Pharmacogenetics of Drug-Resistant Epilepsy (Review of Literature)
Abstract
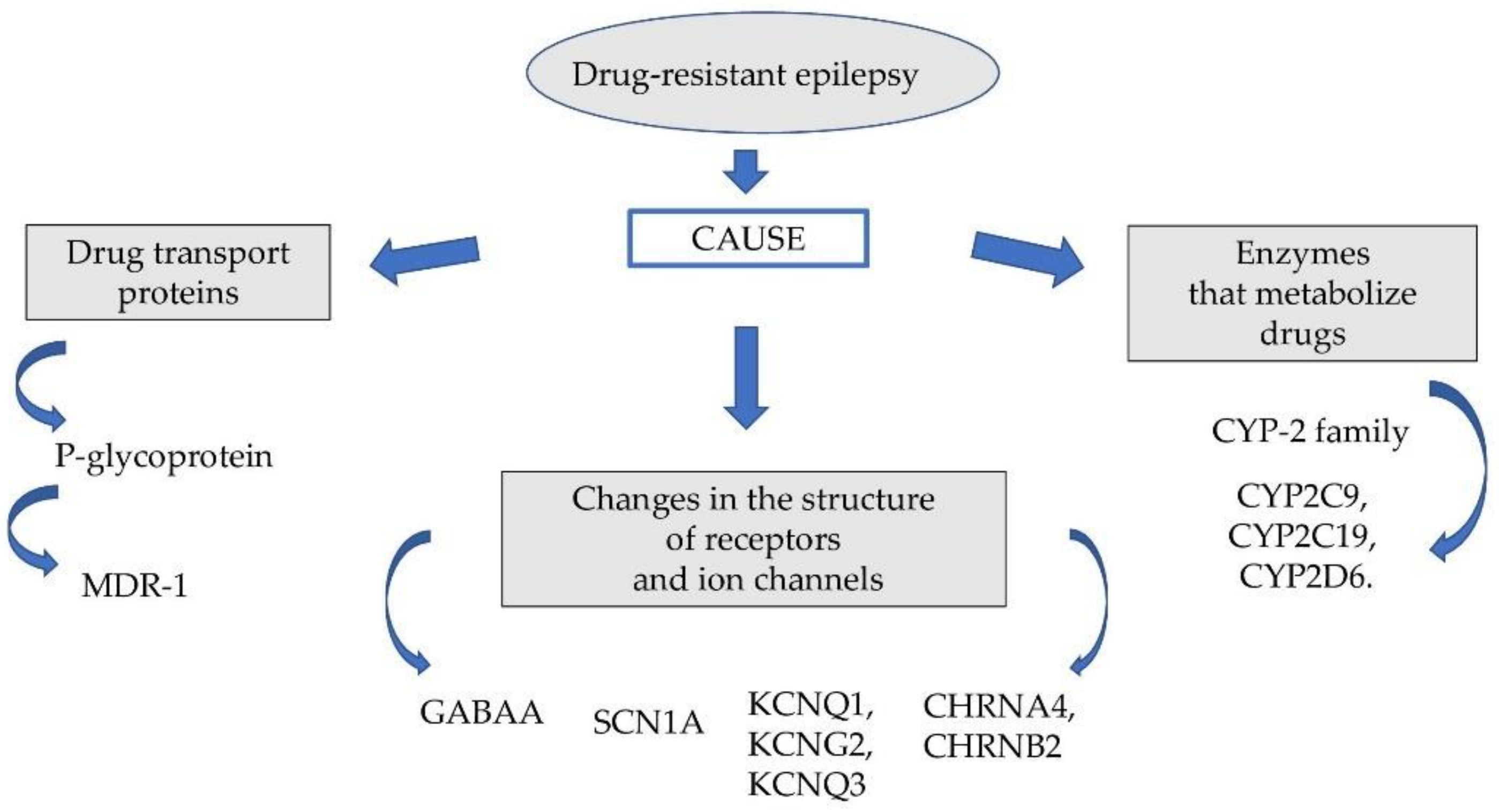
1. Introduction
2. Drug Transport Proteins
3. Enzymes That Metabolize Drugs
4. Changes in the Structure of Receptors and Ion Channels
5. Currently Used Antiepileptic Drugs
6. Conclusive Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vogel, F. Moderne Probleme der Humangenetik. Ergeb. Inn. Med. Kinderheilkd. 1959, 12, 52–125. [Google Scholar]
- Mancinelli, L.; Cronin, M.; Sadée, W. Pharmacogenomics: The promise of personalized medicine. AAPS PharmSci. 2000, 2, E4. [Google Scholar] [CrossRef] [PubMed]
- Klimek, A. Farmakogenetyka padaczki. Aktualn. Neurol. 2007, 7, 4–9. [Google Scholar]
- Białecka, M.; Kłodowska-Duda, G.; Kurzawski, M.; Droździk, M. Farmakogenetyka—Nowe podejście do leczenia choroby Parkinsona. Neurol. Neurochir. Pol. 2008, 42, 131–138. [Google Scholar]
- Hauser, J.; Leszczyńska-Rodziewicz, A. Farmakogenetyka leków przeciwpsychotycznych. Psychiatria 2004, 1, 81–89. [Google Scholar]
- Szoeke, C.E.; Newton, M.; Wood, J.M.; Goldstein, D.; Berkovic, S.F.; Obrien, T.J.; Sheffield, L.J. Update on pharmacogenetics in epilepsy: A brief review. Lancet Neurol. 2006, 5, 189–196. [Google Scholar] [CrossRef]
- Margineanu, D.G.; Klitgaard, H. Mechanisms of drug resistance in epilepsy: Relevance for antiepileptic drug discovery. Expert Opin. Drug Discov. 2009, 4, 23–32. [Google Scholar] [CrossRef]
- Kwan, P.; Arzimanoglou, A.; Berg, A.T.; Brodie, M.J.; Allen Hauser, W.; Mathern, G.; Moshé, S.L.; Perucca, E.; Wiebe, S.; French, J. Definition of drug resistant epilepsy: Consensus proposal by the ad hoc Task Force of the ILAE Commission on Therapeutic Strategies. Epilepsia 2010, 51, 1069–1077. [Google Scholar] [CrossRef] [PubMed]
- Regesta, G.; Tanganelli, P. Clinical aspects and biological bases of drug-resistant epilepsies. Epilepsy Res. 1999, 34, 109–122. [Google Scholar] [CrossRef]
- Spear, B.B. Pharmacogenetics and antiepileptic drugs. Epilepsia 2001, 42, 31–34. [Google Scholar] [CrossRef]
- Löscher, W.; Potschka, H. Role of multidrug transporters in pharmacoresistance to antiepileptic drugs. J. Pharmacol. Exp. Ther. 2002, 301, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Juliano, R.L.; Ling, V. A surface glycoprotein modulating drug permeability in Chinese hamster cells ovary cell mutants. Biochem. Biophys. Acta 1976, 455, 152–162. [Google Scholar] [CrossRef]
- Rao, V.V.; Dahlheimer, J.L.; Bardgett, M.E.; Snyder, A.Z.; Finch, R.A.; Sartorelli, A.C.; Piwnica-Worms, D. Choroid plexus epithelial expression of MDR1 P glycoprotein and multidrug resistance-associated protein contribute to the blood-cerebrospinal-fluid drug-permeability barrier. Proc. Natl. Acad. Sci. USA 1999, 96, 3900–3905. [Google Scholar] [CrossRef] [PubMed]
- Cordon-Cardo, C.; O’Brien, J.P.; Casals, D.; Rittman-Grauer, L.; Biedler, J.L.; Melamed, M.R.; Bertino, J.R. Multidrug-resistance gene (P-glyco-protein) is expressed by endothelial cells at blood-brain barrier sites. Proc. Natl. Acad. Sci. USA 1989, 86, 695–698. [Google Scholar] [CrossRef]
- Johnstone, R.W.; Ruefli, A.A.; Smyth, M.J. Multiple physiological functions for multidrug transporter P-glycoprotein? Trends Biochem. Sci. 2000, 25, 1–6. [Google Scholar] [CrossRef]
- Sakurai, A.; Tamura, A.; Onishi, Y.; Ishikawa, T. Genetic polymorphisms of ATP-binding cassette transporters ABCB1 and ABCG2: Therapeutic implications. Expert Opin. Pharmacother. 2005, 6, 2455–2473. [Google Scholar] [CrossRef] [PubMed]
- Mayer, U.; Wagenaar, E.; Beijnen, J.H.; Smit, J.W.; Meijer, D.K.; van Asperen, J.; Borst, P.; Schinkel, A.H. Substantial excretion of digoxin via the intestinal mucosa and prevention of long-term digoxin accumulation in the brain by the mdr 1a P-glycoprotein. Br. J. Pharmacol. 1996, 119, 1038–1044. [Google Scholar] [CrossRef] [PubMed]
- Mayer, U.; Wagenaar, E.; Dorobek, B.; Beijnen, J.H.; Borst, P.; Schinkel, A.H. Full blockade of intestinal P-glycoprotein and extensive inhibition of blood-brain barrier P-glycoprotein by oral treatment of mice with PSC833. J. Clin. Investig. 1997, 100, 2430–2436. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.S.; Bauer, B.; Hartz, A.M.B. Modulation of P-glycoprotein at the blood-brain barrier: Opportunities to improve central nervous system pharmacotherapy. Pharmacol. Rev. 2008, 60, 196–209. [Google Scholar] [CrossRef] [PubMed]
- Kortekaas, R.; Leenders, K.L.; van Oostrom, J.C.; Vaalburg, W.; Bart, J.; Willemsen, A.T.; Hendrikse, N.H. Blood-brain barrier dysfunction in parkinsonian midbrain in vivo. Ann. Neurol. 2005, 57, 176–179. [Google Scholar] [CrossRef]
- Rizzi, M.; Caccia, S.; Guiso, G.; Richichi, C.; Gorter, J.A.; Aronica, E.; Aliprandi, M.; Bagnati, R.; Fanelli, R.; D’Incalci, M. Limbic seizures induce P-glycoprotein in rodent brain: Functional implications for pharmacoresistance. J. Neurosci. 2002, 22, 5833–5839. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, E.A.; van Schaik, R.; Edelbroek, P.M.; Voskuyl, R.A.; Redeker, S.; Aronica, E.; Wadman, W.J.; Gorter, J.A. Region-specific overexpression of P-glycoprotein at the blood-brain barrier affects brain uptake of phenytoin in epileptic rats. J. Pharmacol. Exp. Ther. 2007, 322, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, E.A.; van Schaik, R.; Edelbroek, P.M.; Redeker, S.; Aronica, E.; Wadman, W.J.; Marchi, N.; Vezzani, A.; Gorter, J.A. Inhibition of the multidrug transporter P-glycoproteinimproves seizure control in phenytoin-treated chronic epileptic rats. Epilepsja 2006, 47, 672–680. [Google Scholar] [CrossRef] [PubMed]
- Brandt, C.; Bethmann, K.; Gastens, A.M.; Löscher, W. The multidrug transporter hypothesis of drug resistance in epilepsy: Proof-of-principle in a rat model of temporal lobe epilepsy. Neurobiol. Dis. 2006, 24, 202–211. [Google Scholar] [CrossRef]
- Sisodiya, S.M.; Thom, M. Widespread upregulation of drug-resistance proteins in fatal human status epilepticus. Epilepsja 2003, 44, 261–264. [Google Scholar] [CrossRef]
- Pekcec, A.; Unkrüer, B.; Stein, V.; Bankstahl, J.P.; Soerensen, J.; Tipold, A.; Baumgärtner, W.; Potschka, H. Over-expression of P-glycoprotein in the canine brain following spontaneous status epilepticus. Epilepsy Res. 2009, 83, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Rambeck, B.; Jürgens, U.H.; May, T.W.; Pannek, H.W.; Behne, F.; Ebner, A.; Gorji, A.; Straub, H.; Speckmann, E.J.; Pohlmann-Eden, B. Comparison of brain extracellular fluid, brain tissue, cerebrospinal fluid, and serum concentrations of antiepileptic drugs measured intra-operatively in patients with intractable seizures. Epilepsia 2006, 47, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Marchi, N.; Hallene, K.L.; Kight, K.M.; Cucullo, L.; Moddel, G.; Bingaman, W.; Dini, G.; Vezzani, A.; Janigro, D. Significance of MDR1 and multiple drug resistance in refractory human epi-leptic brain. BMC Med. 2004, 2, 37. [Google Scholar] [CrossRef] [PubMed]
- Tishler, D.M.; Weinberg, K.I.; Hinton, D.R.; Barbaro, N.; Annett, G.M.; Raffel, C. MDR1 gene expression in brain of patients with medically intractable epilepsy. Epilepsia 1995, 36, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Skalski, D.; Smolarz, B.; Wendorff, J. Związek pomiędzy polimorfizmami pojedynczych nukleotydów genu oporności wielolekowej typu 1. a padaczką lekooporną. Neuropsychiatry Neuropsychol. 2011, 2, 79–84. [Google Scholar]
- Hoffmeyer, S.; Burk, O.; von Richter, O.; Brockmöller, J.; Johne, A.; Cascorbi, I.; Gerloff, T.; Roots, I.; Eichelbaum, M.; Brinkmann, U. Functional polymorphisms of the human multidrug-resistance gene: Multiple sequence variations and correlation of the one allele with P-glycoprotein expression and activity in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 3473–3478. [Google Scholar] [CrossRef]
- Dean, M.; Rzhetsky, A.; Allikmets, R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001, 11, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Zimprich, F.; Sunder-Plassmann, R.; Stogmann, E.; Gleiss, A.; Dal-Bianco, A.; Zimprich, A.; Plumer, S.; Baumgartner, C.; Mannhalter, C. Association of an ABCB1 gene haplotype with pharmacoresistance in temporal lobe epilepsy. Neurology 2004, 63, 1087–1089. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Sadée, W. Searching for polymorphisms that affect gene expression and mRNA processing: Example ABCB1 (MDR1). AAPS J. 2006, 8, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, E.A.; Zibell, G.; Pekcec, A.; Schlichtiger, J.; Edelbroek, P.M.; Holtman, L.; Aronica, E.; Gorter, J.A.; Potschka, H. COX-2 inhibition controls P-glycoprotein expression and promotes brain delivery of phenytoin in chronic epileptic rats. Neuropharmacology 2010, 58, 404–412. [Google Scholar] [CrossRef]
- Lazarowski, A.; Czornyj, L. Potential role of multidrug resis-tant proteins in refractory epilepsy and antiepileptic drugs interactions. Drug Metabol. Drug Interact. 2011, 26, 21–26. [Google Scholar] [CrossRef]
- Siddiqui, A.; Kerb, R.; Weale, M.E.; Brinkmann, U.; Smith, A.; Goldstein, D.B.; Wood, N.W.; Sisodiya, S.M. Association of multi-drug resistance in epilepsy with a polymorphism in the drug-transporter gene ABCB1. N. Engl. J. Med. 2003, 348, 1442–1448. [Google Scholar] [CrossRef]
- Soranzo, N.; Goldstein, D.B.; Sisodiya, S.M. The role of com-mon variation in drug transporter genes in refractory epilepsy. Expert Opin. Pharmacother. 2005, 6, 1305–1312. [Google Scholar] [CrossRef]
- Von Stülpnagel, C.; Plischke, H.; Zill, P.; Bäumel, C.; Spiegel, R.; Gruber, R.; Kluger, G. Letter: Lack of association between MDR1 polymorphisms and pharmacore-sistance to anticonvulsive drugs in patients with childhood-onset epilepsy. Epilepsia 2009, 50, 1835–1837. [Google Scholar] [CrossRef]
- Vahab, S.A.; Sen, S.; Ravindran, N.; Mathew, A.; Vijayan, N.; Nayak, G.; Bhaskaranand, N.; Banerjee, M.; Satyamoorthy, K. Analysis of genotype and haplotype effects of ABCB1 (MDR1) polymorphisms in the risk of medically refractory epilepsy in an Indian population. Drug Metab. Pharmacokinet. 2009, 24, 255–260. [Google Scholar] [CrossRef]
- Tan, N.C.; Heron, S.E.; Scheffer, I.E.; Pelekanos, J.T.; McMahon, J.M.; Vears, D.F.; Mulley, J.C.; Berkovic, S.F. Failure to confirm association of a polymorphism in ABCB1 with multidrug-resistant epilepsy. Neurology 2004, 63, 1090–1092. [Google Scholar] [CrossRef] [PubMed]
- Sills, G.J.; Mohanraj, R.; Butler, E.; McCrindle, S.; Collier, L.; Wilson, E.A.; Brodie, M.J. Lack of association between the C3435T polymorphism in the human multidrug resistance (MDR1) gene and response to antiepileptic drug treatment. Epilepsia 2005, 46, 643–647. [Google Scholar] [CrossRef]
- Cascorbi, I.; Gerloff, T.; Johne, A.; Meisel, C.; Hoffmeyer, S.; Schwab, M.; Schaeffeler, E.; Eichelbaum, M.; Brinkmann, U.; Roots, I. Frequency of single nucleotide polymorphisms in the P-glycoprotein drug trans-porter MDR1 gene in white subjects. Clin. Pharmacol. Ther. 2001, 69, 169–174. [Google Scholar] [CrossRef]
- Mosyagin, I.; Runge, U.; Schroeder, H.W.; Dazert, E.; Vogelgesang, S.; Siegmund, W.; Warzok, R.W.; Cascorbi, I. Association of ABCB1 genetic variants 3435C>T and 2677G>T to ABCB1 mRNA and protein expression in brain tissue from refractory epilepsy patients. Epilepsia 2008, 49, 1555–1561. [Google Scholar] [CrossRef]
- Alpman, A.; Ozkinay, F.; Tekgul, H.; Gokben, S.; Pehlivan, S.; Schalling, M.; Ozkinay, C. Multidrug resistance 1 (MDR1) gene polymorphisms in childhood drug-resistant epilepsy. J. Child. Neurol. 2010, 25, 1485–1490. [Google Scholar] [CrossRef] [PubMed]
- Stasiołek, M.; Romanowicz, H.; Połatyńska, K.; Chamielec, M.; Skalski, D.; Makowska, M.; Smolarz, B. Association between C3435T polymorphism of MDR1 gene and the incidence of drug-resistant epilepsy in the population of Polish children. Behav. Brain Funct. 2016, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Smolarz, B.; Skalski, D.; Rysz, A.; Marchel, A.; Romanowicz, H.; Makowska, M. Polymorphism of the multidrug resistance 1 gene MDR1 G2677T/A (rs2032582) and the risk of drug-resistant epilepsy in the Polish adult population. Acta Neurol. Belg. 2017, 117, 849–855. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Yu, J.; Wang, T.T.; Feng, J.; Zhao, W.B.; Sun, L.; Yu, L.H.; Li, H.J.; Sun, Y. Impact of ABCB1 Polymorphism on Levetiracetam Serum Concentrations in Epileptic Uygur Children in China. Ther. Drug. Monit. 2020, 42, 886–892. [Google Scholar] [CrossRef]
- Gao, L.; Yin, X.; Li, Y.; Xiao, H.; Yang, L.; Fan, H.; Qi, H.; Zhang, J.; Feng, J.; Zheng, F. Association of MDR1 gene polymorphisms with refractory epilepsy in children. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2019, 36, 1073–1076. [Google Scholar] [PubMed]
- Heinrich, A.; Zhong, X.B.; Rasmussen, T.P. Variability in expression of the human MDR1 drug efflux transporter and genetic variation of the ABCB1 gene: Implications for drug-resistant epilepsy. Curr. Opin. Toxicol. 2018, 11–12, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Lv, R.J.; Shao, X.Q.; Cui, T.; Wang, Q. Significance of MDR1 gene C3435T polymorphism in predicting childhood refractory epilepsy. Epilepsy Res. 2017, 132, 21–28. [Google Scholar] [CrossRef]
- Tamimi, D.E.; Abduljabbar, R.; Yousef, A.M.; Saeed, R.M.; Zawiah, M. Association between ABCB1 polymorphisms and response to antiepileptic drugs among Jordanian epileptic patients. Neurol. Res. 2021, 43, 724–735. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Sun, S.; Zhang, W.; Shi, Z. Polymorphisms of ABCG2 and its impact on clinical relevance. Biochem. Biophys. Res. Commun. 2018, 503, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yang, Y.; Dong, Z.; Shi, Z.; Zhang, J.T. Single-nucleotide polymorphisms in a short basic motif in the ABC transporter ABCG2 disable its trafficking out of endoplasmic reticulum and reduce cell resistance to anticancer drugs. J. Biol. Chem. 2019, 294, 20222–20232. [Google Scholar] [CrossRef]
- Chen, J.; Su, Q.; Qin, J.; Zhou, Y.; Ruan, H.; Chen, Z.; Chen, Z.; Li, H.; Zhou, Y.; Zhou, S. Correlation of MCT1 and ABCC2 gene polymorphisms with valproic acid resistance in patients with epilepsy on valproic acid monotherapy. Drug. Metab. Pharmacokinet. 2019, 34, 165–171. [Google Scholar] [CrossRef]
- Al-Eitan, L.N.; Al-Dalalah, I.M.; Mustafa, M.M.; Alghamdi, M.A.; Elshammari, A.K.; Khreisat, W.H.; Aljamal, H.A. Effects of MTHFR and ABCC2 gene polymorphisms on antiepileptic drug responsiveness in Jordanian epileptic patients. Pharmacogenomics Pers. Med. 2019, 12, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yan, Y.; Fang, S.; Zeng, S.; Ma, H.; Qian, L.; Chen, X.; Wei, J.; Gong, Z.; Xu, Z. Comparison of oxcarbazepine efficacy and MHD concentrations relative to age and BMI: Associations among ABCB1, ABCC2, UGT2B7, and SCN2A polymorphisms. Medicine 2019, 98, e14908. [Google Scholar] [CrossRef]
- Shen, C.H.; Zhang, Y.X.; Lu, r.Y.; Jin, B.; Wang, S.; Liu, Z.R.; Tang, Y.L.; Ding, M.P. Specific OCT1 and ABCG2 polymorphisms are associated with Lamotrigine concentrations in Chinese patients with epilepsy. Epilepsy Res. 2016, 127, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Zan, X.; Yue, G.; Hao, Y.; Sima, X. A systematic review and meta-analysis of the association of ABCC2/ABCG2 polymorphisms with antiepileptic drug responses in epileptic patients. Epilepsy Res. 2021, 175, 106678. [Google Scholar] [CrossRef]
- Awasthi, S.; Hallene, K.L.; Fazio, V.; Singhal, S.S.; Cucullo, L.; Awasthi, Y.C.; Dini, G.; Janigro, D. RLIP76 a non-ABC transporter and drug resistance in epilepsy. BMC Neurosci. 2005, 6, 61. [Google Scholar] [CrossRef]
- Soranzo, N.; Kelly, L.; Martinian, L.; Burley, M.W.; Thom, M.; Sali, A.; Kroetz, D.L.; Goldstein, D.B.; Sisodiya, S.M. Lack of support for a role for RLIP76 (RALBP1) in response to treatment or predisposition to epilepsy. Epilepsia 2007, 48, 674–683. [Google Scholar] [CrossRef]
- Leschziner, G.D.; Jorgensen, A.L.; Andrew, T.; Williamson, P.R.; Marson, A.G.; Coffey, A.J.; Middleditch, C.; Balding, D.J.; Rogers, J.; Bentley, D.R. The association between polymorphisms in RLIP76 and drug response in epilepsy. Pharmacogenomics 2007, 8, 1715–1722. [Google Scholar] [CrossRef]
- Lin, J.H.; Lu, A.Y. Role of pharmacokinetics and metabolism in drug discovery and development. Pharmacol. Rev. 1997, 49, 403–449. [Google Scholar]
- Pierzchała, K. Padaczka oporna na leczenie—Epidemiologia i aktualny stan badań. Neurol. Neurochir. Pol. 2010, 44, 285–290. [Google Scholar] [CrossRef]
- Brandolese, R.; Scordo, M.G.; Spina, E.; Gusella, M.; Padrini, R. Severe phenytoin intoxication in a subject homozygous for CYP2C9*3. Clin. Pharmacol. Ther. 2001, 70, 391–394. [Google Scholar] [CrossRef]
- Van der Weide, J.; Steijns, L.S.; van Weelden, M.J.; de Haan, K. The effect of genetic polymorphism of cytochrome P450 CYP2C9 on phenytoin dose requirement. Pharmacogenetics 2001, 11, 287–291. [Google Scholar] [CrossRef]
- Kurkowska-Jastrzębska, I.; Pilip, S.; Niedzielska, I.; Barańska-Gieruszczak, M. Padaczka lekooporna a czynniki genetyczne. Farmakoter. Psych. Neurol. 2005, 1, 25–31. [Google Scholar]
- López-García, M.A.; Feria-Romero, I.A.; Serrano, H.; Rayo-Mares, D.; Fagiolino, P.; Vázquez, M.; Escamilla-Núñez, C.; Grijalva, I.; Escalante-Santiago, D.; Orozco-Suarez, S. Influence of genetic variants of CYP2D6, CYP2C9, CYP2C19 and CYP3A4 on antiepileptic drug metabolism in pediatric patients with refractory epilepsy. Pharmacol. Rep. 2017, 69, 504–511. [Google Scholar] [CrossRef]
- Makowska, M.; Smolarz, B.; Bryś, M.; Forma, E.; Romanowicz, H. An association between the rs1799853 and rs1057910 polymorphisms of CYP2C9, the rs4244285 polymorphism of CYP2C19 and the prevalence rates of drug-resistant epilepsy in children. Int. J. Neurosci. 2020, 30, 1–8. [Google Scholar] [CrossRef]
- Emich-Widera, E.; Likus, W.; Kazek, B.; Niemiec, P.; Balcerzyk, A.; Sieroń, A.L.; Zak, I. CYP3A5*3 and C3435T MDR1 polymorphisms in prognostication of drug-resistant epilepsy in children and adolescents. Biomed. Res. Int. 2013, 2013, 526837. [Google Scholar] [CrossRef] [PubMed]
- Sigel, E.; Steinmann, M.E. Structure, function, and modulation of GABA(A) receptors. J. Biol. Chem. 2012, 287, 40224–40231. [Google Scholar] [CrossRef]
- Jóźwiak, S. Współczesne poglądy na klasyfikację, patogenezę i postępowanie w padaczce lekoopornej. Wiad. Lek. 2007, 60, 258–264. [Google Scholar] [PubMed]
- Gambardella, A.; Manne, J.; Labate, A.; Chifari, R.; La Russa, A.; Serra, P.; Cittadella, R.; Bonavita, S.; Andreoli, V.; LePiane, E. GABA(B) receptor 1 polymorphism (G1465A) is associated with temporal lobe epilepsy. Neurology 2003, 60, 560–563. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Abou-Khalil, B.; Sutcliffe, J.S.; Haines, J.L.; Hedera, P. The GABBR1 locus and the G1465A variant is not associated with temporal lobe epilepsy preceded by febrile seizures. BMC Med. Genet. 2005, 6, 13. [Google Scholar] [CrossRef]
- Salzmann, A.; Moulard, B.; Crespel, A.; Baldy-Moulinier, M.; Buresi, C.; Malafosse, A. GABA receptor 1 polymorphism (G1465A) and temporal lobe epilepsy. Epilepsia 2005, 46, 931–933. [Google Scholar] [CrossRef]
- Jóźwiak, S. Postępy w neurologii dziecięcej w 2005 roku. Med. Prakt. Pediatr. 2006, 4, 80–82. [Google Scholar]
- Różycka, A.; Dorszewska, J.; Jagodziński, P.P. Ion channel dysfunction in pathogenesis of idiopathic epilepsies. Neurol. Neurochir. Pol. 2011, 45, 42–56. [Google Scholar] [CrossRef]
- Goldin, A.L. Mechanisms of sodium channel inactivation. Curr. Opin. Neurobiol. 2003, 3, 284–290. [Google Scholar] [CrossRef]
- Bisulli, F.; Licchetta, L.; Tinuper, P. Sleep related hyper motor epilepsy (SHE): A unique syndrome with heterogeneous genetic etiologies. Sleep Sci. Pract. 2019, 3, 3. [Google Scholar] [CrossRef]
- Remy, S.; Urban, B.W.; Elger, C.E.; Beck, H. Anticonvulsant pharmacology of voltage-gated Na+ channels in hippocampal neurons of control and chronically epileptic rats. Eur. J. Neurosci. 2003, 17, 2648–2658. [Google Scholar] [CrossRef]
- Marini, C.; Guerrini, R. The role of the nicotinic acetylcholine receptors in sleep-related epilepsy. Biochem. Pharmacol. 2007, 74, 1308–1314. [Google Scholar] [CrossRef]
- Tinuper, P.; Bisulli, F. From nocturnal frontal lobe epilepsy to Sleep-Related Hypermotor Epilepsy: A 35-year diagnostic challenge. Seizure 2017, 44, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Steinlein, O.K. Genetic heterogeneity in familial nocturnal frontal lobe epilepsy. Prog. Brain Res. 2014, 213, 1–15. [Google Scholar] [PubMed]
- Steinlein, O.K.; Mulley, J.C.; Propping, P.; Wallace, R.H.; Phillips, H.A.; Sutherland, G.R.; Scheffer, I.E.; Berkovic, S.F. A missense mutation in the neuronal nicotinic acethylcholine receptor a4 subunit is associated with autosomal dominant nocturnal frontal lobe epilepsy. Nat. Genet. 1995, 11, 201–203. [Google Scholar] [CrossRef]
- Phillips, H.A.; Marini, C.; Scheffer, I.E.; Sutherland, G.R.; Mulley, J.C.; Berkovic, S.F. A de novomutation in sporadic nocturnal frontal lobe epilepsy. Ann. Neurol. 2000, 48, 264–267. [Google Scholar] [CrossRef]
- Gu, W.; Bertrand, D.; Steinlein, O.K. A major role of the nicotinic acetylcholine receptor gene CHRNA2 in autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE) is unlikely. Neurosci. Lett. 2007, 422, 74–76. [Google Scholar] [CrossRef]
- De Fusco, M.D.; Becchetti, A.; Patrignani, A.; Annesi, G.; Gambardella, A.; Quattrone, A.; Ballabio, A.; Wanke, E.; Casari, G. The nicotinic receptor beta2 subunit is mutant in nocturnal frontal lobe epilepsy. Nat. Genet. 2000, 26, 275–276. [Google Scholar] [CrossRef]
- Aridon, P.; Marini, C.; Di Resta, C.; Brilli, E.; De Fusco, M.; Politi, F.; Parrini, E.; Manfredi, I.; Pisano, T.; Pruna, D. Increased sensitivity of the neuronal nicotinic receptor a2 subunit causes familial epilepsy with nocturnal wandering and ictal fear. Am. J. Hum. Genet. 2006, 79, 342–350. [Google Scholar] [CrossRef] [PubMed]
- McLellan, A.; Phillips, H.A.; Rittey, C.; Kirkpatrick, M.; Mulley, J.C.; Goudie, D.; Stephenson, J.B.; Tolmie, J.; Scheffer, I.E.; Berkovic, S.F. Phenotypic comparison of two Scottish families with mutations in different genes causing autosomal dominant nocturnal frontal lobe epilepsy. Epilepsia 2003, 44, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Steinlein, O.K.; Kaneko, S.; Hirose, S. Nicotinic acetylcholine receptor mutations. In Jasper’s Basic Mechanisms of the Epilepsies [Internet], 4th ed.; Noebels, J.L., Avoli, M., Rogawski, M.A., Olsen, R.W., Delgado-Escueta, A.V., Eds.; National Center for Biotechnology Information: Bethesda, MD, USA, 2012. [Google Scholar]
- Bertrand, D.; Elmslie, F.; Hughes, E.; Trounce, J.; Sander, T.; Bertrand, S.; Steinlein, O.K. The CHRNB2 mutation I312M is associated with epilepsy and distinct memory deficits. Neurobiol. Dis. 2005, 20, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Combi, R.; Dalprà, L.; Ferini-Strambi, L.; Tenchini, M.L. Frontal lobe epilepsy and mutations of the corticotropin-releasing hormone gene. Ann. Neurol. 2005, 58, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Combi, R.; Ferini-Strambi, L.; Tenchini, M.L. CHRNA2 mutations are rare in the NFLE population: Evaluation of a large cohort of Italian patients. Sleep Med. 2009, 10, 139–142. [Google Scholar] [CrossRef]
- Sansoni, V.; Forcella, M.; Mozzi, A.; Fusi, P.; Ambrosini, R.; Ferini-Strambi, L.; Combi, R. Functional characterization of a CRH missense mutation identified in an ADNFLE family. PLoS ONE 2013, 8, e61306. [Google Scholar] [CrossRef] [PubMed]
- Heron, S.E.; Smith, K.R.; Bahlo, M.; Nobili, L.; Kahana, E.; Licchetta, L.; Oliver, K.L.; Mazarib, A.; Afawi, Z.; Korczyn, A.; et al. Missense mutations in the sodium-gated potassium channel gene KCNT1 cause severe autosomal dominant nocturnal frontal lobe epilepsy. Nat. Genet. 2012, 44, 1188–1190. [Google Scholar] [CrossRef]
- Rubboli, G.; Plazzi, G.; Picard, F.; Nobili, L.; Hirsch, E.; Chelly, J.; Prayson, R.A.; Boutonnat, J.; Bramerio, M.; Kahane, P. Mild malformations of cortical development in sleep-related hypermotor epilepsy due to KCNT1 mutations. Ann. Clin. Transl. Neurol. 2018, 6, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Barcia, G.; Fleming, M.R.; Deligniere, A.; Gazula, V.R.; Brown, M.R.; Langouet, M.; Chen, H.; Kronengold, J.; Abhyankar, A.; Cilio, R. De novo gain-of-function KCNT1 channel mutations cause malignant migrating partial seizures of infancy. Nat. Genet. 2012, 44, 1255–1259. [Google Scholar] [CrossRef]
- Milligan, C.J.; Li, M.; Gazina, E.V.; Heron, S.E.; Nair, U.; Trager, C.; Reid, C.A.; Venkat, A.; Younkin, D.P.; Dlugos, D.J. KCNT1 gain of function in 2 epilepsy phenotypes is reversed by quinidine. Ann. Neurol. 2014, 75, 581–590. [Google Scholar] [CrossRef]
- Møller, R.S.; Heron, S.E.; Larsen, L.H.; Lim, C.X.; Ricos, M.G.; Bayly, M.A.; van Kempen, M.J.; Klinkenberg, S.; Andrews, I.; Kelley, K. Mutations in KCNT1 cause a spectrum of focal epilepsies. Epilepsia 2015, 56, e114–e120. [Google Scholar] [CrossRef] [PubMed]
- Ohba, C.; Kato, M.; Takahashi, N.; Osaka, H.; Shiihara, T.; Tohyama, J.; Nabatame, S.; Azuma, J.; Fujii, Y.; Hara, M. De novo KCNT1 mutations in early-onset epileptic encephalopathy. Epilepsia 2015, 56, e121–e128. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, F.; Ambrosino, P.; Guacci, A.; Chetta, M.; Marchese, G.; Rocco, T.; Soldovieri, M.V.; Manocchio, L.; Mosca, I.; Casara, G. Characterization of two de novoKCNT1 mutations in children with malignant migrating partial seizures in infancy. Mol. Cell. Neurosci. 2016, 72, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Abdelnour, E.; Gallentine, W.; McDonald, M.; Sachdev, M.; Jiang, Y.H.; Mikati, M.A. Does age affect response to quinidine in patients with KCNT1 mutations? Report of three new cases and review of the literature. Seizure 2018, 55, 1–3. [Google Scholar] [CrossRef]
- Dibbens, L.M.; de Vries, B.; Donatello, S.; Heron, S.E.; Hodgson, B.L.; Chintawar, S.; Crompton, D.E.; Hughes, J.N.; Bellows, S.T.; Klein, K.M. Mutations in DEPDC5 cause familial focal epilepsy with variable foci. Nat. Genet. 2013, 45, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Picard, F.; Makrythanasis, P.; Navarro, V.; Ishida, S.; de Bellescize, J.; Ville, D.; Weckhuysen, S.; Fosselle, E.; Suls, A.; De Jonghe, P. DEPDC5 mutations in families presenting as autosomal dominant nocturnal frontal lobe epilepsy. Neurology 2014, 82, 2101–2106. [Google Scholar] [CrossRef]
- Scheffer, I.E.; Bhatia, K.P.; Lopes-Cendes, I.; Fish, D.R.; Marsden, C.D.; Andermann, F.; Andermann, E.; Desbiens, R.; Cendes, F.; Manson, J.I. Autosomal dominant frontal epilepsy misdiagnosed as sleep disorder. Lancet 1994, 343, 515–517. [Google Scholar] [CrossRef]
- Bar-Peled, L.; Chantranupong, L.; Cherniack, A.D.; Chen, W.W.; Ottina, K.A.; Grabiner, B.C.; Spear, E.D.; Carter, S.L.; Meyerson, M.; Sabatini, D.M. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 2013, 340, 1100–1106. [Google Scholar] [CrossRef]
- Van Kranenburg, M.; Hoogeveen-Westerveld, M.; Nellist, M. Preliminary functional assessment and classification of DEPDC5 variants associated with focal epilepsy. Hum. Mutat. 2015, 36, 200–209. [Google Scholar] [CrossRef]
- Citraro, R.; Leo, A.; Constanti, A.; Russo, E.; De Sarro, G. mTOR pathway inhibition as a new therapeutic strategy in epilepsy and epileptogenesis. Pharmacol. Res. 2016, 107, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Scerri, T.; Riseley, J.R.; Gillies, G.; Pope, K.; Burgess, R.; Mandelstam, S.A.; Dibbens, L.; Chow, C.W.; Maixner, W.; Harvey, A.S. Familial cortical dysplasia type IIA caused by a germline mutation in DEPDC5. Ann. Clin. Transl. Neurol. 2015, 2, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Baulac, S. mTOR signaling pathway genes in focal epilepsies. Prog. Brain Res. 2016, 226, 61–79. [Google Scholar]
- Ricos, M.G.; Hodgson, B.L.; Pippucci, T.; Saidin, A.; Ong, Y.S.; Heron, S.E.; Licchetta, L.; Bisulli, F.; Bayly, M.A.; Hughes, J.; et al. Mutations in the mammalian target of rapamycin pathway regulators NPRL2 and NPRL3 cause focal epilepsy. Ann. Neurol. 2016, 79, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Korenke, G.C.; Eggert, M.; Thiele, H.; Nürnberg, P.; Sander, T.; Steinlein, O.K. Nocturnal frontal lobe epilepsy caused by a mutation in the GATOR1 complex gene NPRL3. Epilepsia 2016, 57, e60–e63. [Google Scholar] [CrossRef]
- Hildebrand, M.S.; Tankard, R.; Gazina, E.V.; Damiano, J.A.; Lawrence, K.M.; Dahl, H.H.; Regan, B.M.; Shearer, A.E.; Smith, R.J.; Marini, C.; et al. PRIMA1 mutation: A new cause of nocturnal frontal lobe epilepsy. Ann. Clin. Transl. Neurol. 2015, 2, 821–830. [Google Scholar] [CrossRef]
- Chen, Z.H.; Wang, C.; Zhuo, M.Q.; Zhai, Q.X.; Chen, Q.; Guo, Y.X.; Zhang, Y.X.; Gui, J.; Tang, Z.H.; Zeng, X.L. Exome sequencing identified a novel missense mutation c.464G>A (p.G155D) in Ca2+-binding protein 4 (CABP4) in a Chinese pedigree with autosomal dominant nocturnal frontal lobe epilepsy. Oncotarget 2017, 8, 78940–78947. [Google Scholar] [CrossRef]
- Simonato, M.; Löscher, W.; Cole, A.J.; Dudek, F.E.; Engel, J.; Kaminski, R.M.; Klitgaard, H. Finding a better drug for epilepsy: Preclinical screening strategies and experimental trial design. Epilepsia 2012, 53, 1860–1867. [Google Scholar] [CrossRef] [PubMed]
- Brooks-Kayal, A.R.; Bath, K.G.; Berg, A.T.; Galanopoulou, A.S.; Holmes, G.L.; Jensen, F.E.; Kanner, A.M.; O’Brien, T.J.; Whittemore, V.H.; Winawer, M.R. Issues related to symptomatic and disease-modifying treatments affecting cognitive and neuropsychiatric comorbidities of epilepsy. Epilepsia 2013, 54, 44–60. [Google Scholar] [CrossRef] [PubMed]
- Bialer, M.; White, H. Key factors in the discovery and development of new antiepileptic drugs. Nat. Rev. Drug. Discov. 2010, 9, 68–82. [Google Scholar] [CrossRef]
- Post, J.M.; Loch, S.; Lerner, R.; Remmers, F.; Lomazzo, E.; Lutz, B.; Bindila, L. Antiepileptogenic effect of subchronic palmitoylethanolamide treatment in a mouse model of acute epilepsy. Front. Mol. Neurosci. 2018, 11, 67. [Google Scholar] [CrossRef] [PubMed]
- Davoudi, M.; Shojaei, A.; Palizvan, M.R.; Javan, M.; Mirnajafi-Zadeh, J. Comparison between standard protocol and a novel window protocol for induction of pentylenetetrazol kindled seizures in the rat. Epilepsy Res. 2013, 106, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Akdogan, I.; Yonguc, N.G. Experimental epilepsy models and morphologic alterations of experimental epilepsy models in brain and hippocampus. In Underlying Mechanisms of Epilepsy; Kaneez, F.S., Ed.; InTech: Rijeka, Croatia, 2011; pp. 269–282. [Google Scholar]
- Kandratavicius, L.; Balista, P.A.; Lopes-Aguiar, C.; Ruggiero, R.N.; Umeoka, E.H.; Garcia-Cairasco, N.; Bueno-Junior, L.S.; Leite, J.P. Animal models of epilepsy: Use and limitations. Neuropsychiatr. Dis. Treat. 2014, 10, 1693–1705. [Google Scholar] [CrossRef]
- Gunthorpe, M.J.; Large, C.H.; Sankar, R. The mechanism of action of retigabine (ezogabine), a first-in-class K+ channel opener for the treatment of epilepsy. Epilepsia 2012, 53, 412–424. [Google Scholar] [CrossRef]
- Brodie, M.J.; Lerche, H.; Gil-Nagel, A.; Elger, C.; Hall, S.; Shin, P.; Nohria, V.; Mansbach, H.; RESTORE 2 Study Group. Efficacy and safety of adjunctive ezogabine (retigabine) in refractory partial epilepsy. Neurology 2010, 75, 1817–1824. [Google Scholar] [CrossRef] [PubMed]
- French, J.; Abou-Khalil, B.; Leroy, R.; Yacubian, E.M.; Shin, P.; Hall, S.; Mansbach, H.; Nohria, V.; RESTORE 1/Study 301 Investigators. Randomized, double-blind, placebo-controlled trial of ezogabine (retigabine) in partial epilepsy. Neurology 2011, 76, 1555–1563. [Google Scholar] [CrossRef] [PubMed]
- Locharernkul, C.; Loplumlert, J.; Limotai, C.; Korkij, W.; Desudchit, T.; Tongkobpetch, S.; Kangwanshiratada, O.; Hirankarn, N.; Suphapeetiporn, K.; Shotelersuk, V. Carbamazepine and phenytoin induced Stevens-Johnson syndrome is associated with HLA-B*1502 allele in Thai population. Epilepsia 2008, 49, 2087–2091. [Google Scholar] [CrossRef] [PubMed]
- Alfirevic, A.; Jorgensen, A.L.; Williamson, P.R.; Chadwick, D.W.; Park, B.K.; Pirmohamed, M. HLA-B locus in Caucasian patients with carbamazepine hypersensitivity. Pharmacogenomics 2006, 7, 813–818. [Google Scholar] [CrossRef] [PubMed]
- Hung, S.I.; Chung, W.H.; Liu, Z.S.; Chen, C.H.; Hsih, M.S.; Hui, R.C.; Chu, C.Y.; Chen, Y.T. Common risk allele in aromatic antiepileptic-drug induced Stevens-Johnson syndrome and toxic epidermal necrolysis in Han Chinese. Pharmacogenomics 2010, 11, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, J.; Ye, J. Association between SCN1A polymorphism and carbamazepine responsiveness in epilepsy: A meta-analysis. Epilepsy Res. 2021, 176, 106627. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.C.; Ma, C.L.; Lang, L.Q.; Zhong, M.K. Association of xenobiotic receptor polymorphisms with carbamazepine response in epilepsy patients. Gene 2021, 771, 145359. [Google Scholar] [CrossRef] [PubMed]
- Comfere, N.I.; Sartori-Valinotti, J.C.; Bruce, A.J.; Drage, L.A. Successful treatment of lamotrigine-associated drug hyper-sensitivity syndrome with intravenous IgG. J. Am. Acad. Dermatol. 2012, 66, 249–250. [Google Scholar] [CrossRef] [PubMed]
- Biton, V.; Berkovic, S.F.; Abou-Khalil, B.; Sperling, M.R.; Johnson, M.E.; Lu, S. Brivaracetam as adjunctive treatment for uncontrolled partial epilepsy in adults: A phase III randomized, double-blind, placebo-controlled trial. Epilepsia 2014, 55, 57–66. [Google Scholar] [CrossRef]
- Klein, P.; Schiemann, J.; Sperling, M.R.; Whitesides, J.; Liang, W.; Stalvey, T.; Brandt, C.; Kwan, P. A randomized, double-blind, placebo-controlled, multicenter, parallelgroup study to evaluate the efficacy and safety of adjunctive brivaracetam in adult patients with uncontrolled partial-onset seizures. Epilepsia 2015, 56, 1890–1898. [Google Scholar] [CrossRef] [PubMed]
- Kwan, P.; Trinka, E.; Van Paesschen, W.; Rektor, I.; Johnson, M.E.; Lu, S. Adjunctive brivaracetam for uncontrolled focal and generalized epilepsies: Results of a phase III, double-blind, randomized, placebo-controlled, flexible-dose trial. Epilepsia 2014, 55, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Androsova, G.; Krause, R.; Borghei, M.; Wassenaar, M.; Auce, P.; Avbersek, A.; Becker, F.; Berghuis, B.; Campbell, E.; Coppola, A. EpiPGX Consortium. Comparative effectiveness of antiepileptic drugs in patients with mesial temporal lobe epilepsy with hippocampal sclerosis. Epilepsia 2017, 58, 1734–1741. [Google Scholar] [CrossRef]
- Grosso, S.; Franzoni, E.; Iannetti, P.; Incorpora, G.; Cardinali, C.; Toldo, I.; Verrotti, A.; Caterina Moscano, F.; Lo Faro, V.; Mazzone, L. Efficacy and safety of topiramate in refractory epilepsy of childhood: Longterm follow-up study. J. Child. Neurol. 2005, 20, 893–897. [Google Scholar] [CrossRef]
- Elger, C.; Halasz, P.; Maia, J.; Almeida, L.; Soares-da-Silva, P.; Group, B.I.A.I.S. Efficacy and safety of eslicarbazepine acetate as adjunctive treatment in adults with refractory partial-onset seizures: A randomized, double-blind, placebo-controlled, parallel-group phase III study. Epilepsia 2009, 50, 454–463. [Google Scholar] [CrossRef]
- Gil-Nagel, A.; Lopes-Lima, J.; Almeida, L.; Maia, J.; Soares-da-Silva, P.; Group, B.I.A.I.S. Efficacy and safety of 800 and 1200 mg eslicarbazepine acetate as adjunctive treatment in adults with refractory partial-onset seizures. Acta Neurol. Scand. 2009, 120, 281–287. [Google Scholar] [CrossRef]
- Sperling, M.R.; Abou-Khalil, B.; Harvey, J.; Rogin, J.B.; Biraben, A.; Galimberti, C.A.; Kowacs, P.A.; Hong, S.B.; Cheng, H.; Blum, D.; et al. Eslicarbazepine acetate as adjunctive therapy in patients with uncontrolled partial-onset seizures: Results of a phase III, double-blind, randomized, placebo-controlled trial. Epilepsia 2015, 56, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Stolarek, I.; Blacklaw, J.; Forrest, G.; Brodie, M.J. Vigabatrin and lamotrigine in refractory epilepsy. J. Neurol. Neurosurg. Psychiatry 1994, 57, 921–924. [Google Scholar] [CrossRef]
- Biro, A.; Stephani, U.; Tarallo, T.; Bast, T.; Schlachter, K.; Fleger, M.; Kurlemann, G.; Fiedler, B.; Leiz, S.; Nikanorova, M. Effectiveness and tolerability of perampanel in children and adolescents with refractory epilepsies: First experiences. Neuropediatrics 2015, 46, 110–116. [Google Scholar] [PubMed]
- French, J.A.; Krauss, G.L.; Steinhoff, B.J.; Squillacote, D.; Yang, H.; Kumar, D.; Laurenza, A. Evaluation of adjunctive perampanel in patients with refractory partial-onset seizures: Results of randomized global phase III study 305. Epilepsia 2013, 54, 117–125. [Google Scholar] [CrossRef]
- Zubiaur, P.; Del Peso-Casado, M.; Ochoa, D.; Enrique-Benedito, T.; Mejía-Abril, G.; Navares, M.; Villapalos-García, G.; Román, M.; Abad-Santos, F.; Ovejero-Benito, M.C. ABCB1 C3435T, G2677T/A and C1236T variants have no effect in eslicarbazepine pharmacokinetics. Biomed. Pharmacother. 2021, 142, 112083. [Google Scholar] [CrossRef] [PubMed]
- Wolking, S.; Campbell, C.; Stapleton, C.; McCormack, M.; Delanty, N.; Depondt, C.; Johnson, M.R.; Koeleman, B.P.C.; Krause, R.; Kunz, W.S. Role of Common Genetic Variants for Drug-Resistance to Specific Anti-Seizure Medications. Front. Pharmacol. 2021, 12, 688386. [Google Scholar] [CrossRef]

| Voltage-Gated Ion Channels | Chromosome (Locus) | Gene | Gene Product | Syndrome |
|---|---|---|---|---|
| Potassium channel | 20q13 | KCNQ2 | potassium channel KCNQ2 | mild familial seizures of newborns |
| 8q24 | KCNQ3 | potassium channel KCNQ3 | ||
| Sodium channel | 2q22–23 | SCN2A | sodium channel subunits α2 | benign familial neonatal-infantile seizures (BFNIS) |
| 19q13 | SCN1B | regulatory subunit β of sodium channel | generalized epilepsy with febrile seizures (GEFS+1) | |
| 2q24 | SCN1A | sodium channel subunits α1 | generalized epilepsy with febrile seizures (GEFS+2), Dravet syndrome (severe myoclonic epilepsy in infancy, SMEI) | |
| 2q22–23 | SCN2A | sodium channel subunits α2 | generalized epilepsy with febrile seizures (GEFS+4) | |
| Chloride channel | 3q27–28 | CLNC2 | chloride channels CLC-2 | child absence epilepsy (CAE) juvenile absence epilepsy (JAE) juvenile myoclonic epilepsy (JME) |
| Calcium channel | 6p12 | EFHC1 | regulator of voltage-dependent ion channels | JME |
| 16p13 | CACNA1H | T-type calcium channel | CAE | |
| 2q22–23 | CACNB4 | subunit β2 of calcium channel | JME |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smolarz, B.; Makowska, M.; Romanowicz, H. Pharmacogenetics of Drug-Resistant Epilepsy (Review of Literature). Int. J. Mol. Sci. 2021, 22, 11696. https://doi.org/10.3390/ijms222111696
Smolarz B, Makowska M, Romanowicz H. Pharmacogenetics of Drug-Resistant Epilepsy (Review of Literature). International Journal of Molecular Sciences. 2021; 22(21):11696. https://doi.org/10.3390/ijms222111696
Chicago/Turabian StyleSmolarz, Beata, Marianna Makowska, and Hanna Romanowicz. 2021. "Pharmacogenetics of Drug-Resistant Epilepsy (Review of Literature)" International Journal of Molecular Sciences 22, no. 21: 11696. https://doi.org/10.3390/ijms222111696
APA StyleSmolarz, B., Makowska, M., & Romanowicz, H. (2021). Pharmacogenetics of Drug-Resistant Epilepsy (Review of Literature). International Journal of Molecular Sciences, 22(21), 11696. https://doi.org/10.3390/ijms222111696






