Genomic Identification and Functional Analysis of JHAMTs in the Pond Wolf Spider, Pardosa pseudoannulata
Abstract
1. Introduction
2. Results
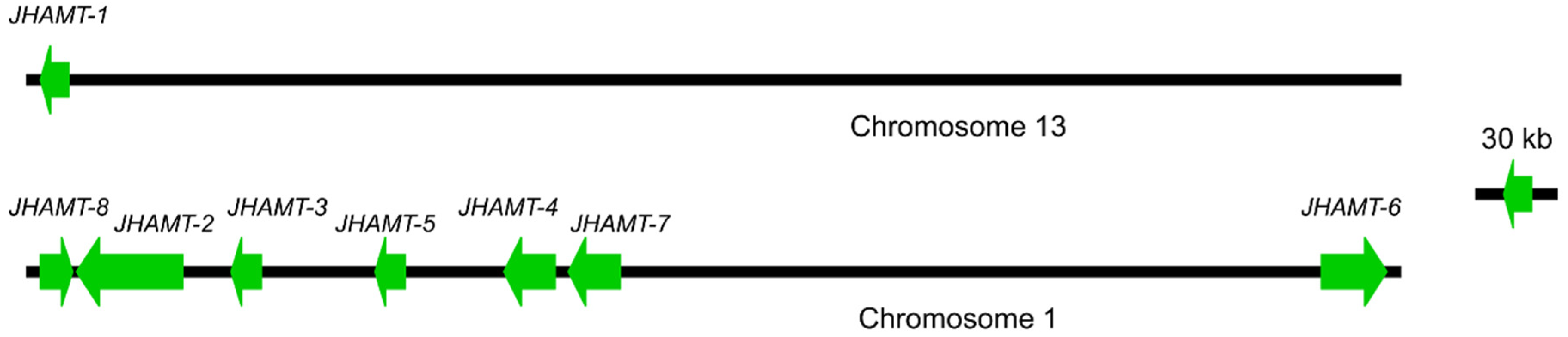
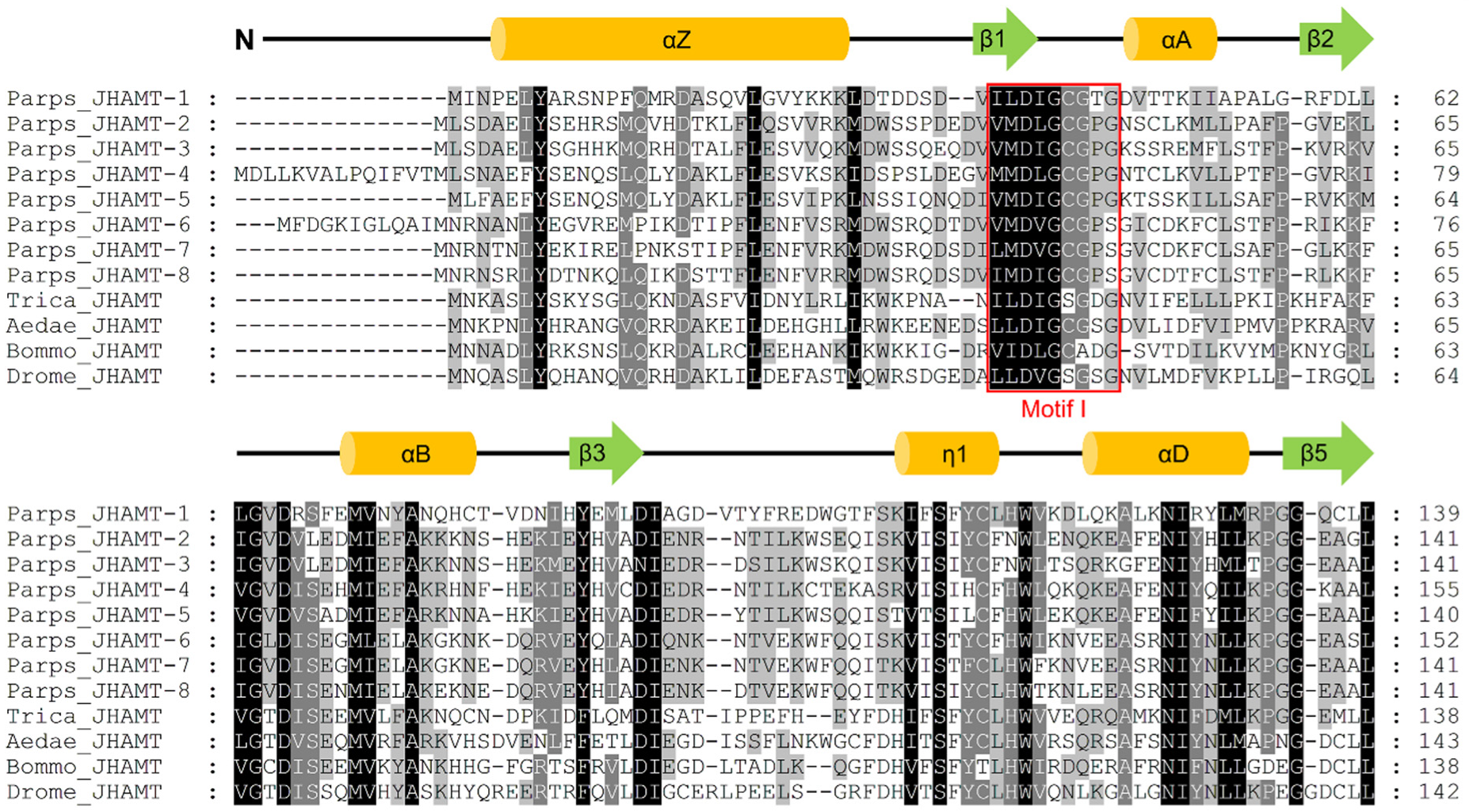
2.1. Characterization of JHAMTs
2.2. Phylogenetic Analysis
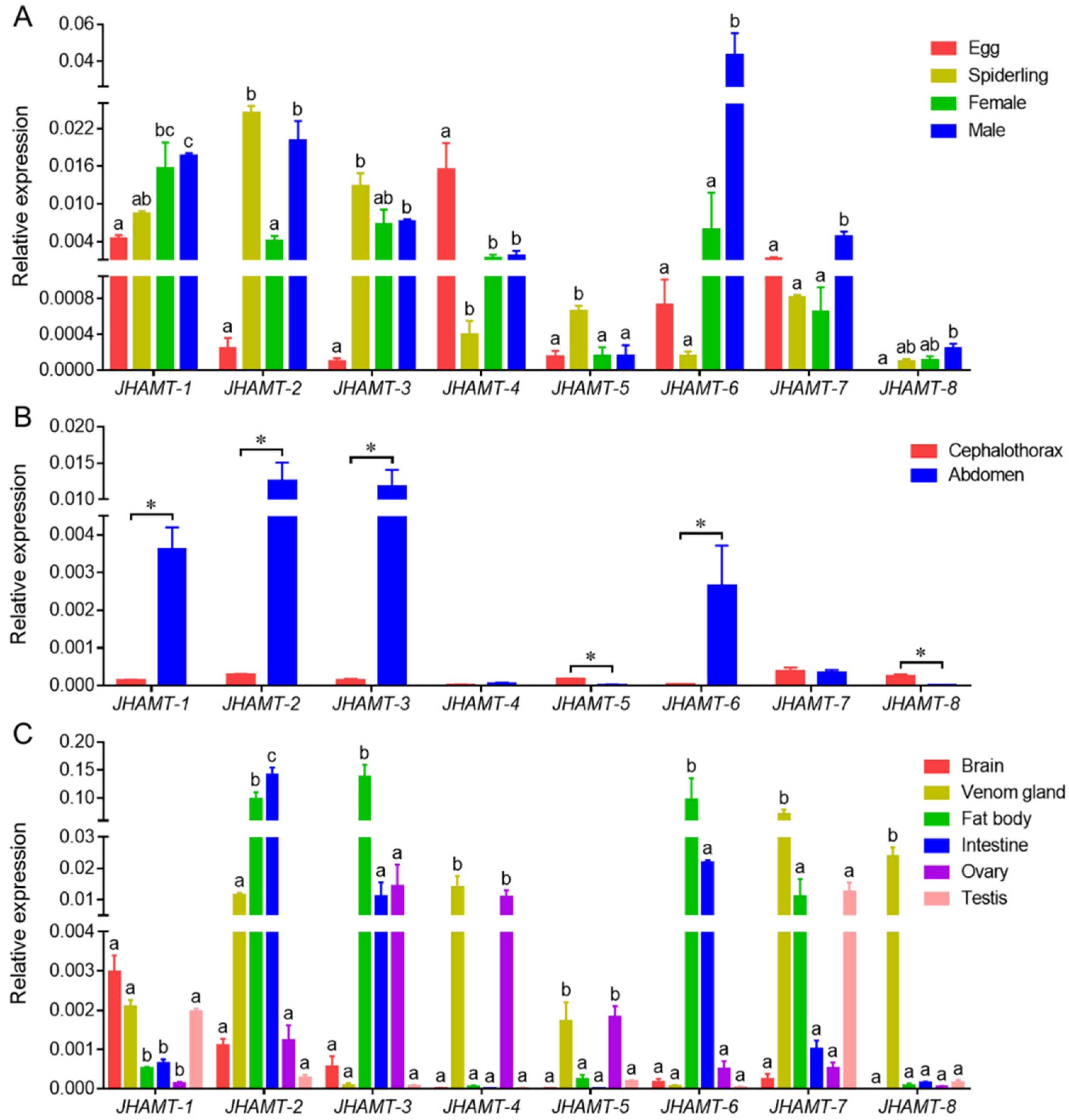
2.3. Spatiotemporal Expression Profile
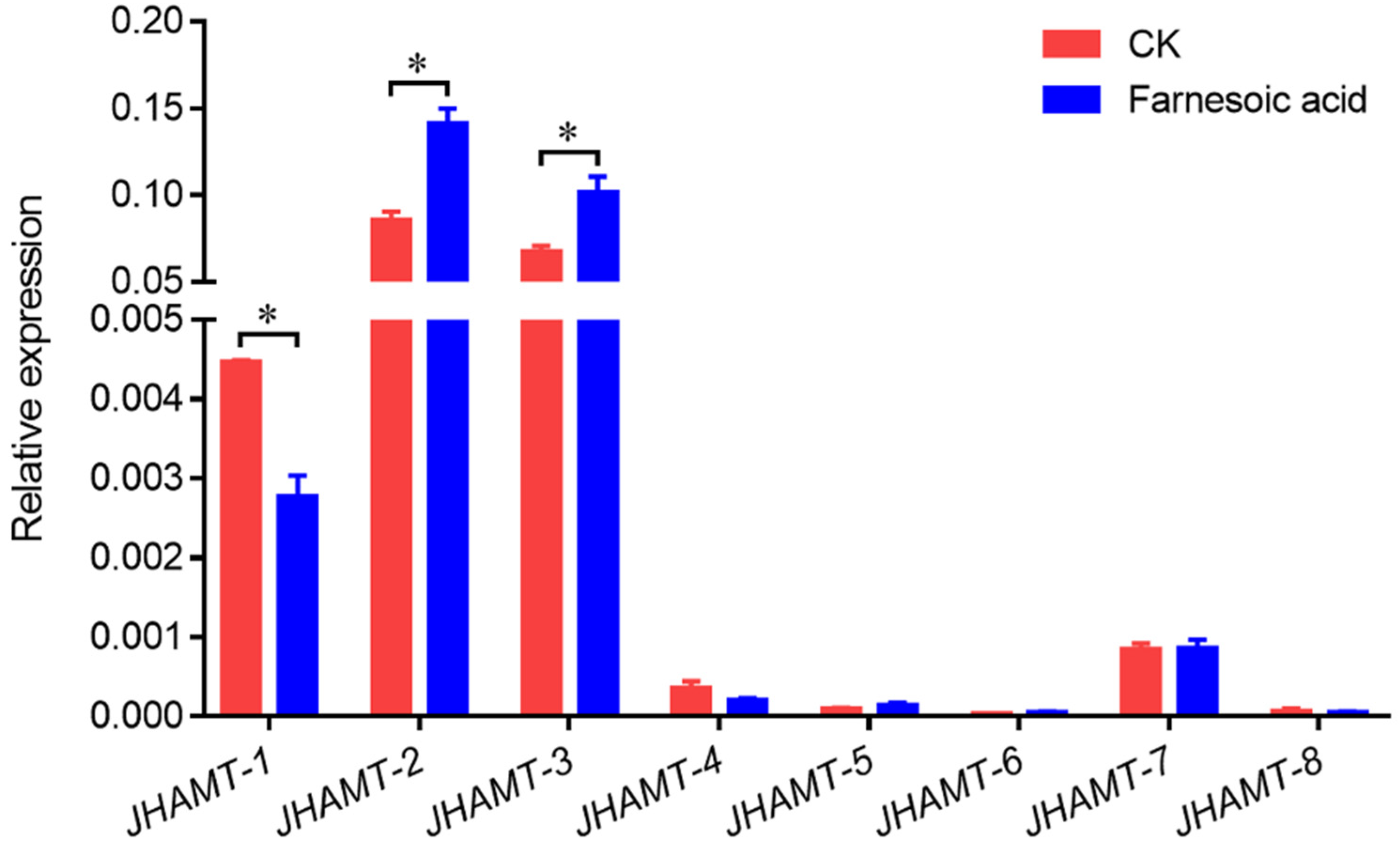
2.4. Effect of Farnesoic Acid Administration
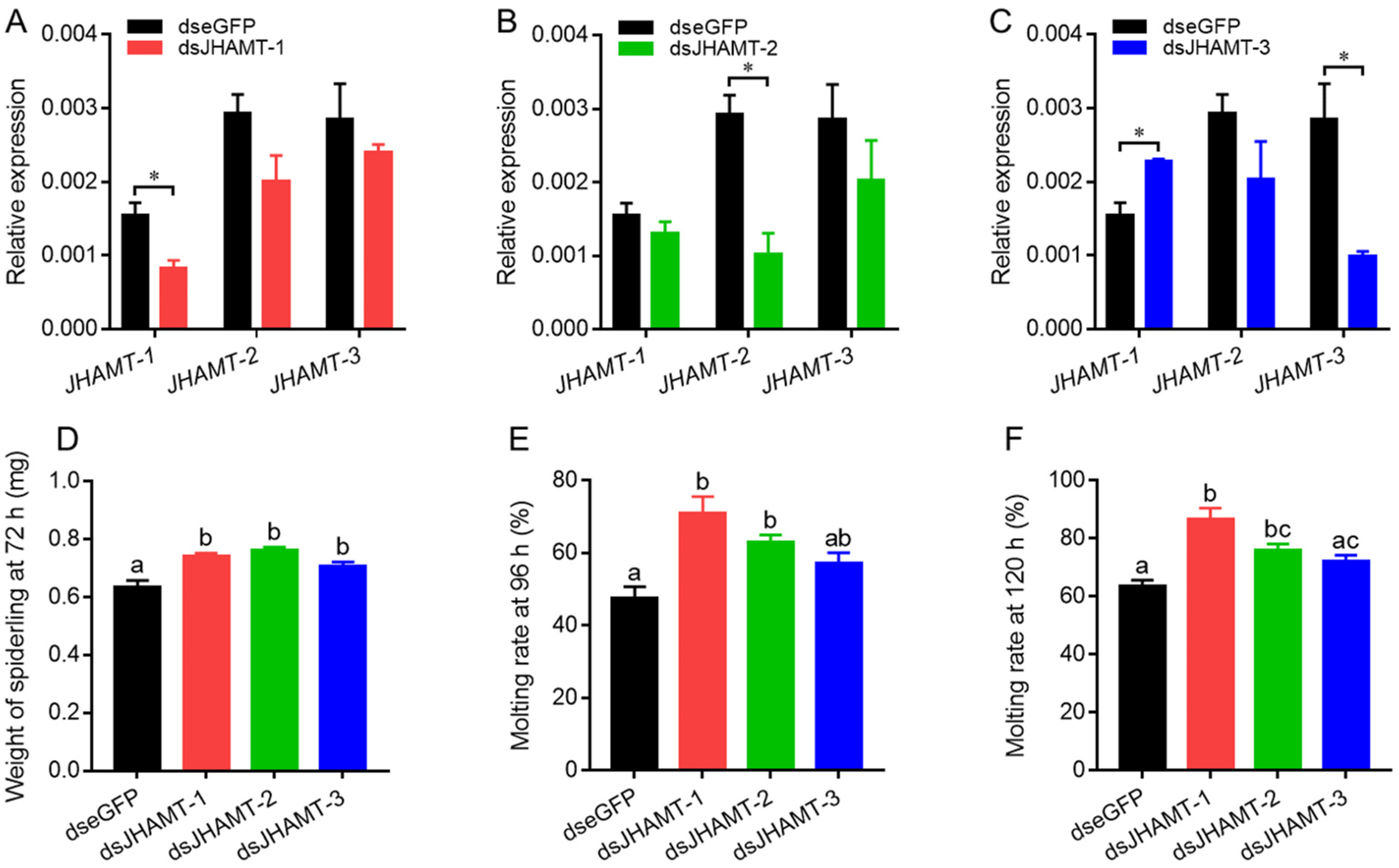
2.5. Effect of JHAMT Silencing
3. Discussion
4. Materials and Methods
4.1. Identification and Characterization of JHAMTs
4.2. Spiders
4.3. Farnesoic Acid Treatment
4.4. RNA Interference
4.5. Real-Time Quantitative PCR
4.6. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Goodman, W.G.; Granger, N.A. The juvenile hormones. In Comprehensive Molecular Insect Science; Gilbert, L.I., Iatrou, K., Gill, S.S., Eds.; Elsevier: Oxford, UK, 2005; Volume 3, pp. 319–408. [Google Scholar]
- Applebaum, S.W.; Avisar, E.; Heifetz, Y. Juvenile hormone and locust phase. Arch. Insect Biochem. Physiol. 1997, 35, 375–391. [Google Scholar] [CrossRef]
- Hartfelder, K. Insect juvenile hormone: From “status quo” to high society. Braz. J. Med. Biol. Res. 2000, 33, 157–177. [Google Scholar] [CrossRef]
- Wyatt, G.R.; Davey, K.G. Cellular and molecular actions of juvenile hormone II. Roles of juvenile hormone in adult insects. Adv. Insect Physiol. 1996, 26, 1–155. [Google Scholar]
- Gilbert, L.I.; Granger, N.A.; Roe, R.M. The juvenile hormones: Historical facts and speculations on future research directions. Insect Biochem. Mol. Biol. 2000, 30, 617–644. [Google Scholar] [CrossRef]
- Riddiford, L.M. Cellular and molecular actions of juvenile hormone I. General considerations and premetamorphic actions. Adv. Insect Physiol. 1994, 24, 213–274. [Google Scholar]
- Shinoda, T.; Itoyama, K. Juvenile hormone acid methyltransferase: A key regulatory enzyme for insect metamorphosis. Proc. Natl. Acad. Sci. USA 2003, 100, 11986–11991. [Google Scholar] [CrossRef]
- Martin, J.L.; McMillan, F.M. SAM (dependent) I AM: The S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002, 12, 783–793. [Google Scholar] [CrossRef]
- Niwa, R.; Niimi, T.; Honda, N.; Yoshiyama, M.; Itoyama, K.; Kataoka, H.; Shinoda, T. Juvenile hormone acid O-methyltransferase in Drosophila melanogaster. Insect Biochem. Mol. Biol. 2008, 38, 714–720. [Google Scholar] [CrossRef]
- Minakuchi, C.; Namiki, T.; Yoshiyama, M.; Shinoda, T. RNAi-mediated knockdown of juvenile hormone acid O-methyltransferase gene causes precocious metamorphosis in the red flour beetle Tribolium castaneum. FEBS J. 2008, 275, 2919–2931. [Google Scholar] [CrossRef]
- Mayoral, J.G.; Nouzova, M.; Yoshiyama, M.; Shinoda, T.; Hernandez-Martinez, S.; Dolghih, E.; Turjanski, A.G.; Roitberg, A.E.; Priestap, H.; Perez, M.; et al. Molecular and functional characterization of a juvenile hormone acid methyltransferase expressed in the corpora allata of mosquitoes. Insect Biochem. Mol. Biol. 2009, 39, 31–37. [Google Scholar] [CrossRef]
- Ollivier, M.; Legeai, F.; Rispe, C. Comparative analysis of the Acyrthosiphon pisum genome and expressed sequence tag-based gene sets from other aphid species. Insect Mol. Biol. 2010, 19, 33–45. [Google Scholar] [CrossRef]
- Li, W.; Huang, Z.Y.; Liu, F.; Li, Z.; Yan, L.; Zhang, S.; Chen, S.; Zhong, B.; Su, S. Molecular cloning and characterization of juvenile hormone acid methyltransferase in the honey bee, Apis mellifera, and its differential expression during caste differentiation. PLoS ONE 2013, 8, e68544. [Google Scholar] [CrossRef]
- Fu, K.-Y.; Li, Q.; Zhou, L.-T.; Meng, Q.-W.; Lu, F.-G.; Guo, W.-C.; Li, G.-Q. Knockdown of juvenile hormone acid methyl transferase severely affects the performance of Leptinotarsa decemlineata (Say) larvae and adults. Pest Manag. Sci. 2016, 72, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.V.; Maestro, J.L. Expression of juvenile hormone acid O-methyltransferase and juvenile hormone synthesis in Blattella germanica. Insect Sci. 2018, 25, 787–796. [Google Scholar] [CrossRef]
- Grbic, M.; Van Leeuwen, T.; Clark, R.M.; Rombauts, S.; Rouze, P.; Grbic, V.; Osborne, E.J.; Dermauw, W.; Ngoc, P.C.; Ortego, F.; et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 2011, 479, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Sun, Q.-Z.; Liu, X.-Y.; Zhang, J.; Dou, W.; Niu, J.-Z.; Wang, J.-J. Expression dynamics of key ecdysteroid and juvenile hormone biosynthesis genes imply a coordinated regulation pattern in the molting process of a spider mite, Tetranychus urticae. Exp. Appl. Acarol. 2019, 78, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Yu, Y.; Wu, Y.; Hao, P.; Di, Z.; He, Y.; Chen, Z.; Yang, W.; Shen, Z.; He, X.; et al. The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat. Commun. 2013, 4, 2602. [Google Scholar] [CrossRef] [PubMed]
- Gulia-Nuss, M.; Nuss, A.B.; Meyer, J.M.; Sonenshine, D.E.; Roe, R.M.; Waterhouse, R.M.; Sattelle, D.B.; de la Fuente, J.; Ribeiro, J.M.; Megy, K.; et al. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat. Commun. 2016, 7, 10507. [Google Scholar] [CrossRef]
- Zhu, J.; Khalil, S.M.; Mitchell, R.D.; Bissinger, B.W.; Egekwu, N.; Sonenshine, D.E.; Roe, R.M. Mevalonate-farnesal biosynthesis in ticks: Comparative synganglion transcriptomics and a new perspective. PLoS ONE 2016, 11, e0141084. [Google Scholar] [CrossRef][Green Version]
- Cheng, D.; Meng, M.; Peng, J.; Qian, W.; Kang, L.; Xia, Q. Genome-wide comparison of genes involved in the biosynthesis, metabolism, and signaling of juvenile hormone between silkworm and other insects. Genet. Mol. Biol. 2014, 37, 444–459. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, A.; Ogawa, K.; Gotoh, H.; Walsh, T.K.; Tagu, D.; Brisson, J.A.; Rispe, C.; Jaubert-Possamai, S.; Kanbe, T.; Tsubota, T.; et al. Juvenile hormone titre and related gene expression during the change of reproductive modes in the pea aphid. Insect Mol. Biol. 2012, 21, 49–60. [Google Scholar] [CrossRef]
- Tobe, S.S.; Stay, B. Structure and regulation of the corpus allatum. Adv. Insect Physiol. 1985, 18, 305–432. [Google Scholar]
- Yang, Z.-M.; Wu, Y.; Xu, G.-M.; Yu, N.; Liu, Z.-W. Methyl farnesoate acts as the endogenous juvenile hormone in spider moulting. Insect Mol. Biol. 2021, submitted. [Google Scholar]
- Slater, G.S.; Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 2005, 6, 31. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Nicholas, K.B.; Nicholas, H.B., Jr.; Deerfield, D.W., II. GeneDoc: Analysis and visualization of genetic variation. EMBNEW News 1997, 4, 14. [Google Scholar]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Yachdav, G.; Kloppmann, E.; Kajan, L.; Hecht, M.; Goldberg, T.; Hamp, T.; Honigschmid, P.; Schafferhans, A.; Roos, M.; Bernhofer, M.; et al. PredictProtein-an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014, 42, W337–W343. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef]
- Meng, X.; Li, C.; Bao, H.; Fang, J.; Liu, Z.; Zhang, Y. Validating the importance of two acetylcholinesterases in insecticide sensitivities by RNAi in Pardosa pseudoannulata, an important predatory enemy against several insect pests. Pest. Biochem. Physiol. 2015, 125, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Swift, M.L. GraphPad prism, data analysis, and scientific graphing. J. Chem. Inf. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Z.-M.; Wu, Y.; Li, F.-F.; Zhou, Z.-J.; Yu, N.; Liu, Z.-W. Genomic Identification and Functional Analysis of JHAMTs in the Pond Wolf Spider, Pardosa pseudoannulata. Int. J. Mol. Sci. 2021, 22, 11721. https://doi.org/10.3390/ijms222111721
Yang Z-M, Wu Y, Li F-F, Zhou Z-J, Yu N, Liu Z-W. Genomic Identification and Functional Analysis of JHAMTs in the Pond Wolf Spider, Pardosa pseudoannulata. International Journal of Molecular Sciences. 2021; 22(21):11721. https://doi.org/10.3390/ijms222111721
Chicago/Turabian StyleYang, Zhi-Ming, Yong Wu, Fang-Fang Li, Zhang-Jin Zhou, Na Yu, and Ze-Wen Liu. 2021. "Genomic Identification and Functional Analysis of JHAMTs in the Pond Wolf Spider, Pardosa pseudoannulata" International Journal of Molecular Sciences 22, no. 21: 11721. https://doi.org/10.3390/ijms222111721
APA StyleYang, Z.-M., Wu, Y., Li, F.-F., Zhou, Z.-J., Yu, N., & Liu, Z.-W. (2021). Genomic Identification and Functional Analysis of JHAMTs in the Pond Wolf Spider, Pardosa pseudoannulata. International Journal of Molecular Sciences, 22(21), 11721. https://doi.org/10.3390/ijms222111721





