Novel Hybrid Peptide Cathelicidin 2 (1-13)-Thymopentin (TP5) and Its Derived Peptides with Effective Antibacterial, Antibiofilm, and Anti-Adhesion Activities
Abstract
:1. Introduction
2. Results
2.1. Physiochemical Properties of Peptides
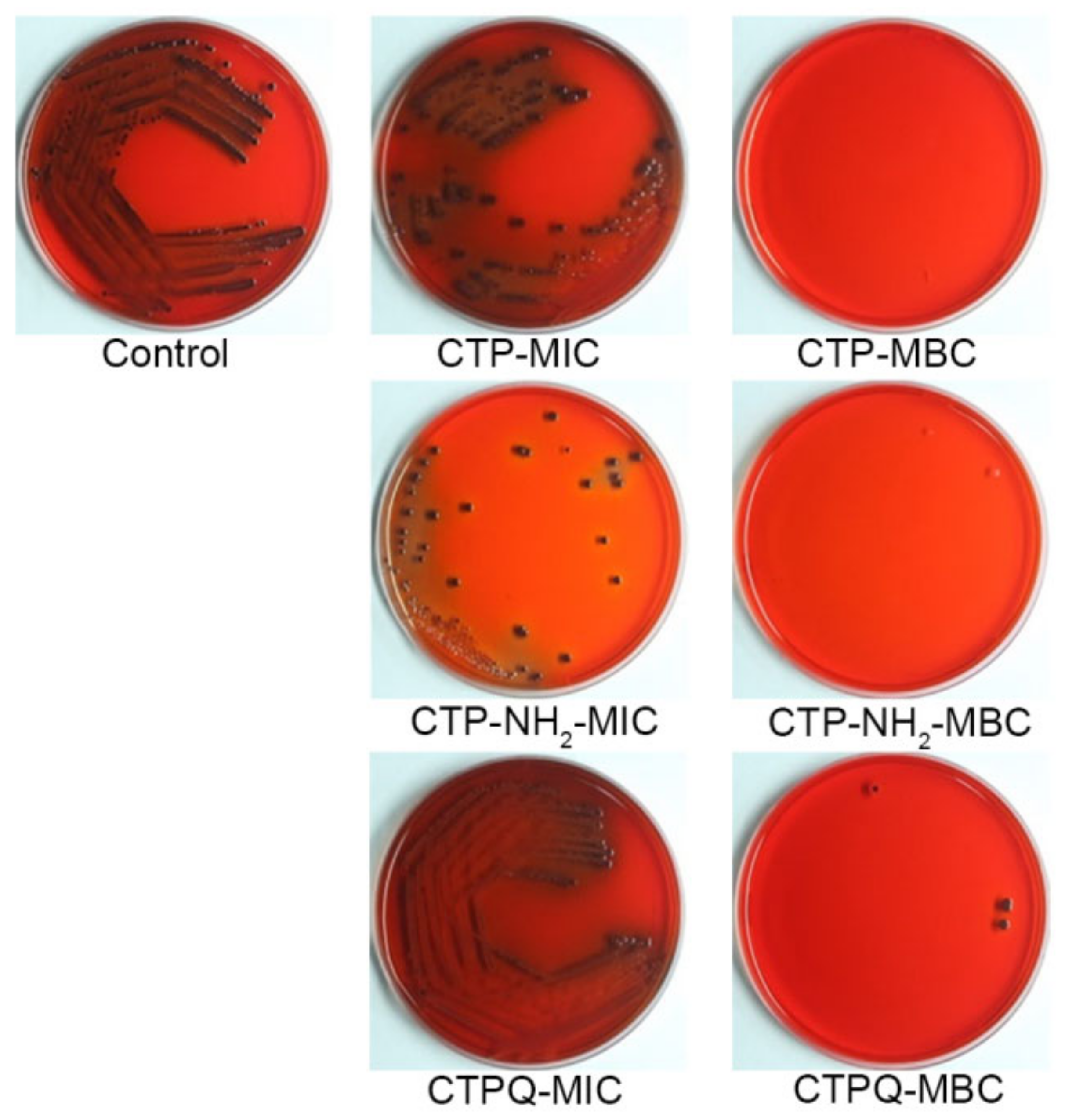
2.2. Determination of Minimal Inhibitory Concentrations (MICs) and Minimal Bactericidal Concentrations (MBCs)
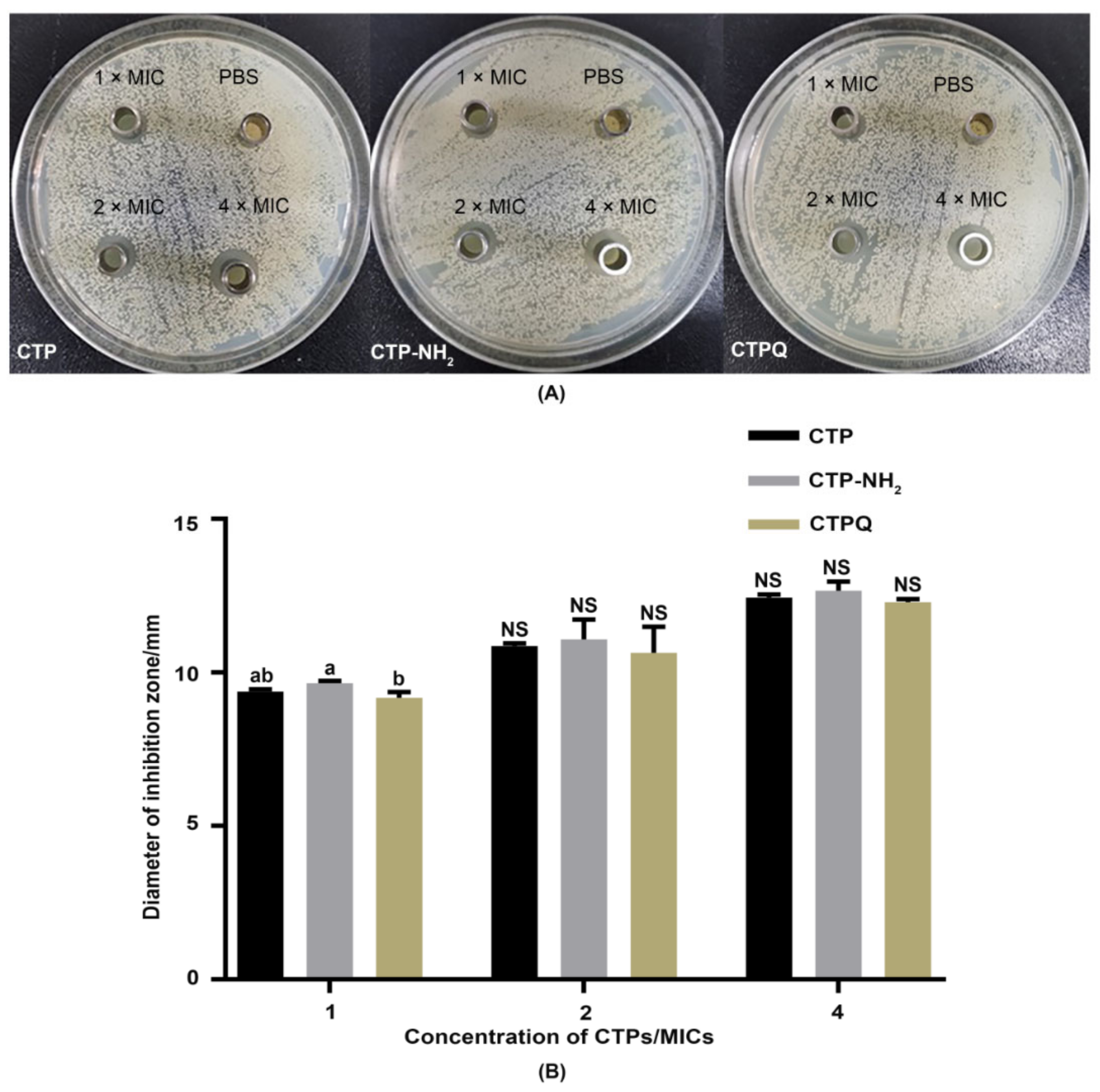
2.3. Inhibited Zone Assay
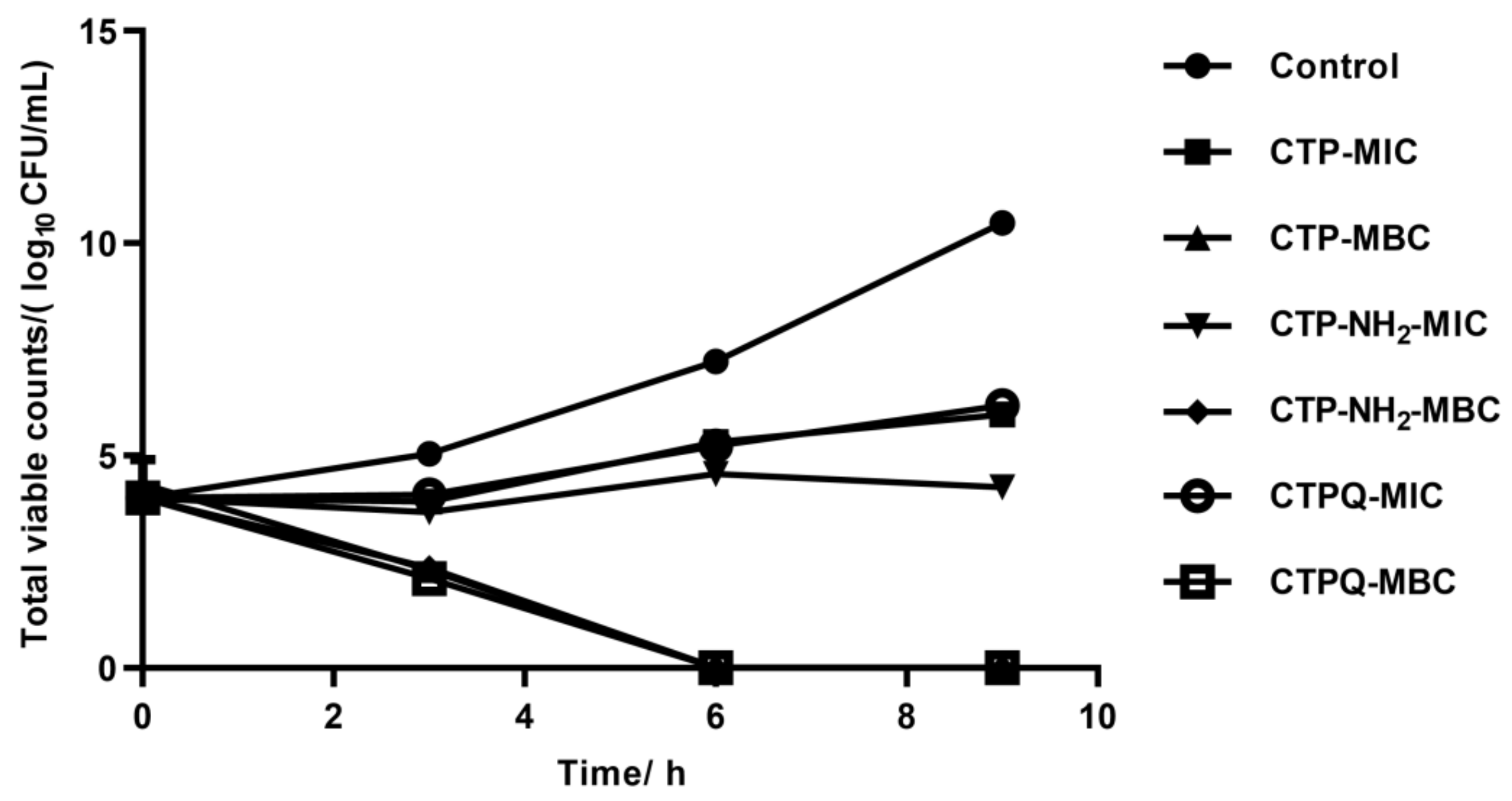
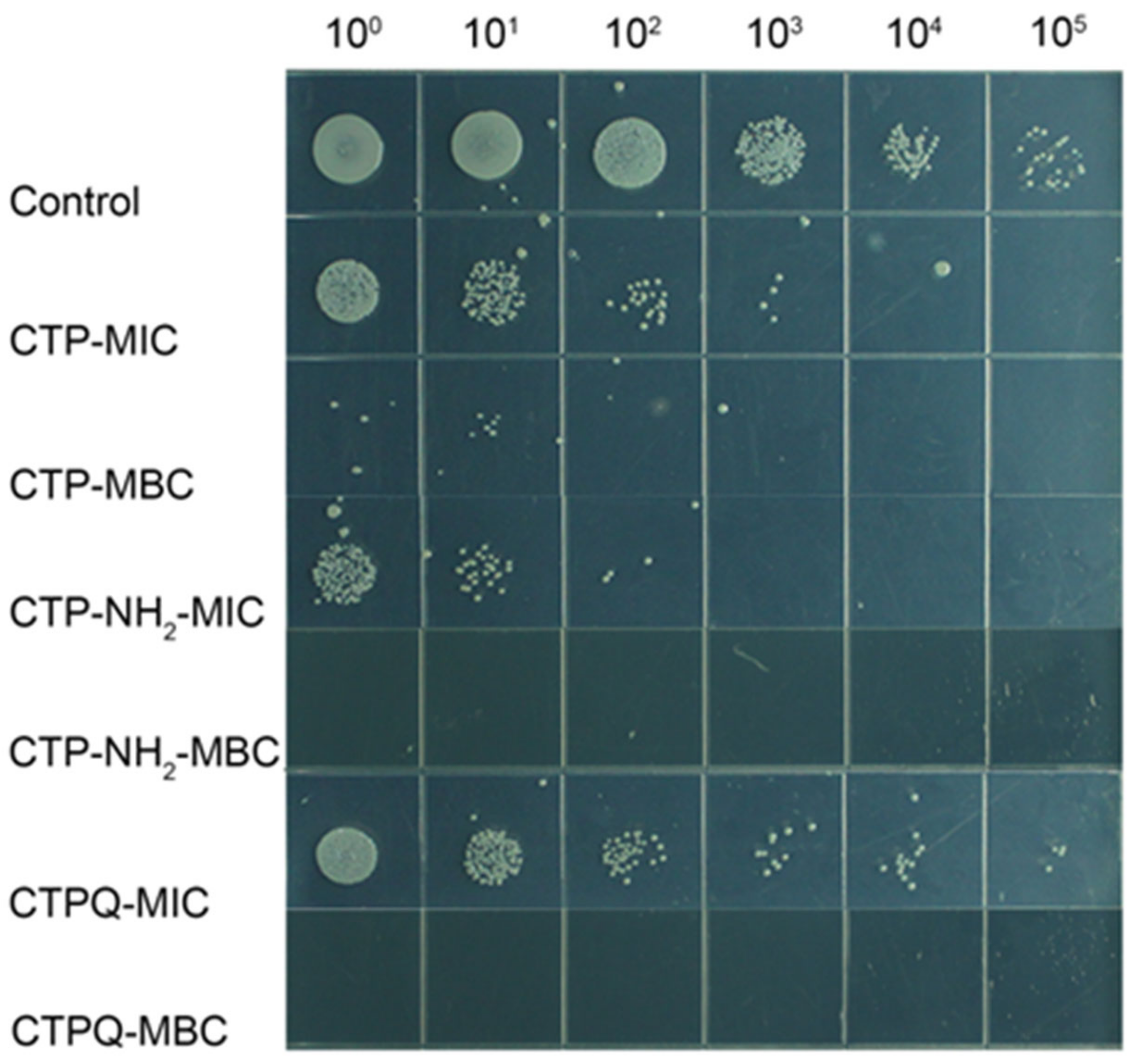
2.4. In Vitro Time-Kill Curve Assay
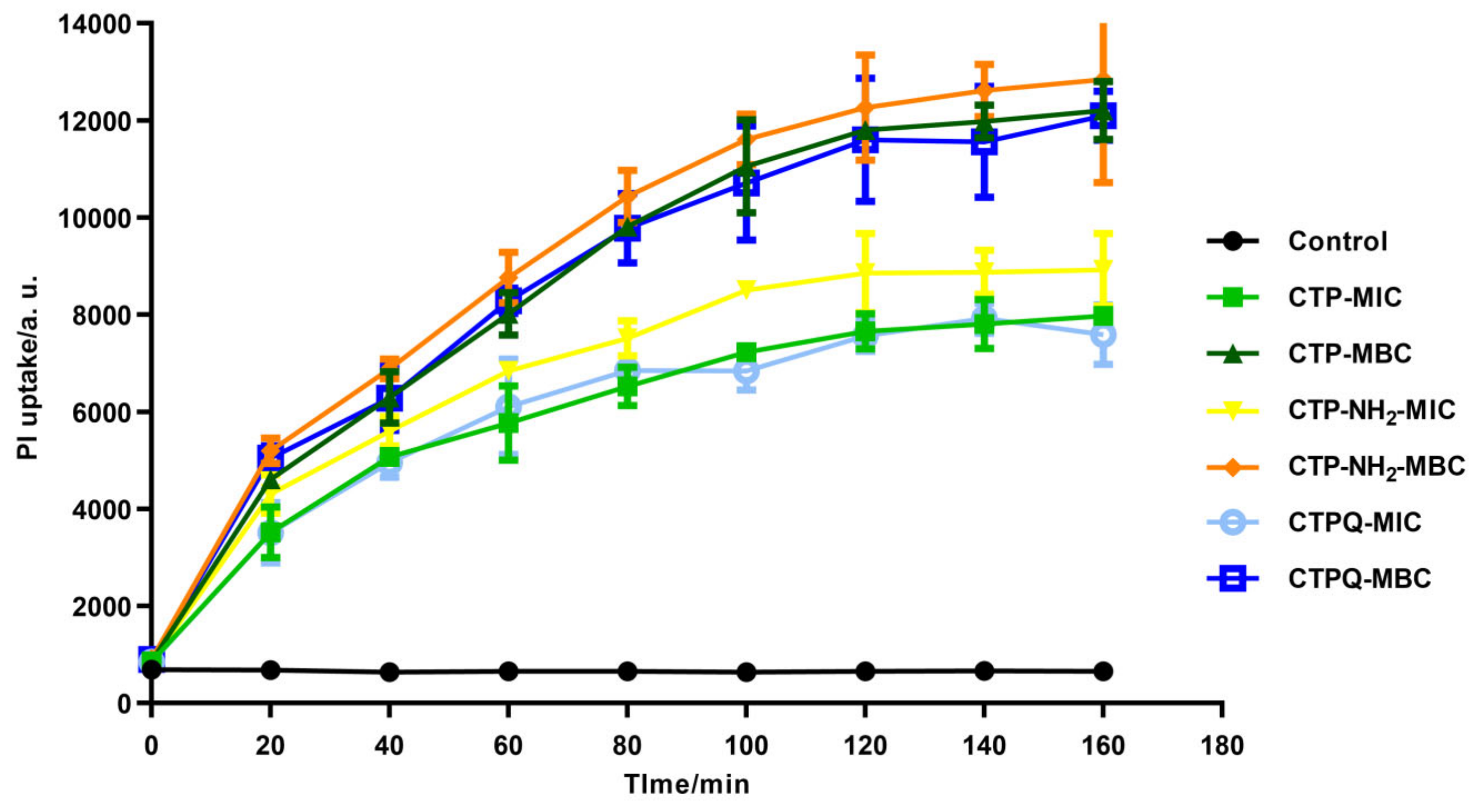
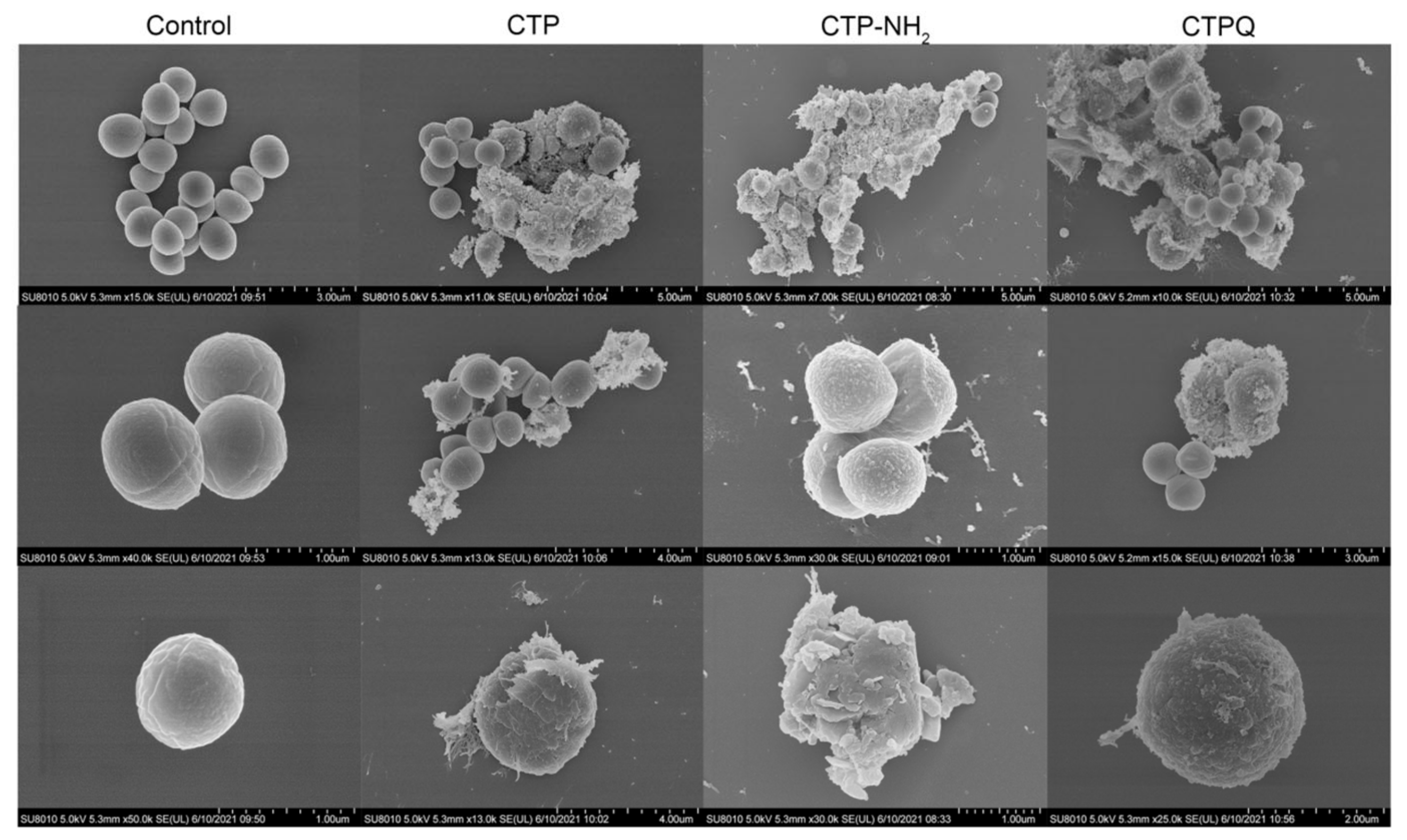
2.5. Cell Membrane Integrity Evaluation and Morphological Observation of S. aureus
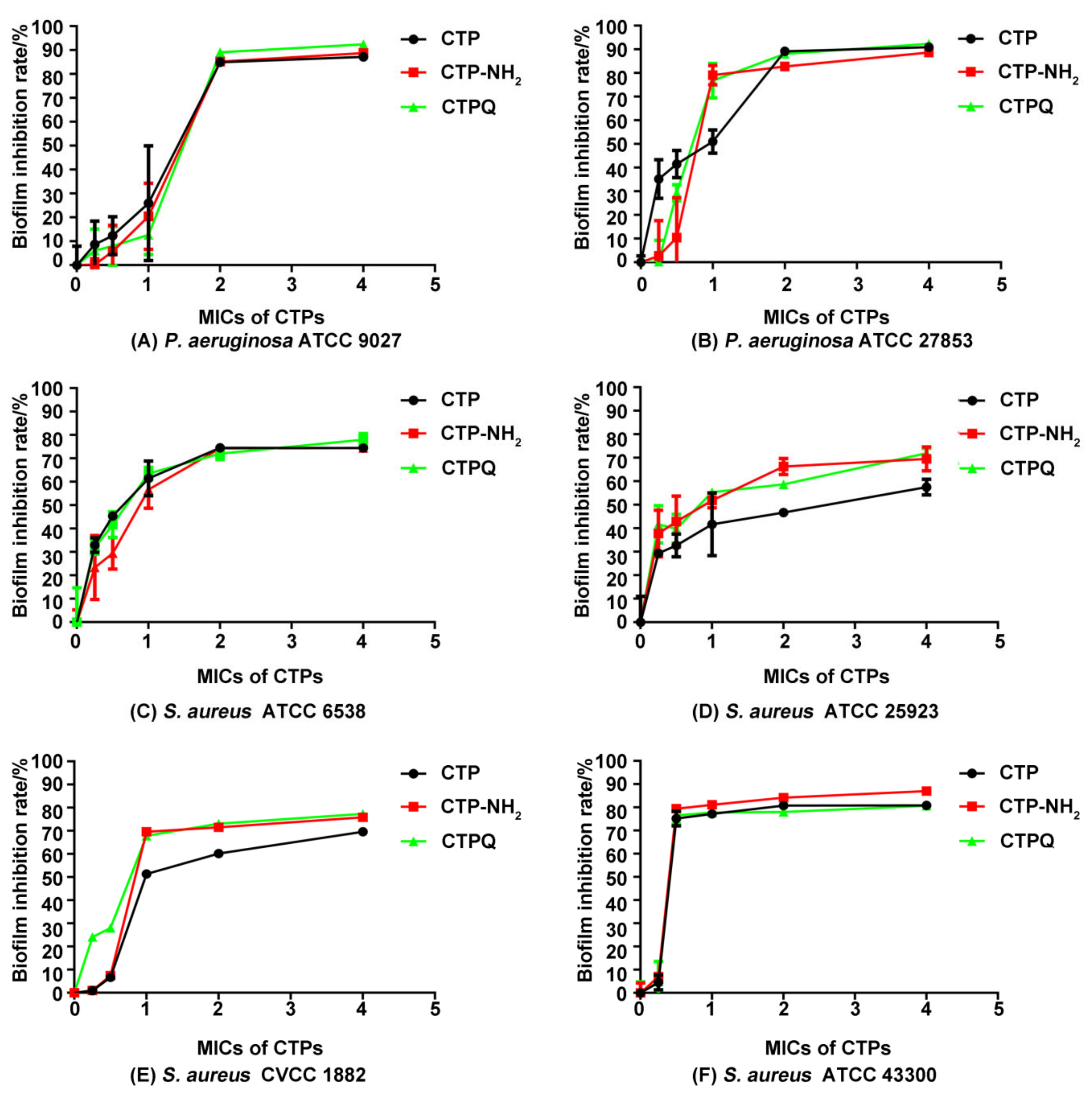
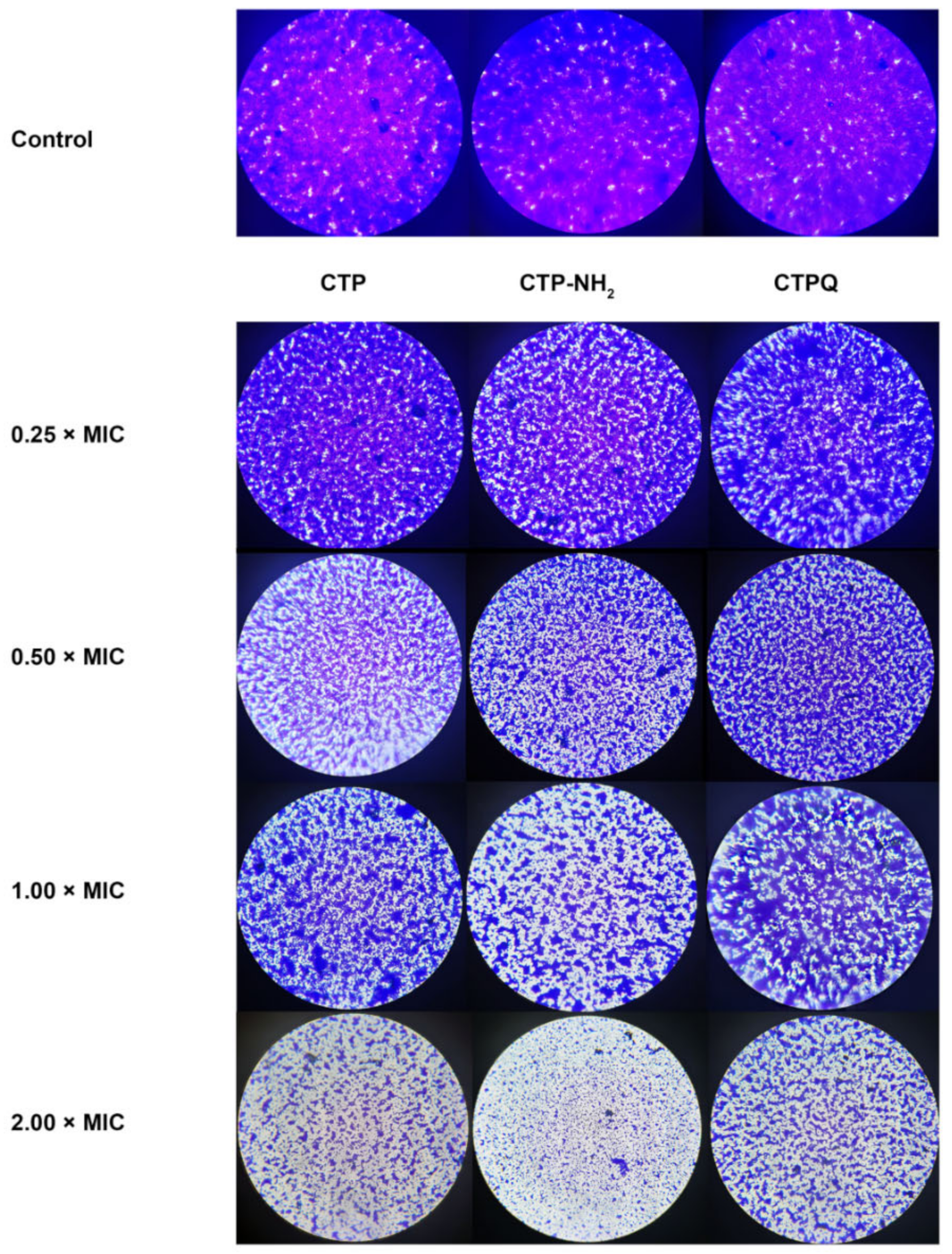
2.6. Antibiofilm Activity Assay
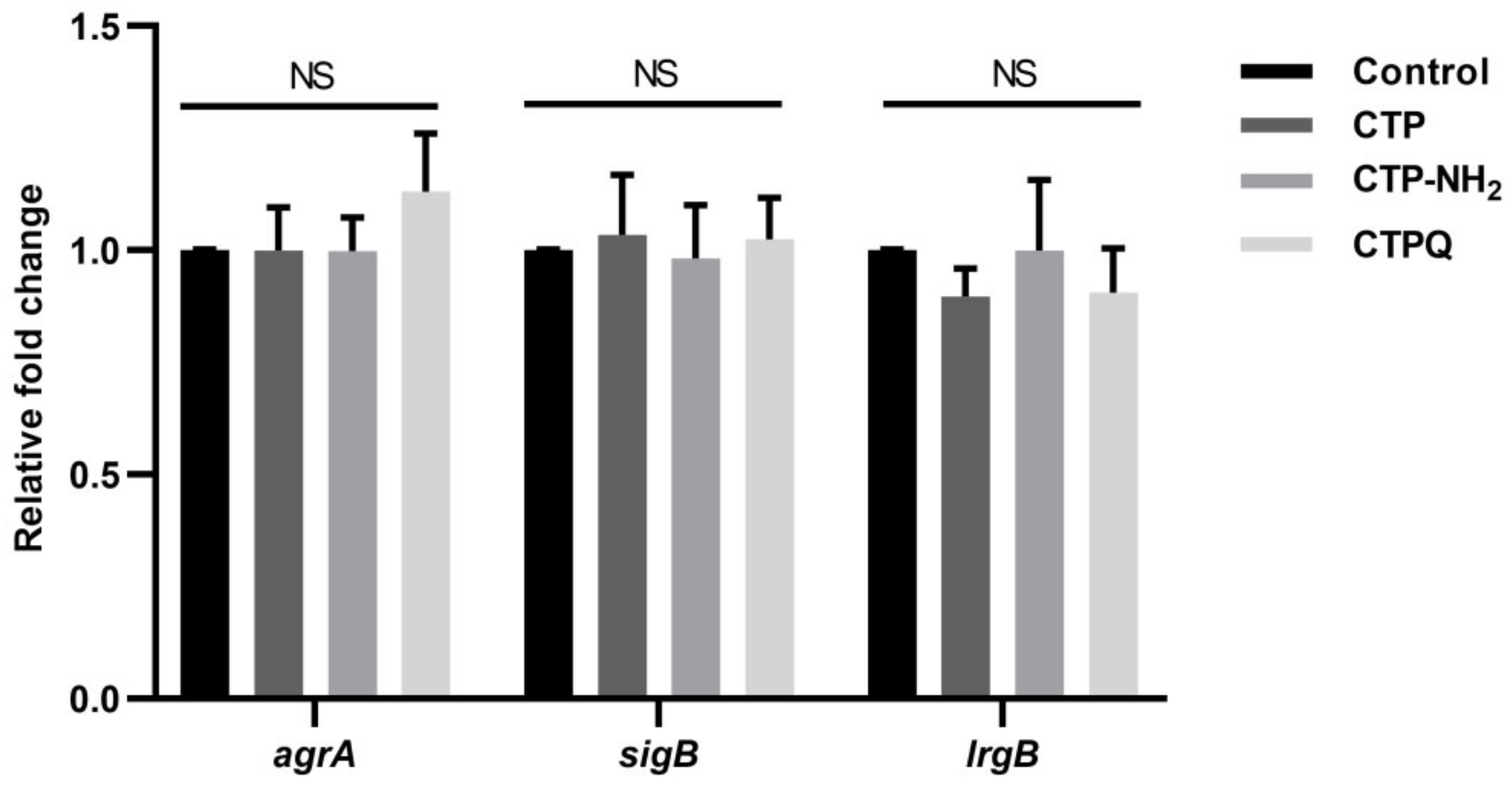
2.7. Confocal Laser Scanning Microscope (CLSM) Assay and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Results of Biofilm-Associated Genes
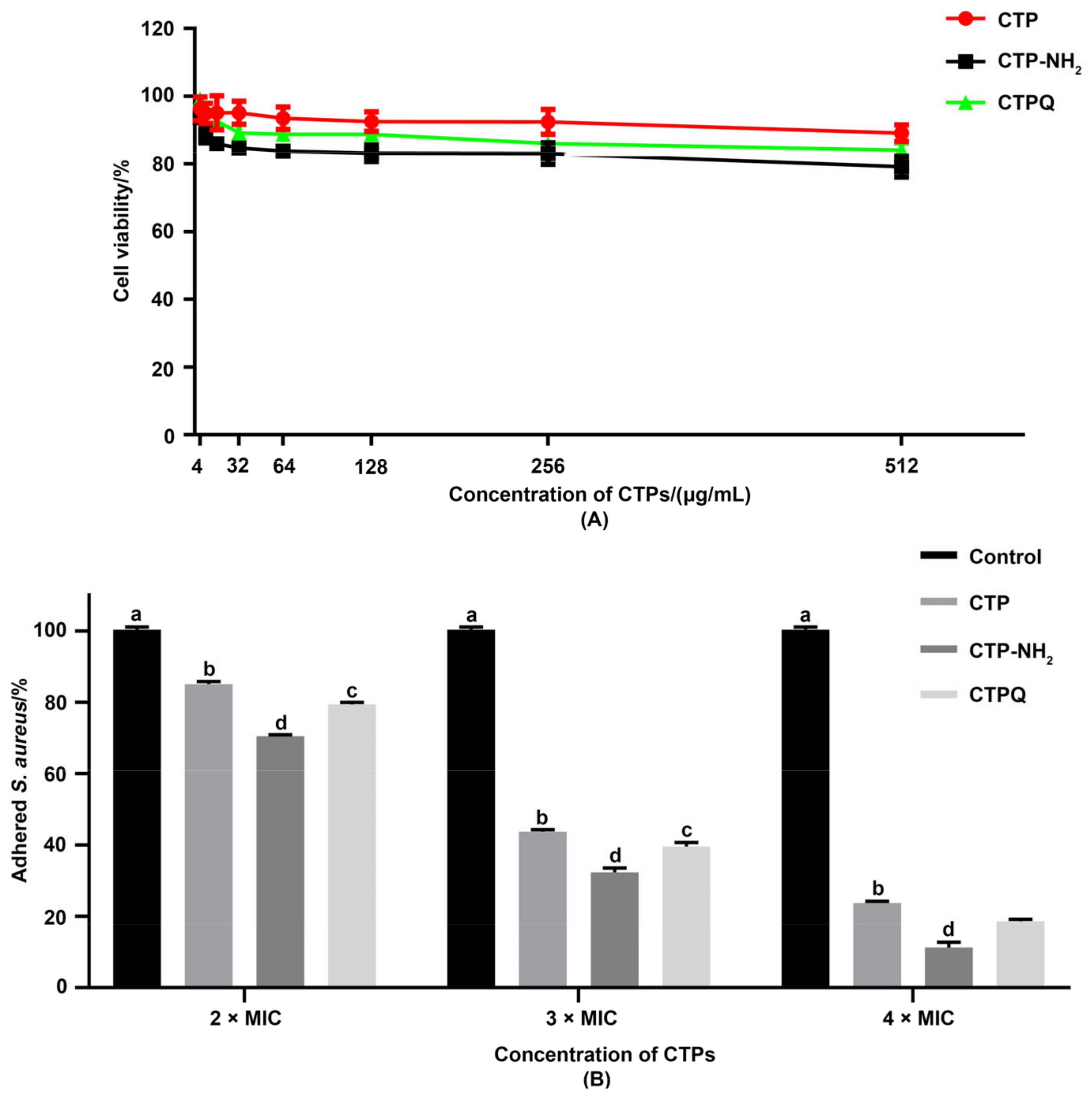
2.8. Cell Viability and Anti-Adhesion Activity Tests
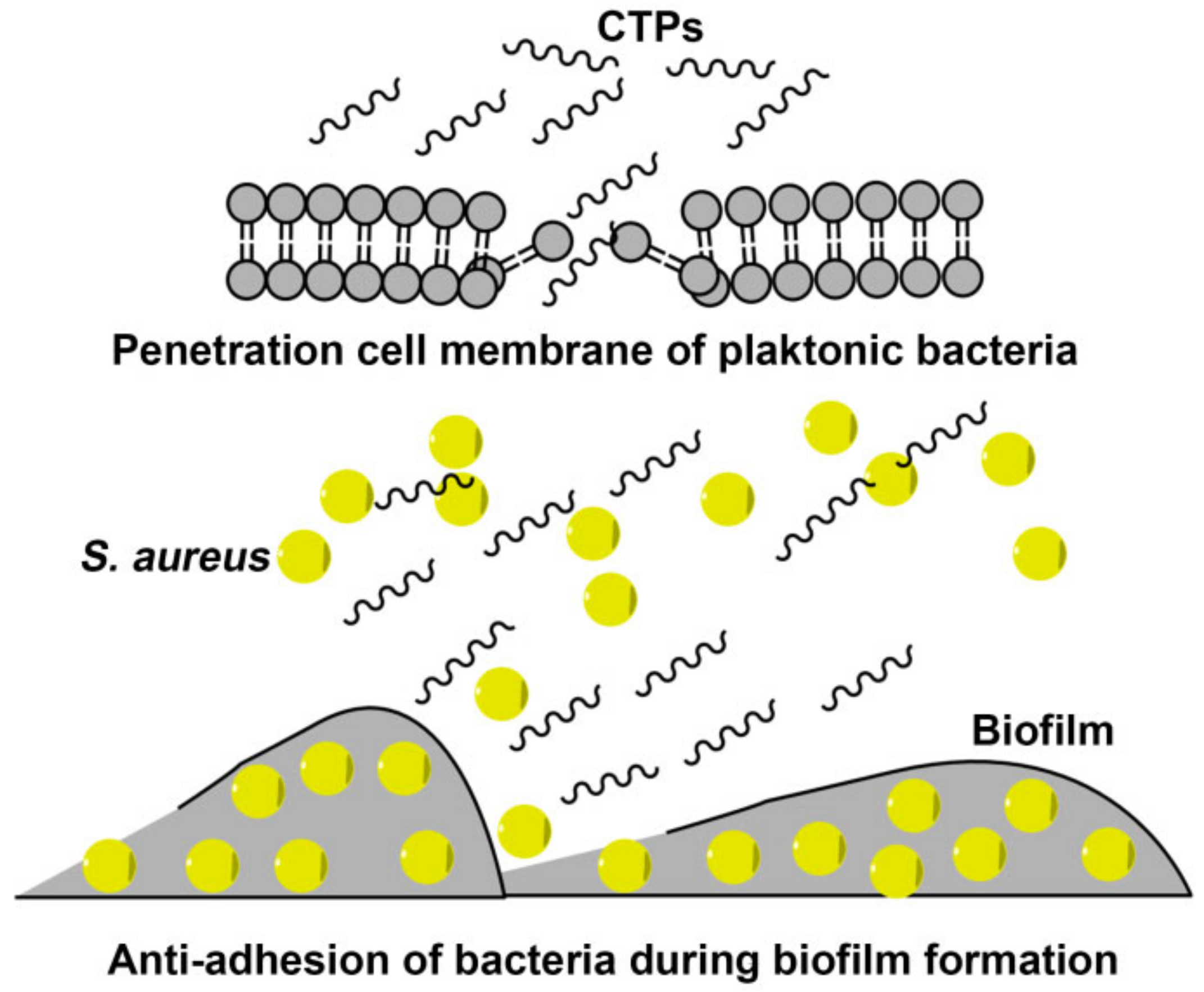
3. Discussion
4. Materials and Methods
4.1. Strains and Peptides
4.2. Physicochemical Properties of Peptides
4.3. Determination of MICs and MBCs
4.4. Inhibition Zones Assay
4.5. Time-Kill Curve Assay
4.6. Enumeration of Viable S. aureus
4.7. CRA Plate Assay of S. aureus
4.8. Assessment of S. aureus Cell Membrane Integrity
4.9. SEM Assay of S. aureus
4.10. TEM Assay of S. aureus
4.11. Antibiofilm Activity Assay
4.12. Optical Microscope Observation of Biofilm
4.13. CLSM Assay of Biofilm
4.14. Biofilm-Related Gene Expression Assays
4.15. Cell Culture
4.16. Cell Viability Assay
4.17. Bacterial Adherence Assays
4.18. Statistics
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus Aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [Green Version]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as Complex Differentiated Communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costerton, J.W.; Geesey, G.G.; Cheng, K.J. How Bacteria Stick. Sci. Am. 1978, 238, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Schilcher, K.; Horswill, A.R. Staphylococcal Biofilm Development: Structure, Regulation, and Treatment Strategies. Microbiol. Mol. Biol. Rev. 2020, 84, e00026-19. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, P.; Krishna, V.P.; Bharath, D.; Lavanya, R.; Vairaprakash, P.; Princy, S.A. Staphylococcus aureus Quorum Regulator SarA Targeted Compound, 2-[(Methylamino) methyl] phenol Inhibits Biofilm and Down-Regulates Virulence Genes. Front. Microbiol. 2017, 8, 1290. [Google Scholar] [CrossRef]
- Scherr, T.D.; Heim, C.E.; Morrison, J.M.; Kielian, T. Hiding in Plain Sight: Interplay between Staphylococcal Biofilms and Host Immunity. Front. Immunol. 2014, 5, 37. [Google Scholar] [CrossRef] [Green Version]
- Hoiby, N.; Johansen, H.K.; Moser, C.; Song, Z.J.; Ciofu, O.; Kharazmi, A. Pseudomonas aeruginosa and the in vitro and In vivo Biofilm Mode of Growth. Microbes Infect. 2001, 3, 23–35. [Google Scholar] [CrossRef]
- Lebeaux, D.; Ghigo, J.; Beloin, C. Biofilm-Related Infections: Bridging the Gap between Clinical Management and Fundamental Aspects of Recalcitrance toward Antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef] [Green Version]
- Hoiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic Resistance of Bacterial Biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Yang, Y.; Liu, L.; Yu, T.; Zhao, J.; Liu, L.; Li, C. Anti-Adhesive Effects of Sialic Acid and Lactobacillus plantarumon on Staphylococcus aureus in vitro. J. Food Saf. 2021, 41, e12875. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Alford, M.A.; Haney, E.F. Antibiofilm Activity of Host Defence Peptides: Complexity Provides Opportunities. Nat. Rev. Microbiol. 2021. [Google Scholar] [CrossRef]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.W.; Schneider, G. Designing Antimicrobial Peptides: Form Follows Function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef] [PubMed]
- Kosciuczuk, E.M.; Lisowski, P.; Jarczak, J.; Strzalkowska, N.; Jozwik, A.; Horbanczuk, J.; Krzyzewski, J.; Zwierzchowski, L.; Bagnicka, E. Cathelicidins: Family of Antimicrobial Peptides, a Review. Mol. Biol. Rep. 2012, 39, 10957–10970. [Google Scholar] [CrossRef] [Green Version]
- Mardirossian, M.; Grzela, R.; Giglione, C.; Meinnel, T.; Gennaro, R.; Mergaert, P.; Scocchi, M. The Host Antimicrobial Peptide Bac7(1-35) Binds to Bacterial Ribosomal Proteins and Inhibits Protein Synthesis. Chem. Biol. 2014, 21, 1639–1647. [Google Scholar] [CrossRef] [Green Version]
- Mardirossian, M.; Perebaskine, N.; Benincasa, M.; Gambato, S.; Hofmann, S.; Huter, P.; Mueller, C.; Hilpert, K.; Innis, C.A.; Tossi, A.; et al. The Dolphin Proline-Rich Antimicrobial Peptide Tur1A Inhibits Protein Synthesis by Targeting the Bacterial Ribosome. Cell Chem. Biol. 2018, 25, 530–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wronska, A.K.; Bogus, M.I. Heat Shock Proteins (HSP 90, 70, 60, and 27) in Galleria mellonella (Lepidoptera) Hemolymph are Affected by Infection with Conidiobolus coronatus (Entomophthorales). PLoS ONE 2020, 15, e02285562. [Google Scholar] [CrossRef]
- Li, L.; Sun, J.; Xia, S.; Tian, X.; Cheserek, M.J.; Le, G. Mechanism of Antifungal Activity of Antimicrobial Peptide APP, a Cell-Penetrating Peptide Derivative, against Candida albicans: Intracellular DNA Binding and Cell Cycle Arrest. Appl. Microbiol. Biot. 2016, 100, 3245–3253. [Google Scholar] [CrossRef] [PubMed]
- Cruz, G.F.; de Araujo, I.; Torres, M.D.T.; de la Fuente-Nunez, C.; Oliveira, V.X., Jr.; Ambrosio, F.N.; Lombello, C.B.; Almeida, D.V.; Silva, F.D.; Garcia, W. Photochemically-Generated Silver Chloride Nanoparticles Stabilized by a Peptide Inhibitor of Cell Division and its Antimicrobial Properties. J. Inorg. Organomet. Polym. Mater. 2020, 30, 2464–2474. [Google Scholar] [CrossRef]
- Okuda, K.; Zendo, T.; Sugimoto, S.; Iwase, T.; Tajima, A.; Yamada, S.; Sonomoto, K.; Mizunoe, Y. Effects of Bacteriocins on Methicillin-Resistant Staphylococcus aureus Biofilm. Antimicrob. Agents Chemother. 2013, 57, 5572–5579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Ye, X.; Sun, P.; Xu, P.; Wang, L.; Liu, Z.; Huang, X.; Bai, Z.; Zhou, C. Antimicrobial and Antibiofilm Activity of the EeCentrocin 1 Derived Peptide EC1-17KV via Membrane Disruption. Ebiomedicine 2020, 55, 102775. [Google Scholar] [CrossRef]
- Abdel-Hamid, M.; Romeih, E.; Saporito, P.; Osman, A.; Mateiu, R.V.; Mojsoska, B.; Jenssen, H. Camel Milk Whey Hydrolysate Inhibits Growth and Biofilm Formation of Pseudomonas aeruginosa PAO1 and Methicillin-Resistant Staphylococcus aureus. Food Control 2020, 111, 107056. [Google Scholar] [CrossRef]
- Brown, M.M.; Kwiecinski, J.M.; Cruz, L.M.; Shahbandi, A.; Todd, D.A.; Cech, N.B.; Horswill, A.R. Novel Peptide from Commensal Staphylococcus simulans Blocks Methicillin-Resistant Staphylococcus aureus Quorum Sensing and Protects Host Skin from Damage. Antimicrob. Agents Chemother. 2020, 64, e00172-20. [Google Scholar] [CrossRef]
- Karathanasi, G.; Bojer, M.S.; Baldry, M.; Johannessen, B.A.; Wolff, S.; Greco, I.; Kilstrup, M.; Hansen, P.R.; Ingmer, H. Linear Peptidomimetics as Potent Antagonists of Staphylococcus aureus Agr Quorum Sensing. Sci. Rep. 2018, 8, 3562. [Google Scholar] [CrossRef]
- Shang, D.; Han, X.; Du, W.; Kou, Z.; Jiang, F. Trp-Containing Antibacterial Peptides Impair Quorum Sensing and Biofilm Development in Multidrug-Resistant Pseudomonas aeruginosa and Exhibit Synergistic Effects with Antibiotics. Front. Microbiol. 2021, 12, 611009. [Google Scholar] [CrossRef]
- Hocquellet, A.; le Senechal, C.; Garbay, B. Importance of the Disulfide Bridges in the Antibacterial Activity of Human Hepcidin. Peptides 2012, 36, 303–307. [Google Scholar] [CrossRef]
- de la Fuente-Nunez, C.; Reffuveille, F.; Haney, E.F.; Straus, S.K.; Hancock, R.E.W. Broad-Spectrum Anti-Biofilm Peptide that Targets a Cellular Stress Response. PLoS Pathog. 2014, 10, e1004152. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wei, X.; Zhang, R.; Koci, M.; Si, D.; Ahmad, B.; Guo, H.; Hou, Y. C-Terminal Amination of a Cationic Anti-Inflammatory Peptide Improves Bioavailability and Inhibitory Activity against LPS-Induced Inflammation. Front. Immunol. 2021, 11, 618312. [Google Scholar] [CrossRef]
- Cao, W.; Zhou, Y.; Ma, Y.; Luo, Q.; Wei, D. Expression and Purification of Antimicrobial Peptide Adenoregulin with C-Amidated Terminus in Escherichia coli. Protein Expr. Purif. 2005, 40, 404–410. [Google Scholar] [CrossRef]
- Zhao, C.Q.; Nguyen, T.; Boo, L.M.; Hong, T.; Espiritu, C.; Orlov, D.; Wang, W.; Waring, A.; Lehrer, R.I. RL-37, an Alpha-Helical Antimicrobial Peptide of the Rhesus Monkey. Antimicrob. Agents Chemother. 2001, 45, 2695–2702. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Lai, Y.; Chang, C.; Tsai, Y.; Tang, C.; Jan, J. Star-Shaped Polypeptides Exhibit Potent Antibacterial Activities. Nanoscale 2019, 11, 11696–11708. [Google Scholar] [CrossRef]
- Cruz, J.; Ortiz, C.; Guzman, F.; Cardenas, C.; Fernandez-Lafuente, R.; Torres, R. Design and Activity of Novel Lactoferrampin Analogues against O157:H7 Enterohemorrhagic Escherichia Coli. Biopolymers 2014, 101, 319–328. [Google Scholar] [CrossRef]
- Yu, H.; Ding, X.; Shang, L.; Zeng, X.; Liu, H.; Li, N.; Huang, S.; Wang, Y.; Wang, G.; Cai, S.; et al. Protective Ability of Biogenic Antimicrobial Peptide Microcin J25 Against Enterotoxigenic Escherichia Coli-Induced Intestinal Epithelial Dysfunction and Inflammatory Responses IPEC-J2 Cells. Front. Cell. Infect. Microbiol. 2018, 8, 242. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.; Hua, K.; Yu, Y.; Cheng, Y.; Cheng, T.; Huang, Y.; Chang, H.; Chen, W. Antibacterial and Antibiofilm Activities of Novel Antimicrobial Peptides against Multidrug-Resistant Enterotoxigenic Escherichia Coli. Int. J. Mol. Sci. 2021, 22, 3926. [Google Scholar] [CrossRef]
- Kannappan, A.; Gowrishankar, S.; Srinivasan, R.; Pandian, S.K.; Ravi, A.V. Antibiofilm Activity of Vetiveria zizanioides Root Extract against Methicillin-Resistant Staphylococcus aureus. Microb. Pathog. 2017, 110, 313–324. [Google Scholar] [CrossRef]
- Naskar, A.; Lee, S.; Kim, K. Antibacterial Potential of Ni-Doped Zinc Oxide Nanostructure: Comparatively More Effective against Gram-Negative Bacteria Including Multi-Drug Resistant Strains. RSC Adv. 2020, 1, 1232–1242. [Google Scholar] [CrossRef] [Green Version]
- McInnes, R.S.; McCallum, G.E.; Lamberte, L.E.; van Schaik, W. Horizontal Transfer of Antibiotic Resistance Genes in the Human Gut Microbiome. Curr. Opin. Microbiol. 2020, 53, 35–43. [Google Scholar] [CrossRef]
- Guilhelmelli, F.; Vilela, N.; Albuquerque, P.; Derengowski, L.D.S.; Silva-Pereira, I.; Kyaw, C.M. Antibiotic Development Challenges: The Various Mechanisms of Action of Antimicrobial Peptides and of Bacterial Resistance. Front. Microbiol. 2013, 4, 353. [Google Scholar] [CrossRef] [Green Version]
- Chou, H.; Kuo, T.; Chiang, J.; Pei, M.; Yang, W.; Yu, H.; Lin, S.; Chen, W. Design and Synthesis of Cationic Antimicrobial Peptides with Improved Activity and Selectivity Against Vibrio Spp. Int. J. Antimicrob. Agents 2008, 32, 130–138. [Google Scholar] [CrossRef]
- Nam, B.; Moon, J.; Park, E.; Kong, H.; Kim, Y.; Kim, D.; Kim, W.; An, C.; Seo, J. Antimicrobial and Antitumor Activities of Novel Peptides Derived from the Lipopolysaccharide- and β-1,3-Glucan Binding Protein of the Pacific Abalone Haliotis discus hannai. Mar. Drugs 2016, 14, 227. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Li, W.; O’Brien-Simpson, N.; Separovic, F.; Sani, M. C-Terminus Amidation Influences Biological Activity and Membrane Interaction of Maculatin 1. Amino Acids 2021, 53, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Mahlapuu, M.; Hakansson, J.; Ringstad, L.; Bjorn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [Green Version]
- Malmsten, M. Interactions of Antimicrobial Peptides with Bacterial Membranes and Membrane Components. Curr. Top. Med. Chem. 2016, 16, 16–24. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The Expanding Scope of Antimicrobial Peptide Structures and their Modes of Action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Luo, L.; Chen, J.; Sun, H.; Wang, X.; Yi, Y.; Lv, X. Cell Wall and DNA Damage of Staphylococcus aureus by Bacteriocin BM1157. LWT-Food Sci. Technol. 2020, 134, 109842. [Google Scholar] [CrossRef]
- Kwon, J.Y.; Kim, M.K.; Mereuta, L.; Seo, C.H.; Luchian, T.; Park, Y. Mechanism of Action of Antimicrobial Peptide P5 Truncations against Pseudomonas aeruginosa and Staphylococcus aureus. AMB Express 2019, 9, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, X.; Wu, R.; Si, D.; Liao, X.; Zhang, L.; Zhang, R. Novel Hybrid Peptide Cecropin A (1–8)-LL37 (17–30) with Potential Antibacterial Activity. Int. J. Mol. Sci. 2016, 17, 983. [Google Scholar] [CrossRef]
- Tan, T.; Wu, D.; Li, W.; Zheng, X.; Li, W.; Shan, A. High Specific Selectivity and Membrane-Active Mechanism of Synthetic Cationic Hybrid Antimicrobial Peptides Based on the Peptide FV7. Int. J. Mol. Sci. 2017, 18, 339. [Google Scholar] [CrossRef] [Green Version]
- Jiale, Z.; Jian, J.; Xinyi, T.; Haoji, X.; Xueqin, H.; Xiao, W. Design of a Novel Antimicrobial Peptide 1018M Targeted ppGpp to Inhibit MRSA Biofilm Formation. AMB Express 2021, 11, 49. [Google Scholar] [CrossRef]
- Qi, R.; Zhang, N.; Zhang, P.; Zhao, H.; Liu, J.; Cui, J.; Xiang, J.; Han, Y.; Wang, S.; Wang, Y. Gemini Peptide Amphiphiles with Broad-Spectrum Antimicrobial Activity and Potent Antibiofilm Capacity. ACS Appl. Mater. Interfaces 2020, 12, 17220–17229. [Google Scholar] [CrossRef]
- Shyla, G.; Vineethkumar, T.V.; Arun, V.; Divya, M.P.; Thomas, S.; George, S. Functional Characterization of two Novel Peptides and their Analogs Identified from the Skin Secretion of Indosylvirana aurantiaca, an Endemic Frog Species of Western Ghats, India. Chemoecology 2019, 29, 179–187. [Google Scholar] [CrossRef]
- Choudhary, K.S.; Mih, N.; Monk, J.; Kavvas, E.; Yurkovich, J.T.; Sakoulas, G.; Palsson, B.O. The Staphylococcus aureus Two-Component System AgrAC Displays Four Distinct Genomic Arrangements that Delineate Genomic Virulence Factor Signatures. Front. Microbiol. 2018, 9, 1082. [Google Scholar] [CrossRef] [PubMed]
- Archer, N.K.; Mazaitis, M.J.; Costerton, J.W.; Leid, J.G.; Powers, M.E.; Shirtliff, M.E. Staphylococcus aureus Biofilms Properties, Regulation and Roles in Human Disease. Virulence 2011, 2, 445–459. [Google Scholar] [CrossRef] [Green Version]
- Vollaro, A.; Esposito, A.; Esposito, E.P.; Zarrilli, R.; Guaragna, A.; de Gregorio, E. PYED-1 Inhibits Biofilm Formation and Disrupts the Preformed Biofilm of Staphylococcus aureus. Antibiotics 2020, 9, 240. [Google Scholar] [CrossRef]
- Ren, D.; Li, C.; Qin, Y.; Yin, R.; Li, X.; Tian, M.; Du, S.; Guo, H.; Liu, C.; Zhu, N.; et al. Inhibition of Staphylococcus aureus Adherence to Caco-2 Cells by lactobacilli and Cell Surface Properties that Influence Attachment. Anaerobe 2012, 18, 508–515. [Google Scholar] [CrossRef]
- Jaglic, Z.; Desvaux, M.; Weiss, A.; Nesse, L.L.; Meyer, R.L.; Demnerova, K.; Schmidt, H.; Giaouris, E.; Sipailiene, A.; Teixeira, P.; et al. Adhesins and Exopolymers of Selected Foodborne Pathogens. Microbiology 2014, 160, 2561–2582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andjelkovic, M.; Tsilia, V.; Rajkovic, A.; De Cremer, K.; Van Loco, J. Application of LC-MS/MS MRM to Determine Staphylococcal Enterotoxins (SEB and SEA) in Milk. Toxins 2016, 8, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, S.; Ji, Y.; Chen, Q.; Mei, Z.; Jiang, Y.; Cui, W.; Zhou, H.; Wang, L.; Qiao, X.; Xu, Y.; et al. Antibacterial Activity of the Recombinant Bovine Lactoferrin Peptide Expressed by Lactococcus lactis. Acta Microbiol. Sin. 2021, 61, 428–443. [Google Scholar]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard-Ninth Edition. Clinical and Laboratory Standards Institute 2012, M07-A9, Volume 32. Available online: https://www.coursehero.com/file/59636428/03-CLSI-M07-A9-2012-1pdf/ (accessed on 29 September 2021).
- Li, Z.; Cheng, Q.; Guo, H.; Zhang, R.; Si, D. Expression of Hybrid Peptide EF-1 in Pichia pastoris, its Purification, and Antimicrobial Characterization. Molecules 2020, 25, 5538. [Google Scholar] [CrossRef]
- Shu, Q.; Lou, H.; Wei, T.; Zhang, X.; Chen, Q. Synergistic Antibacterial and Antibiofilm Effects of Ultrasound and MEL-A against Methicillin-Resistant Staphylococcus aureus. Ultrason. Sonochem. 2021, 72, 105452. [Google Scholar] [CrossRef]
- Bai, J.; Zhong, K.; Wu, Y.; Elena, G.; Gao, H. Antibiofilm Activity of Shikimic Acid against Staphylococcus aureus. Food Control 2019, 95, 327–333. [Google Scholar] [CrossRef]
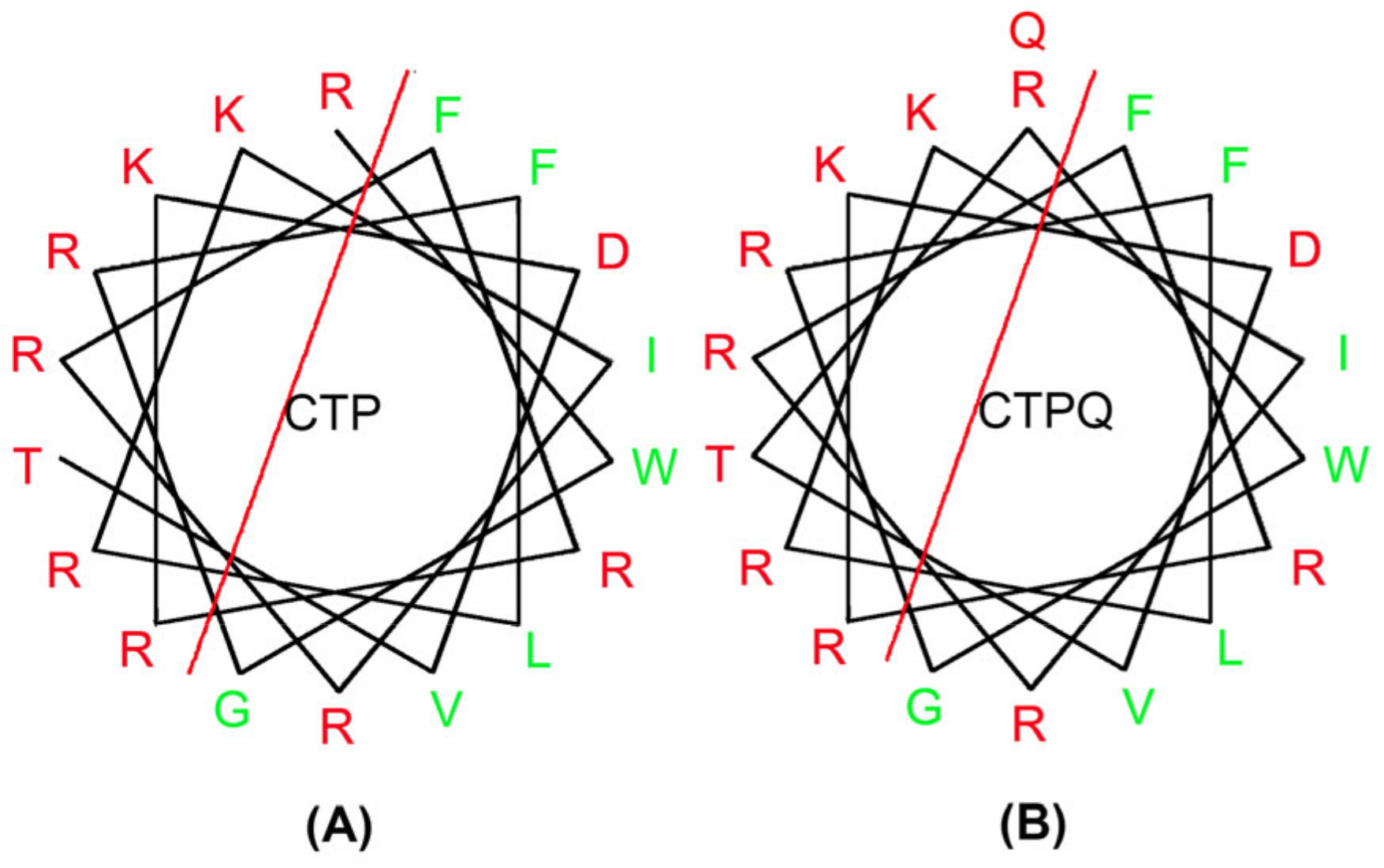


| Name of Peptide | Normalized Hydrophobic Moment | Normalized Hydrophobicity | Net Charge | Amphiphilicity Index |
|---|---|---|---|---|
| CTP | 1.11 | 0.34 | 8.00 | 1.75 |
| CTPQ | 1.07 | 0.40 | 8.00 | 1.72 |
| Strains | Peptides | MICs (μg/mL) | MBCs (μg/mL) |
|---|---|---|---|
| S. aureus ATCC 6385 | CTP | 16 | 32 |
| CTPQ | 16 | 32 | |
| CTP-NH2 | 8 | 16 | |
| S. aureus ATCC 43300 | CTP | 32 | 64 |
| CTPQ | 32 | 64 | |
| CTP-NH2 | 32 | 64 | |
| S. aureus CVCC 1882 | CTP | 4 | 4 |
| CTPQ | 4 | 4 | |
| CTP-NH2 | 2 | 8 | |
| S. aureus ATCC 25923 | CTP | 64 | 512 |
| CTPQ | 64 | 512 | |
| CTP-NH2 | 32 | 512 | |
| S. castellani CMCC 51592 | CTP | 32 | 128 |
| CTPQ | 32 | 128 | |
| CTP-NH2 | 32 | 64 | |
| S. typhimurium ATCC 14028 | CTP | 32 | 128 |
| CTPQ | 32 | 512 | |
| CTP-NH2 | 32 | 64 | |
| S. pullorum CVCC 519 | CTP | 8 | 64 |
| CTPQ | 8 | 64 | |
| CTP-NH2 | 8 | 32 | |
| E. coli ATCC K99 | CTP | 32 | 256 |
| CTPQ | 32 | 128 | |
| CTP-NH2 | 16 | 64 | |
| EHEC O157 H7 | CTP | 16 | 256 |
| CTPQ | 16 | 128 | |
| CTP-NH2 | 8 | 128 | |
| P. aeruginosa ATCC 27853 | CTP | 64 | 128 |
| CTPQ | 32 | 64 | |
| CTP-NH2 | 16 | 64 | |
| P. aeruginosa ATCC 9027 | CTP | 64 | 256 |
| CTPQ | 64 | 128 | |
| CTP-NH2 | 32 | 128 | |
| P. aeruginosa CGMCC 1.10712 | CTP | 64 | 128 |
| CTPQ | 32 | 64 | |
| CTP-NH2 | 8 | 64 |
| Gene Name | Primer Sequence | ATCC Genome Locus_Tag 1 |
|---|---|---|
| agrA | F: 5′-CTGATAATCCTTATGAGGTGCTTGA-3′ | KNNFDEDG_02655 |
| R: 5′-CGTAAGTTCACTGTGACTCGTAACG-3′ | ||
| lrgB | F: 5′-ACTACAGCGATTGCGTTACCA-3′ | KNNFDEDG_01697 |
| R: 5′-CTTGCCATTGATTCTTCTACAGGT-3′ | ||
| sigB | F: 5′-TTGACCATTCCATTGAAGCTG-3′ | KNNFDEDG_02624 |
| R: 5′-AACCGATACGCTCACCTGTC-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, H.-N.; Tong, Y.-C.; Wang, H.-L.; Zhang, J.; Li, Z.-X.; Abbas, Z.; Yang, T.-T.; Liu, M.-Y.; Chen, P.-Y.; Hua, Z.-C.; et al. Novel Hybrid Peptide Cathelicidin 2 (1-13)-Thymopentin (TP5) and Its Derived Peptides with Effective Antibacterial, Antibiofilm, and Anti-Adhesion Activities. Int. J. Mol. Sci. 2021, 22, 11681. https://doi.org/10.3390/ijms222111681
Guo H-N, Tong Y-C, Wang H-L, Zhang J, Li Z-X, Abbas Z, Yang T-T, Liu M-Y, Chen P-Y, Hua Z-C, et al. Novel Hybrid Peptide Cathelicidin 2 (1-13)-Thymopentin (TP5) and Its Derived Peptides with Effective Antibacterial, Antibiofilm, and Anti-Adhesion Activities. International Journal of Molecular Sciences. 2021; 22(21):11681. https://doi.org/10.3390/ijms222111681
Chicago/Turabian StyleGuo, He-Nan, Yu-Cui Tong, Hui-Li Wang, Jing Zhang, Zhong-Xuan Li, Zaheer Abbas, Tian-Tian Yang, Meng-Yao Liu, Pei-Yao Chen, Zheng-Chang Hua, and et al. 2021. "Novel Hybrid Peptide Cathelicidin 2 (1-13)-Thymopentin (TP5) and Its Derived Peptides with Effective Antibacterial, Antibiofilm, and Anti-Adhesion Activities" International Journal of Molecular Sciences 22, no. 21: 11681. https://doi.org/10.3390/ijms222111681
APA StyleGuo, H.-N., Tong, Y.-C., Wang, H.-L., Zhang, J., Li, Z.-X., Abbas, Z., Yang, T.-T., Liu, M.-Y., Chen, P.-Y., Hua, Z.-C., Yan, X.-N., Cheng, Q., Ahmat, M., Wang, J.-Y., Zhang, L.-L., Wei, X.-B., Liao, X.-D., & Zhang, R.-J. (2021). Novel Hybrid Peptide Cathelicidin 2 (1-13)-Thymopentin (TP5) and Its Derived Peptides with Effective Antibacterial, Antibiofilm, and Anti-Adhesion Activities. International Journal of Molecular Sciences, 22(21), 11681. https://doi.org/10.3390/ijms222111681







