.; Talarn, C.; et al. Common variants in NLRP2 and NLRP3 genes are strong prognostic factors for the outcome of HLA-identical sibling allogeneic stem cell transplantation. Blood 2008, 112, 4337–4342. [Google Scholar] [CrossRef]
- Takami, A.; Espinoza, J.L.; Onizuka, M.; Ishiyama, K.; Kawase, T.; Kanda, Y.; Sao, H.; Akiyama, H.; Miyamura, K.; Okamoto, S.; et al. A single-nucleotide polymorphism of the Fcgamma receptor type IIIA gene in the recipient predicts transplant outcomes after HLA fully matched unrelated BMT for myeloid malignancies. Bone Marrow Transplant. 2010, 46, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Damsgaard, C.T.; Lauritzen, L.; Calder, P.C.; Kjaer, T.M.; Frøkiaer, H. Whole-blood culture is a valid low-cost method to measure monocytic cytokines—A comparison of cytokine production in cultures of human whole-blood, mononuclear cells and monocytes. J. Immunol. Methods 2009, 340, 95–101. [Google Scholar] [CrossRef]
- He, Y.; Hara, H.; Núñez, G. Mechanism and Regulation of NLRP3 Inflammasome Activation. Trends Biochem. Sci. 2016, 41, 1012–1021. [Google Scholar] [CrossRef]
- Takahashi, H.; Okayama, N.; Yamaguchi, N.; Miyahara, Y.; Morishima, Y.; Suehiro, Y.; Yamasaki, T.; Tamada, K.; Takahashi, S.; Tojo, A.; et al. Associations of interactions between NLRP3 SNPs and HLA mismatch with acute and extensive chronic graft-versus-host diseases. Sci. Rep. 2017, 7, 13097. [Google Scholar] [CrossRef]
- Espinoza, J.L.; Takami, A.; Onizuka, M.; Morishima, Y.; Fukuda, T.; Kodera, Y.; Akiyama, H.; Miyamura, K.; Mori, T.; Nakao, S. Recipient PTPN22 -1123 C/C Genotype Predicts Acute Graft-versus-Host Disease after HLA Fully Matched Unrelated Bone Marrow Transplantation for Hematologic Malignancies. Biol. Blood Marrow Transplant. 2013, 19, 240–246. [Google Scholar] [CrossRef]
- Lu, F.; Chen, H.; Hong, Y.; Lin, Y.; Liu, L.; Wei, N.; Wu, Q.; Liao, S.; Yang, S.; He, J.; et al. A gain-of-function NLRP3 3’-UTR polymorphism causes miR-146a-mediated suppression of NLRP3 expression and confers protection against sepsis progression. Sci. Rep. 2021, 11, 13300. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Blocking interleukin-1β in acute and chronic autoinflammatory diseases. J. Intern. Med. 2011, 269, 16–28. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Huang, Y.; Wang, Y.; Wang, P.; Song, H.; Wang, F. Antibiotics induced intestinal tight junction barrier dysfunction is associated with microbiota dysbiosis, activated NLRP3 inflammasome and autophagy. PLoS ONE 2019, 14, e0218384. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.; Li, B.; Luo, Y.; Gong, Y.; Jin, X.; Zhang, J.; Zhou, Y.; Zhuo, X.; Wang, Z.; et al. Gut microbiota dysbiosis promotes age-related atrial fibrillation by lipopolysaccharide and glucose-induced activation of NLRP3-inflammasome. Cardiovasc. Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood 2011, 117, 3720–3732. [Google Scholar] [CrossRef]
- Park, M.J.; Lee, S.H.; Lee, E.J.; Kim, E.K.; Choi, J.Y.; Cho, M.L. IL-1 Receptor Blockade Alleviates Graft-versus-Host Disease through Downregulation of an Interleukin-1β-Dependent Glycolytic Pathway in Th17 Cells. Mediat. Inflamm. 2015, 2015, 631384. [Google Scholar] [CrossRef] [PubMed]
- Antin, J.H.; Weisdorf, D.; Neuberg, D.; Nicklow, R.; Clouthier, S.; Lee, S.J.; Alyea, E.; McGarigle, C.; Blazar, B.R.; Sonis, S.; et al. Interleukin-1 blockade does not prevent acute graft-versus-host disease: Results of a randomized, double-blind, placebo-controlled trial of interleukin-1 receptor antagonist in allogeneic bone marrow transplantation. Blood 2002, 100, 3479–3482. [Google Scholar] [CrossRef] [PubMed]
- de Mooij, C.E.M.; Netea, M.G.; van der Velden, W.J.F.M.; Blijlevens, N.M.A. Targeting the interleukin-1 pathway in patients with hematological disorders. Blood 2017, 129, 3155–3164. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Espinoza, J.L.; Kamio, K.; Lam, V.Q.; Takami, A. The Impact of NLRP3 Activation on Hematopoietic Stem Cell Transplantation. Int. J. Mol. Sci. 2021, 22, 11845. https://doi.org/10.3390/ijms222111845
Espinoza JL, Kamio K, Lam VQ, Takami A. The Impact of NLRP3 Activation on Hematopoietic Stem Cell Transplantation. International Journal of Molecular Sciences. 2021; 22(21):11845. https://doi.org/10.3390/ijms222111845
Chicago/Turabian StyleEspinoza, J. Luis, Kosuke Kamio, Vu Quang Lam, and Akiyoshi Takami. 2021. "The Impact of NLRP3 Activation on Hematopoietic Stem Cell Transplantation" International Journal of Molecular Sciences 22, no. 21: 11845. https://doi.org/10.3390/ijms222111845
APA StyleEspinoza, J. L., Kamio, K., Lam, V. Q., & Takami, A. (2021). The Impact of NLRP3 Activation on Hematopoietic Stem Cell Transplantation. International Journal of Molecular Sciences, 22(21), 11845. https://doi.org/10.3390/ijms222111845







