SARS-CoV-2 Exposed Mesenchymal Stromal Cell from Congenital Pulmonary Airway Malformations: Transcriptomic Analysis and the Expression of Immunomodulatory Genes
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Isolation, Culture, and Characterization of MSCs
4.3. Virus
4.4. In Vitro MSCs SARS-CoV-2 Infection Assay
4.5. MSC-Lung External Control Group
4.6. RNA-Seq Analysis
4.7. Cytokine and Chemokine Measurement by Multiplex Assay
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pal, M.; Berhanu, G.; Desalegn, C.; Kandi, V. Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2): An Update. Cureus 2020, 12, e7423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Attaway, A.H.; Scheraga, R.G.; Bhimraj, A.; Biehl, M.; Hatipoğlu, U. Severe covid-19 pneumonia: Pathogenesis and clinical management. BMJ Clin. Res. Ed. 2021, 372, n436. [Google Scholar] [CrossRef]
- Lou, S.; Duan, Y.; Nie, H.; Cui, X.; Du, J.; Yao, Y. Mesenchymal stem cells: Biological characteristics and application in disease therapy. Biochimie 2021, 185, 9–21. [Google Scholar] [CrossRef]
- Häberle, H.; Magunia, H.; Lang, P.; Gloeckner, H.; Körner, A.; Koeppen, M.; Backchoul, T.; Malek, N.; Handgretinger, R.; Rosenberger, P.; et al. Mesenchymal Stem Cell Therapy for Severe COVID-19 ARDS. J. Intensive Care Med. 2021, 36, 681–688. [Google Scholar] [CrossRef]
- Hashemian, S.R.; Aliannejad, R.; Zarrabi, M.; Soleimani, M.; Vosough, M.; Hosseini, S.E.; Hossieni, H.; Keshel, S.H.; Naderpour, Z.; Hajizadeh-Saffar, E.; et al. Mesenchymal stem cells derived from perinatal tissues for treatment of critically ill COVID-19-induced ARDS patients: A case series. Stem Cell Res. Ther. 2021, 12, 91. [Google Scholar] [CrossRef]
- Can, A.; Coskun, H. The rationale of using mesenchymal stem cells in patients with COVID-19-related acute respiratory distress syndrome: What to expect. Stem Cells Transl. Med. 2020, 9, 1287–1302. [Google Scholar] [CrossRef] [PubMed]
- Leng, Z.; Zhu, R.; Hou, W.; Feng, Y.; Yang, Y.; Han, Q.; Shan, G.; Meng, F.; Du, D.; Wang, S.; et al. Transplantation of ACE2(-) Mesenchymal Stem Cells Improves the Outcome of Patients with COVID-19 Pneumonia. Aging Dis. 2020, 11, 216–228. [Google Scholar] [CrossRef] [Green Version]
- Tong, Y.; Zuo, J.; Yue, D. Application Prospects of Mesenchymal Stem Cell Therapy for Bronchopulmonary Dysplasia and the Challenges Encountered. Biomed. Res. Int. 2021, 2021, 9983664. [Google Scholar] [CrossRef]
- Pelizzo, G.; Avanzini, M.A.; Lenta, E.; Mantelli, M.; Croce, S.; Catenacci, L.; Acquafredda, G.; Ferraro, A.L.; Giambanco, C.; D′Amelio, L.; et al. Allogeneic mesenchymal stromal cells: Novel therapeutic option for mutated FLNA-associated respiratory failure in the pediatric setting. Pediatric Pulmonol. 2020, 55, 190–197. [Google Scholar] [CrossRef]
- Stocker, J. Congenital pulmonary airway malformation: A new name for and an expanded classification of congenital cystic adenomatoid malformation of the lung. Histopathology 2002, 41, 424–430. [Google Scholar]
- Pelizzo, G.; Costanzo, F.; Andreatta, E.; Calcaterra, V. Congenital pulmonary airway malformations: From prenatal diagnosis to postnatal outcome. Minerva Pediatrica 2016, 68, 299–311. [Google Scholar] [PubMed]
- Priest, J.R.; Williams, G.M.; Hill, D.A.; Dehner, L.P.; Jaffé, A. Pulmonary cysts in early childhood and the risk of malignancy. Pediatr. Pulmonol. 2009, 44, 14–30. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef]
- Pelizzo, G.; Avanzini, M.A.; Folini, M.; Bussani, R.; Mantelli, M.; Croce, S.; Acquafredda, G.; Travaglino, P.; Cimino-Reale, G.; Boni, M.; et al. CPAM type 2-derived mesenchymal stem cells: Malignancy risk study in a 14-month-old boy. Pediatr. Pulmonol. 2017, 52, 990–999. [Google Scholar] [CrossRef]
- Stingi, A.; Cirillo, L. SARS-CoV-2 infection and cancer. BioEssays 2021, 43, 2000289. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Valeri, A.; Chiricosta, L.; Calcaterra, V.; Biasin, M.; Cappelletti, G.; Carelli, S.; Zuccotti, G.V.; Bramanti, P.; Pelizzo, G.; Mazzon, E.; et al. Transcriptomic Analysis of HCN-2 Cells Suggests Connection among Oxidative Stress, Senescence, and Neuron Death after SARS-CoV-2 Infection. Cells 2021, 10, 2189. [Google Scholar] [CrossRef] [PubMed]
- Leblanc, C.; Baron, M.; Desselas, E.; Phan, M.H.; Rybak, A.; Thouvenin, G.; Lauby, C.; Irtan, S. Congenital pulmonary airway malformations: State-of-the-art review for pediatrician’s use. Eur. J. Pediatrics 2017, 176, 1559–1571. [Google Scholar] [CrossRef]
- Gajewska-Knapik, K.; Impey, L. Congenital lung lesions: Prenatal diagnosis and intervention. Semin. Pediatric Surg. 2015, 24, 156–159. [Google Scholar] [CrossRef]
- Adzick, N.S. Management of fetal lung lesions. Clin. Perinatol. 2009, 36, 363–376. [Google Scholar] [CrossRef]
- Peters, R.T.; Burge, D.M.; Marven, S.S. Congenital lung malformations: An ongoing controversy. Ann. R. Coll. Surg. Engl. 2013, 95, 144–147. [Google Scholar] [CrossRef] [Green Version]
- Aslan, A.T.; Yalcin, E.; Soyer, T.; Dogru, D.; Talim, B.; Ciftci, A.O.; Ozcelik, U.; Kiper, N. Prenatal period to adolescence: The variable presentations of congenital cystic adenomatoid malformation. Pediatrics Int. 2006, 48, 626–630. [Google Scholar] [CrossRef]
- Pelizzo, G.; Barbi, E.; Codrich, D.; Lembo, M.A.; Zennaro, F.; Bussani, R.; Schleef, J. Chronic inflammation in congenital cystic adenomatoid malformations. An underestimated risk factor? J. Pediatric Surg. 2009, 44, 616–619. [Google Scholar] [CrossRef]
- Shyam, H.; Singh, S.K.; Kant, R.; Saxena, S.K. Mesenchymal stem cells in regenerative medicine: A new paradigm for degenerative bone diseases. Regen. Med. 2017, 12, 111–114. [Google Scholar] [CrossRef] [Green Version]
- Allison, S. Soluble ACE2 in SARS-CoV-2 infection. Nat. Rev. Nephrol. 2021, 17, 297. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Schäfer, R.; Spohn, G.; Bechtel, M.; Bojkova, D.; Baer, P.C.; Kuçi, S.; Seifried, E.; Ciesek, S.; Cinatl, J. Human Mesenchymal Stromal Cells Are Resistant to SARS-CoV-2 Infection under Steady-State, Inflammatory Conditions and in the Presence of SARS-CoV-2-Infected Cells. Stem Cell Rep. 2021, 16, 419–427. [Google Scholar] [CrossRef]
- Gugliandolo, A.; Chiricosta, L.; Calcaterra, V.; Biasin, M.; Cappelletti, G.; Carelli, S.; Zuccotti, G.; Avanzini, M.A.; Bramanti, P.; Pelizzo, G.; et al. SARS-CoV-2 Infected Pediatric Cerebral Cortical Neurons: Transcriptomic Analysis and Potential Role of Toll-like Receptors in Pathogenesis. Int. J. Mol. Sci. 2021, 22, 8059. [Google Scholar] [CrossRef]
- Gadanec, L.K.; McSweeney, K.R.; Qaradakhi, T.; Ali, B.; Zulli, A.; Apostolopoulos, V. Can SARS-CoV-2 Virus Use Multiple Receptors to Enter Host Cells? Int. J. Mol. Sci. 2021, 22, 992. [Google Scholar] [CrossRef]
- Aguilar-Briseño, J.A.; Upasani, V.; Ellen, B.M.t.; Moser, J.; Pauzuolis, M.; Ruiz-Silva, M.; Heng, S.; Laurent, D.; Choeung, R.; Dussart, P.; et al. TLR2 on blood monocytes senses dengue virus infection and its expression correlates with disease pathogenesis. Nat. Commun. 2020, 11, 3177. [Google Scholar] [CrossRef]
- Costela-Ruiz, V.J.; Illescas-Montes, R.; Puerta-Puerta, J.M.; Ruiz, C.; Melguizo-Rodríguez, L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020, 54, 62–75. [Google Scholar] [CrossRef]
- Hirano, T. IL-6 in inflammation, autoimmunity and cancer. Int. Immunol. 2020, 33, 127–148. [Google Scholar] [CrossRef]
- Kaneko, N.; Kurata, M.; Yamamoto, T.; Morikawa, S.; Masumoto, J. The role of interleukin-1 in general pathology. Inflamm. Regen. 2019, 39, 12. [Google Scholar] [CrossRef] [Green Version]
- Qazi, B.S.; Tang, K.; Qazi, A. Recent Advances in Underlying Pathologies Provide Insight into Interleukin-8 Expression-Mediated Inflammation and Angiogenesis. Int. J. Inflamm. 2011, 2011, 908468. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Guo, S.; Hibbert, J.M.; Jain, V.; Singh, N.; Wilson, N.O.; Stiles, J.K. CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev. 2011, 22, 121–130. [Google Scholar] [CrossRef]
- Yamada, T.; Sato, S.; Sotoyama, Y.; Orba, Y.; Sawa, H.; Yamauchi, H.; Sasaki, M.; Takaoka, A. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat. Immunol. 2021, 22, 820–828. [Google Scholar] [CrossRef]
- Goodman, A.G.; Smith, J.A.; Balachandran, S.; Perwitasari, O.; Proll, S.C.; Thomas, M.J.; Korth, M.J.; Barber, G.N.; Schiff, L.A.; Katze, M.G. The Cellular Protein P58IPK Regulates Influenza Virus mRNA Translation and Replication through a PKR-Mediated Mechanism. J. Virol. 2007, 81, 2221–2230. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.W.; Wilusz, J.; Katze, M.G. Regulation of eukaryotic protein synthesis: Selective influenza viral mRNA translation is mediated by the cellular RNA-binding protein GRSF-1. Proc. Natl. Acad. Sci. USA 1999, 96, 6694–6699. [Google Scholar] [CrossRef] [Green Version]
- Hottiger, M.O.; Nabel, G.J. Viral replication and the coactivators p300 and CBP. Trends Microbiol. 2000, 8, 560–565. [Google Scholar] [CrossRef]
- Marzio, G.; Tyagi, M.; Gutierrez, M.I.; Giacca, M. HIV-1 Tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc. Natl. Acad. Sci. USA 1998, 95, 13519–13524. [Google Scholar] [CrossRef] [Green Version]
- Maeda, Y.; Davé, V.; Whitsett, J.A. Transcriptional Control of Lung Morphogenesis. Physiol. Rev. 2007, 87, 219–244. [Google Scholar] [CrossRef]
- Zhang, Y.; Mao, D.; Roswit, W.T.; Jin, X.; Patel, A.C.; Patel, D.A.; Agapov, E.; Wang, Z.; Tidwell, R.M.; Atkinson, J.J.; et al. PARP9-DTX3L ubiquitin ligase targets host histone H2BJ and viral 3C protease to enhance interferon signaling and control viral infection. Nat. Immunol. 2015, 16, 1215–1227. [Google Scholar] [CrossRef] [PubMed]
- Swarnkar, G.; Chen, T.H.-P.; Arra, M.; Nasir, A.M.; Mbalaviele, G.; Abu-Amer, Y. NUMBL Interacts with TAK1, TRAF6 and NEMO to Negatively Regulate NF-κB Signaling During Osteoclastogenesis. Sci. Rep. 2017, 7, 12600. [Google Scholar] [CrossRef]
- Ruiz-García, A.; López-López, S.; García-Ramírez, J.J.; Baladrón, V.; Ruiz-Hidalgo, M.J.; López-Sanz, L.; Ballesteros, Á.; Laborda, J.; Monsalve, E.M.; Díaz-Guerra, M.J.M. The Tetraspanin TSPAN33 Controls TLR-Triggered Macrophage Activation through Modulation of NOTCH Signaling. J. Immunol. 2016, 197, 1600421. [Google Scholar] [CrossRef]
- Groot, A.J.; Vooijs, M.A. The Role of Adams in Notch Signaling. In Notch Signaling in Embryology and Cancer; Reichrath, J., Reichrath, S., Eds.; Springer: New York, NY, USA, 2012; pp. 15–36. [Google Scholar] [CrossRef] [Green Version]
- Pabois, A.; Devalliere, J.; Quillard, T.; Coulon, F.; Gerard, N.; Laboisse, C.; Toquet, C.; Charreau, B. The disintegrin and metalloproteinase ADAM10 mediates a canonical Notch-dependent regulation of IL-6 through Dll4 in human endothelial cells. Biochem. Pharmacol. 2014, 91, 510–521. [Google Scholar] [CrossRef]
- Chen, G.Q.; Wang, Q.M.; Yu, M.; Cheng, Y.D.; Zhang, Z.C.; Wang, W.S.; Qiu, Y.; Sun, L.H.; Peng, K.; Yang, H. Notch signaling is involved in regulation of LPS-induced macrophage apoptosis through JNK/NF-kB signaling pathway. J. Biol. Regul. Homeost. Agents 2020, 34, 283. [Google Scholar] [CrossRef]
- Palaga, T.; Miele, L.; Golde, T.E.; Osborne, B.A. TCR-mediated Notch signaling regulates proliferation and IFN-gamma production in peripheral T cells. J. Immunol. 2003, 171, 3019–3024. [Google Scholar] [CrossRef] [PubMed]
- López-López, S.; Monsalve, E.M.; de Ávila, M.J.R.; González-Gómez, J.; de León, N.H.; Ruiz-Marcos, F.; Baladrón, V.; Nueda, M.L.; García-León, M.J.; Screpanti, I.; et al. NOTCH3 signaling is essential for NF-κB activation in TLR-activated macrophages. Sci. Rep. 2020, 10, 14839. [Google Scholar] [CrossRef]
- Wei, Y.; Ma, D.; Gao, Y.; Zhang, C.; Wang, L.; Liu, F. Ncor2 is required for hematopoietic stem cell emergence by inhibiting Fos signaling in zebrafish. Blood 2014, 124, 1578–1585. [Google Scholar] [CrossRef]
- Luo, Z.; Mu, L.; Zheng, Y.; Shen, W.; Li, J.; Xu, L.; Zhong, B.; Liu, Y.; Zhou, Y. NUMB enhances Notch signaling by repressing ubiquitination of NOTCH1 intracellular domain. J. Mol. Cell Biol. 2020, 12, 345–358. [Google Scholar] [CrossRef]
- Joshi, I.; Minter, L.M.; Telfer, J.; Demarest, R.M.; Capobianco, A.J.; Aster, J.C.; Sicinski, P.; Fauq, A.; Golde, T.E.; Osborne, B.A. Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood 2009, 113, 1689–1698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.Y.; Reichlin, A.; Santana, A.; Sokol, K.A.; Nussenzweig, M.C.; Choi, Y. TRAF2 Is Essential for JNK but Not NF-κB Activation and Regulates Lymphocyte Proliferation and Survival. Immunity 1997, 7, 703–713. [Google Scholar] [CrossRef] [Green Version]
- Milhas, D.; Cuvillier, O.; Therville, N.; Clavé, P.; Thomsen, M.; Levade, T.; Benoist, H.; Ségui, B. Caspase-10 Triggers Bid Cleavage and Caspase Cascade Activation in FasL-induced Apoptosis. J. Biol. Chem. 2005, 280, 19836–19842. [Google Scholar] [CrossRef] [Green Version]
- Cai, J.; Wang, D.; Zhang, G.; Guo, X. The Role Of PD-1/PD-L1 Axis In Treg Development And Function: Implications For Cancer Immunotherapy. Oncol. Targets Ther. 2019, 12, 8437–8445. [Google Scholar] [CrossRef] [Green Version]
- Basit, A.; Reutershan, J.; Morris, M.A.; Solga, M.C.; Edward, R.J.; Ley, K. ICAM-1 and LFA-1 play critical roles in LPS-induced neutrophil recruitment into the alveolar space. Am. J. Physiol. -Lung Cell. Mol. Physiol. 2006, 291, L200–L207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, S.; Schaller, M.A.; Neupane, R.; Kunkel, S.L.; Lukacs, N.W. Regulation of T Cell Activation by Notch Ligand, DLL4, Promotes IL-17 Production and Rorc Activation. J. Immunol. 2009, 182, 7381–7388. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Zuleta, W.G.; Sanchez, E. IL-9: Function, Sources, and Detection. Methods Mol. Biol. 2017, 1585, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Dunn, A.; Ward, A. G-CSF: Function and modes of action (Review). Int. J. Mol. Med. 2002, 10, 3–10. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, C.H.; Roberts, A.I.; Das, J.; Xu, G.; Ren, G.; Zhang, Y.; Zhang, L.; Yuan, Z.R.; Tan, H.S.W.; et al. Granulocyte-macrophage colony-stimulating factor (GM-CSF) and T-cell responses: What we do and don’t know. Cell Res. 2006, 16, 126–133. [Google Scholar] [CrossRef]
- De la Fuente López, M.; Landskron, G.; Parada, D.; Dubois-Camacho, K.; Simian, D.; Martinez, M.; Romero, D.; Roa, J.C.; Chahuán, I.; Gutiérrez, R.; et al. The relationship between chemokines CCL2, CCL3, and CCL4 with the tumor microenvironment and tumor-associated macrophage markers in colorectal cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2018, 40, 1010428318810059. [Google Scholar] [CrossRef] [Green Version]
- Pelizzo, G.; Veschi, V.; Mantelli, M.; Croce, S.; Di Benedetto, V.; D’Angelo, P.; Maltese, A.; Catenacci, L.; Apuzzo, T.; Scavo, E.; et al. Microenvironment in neuroblastoma: Isolation and characterization of tumor-derived mesenchymal stromal cells. BMC Cancer 2018, 18, 1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrow, K.A.; Rich, L.M.; Vanderwall, E.R.; Reeves, S.R.; Rathe, J.A.; White, M.P.; Debley, J.S. Inactivation of Material from SARS-CoV-2-Infected Primary Airway Epithelial Cell Cultures. Methods Protoc. 2021, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Spadafora, R.; Lu, J.; Khetani, R.S.; Zhang, C.; Iberg, A.; Li, H.; Shi, Y.; Lerou, P.H. Lung-Resident Mesenchymal Stromal Cells Reveal Transcriptional Dynamics of Lung Development in Preterm Infants. Am. J. Respir. Crit. Care Med. 2018, 198, 961–964. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coordinators, N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatic 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatic 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatic 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
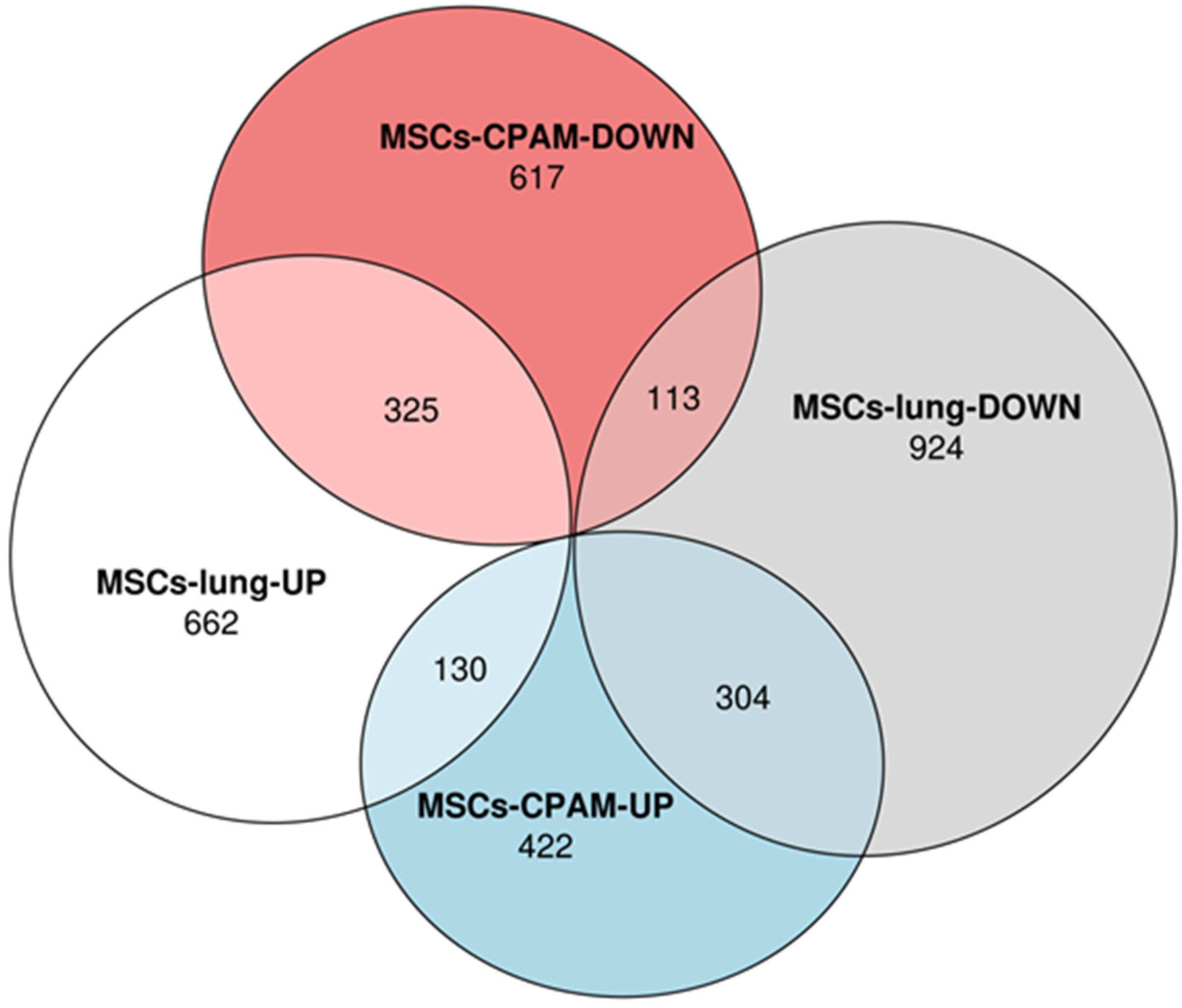

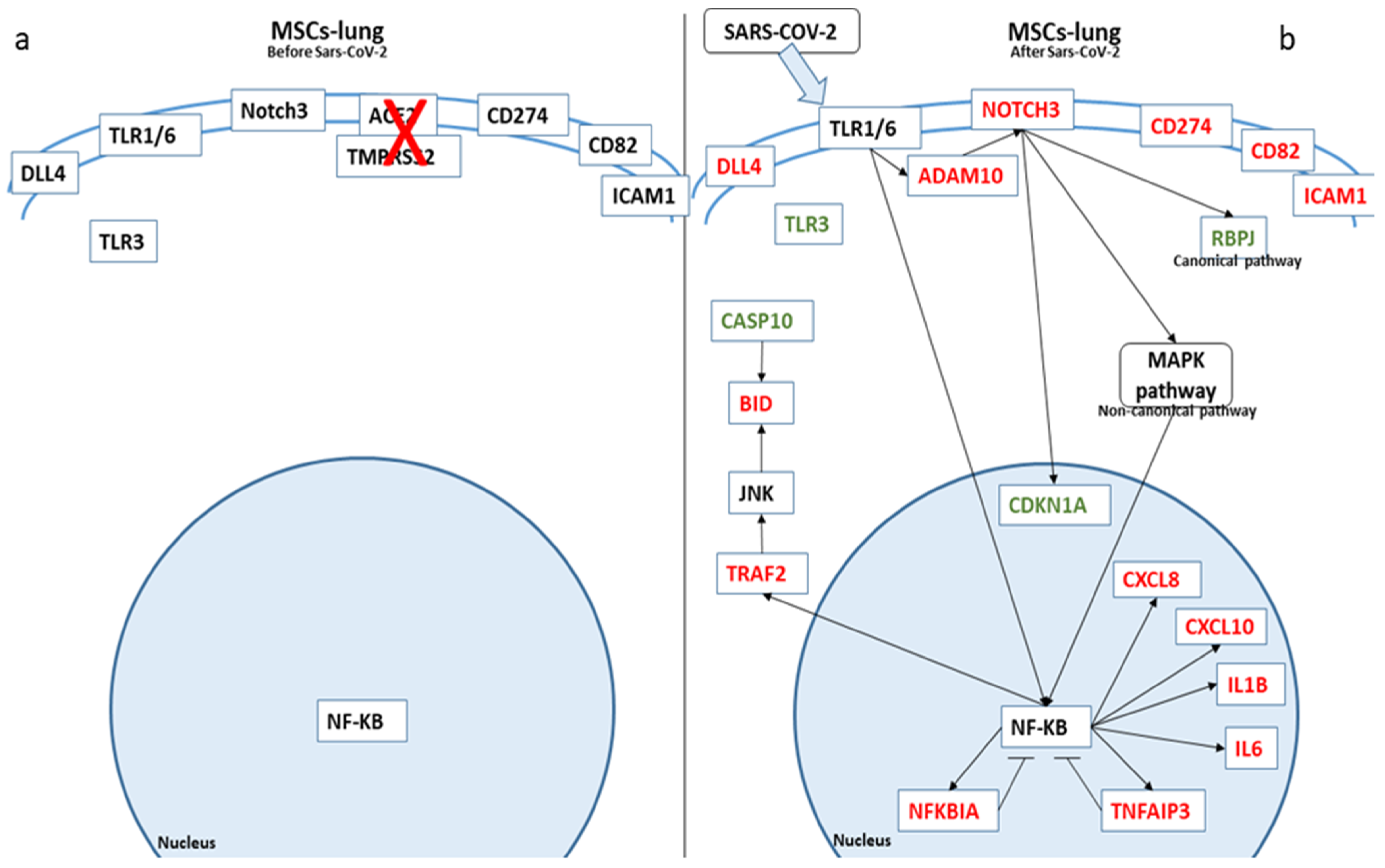

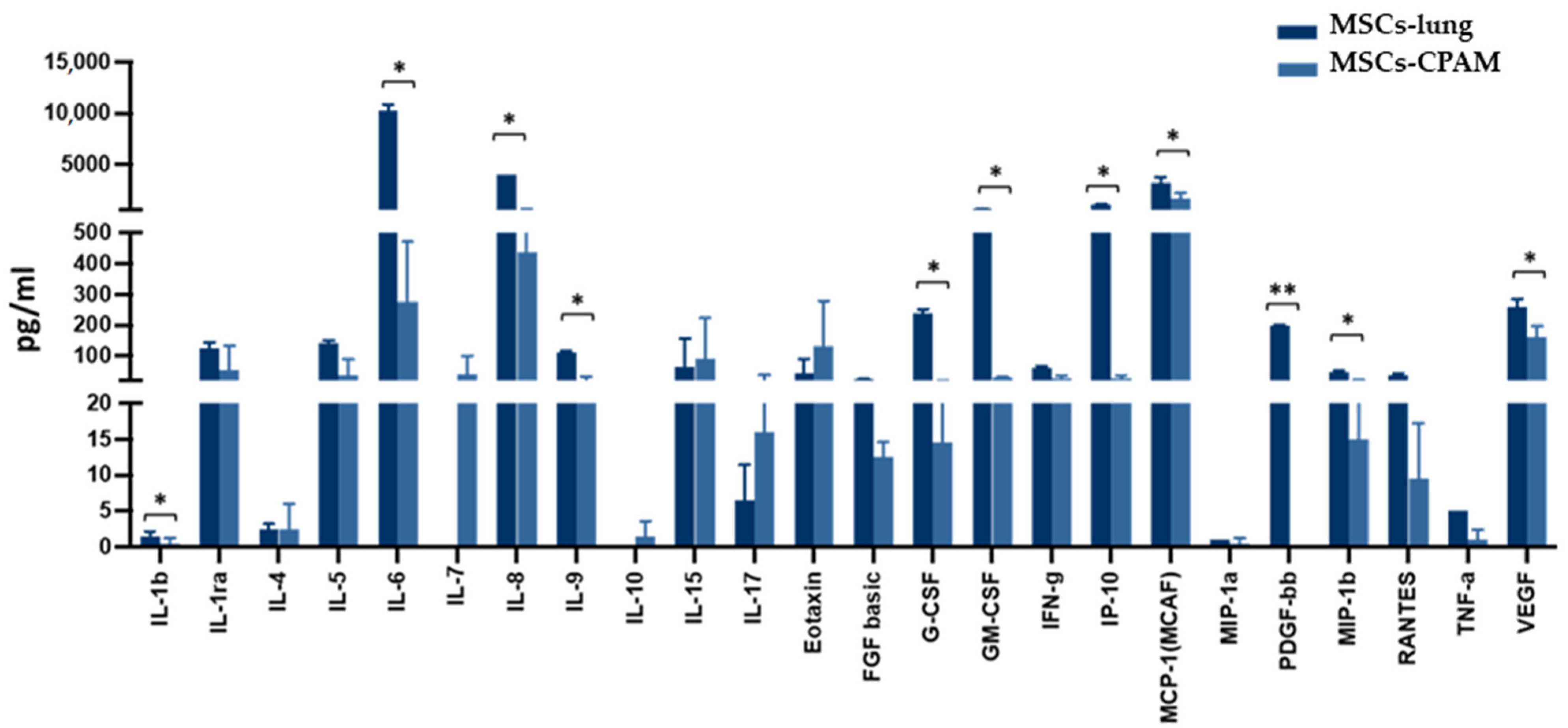
| Genes | MSCs-Lung | MSCs-Lung Infected | MSCs-CPAM | MSCs-CPAM Infected | MSCs-TA |
|---|---|---|---|---|---|
| NT5E | 4041.59 | 4097.82 | 1838.26 | 1560.99 | 4942.13 |
| THY1 | 2641.86 | 2736.23 | 1092.17 | 1094.85 | 2940.66 |
| ENG | 1880.03 | 1728.19 | 1219.84 | 1019.38 | 575.79 |
| HLA-A | 7579.78 | 7565.95 | 3017.68 | 3353.59 | 1914.89 |
| HLA-B | 5677 | 6142.11 | 2011.94 | 2213.41 | 1209.48 |
| HLA-C | 4216.12 | 4170.44 | 1555.12 | 1642.62 | 656.22 |
| Gene | Log2 Fold Change MSCs-TA-Lung Exposed | Log2 Fold Change MSCs-Lung | Log2 Fold Change MSCs-CPAM |
|---|---|---|---|
| ADAM17 | 1.05 | - | −1.56 |
| CREBBP | 3.24 | −0.36 | - |
| DLL4 | - | 5.09 | - |
| DTX3L | - | −1.44 | - |
| DVL1 | - | - | 1.56 |
| DVL2 | 0.8 | −0.65 | - |
| HDAC2 | 0.87 | - | −0.72 |
| JAG1 | 3.33 | 0.4 | 1.59 |
| MAML2 | 1.81 | - | −1.06 |
| MAML3 | 2.76 | 0.73 | - |
| NCOR2 | 1.47 | −0.59 | 0.71 |
| NOTCH3 | 1.97 | 1.33 | - |
| NUMB | - | - | −0.97 |
| NUMBL | 0.85 | −0.75 | 1.61 |
| RBPJ | 0.55 | −1.03 | 1.17 |
| TLE3 | - | - | 1.03 |
| Gene | Log2 Fold Change MSCs-TA-Lung Exposed | Log2 Fold Change MSCs-Lung | Log2 Fold Change MSCs-CPAM |
|---|---|---|---|
| DLL4 | - | 5.09 | - |
| CD274 | −2.38 | 1.77 | - |
| CD82 | 2.48 | 1.56 | - |
| ICAM1 | 1.65 | 1.18 | −1.55 |
| TNFAIP3 | 3.29 | 2.18 | −2.67 |
| NFKB1 | 1.13 | - | - |
| NFKBIA | 3.41 | 0.90 | −1.86 |
| TRAF2 | - | 1.38 | - |
| ADAM10 | 1.53 | 0.35 | −0.49 |
| IL6 | 2.48 | 2.22 | −2.53 |
| IL1B | 5.47 | 3.25 | −10.39 |
| CXCL8 | 6.07 | 6.25 | −7.63 |
| CXCL10 | - | 7.42 | - |
| CASP10 | - | −2.23 | - |
| CDKN1A | - | −0.58 | - |
| BID | - | 1.00 | − |
| DNAJC3 | −1.35 | - | −0.95 |
| GRSF1 | −0.3 | - | −1.02 |
| MAPK10 | - | - | −2.98 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valeri, A.; Chiricosta, L.; Gugliandolo, A.; Biasin, M.; Avanzini, M.A.; Calcaterra, V.; Cappelletti, G.; Carelli, S.; Zuccotti, G.V.; Silvestro, S.; et al. SARS-CoV-2 Exposed Mesenchymal Stromal Cell from Congenital Pulmonary Airway Malformations: Transcriptomic Analysis and the Expression of Immunomodulatory Genes. Int. J. Mol. Sci. 2021, 22, 11814. https://doi.org/10.3390/ijms222111814
Valeri A, Chiricosta L, Gugliandolo A, Biasin M, Avanzini MA, Calcaterra V, Cappelletti G, Carelli S, Zuccotti GV, Silvestro S, et al. SARS-CoV-2 Exposed Mesenchymal Stromal Cell from Congenital Pulmonary Airway Malformations: Transcriptomic Analysis and the Expression of Immunomodulatory Genes. International Journal of Molecular Sciences. 2021; 22(21):11814. https://doi.org/10.3390/ijms222111814
Chicago/Turabian StyleValeri, Andrea, Luigi Chiricosta, Agnese Gugliandolo, Mara Biasin, Maria Antonietta Avanzini, Valeria Calcaterra, Gioia Cappelletti, Stephana Carelli, Gian Vincenzo Zuccotti, Serena Silvestro, and et al. 2021. "SARS-CoV-2 Exposed Mesenchymal Stromal Cell from Congenital Pulmonary Airway Malformations: Transcriptomic Analysis and the Expression of Immunomodulatory Genes" International Journal of Molecular Sciences 22, no. 21: 11814. https://doi.org/10.3390/ijms222111814
APA StyleValeri, A., Chiricosta, L., Gugliandolo, A., Biasin, M., Avanzini, M. A., Calcaterra, V., Cappelletti, G., Carelli, S., Zuccotti, G. V., Silvestro, S., Mazzon, E., & Pelizzo, G. (2021). SARS-CoV-2 Exposed Mesenchymal Stromal Cell from Congenital Pulmonary Airway Malformations: Transcriptomic Analysis and the Expression of Immunomodulatory Genes. International Journal of Molecular Sciences, 22(21), 11814. https://doi.org/10.3390/ijms222111814










