Abstract
Mitogen-activated protein kinases (MAPKs) are essential for proper cell functioning as they regulate many molecular effectors. Careful regulation of MAPKs is therefore required to avoid MAPK pathway dysfunctions and pathologies. The mammalian genome encodes about 200 phosphatases, many of which dephosphorylate the MAPKs and bring them back to an inactive state. In this review, we focus on the normal and pathological functions of dual-specificity phosphatase 9 (DUSP9)/MAP kinase phosphatases-4 (MKP-4). This cytoplasmic phosphatase, which belongs to the threonine/tyrosine dual-specific phosphatase family and was first described in 1997, is known to dephosphorylate ERK1/2, p38, JNK and ASK1, and thereby to control various MAPK pathway cascades. As a consequence, DUSP9 plays a major role in human pathologies and more specifically in cardiac dysfunction, liver metabolic syndromes, diabetes, obesity and cancer including drug response and cell stemness. Here, we recapitulate the mechanism of action of DUSP9 in the cell, its levels of regulation and its roles in the most frequent human diseases, and discuss its potential as a therapeutic target.
1. General Introduction
The mitogen-activated protein kinase (MAPK) signaling pathways are crucial in cell function and homeostasis. MAPKs regulate pathophysiological processes by controlling signal translation and cellular response such as survival, proliferation, differentiation and migration [1,2,3]. They are activated by a double phosphorylation process on tyrosine and threonine residues in a conserved Thr-X-Tyr motif (X being any amino acid) [4,5]. Activation of MAPK pathways triggers multiple intracellular signaling cascades. Each cascade is initiated by a specific signal and leads to the activation of a particular MAPK [6]. Once activated, MAPK can phosphorylate various cytoplasmic and/or nuclear substrates and induce changes in the function of target proteins and gene expression [6]. Spatial localization of MAPKs also determines the target substrates and subsequent cellular effects [7].
In humans, MAPKs are divided into three main families: ERK (kinases regulated by the extracellular signal), p38 (protein kinases activated by stress) and JNK (amino-terminal kinases Jun) [2,3,6]. Each of the MAPK modules involves three sequentially activated kinases: MAPKKK, MAPKK and MAPK [4,5,8]. ERK is the best-studied member of the MAPKs. It is activated by numerous extracellular signals such as growth factors, mitogens, cytokines, viruses, receptor ligands coupled to G proteins, and oncogenes. It thus regulates cell signaling under normal and pathological conditions. When the MAPK pathway is inactive, ERK is localized in the cytoplasm. Once activated, phosphorylated ERK is dimerized and is translocated to the nucleus [4,8]. In the nucleus, phosphorylated ERK binds to transcription factors such as proto-oncogenes c-Fos and c-Jun and induces the expression of many genes, most of which are associated with cell proliferation, motility, stemness and survival [8]. As a consequence, MAPK signaling and ERK kinases play a key role in cancer by regulating proliferation, migration, angiogenesis and metastasis [8]. ERK signaling is deregulated in about a third of human cancers and constitutive ERK activation has been reported in many types of tumors [9]. The ERK cascade involves several upstream kinases including RAS, RAF and MEK1/2, forming the RAS/RAF/MEK/ERK pathway and several downstream MAPKs [8,10]. RAS/RAF/MEK/ERK signaling dysfunction is a major trigger for the development of several cancers and RAS mutations (mainly K-RAS) account for 30% of all analyzed tumors [8,11]. Regarding p38 and JNK, they are mainly linked to stress and cell apoptosis [8]. There are four p38 MAP kinases in mammals: α, β, γ and δ. Of all the p38 MAPK isoforms, p38a is the best characterized and is expressed in most cell types [4].
According to the PhosphoSite database, approximately 17,000 proteins have at least one phosphorylated residue in their peptide sequence [12]. Therefore, very precise regulation of protein phosphorylation is necessary for the proper functioning of cells. Deregulation of MAPKs has been described in various pathologies such as metabolic diseases and malignancies in association with drug resistance [2,3,7,12,13,14]. Under normal conditions, MAPKs are tightly regulated by protein phosphatases. Phosphatases reverse phosphorylation and return MAPKs to an inactive state. The dual specificity phosphatase (DUSP) family belongs to the 199 phosphatases encoded in the human genome. This family is composed of 61 phosphatases capable of downregulating MAPKs by dephosphorylating both tyrosine and serine/threonine residues in a single substrate [12,15]. The phosphorylation of proteins is a reversible process. This prevents the abnormal activation of the signal and fine-tunes its activity and downstream effects [3,12]. The balance between phosphorylation and dephosphorylation controls the expression, function, activity and localization of many proteins [12,16]. Dephosphorylation by DUSPs regulates the duration, intensity and spatiotemporal profile of the MAPK signaling cascade [17]. This dephosphorylation takes place thanks to the highly conserved phosphatase site which contains arginine, cysteine and aspartic acid [3,12,18]. In addition to the active site common to all DUSPs, some DUSPs contain a MAP kinase-binding motif (MKB), also called a kinase-interacting motif (KIM), which interacts with the common docking domain of MAPKs to allow the interaction between the enzyme and the substrate [3,12,18,19]. Ten DUSPs containing the KIM domain are classified as typical DUSPs or MAP kinase phosphatases (MKPs) (Table 1), while those which do not have this domain (16 phosphatases in total) are called atypical DUSPs [3,5,20]. However, there are a few exceptions. DUSP2, DUSP5 and DUSP8 are typical DUSPs and contain the KIM domain but they are not called MKPs. On the other hand, DUSP14 and DUSP26, which are atypical DUSPs and do not contain a KIM domain, are called MKP6 and MKP8, respectively (Table 1) [3,20]. Typical DUSPs are the best characterized within the DUSP family [20] and this comprises the typical DUSP named DUSP9 or MKP4, which was first described in 1997 by Muda and collaborators [18]. This 42-kDa protein dephosphorylates several substrates including JNK, p38, the MAPKKK apoptosis signal-regulating kinase 1 (ASK1) and ERK1/2 with a high specificity for ERK kinases [3,18]. In this review, we discuss recent findings about DUSP9, a cytoplasmic phosphatase encoded by a four-exon gene located on the X chromosome [18,21].

Table 1.
General information and main functions of MAP kinase phosphatases (MKP).
2. General Characteristics of DUSP9 and Mechanisms of Regulation
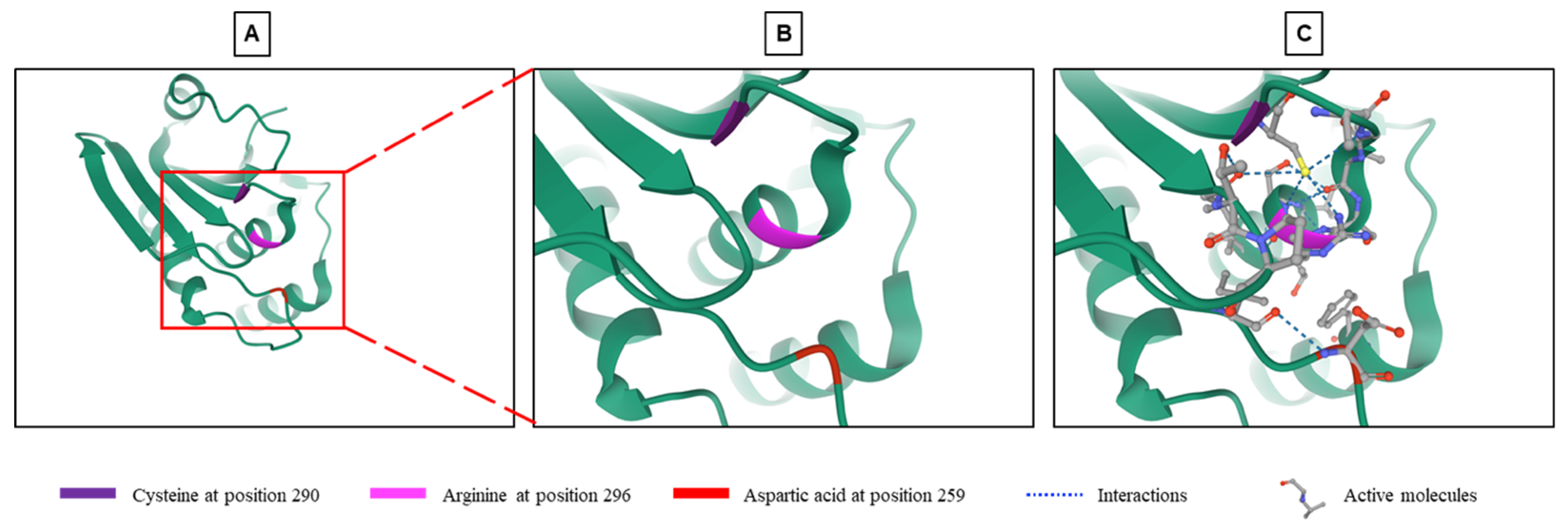
DUSP9 is a typical DUSP characterized by the presence of an MKB/KIM motif and a phosphatase domain, which shares structural homology with other DUSPs [12]. Sequence homology analysis of DUSP9 showed 61% identity with DUSP22/MKP-X, 57% with DUSP6/MKP-3 and 35% with DUSP8 [18]. DUSP9 contains a C-terminal catalytic domain common to all DUSPs. The core of this domain, which consists in residues of arginine, cysteine and aspartic acid (Figure 1), is highly conserved and carries the phosphatase activity [11,18,19,43]. Arginine at position 296 forms hydrogen bonds with a phosphate group on the substrate and stabilizes the transition state. Cysteine at position 290 functions as an active nucleophile site forming a covalent thiol-phosphate intermediate, while aspartic acid in position 259 acts as a catalytic acid to give a proton to the leaving group (Figure 1) [18,43,44].
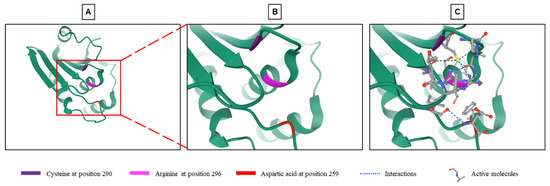
Figure 1.
Structure of the phosphatase active site of DUSP9. (A) Crystallographic representation of the phosphatase site of DUSP9 with a focus on its catalytic part (see region of interest in red box) composed of a cysteine at position 290, an arginine at position 296 and an aspartic acid at position 259 (from RCSB Protein Data Bank: https://www.rcsb.org/3d-view/3LJ8, accessed on 1 September 2021) [45]. (B) Higher magnification of region of interest shown in panel A. (C) Same image than in Panel B with inserted substrates and molecular interactions between the three amino acyls and substrate.
The MKB/KIM motif is composed of two CDC25 homology domains and an intermediate group of basic amino acids mediating the interaction with the common domain of MAPKs [16,18,19,44]. DUSP9 is able to undergo a conformational rearrangement allowing it to alternate between a partially active structure and a fully active structure [43]. A crystallographic representation of the catalytic site of DUSP9 showed a unique structure with significant differences between the catalytic core and several surrounding loops compared to other MKPs. The catalytic site of DUSP9 deviates considerably from the canonical conformation of DUSPs, which may explain the low catalytic activity of this protein in the absence of specific substrates [19,43]. The DUSP9 protein alone has very low catalytic activity but binding to MAPK through the MKB/KIM domain significantly increases its phosphatase activity [18,19]. The binding of the substrate likely triggers a conformational change and thus increases its catalytic efficiency [19,43]. DUSP9 is capable of binding and being activated by different MAPKs. Measurements in the presence of para-nitrophenylphosphate (pNPP) showed DUSP9 binding preference and activation by ERK2, JNK and p38 MAPK [19,34,44].
At a functional level, DUSP9 phosphatase is unique. It plays an important role in the dephosphorylation and inactivation of specific kinases such as ASK1, ERK1/2, p38 and JNK (Figure 2). This results in a negative control of MAPK signal transduction and a fine tuning of their duration and intensity [15,17,32]. The endogenous catalytic activity of DUSP9 was studied for the first time by Muda and collaborators by measuring the hydrolysis of pNPP in the presence of increasing doses of a purified human recombinant protein. DUSP9 displayed a dose-dependent catalytic activity which was directly proportional to the amount of protein added [18]. The DUSP9 protein has a broad specificity for MAPK substrates. It can dephosphorylate ERK-family MAPKs, stress-activated JNK and p38 MAPKs (Figure 2), but its effect is significantly higher and more specific for ERK kinases [18,43]. In hepatic tumor cells, DUSP9 negatively regulates the RAS/RAF/MEK/ERK signal by dephosphorylating ERK1/ERK2 and a low level of DUSP9 is correlated with an elevated level of phospho-ERK1/2 in hepatocellular carcinoma (HCC) samples [15]. Co-incubation of ERK2 with increasing concentrations of DUSP9 results in the dose-dependent blockade of ERK2 target phosphorylation such as stathmin [18]. This catalytic activity is effectively inhibited by sodium vanadate, which is an inhibitor of protein tyrosine phosphatases [18].
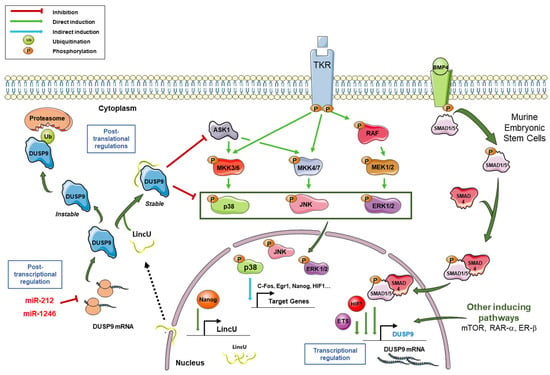
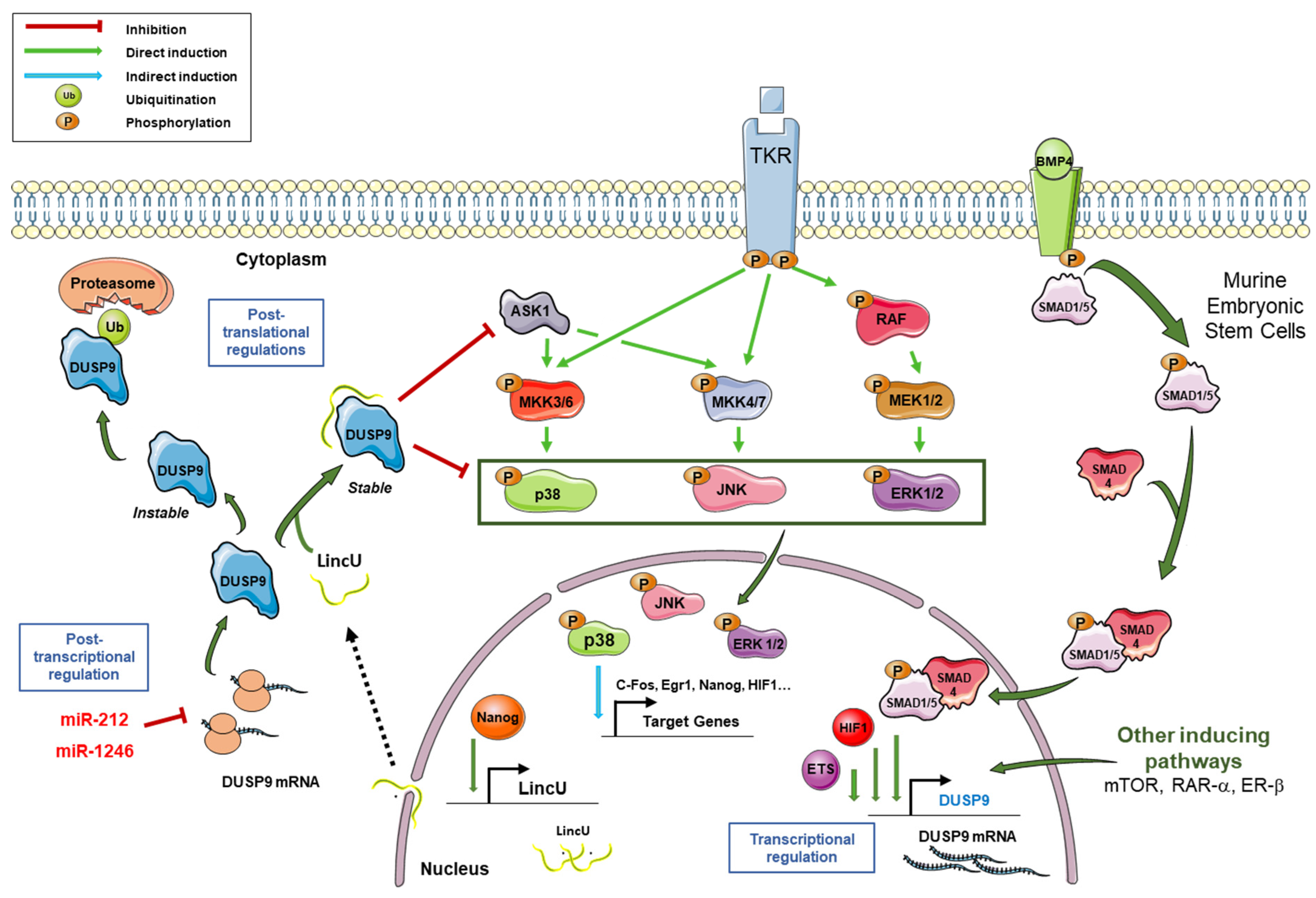
Figure 2.
Regulation of DUSP9 expression and connection with MAPK pathways. Following the activation of tyrosine kinase receptors (TKR), MAPK pathways are activated through the successive phosphorylations of MAPKKKs (among which ASK1), MAPKKs MKK3/6, MKK4/7 and MEK1/2, and MAPKs p38, JNK and ERK. Phosphorylated MAPKs translocate to the nucleus and induce expression of the downstream targets c-FOS, ERG1, NANOG and HIF1, among others. In murine embryonic stem cells, the binding of BMP4 on its receptor induces the phosphorylation of Smad1/5, which then associates with Smad4. The Smad1/5–Smad4 complex translocates to the nucleus, binds the DUSP9 promoter and induces its transcriptional expression. Transcription factors HIF1-α and ETS can also induce DUSP9 expression. Besides the BMP signal, mTOR, RAR-α and ERβ pathways can also potentiate the transcription of the DUSP9 gene. Following NANOG binding on LINCU promoter, the long non-coding RNA LincU is transcribed and exported in the cytoplasm where it associates with DUSP9 and stabilizes it. Stable DUSP9 can dephosphorylate its substrates, including ERK1/2, p38, JNK and ASK1. In the absence of LincU RNA, DUSP9 protein is unstable and is polyubiquitinylated before its degradation by the proteasome. DUSP9 can also be post-transcriptionally regulated by miR-212 and miR-1246, which target the 3′-untranslated region of its mRNA. In summary, DUSP9 expression is tightly regulated by transcriptional, post-transcriptional and post-translational mechanisms.
In normal mature tissues, DUSP9 is mainly expressed in kidney, adipose tissue and placenta, while it is only minimally present in brain, ovary, testis and urinary bladder (National Center for Biotechnology Information: available online: https://www.ncbi.nlm.nih.gov/gene/1852, accessed on 15 July 2021) [32,46,47]. In order to maintain cell homeostasis, the level and activity of DUSP9 has to be tightly regulated at transcriptional, post-transcriptional and post-translational levels (Figure 2) [3]. At the transcriptional level, the expression of DUSP9 can be regulated by various transcription factors such as EFS family members and hypoxia-inducible factor 1 alpha (HIF1α). For instance, genetic depletion by gene knock-down or digoxin-induced pharmacological inhibition of HIF1 blocks the expression of DUSP9 and induces the loss of its inhibitory effect on the ERK signaling pathway in MDA-MB-231 breast cancer cells [17]. Thus, HIF1 positively regulates DUSP9 expression. DUSP9 is also transcriptionally regulated by BMP signaling in mouse embryonic stem cells (mESCs). The bone morphogenetic protein 4 (Bmp4) induces the recruitment of Smad1/5 and Smad4 on the promoter region of DUSP9 and induces DUSP9 expression at the mRNA level, and thus at the protein level. The positive regulation of DUSP9 by Bmp4 is accompanied by inhibition of ERK pathway activity and downregulation of its targets Egr1 and Fos (Figure 2). This DUSP9-mediated dephosphorylation was not observed for p38 and JNK, showing its specificity for ERK1/2 signaling which is crucial for the renewal and differentiation of mESCs [31]. In the same cells, DUSP9 can also undergo post-translational regulation by a long non-coding RNA (lncRNA) called LincU [48]. Following induced expression of LincU by the transcription factor Nanog, this lncRNA directly binds DUSP9 and maintains the phosphatase in an active and stable conformation. As a result, DUSP9, bound to LincU, constantly dephosphorylates ERK1/2 and thus, totally blocks RAS/RAF/MEK/ERK signaling. Interestingly, the phosphorylation level of ERK1/2 was inversely correlated with LincU expression and the amount of DUSP9 protein, while the level of DUSP9 mRNA remained unchanged [48]. Thus, LincU interacts with DUSP9 protein and stabilizes it in an active state, thereby triggering a constitutive dephosphorylation state of DUSP9 kinase substrates (Figure 2) [48]. DUSP9 can also be transcriptionally regulated by the retinoic acid receptor (RAR). Microarray analysis of differential gene expression in Caco-2 cells treated with RAR agonists, Ch55 and Am580 demonstrated dose-dependent induction of DUSP9 expression by RAR signaling. DUSP9 was one of the most induced genes after RAR activation. Induction of DUSP9 mRNA and protein by RAR signaling was confirmed by RT-qPCR and immunolabelling [49]. Similar results were obtained with HT29 and HeLa cells. On the other hand, DUSP9 induction was inhibited in Caco-2 cells expressing the dominant negative form of RARα or treated by the specific RAR antagonist LE540. Chromatin immunoprecipitation analysis showed that RAR induces DUSP9 expression by binding directly to the DUSP9 promoter through an inverted direct repeat separated by 1 (DR1 element). By inducing DUSP9, RAR signaling inactivates ERK during the differentiation of colorectal cancer (CRC) cells [49]. In a more recent paper, an upregulation of DUSP9 was also reported in rat ovaries following Estrogen Receptor-beta signaling induction, supporting the hypothesis that DUSP9 is one of the key genes involved in gonadotrophin-mediated ovarian follicle development [50].
In the cytoplasm, DUSP9 can be degraded by the proteasome following its ubiquitination [3,48]. Pretreatment with the potent proteasome inhibitor MG132 stabilizes DUSP9 protein and increases its level in LincU-deficient mESCs [48]. These data indicate that the ubiquitination-proteasome pathway is involved in the degradation of DUSP9 induced by LincU deficiency. Immunoprecipitation performed after pretreatment with MG132 and immunolabelling with an anti-ubiquitin antibody showed polyubiquitinated DUSP9 bands, suggesting that the knockdown of LincU causes ubiquitination of DUSP9 [48]. This indicates that LincU binds to DUSP9, increases its stability and protects it from ubiquitin-mediated degradation [3,48].
Similar findings were obtained in human embryonic stem cells (hESCs). The overexpression of LincU in hESCs also protected DUSP9 from degradation and inhibited the phosphorylation of ERK1/2, indicating the conserved role of LincU in the stabilization of DUSP9 [48]. Finally, DUSP9 can be regulated by microRNAs (miRNAs), which are small non-coding RNAs that act as post-transcriptional regulators and which are often deregulated in pathological conditions and cancers [10,51,52,53,54]. MiR-1246 and miR-212 can target the 3′-untranslated region (UTR) of the DUSP9 transcript and reduce its expression in CRC cells and hepatoblastoma-derived HepG2 cells, respectively [55,56]. Moreover, Chang and collaborators previously reported a link between DUSP9 expression and miR-133b and miR-4458 in CRC [57]. In our data, we noticed a decreased level of DUSP9 protein following ectopic expression of miR-4510 in hepatoma cells Huh7 [10].
3. DUSP9 in Embryonic Stem Cell Pluripotency and Sex Differences
The feeder-free culture of undifferentiated mESCs depends on the cumulative effect of pluripotency factors (i.e., LIF, BMP4) and low activity of commitment-associated pathways, including the MAPK pathway. In 2012, Li and collaborators investigated the role of DUSP9 in cell stemness and pluripotency [31]. They found that, associated to Lif, Bmp4 supports the pluripotency state of mESCs by keeping ERK pathway activity at a low level, independently of the upstream MAPKK MEK kinase. As Bmp4 is strongly connected to Smad signaling, they showed that Bmp4 mediates its inhibitory effect on ERK by inducing the binding of transcriptional Smad1/5+Smad4 complex on the Dusp9 promoter, thereby promoting its transcription and expression. An increase in DUSP9 led to the dephosphorylation of ERK1/2 and the downregulation of the ERK pathway target genes Egr1 and Fos. By comparison, the Bmp4/Smad/DUSP9 axis was inactive in murine somatic cells [31]. Altogether, these data demonstrated that DUSP9 is an important downstream regulator of BMP/Smad signaling and a key factor in maintaining the stemness of mESCs.
The pluripotency of embryonic stem cell lines is also closely associated with the DNA methylation status of the cells. In an attempt to shed light on this link, Choi and collaborators generated isogenic models of male and female mESCs and murine embryonic germ cells (mEGC), and found that sex rather than embryonic cell type determines the DNA methylation status [58]. The methylation pattern in these undifferentiated cells was controlled by the ratio of X chromosomes to autosomes. Comparative transcriptomic analysis in male and female isogenic embryonic stem cell lines identified DUSP9 as an X chromosome-linked gene more abundantly expressed in female than in male mESCs. In line with the lower activity of MAPK pathway in female mESCs compared to male ones and the presence of ERK1/2, p38 and Jnk1/2/3 genes to autosomes, the methylation level of DNA was reduced in cells ectopically expressing DUSP9 [58]. Forced expression of DUSP9 reduced the global methylation levels in male mESCs, while its deletion in female ones led, after several passages, to an elevation of methylation levels. In DUSP9-overexpressing male cells, the MAPK pathway was inhibited and its downstream targets were less expressed, while the naïve pluripotency markers Ror2 and Prdm14 were increased and the primed pluripotency marker Dmnt3B was decreased [58]. These data demonstrated that DUSP9 plays an important role in embryonic stem cell sex differences and methylation status. In female cells, X chromosome-linked DUSP9 expression keeps the MAPK pathway activity and the expression of DNMT3A/3B at a low level, leading to a global hypomethylation by comparison to male cells [58]. These results were corroborated by a more recent study in which the authors showed that the effect of DUSP9 on global hypomethylation and sex differences was uncoupled from the opening of the chromatin, cell growth and the delay to exit pluripotency [59].
In a more recent study using an elaborate multistep screening approach and mESCs, the authors found that DUSP9 is an X-chromosomal gene and is involved in sex differences during early development [60]. In vivo, its expression was higher in females than in males. Forced expression of DUSP9 enhanced the pluripotency of mESCs by dephosphorylating Mek and ERK1/2, thereby decreasing the expression of the pluripotent factors NANOG and PRDM14. The study also confirmed the role of DUSP9 in the regulation of global DNA methylation mediated by MAPK pathways [60].
4. DUSP9 and Metabolic Diseases
Insulin resistance is defined as the failure of many cells to respond to insulin hormone, to mediate insulin signaling and to take up glucose, thereby leading to abnormally high blood sugar levels known as hyperglycemia. This metabolic syndrome is often associated with chronic inflammation, cell stress responses and pathological conditions such as obesity, non-alcoholic fatty liver disease (NAFLD) and type 2 diabetes mellitus [34,47,61]. Molecularly, the normal response of cells to insulin stimulus depends on tyrosine-kinase insulin receptor membrane expression and downstream phosphorylation cascades and effectors, which comprise the pro-mitotic ERK1/2-dependent MAPK pathway, the JNK- and p38 MAPK-dependent SAP pathway response, the pro-metabolic PI3K/PDK1/AKT-pathway and also the fuel-sensing enzyme AMP-activated protein kinase (AMPK) [61,62,63,64]. Thus, insulin resistance is a pathological condition associated with over-activated phosphorylation cascades involving MAPK, PI3K, AMPK and SAP kinases.
The first report of a link between DUSP9 and insulin resistance was published in 2003 [47]. Using an alkaline-phosphatase-based functional screen reproducing the insulin-operated transcriptional shut down of PEPCK gene in target cells, Xu and collaborators identified DUSP9 out of 10,000 cDNA clones as a potent inhibitor of insulin signal transduction [47]. In physiological conditions, DUSP9 is expressed in mature brain, kidney, testis, white adipose tissue, embryonic liver and placental trophoblast giant cells [18,46,47,65]. By comparing the expression of DUSP9 in different murine models, Xu and collaborators found that it becomes detectable in brown adipose tissue, liver and muscle of obese mice, while being undetectable in these tissues in mice fed on a regular diet [47]. DUSP9 expression remained unchanged in kidneys and testis in the different tested murine models. Next, they showed that DUSP9 expression increases during insulin-induced adipogenesis and culminates in mature adipocytes. Interestingly, forced DUSP9 expression impedes insulin-mediated adipocyte differentiation and limits the uptake of glucose in mature adipocytes [47]. Overall, these data identified DUSP9 as a key regulator of insulin signaling and highlighted its potential role in insulin resistance and metabolic diseases by dephosphorylating kinases involved in metabolic processes, glucose uptake and storage.
In 2004, Bazuine and collaborators demonstrated the negative role of DUSP9 in induced insulin resistance through an alternative phosphorylation cascade governed by p38 MAPK [66]. They also showed that dexamethasone, which inhibits insulin signaling and lowers glucose uptake, induces DUSP9 and DUSP1/MKP-1 expression in adipocytes through a direct or indirect mechanism involving the glucocorticoid receptor. Here again, the expression of DUSP9 decreased glucose uptake and led to ERK dephosphorylation, mimicking the action of dexamethasone. However, unlike Xu and collaborators [47], Bazuine and collaborators showed that DUSP9 attenuates arsenite-mediated but not insulin-mediated p38 MAPK dephosphorylation [66]. These discrepancies can be explained by the pleiotropic effect of DUSP9 on multiple signaling pathways, as it can dephosphorylate and inactivate either ERK1/2, JNK, p38 and/or ASK1 kinases depending on the tissue and biological conditions [34,66,67]. Nevertheless, the insulin receptor substrate-1 (IRS1), which is the target of many of these kinases [61], is phosphorylated on Ser-307. This phosphorylation blocks the subsequent tyrosine phosphorylation of IRS1 by the kinase domain of the insulin receptor after insulin binding. Once phosphorylated on ser-307, IRS-1 dissociates from p85 PI3K and impairs downstream PDK1/AKT signaling [67]. By dephosphorylating and inactivating ERK1/2, JNK, p38 MAPK and/or ASK1 kinases, DUSP9 restores the tyrosine phosphorylation level of IRS-1 and its capacity to interact with p85-PI3K and mediate insulin signal transduction. Moreover, DUSP9 can also impair the action of extracellular mediators and stress inducers (i.e., proinflammatory cytokines, oxidative compounds, endoplasmic reticulum stress effectors), which can induce insulin resistance by abnormally activating MAPK or SAP pathways [34,67]. Importantly, DUSP9 can impede insulin resistance in vivo in the ob/ob murine model by lowering blood glucose levels and by restoring the normal level of enzymes involved in gluconeogenesis and lipogenesis such as fructose-1,6-biphosphatase, SREBP1C, SCD1 and ACC [34,67]. As a result, hyperglycemia, the synthesis of lipids and the accumulation of triglycerides in liver decrease, thereby attenuates the impact of overall hepatic steatosis. On the other hand, DUSP9 expression can be modulated in insulin-sensitive tissues such as white adipocyte tissue, liver and muscle in normal or pathological conditions, depending on both the type of feeding diet and tissue [67].
In agreement with the two previous studies, Ye and collaborators reported the protective effect of the ectopic expression of DUSP9 against nonalcoholic steatohepatitis (NASH), the most frequent and pathophysiological form of NAFLD [34]. NASH is usually associated with type 2 diabetes and obesity, and is characterized by chronic inflammation, insulin resistance and lipid accumulation. Using liver-specific murine models, various feeding conditions and genetic gain- and loss-of-function approaches, they showed that the DUSP9 protein level is slowly and steadily decreased in the liver of mice fed on a high fat diet or presenting obesity. Interestingly, this insulin resistance-induced down-regulation of DUSP9 occurred at the protein level but not at the transcript level, suggesting the translational regulation and/or the potential implication of miRNAs [34]. Interestingly, we found a significant down-regulation of DUSP9 in HCC-derived Huh7 cells transfected with miR-4510, a primate-specific miRNA which inactivates Wnt/β-catenin and RAS/RAF/MEK/ERK signals by targeting RAF proto-oncogene serine/threonine-protein kinase RAF1 and glypican-3 [10,52]. In mice, DUSP9 deficiency exacerbated the gain in body weight, liver weight, lipid accumulation and synthesis mediated by a high fat/high-cholesterol diet [34]. However, under the same dietary conditions, hepatocyte-specific DUSP9 transgenic mice were less prone to accumulate lipids and triglycerides and to develop liver injury, fibrosis, inflammation, NAFLD and NASH [34]. Importantly, the hepatic over-expression of DUSP9 directly affected the blood levels of glucose and insulin, glucose tolerance and the sensitivity of cells to insulin [34]. At a molecular level, DUSP9 deficiency increased the phosphorylation of ERK1/2, JNK and p38, decreased the phosphorylation of IRS-1, AKT and GSK3-β proteins and increased the expression of pro-inflammatory cytokines and chemokines in an inhibitor of nuclear factor kappa B kinase subunit beta (IKK-β)/nuclear factor kappa B (NF-κB)-dependent manner. In contrast, over-expression of DUSP9 reduced the phosphorylation of JNK and p38 MAPK in hepatocytes. Interestingly, these authors uncovered a new target of DUSP9 named ASK1 (also known as MAP3K5 or MAPKKK5) and demonstrated the capacity of DUSP9 to control the SAP response downstream p38 MAPK by dephosphorylating ASK1 and decreasing MAPK kinase 4/7 expression. Moreover, they demonstrated the requirement of ASK1 activation in SAP signaling, lipid accumulation and induced-insulin resistance in hepatocytes [34]. Finally, a recent study reported that higher expression of DUSP9 in placental cytotrophoblasts is associated with hyperglycemia in pregnant women suffering from type 2 diabetes mellitus compared to pregnant women not presenting it. This raises the question of the role of DUSP9 as an effector of gestational insulin resistance [32].
5. DUSP9 and Cardiac Disease
A recent paper by Jiang and collaborators reported that DUSP9 is implicated in the pathological process of heart failure [30]. The progression of this cardiac alteration was attributed to both pathological heart tissue remodeling and over-activation of stress responses, involving many signaling pathways including MAPKs. The authors showed that DUSP9 is required to limit pressure overload-mediated cardiac hypertrophy and heart abnormality. The protective effect of DUSP9 on cardiomyocytes was mediated by the binding and dephosphorylation of the MAPKKK ASK1, subsequently preventing the activation of the downstream p38/JNK pathway [30]. In absence of additional reports on the role of DUSP9 in heart pathologies, future studies are required to improve our understanding of the relationship between DUSP9 phosphatase and cardiac diseases.
6. DUSP9 in Cancers
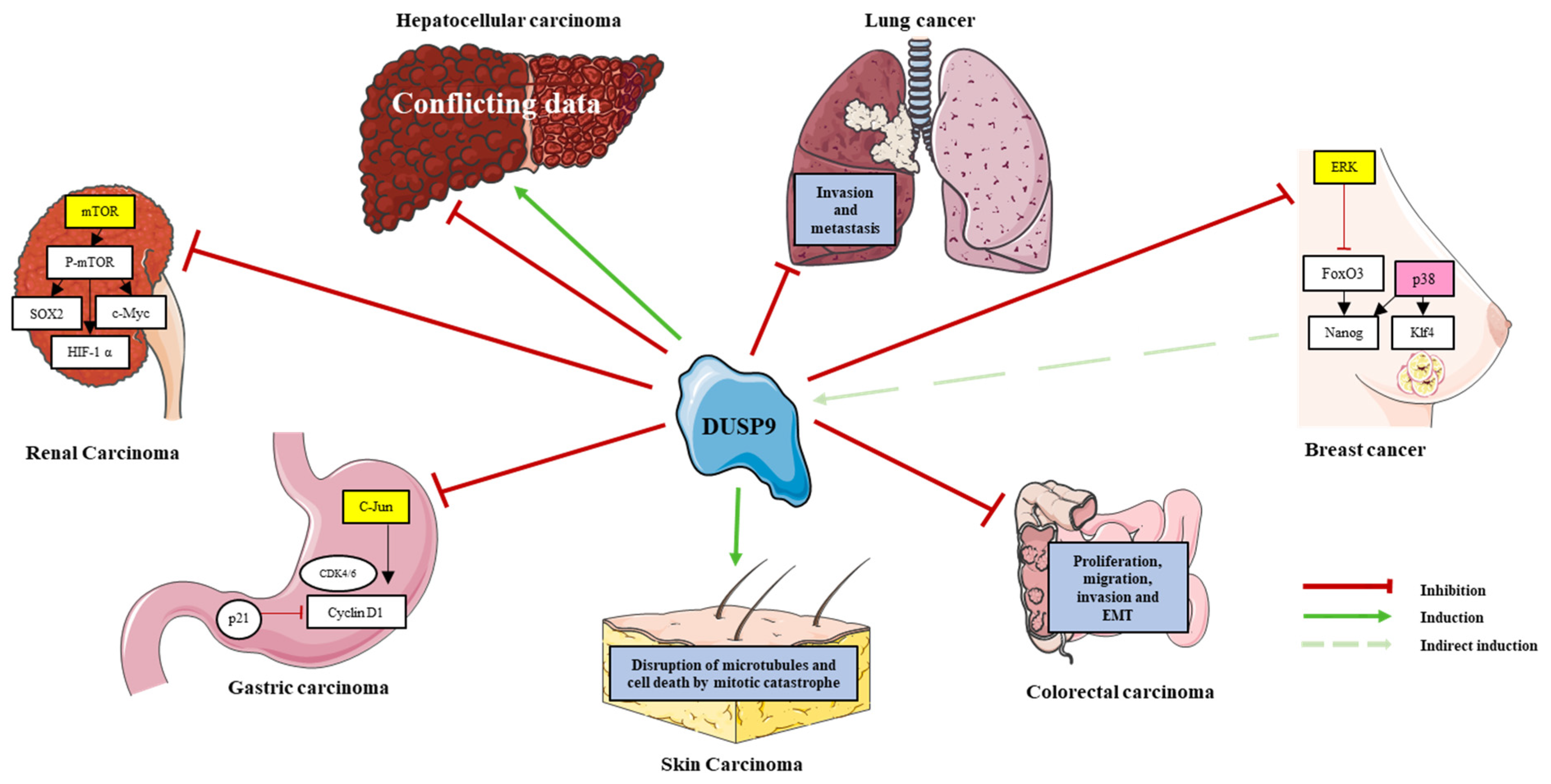
DUSP9 is implicated in many types of cancer (Table 2; Figure 3). It was identified by the Walktrap algorithm as an HCC-associated gene and a potential target for therapeutic research because of its interaction with MAPKs and the proto-oncogenes JUNB and FOSB [68]. However, its mechanistic role in cancer is unclear and seems context-dependent. In this section, we summarize the main data gathered from the literature on the role of DUSP9 in cancer.

Table 2.
Expression of DUSP9 in cancers versus non-tumoral tissues and main results from the literature.
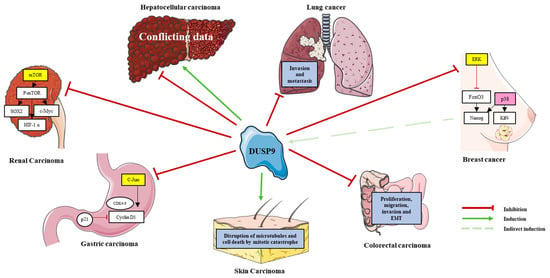
Figure 3.
Role of DUSP9 in various cancers. Positive and negative effect of DUSP9 (see legend at the bottom right) on putative molecular pathways and oncogenic processes in the corresponding cancer. Targets of DUSP9 phosphatase (shown in yellow boxes) are inactivated, and this inactivation induces p38 kinase activity (shown in pink box) in breast cancer. See Section 6 for a fuller description.
6.1. Breast Cancer
Despite progress in highly effective chemotherapies against breast cancer, the triple negative breast cancer (TNBC) subtype remains highly resistant to treatment and presents a high risk of recurrence and mortality [75]. A link between DUPS9 and breast cancer was first described in 2018 by Lu and collaborators who investigated MAPK pathway activity in response to chemotherapy in TNBC [17]. Treatment of MDA-MB-231 cells with paclitaxel or carboplatin inhibited ERK phosphorylation and induced p38 phosphorylation. These effects were lost by the genetic or pharmacological blockade of HIF1α, demonstrating their HIF1α dependency. Using a large cohort of breast cancer samples from the TGCA database, the authors found a strong positive correlation between DUSP9 expression and a 10-gene HIF signature in a TBNC-enriched tumor subtype. Depletion of DUSP9 in MDA-MB-231 cells abolished paclitaxel-mediated ERK inhibition, indicating that ERK is a substrate of DUSP9. MDA-MB-231 cells cultured as mammospheres, which promotes breast cancer stem cell (BCSC) enrichment, expressed a higher amount of DUSP9 protein compared to cells cultured in standard monolayer condition. Additional analysis of chemotherapy-resistant BCSCs in mammospheres showed that paclitaxel or carboplatin induces DUSP9 expression by promoting the direct binding of transcription factor HIF1α to DUSP9 promoter and its transcriptional activation. This leads to ERK inhibition and p38 activation, thereby driving TNBC cells to switch from ERK to p38 signaling, evade chemotherapy and resist the drug through enrichment of BCSCs [17]. DUSP9 is therefore a key factor in chemotherapy-induced BCSC enrichment. In parallel, ERK inhibition caused the upregulation of NANOG and KLF4 mRNAs due to ERK-substrate FoxO3 dephosphorylation, while activated p38 indirectly stabilized NANOG and KLF4 mRNA through ZFP36L1 RNA-binding protein following its phosphorylation by MK2 [17]. Expression of the pluripotency factors NANOG, SOX2 and KLF4 induced by paclitaxel in MDA-MB-231 cells was attenuated by DUSP9 knockdown [17]. Therefore, DUSP9 expression promotes BCSC phenotype specification by inhibiting the ERK pathway and supporting the p38 pathway (Figure 3). In vivo, inactivation of DUSP9 did not significantly decrease the ability of initiating cells to form tumors. This result was consistent with in vitro results showing that DUSP9 knockdown did not significantly decrease the number of mammosphere formations in the absence of paclitaxel [17]. Together, these data suggest that DUSP9 contributes to cancer stemness and drug resistance and thus positively influences the growth and progression of TNBCs. This hypothesis, which is in line with the findings of another study [35], was further supported by an increase in DUSP9 transcript levels in breast cancer patients who received chemotherapy [17]. However, DUSP9 mRNA level was not an independent prognostic factor for relapse-free survival [17]. In 2020, Jimenez and collaborators showed that pERK1/2 is repressed and p38 is upregulated following engraftment of TNBC cells HCC70, HCC1806 and MDA-MB-48 in immune-deficient mice [35], The repression of pERK1/2 was associated with DUSP9 upregulation and high phosphatase activity. The authors also found an inverse relationship between phospho-ERK1/2 activity and DUSP9 expression in TNBC xenografts, as well as in TNBC mammospheres [35], However, in contradiction with the previous study, they showed that the pharmacological or genetic depletion of DUSP9 reduced the growth of TNBC mammospheres in vitro, as well as tumor growth and mass in vivo [35],
6.2. Colorectal Cancer
The first link between DUSP9 and CRC was documented in 2006 by Sansom and collaborators. They showed an upregulation of DUSP9 mRNA in APC-deficient mice intestines, suggesting a connection between DUSP9, the Wnt pathway and the early stage of colon tumorigenesis [76]. In 2015, a quantitative pyrosequencing methylation screening applied to 79 CRC and 22 healthy colon samples evidenced differences in the methylation status of the DUSP9 promoter, although no conclusion could be drawn regarding the role of DUSP9 promoter methylation in colon carcinogenesis [77]. Later, antagonistic interactions between ERK signaling and RAR signaling was reported in CRC cells [49]. Indeed, by increasing DUSP9 expression, RAR signaling counteracts ERK pathway activation. As a result, ERK1/2 kinases were dephosphorylated and CRC cells engaged in differentiation. On the other hand, ERK signaling was shown to inhibit RAR signaling by a transcriptional mechanism mediated by RIP140/histone deacetylase (HDAC) [49]. The upregulation of DUSP9 mediated by RAR signaling in CRC cells was consecutive to the direct binding of RAR to the DUSP9 promoter through a consensus sequence for the RAR/RXR heterodimer corresponding to the DR1 element [49].
Analysis of DUSP9 expression by different experimental approaches in paired CRC tissues showed the downregulation of DUSP9 mRNA and protein in CRC tissues compared to peritumoral tissues [56]. Patients with low DUSP9 transcripts had significantly shorter overall survival and recurrence-free survival than those with high protein levels. In addition, decreased DUSP9 level was closely associated with larger tumors, deeper invasion, and advanced cancer stage, indicating that DUSP9 down-regulation is associated with tumor progression and poor prognosis in CRC [56].
At a mechanistic level, the DUSP9 promoter was hypomethylated in normal intestine compared to CRC, and the treatment of SW480 cells treated with 5-aza-dC, an inhibitor of DNA methyltransferase activity, induced DUSP9 protein level [56]. Interestingly, hypermethylation of DUSP9 promoter was also observed in many other solid tumors including bladder urothelial carcinoma, breast carcinoma, cervical squamous cell carcinoma, endocervical carcinoma, lung carcinoma, and pancreatic carcinoma. Therefore, hypermethylation of the CpG islands near the transcription initiation site of DUSP9 promoter could in part explain DUSP9 downregulation in CRC and other cancers [56]. In addition, Qiu and collaborators showed that miRNAs also participate in DUSP9 downregulation in CRC. They showed that miR-1246 reduces DUSP9 level in CRC cells through an miR-1246-binding site located in the 3′UTR of its transcript and blocks DUSP9-mediated phenotypical changes [56]. Moreover, miR-1246 levels were inversely correlated with DUSP9 mRNA in 30 CRC samples.
At the functional level, forced DUSP9 expression affected the proliferation, migration, invasion and epithelial–mesenchymal transition (EMT) of CRC cells in vitro and in vivo [56]. (Table 2; Figure 3). In vitro, changes in the growth and migration of CRC cells were cell-dependent [56]. For example, DUSP9-silenced SW480 cells produced larger tumors in mice compared to controls. In contrast, tumors in mice with DUSP9-expressing LoVo cells grew more slowly and their size was significantly smaller than in controls [56]. Transcriptome profiling studies performed in DUSP9-silenced SW480 cells reported an enrichment of several signaling pathways related to tumor growth and metastasis, such as ERK, JNK, WNT, AKT/mTOR and ERBB [56]. DUSP9 silencing activated ERK signaling and promoted tumor development [49,56]. Treatment of DUSP9-silenced SW480 cells with PD98059 (a specific inhibitor of the ERK signaling pathway) counteracted their growth advantage, while DUSP9-expressing LoVo cells treated with curcumin (an activator of the ERK signaling pathway) grew significantly faster [56]. Together, these data show that DUSP9 plays an important role in CRC cell tumorigenicity but its role depends heavily on the cellular context and likely on the functional balance between the ERK pathway and the p38 pathway.
6.3. Gastric Cancer
In gastric cancer (GC), the expression level of DUSP9 is low at all stages of the disease and is the lowest in advanced stages [70]. This decreased expression starts very early in tumor progression. DUSP9 down-regulation is detectable in adenomas and also observed in GC cell lines compared to normal gastric cells [70]. Bisulfite sequencing PCR analysis of 30 GCs and matched healthy tissues from gastric mucosa showed high methylation of the DUSP9 promoter at the CpG island in cancerous tissues compared to normal tissues. Treatment of GC-derived MKN-1 cells with the DNA-hypomethylating 5-aza-2′-deoxycytidine (5-aza-dC) drug significantly induced DUSP9 expression [70]. In agreement with another study [56], downregulation of DUSP9 in GC is likely due to DUSP9 promoter hypermethylation and transcription shutdown. Forced expression of DUSP9 in MKN-1 cells inhibited cell growth by approximately 50%, while DUSP9 silencing promoted cell growth by 74%. DUSP9-expressing MKN-1 cells remained blocked in the S-G2/M phases and presented a down-regulation of the cell cycle-associated proteins c-Jun, CCND1, CDK4 and CDK6, while p21 expression was stimulated. These anti-proliferative effects were counteracted by the treatment of DUSP9-expressing cells with the JNK inhibitor SP600125. In vivo, mice xenografted with DUSP9-expressing MKN-1 cells developed larger tumors than the control mice [70].
6.4. Liver Cancer
DUSP9 also appears to play an important role in liver tumors (Table 2, Figure 3). The Walktrap algorithm identified DUSP9 as one of the key genes involved in the onset of HCC [68], likely because of the importance of the ERK pathway in this hepatic malignancy [10,78]. However, its exact functions in liver cancer is a matter of debate [15,21,73].
In 2013, a first analysis of biological samples showed that DUSP9 mRNA and protein were under-expressed in HCCs compared to non-tumoral livers [79]. Using the hepatoblastoma-derived and not the HCC-derived cell line HepG2 [74,80], the authors also showed that the depletion of DUSP9 slightly increased cell proliferation and improved the basal survival of cells in vitro. Analysis of a cohort of 134 HCCs showed that the DUSP9 decrease was a strong independent prognostic factor and was associated with poor overall survival and a high risk of recurrence [79]. The same team published a second paper in 2019 reporting the tumor suppressive role of DUSP9 in liver cancer [15]. They first showed that DUSP9 interacts with ERK1/2 kinases in tumor tissues and hepatoblastoma-derived HepG2 cells. DUSP9 expression was decreased in the human liver tumor cell lines HepG2, SK-Hep1 and SMMC-7721 compared to the human hepatocyte cell line LO2. After the laborious selection of one effective anti-DUSP9 small interfering RNA (one out of six tested), they showed that medium DUSP9 silencing enhanced ERK1/2 phosphorylation and inversely, that a moderate increase in phosphatase level (50% increase) decreased ERK1/2 phosphorylation [15]. The depletion of DUSP9 promoted the growth and clonogenicity of hepatoma cells, and the formation of spheroids, while forced DUSP9 expression produced the opposite effect. Treatment of hepatoma cells with the ERK kinase antagonist PD98059 totally abrogated the proliferative effect induced by the depletion of DUSP9. Unfortunately, this inhibitor was not tested alone and the statistical test used to analyze these data was incorrect, precluding any definitive conclusions [15]. In vivo, forced expression of DUSP9 inhibited the growth of grafted hepatoma cells in mice. Consequently, tumor volume and mass were significantly smaller compared to cells expressing basal or low level of DUSP9 [15]. Inversely, knockdown of DUSP9 promoted tumor development and expansion in mice. Additional analyses showed that the phenotypical changes mediated by DUSP9 were due to enhanced and diminished ERK1/2 dephosphorylation in DUSP9 silenced and DUSP9-expressing tumors, respectively. Together, these data supported the hypothesis that DUSP9 exerts a tumor suppressive role in HCC by inactivating the ERK pathway and lowering the level of the ERK targets MYC and Cyclin D1 [15]. The authors then measured the level of DUSP9, ERK1/2 and phospho-ERK1/2 in eight HCC samples and found, as previously reported [79], that DUSP9 was decreased in HCC, and that ERK1/2 and phospho-ERK1/2 were increased in the same samples. These data were confirmed by immunohistochemistry (IHC) analysis using a cohort of 160 HCCs. Finally authors correlated the expression of DUSP9 with clinicopathological factors and found a significant association between low DUSP9 expression or high ERK1/2 and phospho-ERK1/2 levels and poor overall survival [15].
In another study, again misusing the HepG2 cells as HCC-derived cells, the authors investigated at the cellular and molecular level the benefit of using phenolic compounds in metabolic syndromes [55]. These syndromes may evolve into an HCC [55]. Phenolic compounds enhanced glucose consumption and reduced the inflammatory response of palmitate-treated HepG2 cells. This effect was associated with dephosphorylation of p38 and JNK1, a decrease in miR-212 and a slight increase in DUSP9 [55]. The authors also found that miR-212 could repress DUSP9 by directly binding its 3′UTR. These data further support the role of DUSP9 in the regulation of stress responses related to obesity and insulin resistance (see Section 4).
In 2008, DUSP9 was reported to be part of a 16-gene signature discriminating hepatoblastomas classified as C1 and C2 groups [73]. Hepatoblastoma is an embryonal tumor and a primary liver cancer affecting children below the age of five. Cairo, Armengol and collaborators found that the DUSP9 level was increased in most C2 samples, a group of tumors associated with poor prognosis, an advanced disease stage, high proliferation, chromosomal instability, early fetal origin and MYC signaling [73]. We further found an overexpression of DUSP9 in the poor prognosis C2A group [74]. Therefore, unlike the previous studies [15,79], DUSP9 may play rather an oncogenic role in a subgroup of hepatoblastoma (Table 2), and the early fetal origin of C2 tumors again supports a link between DUSP9 and highly undifferentiated tumoral cells such as cancer stem cells or precursors.
In an attempt to clarify the role of oval cells (also known as intra-hepatic stem cells) in HCC onset, Sanders and collaborators investigated the role of mTOR1 and mTOR1 inhibitor rapamycin in liver cancer initiation using different liver injury models in Fisher rats [81]. Oval cells generated by the choline deficient plus ethionine model expressed high levels of the DUSP9/MKP-4 gene and cyclin D1. This effect was counteracted by the administration of rapamycin. DUSP9 levels were also higher in fetal livers than in adult livers. The authors concluded that DUSP9 is a marker of fetal liver and is more specifically expressed by the very undifferentiated oval cells [81], which is in agreement with the increased expression of DUSP9 in embryonic-derived hepatoblastoma [73,74].
A very recent report from a Canadian team also investigated the role of DUSP9 in HCC [21]. By interrogating GEO datasets, the authors found that DUSP9 transcript (along with Glypican-3 and α-fetoprotein) was overexpressed in HCCs, fetal livers and human hepatoma cell lines compared to their corresponding non-tumoral counterparts. Higher DUSP9 expression was associated with an increased amount of serum α-fetoprotein, more advanced disease stage, microvascular invasion and higher risk of recurrence after surgery, but not with overall survival. DUSP9 protein was also more expressed in hepatoma cell lines than in immortalized hepatic THLE-2 cells, and in HCC samples than in non-tumoral livers [21]. DUSP9 upregulation in HCC was associated with transcriptional regulation mediated by ETS transcription factors. Moreover, DUSP9 mRNA expression positively correlated with expression of ETV4, ELF3, ERF and ETV5 transcripts. In opposition with a previous study [15], Chen and collaborators showed that the forced expression of DUSP9 increased the proliferative capacity of hepatoma cells expressing low level of DUSP9 protein. Inversely, hepatoma cell knockout (KO) for DUSP9 proliferated less, had reduced clonogenic capacity, were more sensitive to doxorubicin and harbored higher ERK pathway activity [21]. In vivo, DUSP9 silenced cells generated smaller tumors and had lower growing capacity. Finally, grafted DUSP9-KO Hep3B cells were unable to form a tumor in comparison with parental cells. DUSP9 silencing was associated with increased MAPK activity and cell survival [21].
Altogether, these data show that the precise role of DUSP9 in liver cancers is still a matter of debate and that additional investigations need to be carried out to determine if DUSP9 acts as an oncogene or a tumor suppressor in liver malignancies.
6.5. Lung Cancer
A transcriptome sequencing analysis in patients with LA identified DUSP9 down-regulation as one factor associated with cell invasion and metastasis [72] (Table 2; Figure 3). Concerning the mechanism, the blockade of miR-1233-3p function by the circular RNA has_circ_0004050 stimulated DUSP9 expression in A549 cells leading to ERK/JNK pathway inactivation and tumor inhibition. This suggested that DUSP9 acts as a tumor suppressor in lung cancer [82].
6.6. Kidney Cancer
The role of DUSP9 in renal cancer has been highlighted in several studies and DUSP9 is regularly found to be down-regulated in renal tumors compared to normal tissues [33,83,84]. In 2010, Zhou and collaborators analyzed 10 clear cell renal cell carcinomas (ccRCC) and matched normal adjacent tissues. They reported for the first time the decreased expression of DUSP9 mRNA in tumor tissues, suggesting the pivotal role of DUSP9 in the development of renal cancer [84]. One year later, the down-regulation of DUSP9 mRNA and protein was further confirmed in ccRCC using real time quantitative RT-PCR and IHC [33]. Additional analysis in a cohort of 211 ccRCC samples showed that the DUSP9 decrease was associated with poor overall survival in patients and higher pathological disease stages (Fuhrman grade) [33]. In 2020, a new study reported the decreased expression of DUSP9 mRNA in approximately 81% of tumor tissues and most ccRCC cell lines [69]. The decrease in DUSP9 mRNA was always accompanied by a decrease in DUSP9 protein [33,69]. The same year, the down-regulation of DUSP9 mRNA in ccRCC was confirmed by a Polish group [83]. Phenotypically, forced expression of DUSP9 in ccRCC cell lines inhibited cell growth and migration in vitro and impeded tumor growth and size in vivo in a xenograft nude mouse model, thus showing for the first time the tumor suppressor role of DUSP9 in renal cancer [69]. This tumor suppressor role was mediated by inhibition of the mTOR pathway, as shown by dephosphorylation of mTOR on serine 2448 and a decrease in its pathway-associated targets HIF-1α, Sox2 and c-Myc (Figure 3) [69].
6.7. Skin Cancer
Using a carcinogenic model of squamous cell carcinoma (SCC) and a transcriptomic approach, Liu and collaborators showed that DUSP9 mRNA is down-regulated in tumor initiated cells compared to non-transformed parental cells, and completely lost in malignant cells, which harbor the typical characteristics of advanced-stage disease [71]. Carcinogenic induction using either UVB light or 7,12-dimethylbenz (a) anthracene compound led to a loss of DUSP9 expression in 100% and 40% of treated mice, respectively [71]. Using a lentiviral approach, forced expression of DUSP9 induced the death of nearly 90% of initiated cells after nine days by inhibiting the phosphorylation of ERK, JNK and p38. Under the same conditions, no phenotypical change was observed with DUSP6/MKP3 ectopic expression. Unexpectedly, DUSP9-expressing cells accumulated in the cell-death associated G2/M phase and were not arrested in the G1 phase, as is normally the case following ERK, JNK and p38 dephosphorylation. Cells in the G2/M phase were larger with condensed chromatin and underwent a mitotic catastrophe. Additional analysis showed that DUSP9 disorganized the microtubules [71]. In newborn BALB/c mice, the graft of DUSP9-expressing initiated cells resulted, on average, in seven-fold smaller tumors than the control cells 30 days after inoculation [71]. Histological analysis showed that tumors resulting from DUSP9-transduced cells did not express DUSP9, suggesting that engrafted cells at the origin of the tumor were likely not infected. Similar in vivo data were obtained with cells conditionally expressing DUSP9 following induction with tetracycline. Histological analysis of the tumor tissues showed a major change in the morphology of cells, which were larger and presented decondensed nuclei, indicating a tumoricidal effect (Table 2, Figure 3). Altogether, these in vitro and in vivo data suggest that DUSP9 plays a role in preventing SCC cell transformation and skin carcinoma expansion.
7. DUSP9 Is the Target for Therapy
Given the central role of DUSP9 in cancer and metabolic diseases, some groups have attempted to identify potent molecules specifically targeting it. Unfortunately, many human protein tyrosine phosphatases share similarities in their active site [43,85,86], thus precluding the development of specific selective drugs targeting the catalytic core of DUSP9.
A first attempt was to manipulate the activity of DUSP9 by targeting the loops surrounding the active phosphatase site and its spatial conformation to increase its efficiency (Figure 1C). To this end, 85,000 compounds with the physicochemical characteristics of drug candidates were selected from the InterBioScreen library and tested by virtual screening using the three-dimensional structure of the DUSP9 active site determined by crystallographic studies and in silico interaction simulations [43]. The 100 molecules with the best score in the virtual screening were selected. The study of the inhibitory activity of these molecules on DUSP9, carried out in vitro using a recombinant human protein, selected five very powerful molecules. Analysis of the molecular mechanism showed that four of the five compounds bound to DUSP9 protein between the catalytic core and the surrounding loops with a higher affinity near the loops. This interaction increased the substrate affinity and specificity of DUSP9 [43]. The loops which surround the catalytic site and participate in the interaction represent a good therapeutic target (Figure 1). The structure of these loops is different from one phosphatase to another, and they are less hydrophilic than the active site. Targeting these loops could therefore lead to the development of specific drugs capable of blocking the movement of loops in the active conformation of DUSP9 [43].
A recent study reported the capacity of the lncRNA LincU to maintain DUSP9 in an active state and block the MAPK pathway [15,48]. This remarkable result suggests that a therapeutic approach aiming at keeping DUSP9 in an active state may be beneficial for the treatment of patients with metabolic syndrome or tumor. This could be particularly useful for the development of specific treatments against type 2 diabetes and tumors under-expressing DUSP9 [43,67,71].
Another study reported that ginkgolic acid, which is extracted from the Ginkgo bilola leaf or seed coat, can inhibit DUSP9 activity [87]. It was selected following the screening of 658 natural compounds by an enzymatic test based on a recombinant purified human DUSP9 protein. In vitro, ginkgolic acid reduced DUSP9 phosphatase activity by 70% [87]. Interestingly, ginkgolic acid was found to be a potent antitumor agent in several solid tumors, suggesting its therapeutic potential for the treatment of cancer [84,88,89] This suggests its putative use in tumors overexpressing DUSP9 such as C2 or C2A hepatoblastomas. However, its use in clinical settings will depend on both the development of synthetic derivatives of the natural compound and the demonstration of their harmlessness in preclinical tumor models.
8. Conclusions
- (1)
- DUSP9 is tightly regulated at transcriptional, post-transcriptional and post-translational levels by transcription factors, promoter methylation, miRNAs, lncRNA and ubiquitination. Its rigorous regulation is therefore necessary to maintain normal cell function and physiological homeostasis.
- (2)
- DUSP9 has a central role in sex differences, metabolic disorders and tumorigenesis. At a pathophysiological level, DUSP9 is strongly involved in the regulation of insulin signaling, and consequently, of the downstream phosphorylation cascades and metabolic processes. Therefore, any therapeutic intervention to increase DUSP9 expression or to control the activity of ERK1/2, JNK, p38 MAPK and/or ASK1 kinases could be beneficial for patients presenting metabolic syndromes such as type 2 diabetes, morbid obesity, liver cirrhosis, NAFLD or its most severe form, NASH.
- (3)
- DUSP9 is central in tumorigenesis and is involved in many adult and pediatric cancers. It clearly acts as a tumor suppressor in kidney cancer, gastric cancer, skin cancer, CRC and lung cancer. Its high expression is associated with poor prognosis in C2A hepatoblastomas and with cancer stemness and drug resistance in breast cancer (Table 2; Figure 3). However, the role of DUSP9 in the adult liver is still a matter of debate and needs further investigations to determine its pro- or anti-tumoral function in hepatic malignancies (Table 2; Figure 3). As NASH can lead to malignant HCC, therapies influencing DUSP9 activity could also be beneficial in patients with liver cancer.
- (4)
- DUSP9 is also an important gene in heart tissue preservation and can counteract the negative effect mediated by pressure overload on cardiac hypertrophy and cardiomyocytes.
- (5)
- Collectively, these data clearly demonstrated the critical role of DUSP9 in both cell physiology and pathologies. It is therefore a promising therapeutic target to fight against the frequent human diseases that are diabetes, heart failure and cancers.
Funding
This study was supported by grants to FZK and CFG from the Fondation Groupama pour la Santé, Groupama Centre-Atlantique, the Région Nouvelle-Aquitaine (N°2018-1R30114), La Fondation ARC pour la Recherche sur le Cancer (contract N° PJA 20191209631) and The Ligue Nationale contre le Cancer (contract n°AAPPEAC2018.LCC/CG). FZK, CFG and the MIRCADE team also received donations from the following charities: Aidons Marina, E.S.CA.P.E., Eva pour la Vie, Les Récoltes de l’Espoir, Pour Emma, Scott & Co and Sphères.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| 3′UTR | 3′-untranslated region |
| ASK1 | MAPKKK apoptosis signal-regulating kinase 1 |
| CRC | colorectal carcinoma |
| DR1 element | inverted direct repeat separated by 1 |
| DUSP | dual specificity phosphatase |
| ERβ | estrogen receptor-beta |
| HCC | hepatocellular carcinoma |
| hESC | human embryonic stem cell |
| HIF1α | hypoxia-inducible factor 1 alpha |
| IHC | immunohistochemistry |
| IRS1 | insulin receptor substrate-1 |
| KIM | kinase-interacting motif |
| KO | knockout |
| MAPK | mitogen-activated protein kinase |
| mEGC | murine embryonic germ cell |
| mESC | mouse embryonic stem cell |
| MKB | MAP kinase-binding motif |
| MKP | MAP kinase phosphatase |
| NASH | non-alcoholic steatohepatitis |
| NAFLD | non-alcoholic fatty liver disease |
| pNPP | para-nitrophenylphosphate |
| RAR | retinoic acid receptor |
References
- Keshet, Y.; Seger, R. The MAP Kinase Signaling Cascades: A System of Hundreds of Components Regulates a Diverse Array of Physiological Functions. In MAP Kinase Signaling Protocols; Seger, R., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2010; Volume 661, pp. 3–38. ISBN 978-1-60761-794-5. [Google Scholar]
- Jiang, L.; Wang, Y.; Liu, G.; Liu, H.; Zhu, F.; Ji, H.; Li, B. C-Phycocyanin Exerts Anti-Cancer Effects via the MAPK Signaling Pathway in MDA-MB-231 Cells. Cancer Cell Int. 2018, 18, 12. [Google Scholar] [CrossRef]
- Chen, H.-F.; Chuang, H.-C.; Tan, T.-H. Regulation of Dual-Specificity Phosphatase (DUSP) Ubiquitination and Protein Stability. Int. J. Mol. Sci. 2019, 20, 2668. [Google Scholar] [CrossRef]
- Cuenda, A.; Rousseau, S. P38 MAP-Kinases Pathway Regulation, Function and Role in Human Diseases. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2007, 1773, 1358–1375. [Google Scholar] [CrossRef]
- Gaestel, M. MAPK-Activated Protein Kinases (MKs): Novel Insights and Challenges. Front. Cell Dev. Biol. 2016, 3, 88. [Google Scholar] [CrossRef]
- Morrison, D.K. MAP Kinase Pathways. Cold Spring Harb. Perspect. Biol. 2012, 4, a011254. [Google Scholar] [CrossRef]
- Burotto, M.; Chiou, V.L.; Lee, J.-M.; Kohn, E.C. The MAPK Pathway across Different Malignancies: A New Perspective: Tissue-Specific MAPK Signaling. Cancer 2014, 120, 3446–3456. [Google Scholar] [CrossRef]
- Guo, Y.; Pan, W.; Liu, S.; Shen, Z.; Xu, Y.; Hu, L. ERK/MAPK Signalling Pathway and Tumorigenesis (Review). Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, A.S.; Hagan, S.; Rath, O.; Kolch, W. MAP Kinase Signalling Pathways in Cancer. Oncogene 2007, 26, 3279–3290. [Google Scholar] [CrossRef] [PubMed]
- Ghousein, A.; Mosca, N.; Cartier, F.; Charpentier, J.; Dupuy, J.; Raymond, A.; Bioulac-Sage, P.; Grosset, C.F. MiR-4510 Blocks Hepatocellular Carcinoma Development through RAF1 Targeting and RAS/RAF/MEK/ERK Signalling Inactivation. Liver Int. 2020, 40, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Low, H.B.; Zhang, Y. Regulatory Roles of MAPK Phosphatases in Cancer. Immune Netw. 2016, 16, 85–98. [Google Scholar] [CrossRef]
- Nayak, J.; Gastonguay, A.J.; Talipov, M.R.; Vakeel, P.; Span, E.A.; Kalous, K.S.; Kutty, R.G.; Jensen, D.R.; Pokkuluri, P.R.; Sem, D.S.; et al. Protein Expression, Characterization and Activity Comparisons of Wild Type and Mutant DUSP5 Proteins. BMC Biochem. 2014, 15, 27. [Google Scholar] [CrossRef]
- Kutty, R.G. Dual Specificity Phosphatase 5-Substrate Interaction: A Mechanistic Perspective. Compr. Physiol. 2017, 7, 1449–1461. [Google Scholar] [PubMed]
- Seternes, O.-M.; Kidger, A.M.; Keyse, S.M. Dual-Specificity MAP Kinase Phosphatases in Health and Disease. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2019, 1866, 124–143. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Zhang, C.; Qu, L.; Lu, C.; Xiao, M.; Ni, R.; Liu, J. MKP-4 Suppresses Hepatocarcinogenesis by Targeting ERK1/2 Pathway. Cancer Cell Int. 2019, 19, 61. [Google Scholar] [CrossRef]
- Huang, C.-Y.; Tan, T.-H. DUSPs, to MAP Kinases and Beyond. Cell Biosci. 2012, 2, 24. [Google Scholar] [CrossRef]
- Lu, H.; Tran, L.; Park, Y.; Chen, I.; Lan, J.; Xie, Y.; Semenza, G.L. Reciprocal Regulation of DUSP9 and DUSP16 Expression by HIF1 Controls ERK and P38 MAP Kinase Activity and Mediates Chemotherapy-Induced Breast Cancer Stem Cell Enrichment. Cancer Res. 2018, 78, 4191–4202. [Google Scholar] [CrossRef]
- Muda, M.; Boschert, U.; Smith, A.; Antonsson, B.; Gillieron, C.; Chabert, C.; Camps, M.; Martinou, I.; Ashworth, A.; Arkinstall, S. Molecular Cloning and Functional Characterization of a Novel Mitogen-Activated Protein Kinase Phosphatase, MKP-4. J. Biol. Chem. 1997, 272, 5141–5151. [Google Scholar] [CrossRef]
- Camps, M.; Nichols, A.; Gillieron, C.; Antonsson, B.; Muda, M.; Chabert, C.; Boschert, U.; Arkinstall, S. Catalytic Activation of the Phosphatase MKP-3 by ERK2 Mitogen-Activated Protein Kinase. Science 1998, 280, 1262–1265. [Google Scholar] [CrossRef]
- Buffet, C. Anomalies Moléculaires de la Voie MAPK et Cancer Papillaire de la Thyroïde: Étude de Deux Phosphatases Spécifiques de ERK, DUSP5 et DUSP6. Ph.D. Thesis, Université René Descartes, Paris, France, 2015. [Google Scholar]
- Chen, K.; Gorgen, A.; Ding, A.; Du, L.; Jiang, K.; Ding, Y.; Sapisochin, G.; Ghanekar, A. Dual-Specificity Phosphatase 9 Regulates Cellular Proliferation and Predicts Recurrence After Surgery in Hepatocellular Carcinoma. Hepatol. Commun. 2021, 5, 1310–1328. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhang, Y.; Yu, H.; Shen, B.; Liang, Y.; Jin, R.; Liu, X.; Shi, L.; Cai, X. Role of DUSP1/MKP1 in Tumorigenesis, Tumor Progression and Therapy. Cancer Med. 2016, 5, 2061–2068. [Google Scholar] [CrossRef]
- Wei, W.; Jiao, Y.; Postlethwaite, A.; Stuart, J.M.; Wang, Y.; Sun, D.; Gu, W. Dual-Specificity Phosphatases 2: Surprising Positive Effect at the Molecular Level and a Potential Biomarker of Diseases. Genes Immun. 2013, 14, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, W.-Y.; Lin, Y.-C.; Liao, F.-H.; Chan, Y.-C.; Huang, C.-Y. Dual-Specificity Phosphatase 4 Regulates STAT5 Protein Stability and Helper T Cell Polarization*. PLoS ONE 2015, 10, e0145880. [Google Scholar] [CrossRef] [PubMed]
- Menyhart, O.; Budczies, J.; Munkácsy, G.; Esteva, F.J.; Szabó, A.; Miquel, T.P.; Győrffy, B. DUSP4 Is Associated with Increased Resistance against Anti-HER2 Therapy in Breast Cancer. Oncotarget 2017, 8, 77207–77218. [Google Scholar] [CrossRef]
- Seo, H.; Cho, Y.-C.; Ju, A.; Lee, S.; Park, B.C.; Park, S.G.; Kim, J.-H.; Kim, K.; Cho, S. Dual-Specificity Phosphatase 5 Acts as an Anti-Inflammatory Regulator by Inhibiting the ERK and NF-ΚB Signaling Pathways. Sci. Rep. 2017, 7, 17348. [Google Scholar] [CrossRef]
- Muhammad, K.A.; Nur, A.A.; Nurul, H.S.; Narazah, M.Y.; Siti, R.A.R. Dual-Specificity Phosphatase 6 (DUSP6): A Review of Its Molecular Characteristics and Clinical Relevance in Cancer. Cancer Biol. Med. 2018, 15, 14. [Google Scholar] [CrossRef]
- Luan, T.; Zhang, X.; Wang, S.; Song, Y.; Zhou, S.; Lin, J.; An, W.; Yuan, W.; Yang, Y.; Cai, H.; et al. Long Non-Coding RNA MIAT Promotes Breast Cancer Progression and Functions as CeRNA to Regulate DUSP7 Expression by Sponging MiR-155-5p. Oncotarget 2017, 8, 76153–76164. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Zhou, Y.; Long, R.; Chen, C.; Zhao, J.; Cui, P.; Guo, M.; Liang, G.; Xu, L. DUSP8 Phosphatase: Structure, Functions, Expression Regulation and the Role in Human Diseases. Cell Biosci. 2019, 9, 70. [Google Scholar] [CrossRef]
- Jiang, L.; Ren, L.; Guo, X.; Zhao, J.; Zhang, H.; Chen, S.; Le, S.; Liu, H.; Ye, P.; Chen, M.; et al. Dual-Specificity Phosphatase 9 Protects against Cardiac Hypertrophy by Targeting ASK1. Int. J. Biol. Sci. 2021, 17, 2193–2204. [Google Scholar] [CrossRef]
- Li, Z.; Fei, T.; Zhang, J.; Zhu, G.; Wang, L.; Lu, D.; Chi, X.; Teng, Y.; Hou, N.; Yang, X.; et al. BMP4 Signaling Acts via Dual-Specificity Phosphatase 9 to Control ERK Activity in Mouse Embryonic Stem Cells. Cell Stem Cell 2012, 10, 171–182. [Google Scholar] [CrossRef]
- Wei, Q.; Pu, X.; Zhang, L.; Xu, Y.; Duan, M.; Wang, Y. Expression of Dual-Specificity Phosphatase 9 in Placenta and Its Relationship with Gestational Diabetes Mellitus. J. Diabetes Res. 2019, 2019, 1–7. [Google Scholar] [CrossRef]
- Wu, S.; Wang, Y.; Sun, L.; Zhang, Z.; Jiang, Z.; Qin, Z.; Han, H.; Liu, Z.; Li, X.; Tang, A.; et al. Decreased Expression of Dual-Specificity Phosphatase 9 Is Associated with Poor Prognosis in Clear Cell Renal Cell Carcinoma. BMC Cancer 2011, 11, 413. [Google Scholar] [CrossRef]
- Ye, P.; Xiang, M.; Liao, H.; Liu, J.; Luo, H.; Wang, Y.; Huang, L.; Chen, M.; Xia, J. Dual-Specificity Phosphatase 9 Protects Against Nonalcoholic Fatty Liver Disease in Mice through ASK1 Suppression: Steatohepatitis/Metabolic Liver Disease. Hepatology 2019, 69, 76–93. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, T.; Barrios, A.; Tucker, A.; Collazo, J.; Arias, N.; Fazel, S.; Halim, M.; Huynh, T.; Singh, R.; Pervin, S. DUSP9-Mediated Reduction of PERK1/2 Supports Cancer Stem Cell-like Traits and Promotes Triple Negative Breast Cancer. Am. J. Cancer Res. 2020, 10, 3487–3506. [Google Scholar]
- Wu, Y.-K.; Hu, L.-F.; Lou, D.-S.; Wang, B.-C.; Tan, J. Targeting DUSP16/TAK1 Signaling Alleviates Hepatic Dyslipidemia and Inflammation in High Fat Diet (HFD)-Challenged Mice through Suppressing JNK MAPK. Biochem. Biophys. Res. Commun. 2020, 524, 142–149. [Google Scholar] [CrossRef]
- Yang, C.-Y.; Li, J.-P.; Chiu, L.-L.; Lan, J.-L.; Chen, D.-Y.; Chuang, H.-C.; Huang, C.-Y.; Tan, T.-H. Dual-Specificity Phosphatase 14 (DUSP14/MKP6) Negatively Regulates TCR Signaling by Inhibiting TAB1 Activation. J. Immunol. 2014, 192, 1547–1557. [Google Scholar] [CrossRef]
- Jung, S.; Nah, J.; Han, J.; Choi, S.-G.; Kim, H.; Park, J.; Pyo, H.-K.; Jung, Y.-K. Dual-Specificity Phosphatase 26 (DUSP26) Stimulates Aβ42 Generation by Promoting Amyloid Precursor Protein Axonal Transport during Hypoxia. J. Neurochem. 2016, 137, 770–781. [Google Scholar] [CrossRef] [PubMed]
- Keyse, S.M. Dual-Specificity MAP Kinase Phosphatases (MKPs) and Cancer. Cancer Metastasis Rev. 2008, 27, 253–261. [Google Scholar] [CrossRef]
- Lang, R.; Hammer, M.; Mages, J. DUSP Meet Immunology: Dual Specificity MAPK Phosphatases in Control of the Inflammatory Response. J. Immunol. 2006, 177, 7497–7504. [Google Scholar] [CrossRef] [PubMed]
- Lang, R.; Raffi, F.A.M. Dual-Specificity Phosphatases in Immunity and Infection: An Update. Int. J. Mol. Sci. 2019, 20, 2710. [Google Scholar] [CrossRef]
- Theodosiou, A.; Ashworth, A. MAP Kinase Phosphatases. Genome Biol. 2002, 3, REVIEWS3009. [Google Scholar] [CrossRef]
- Jeong, D.G.; Yoon, T.S.; Jung, S.K.; Park, B.C.; Park, H.; Ryu, S.E.; Kim, S.J. Exploring Binding Sites Other than the Catalytic Core in the Crystal Structure of the Catalytic Domain of MKP-4. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 25–31. [Google Scholar] [CrossRef]
- Hong, S.B.; Lubben, T.H.; Dolliver, C.M.; Petrolonis, A.J.; Roy, R.A.; Li, Z.; Parsons, T.F.; Li, P.; Xu, H.; Reilly, R.M.; et al. Expression, Purification, and Enzymatic Characterization of the Dual Specificity Mitogen-Activated Protein Kinase Phosphatase, MKP-4. Bioorg. Chem. 2005, 33, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: Modern Web App for 3D Visualization and Analysis of Large Biomolecular Structures. Nucleic Acids Res. 2021, 49, W431–W437. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Björling, E.; Agaton, C.; Szigyarto, C.A.-K.; Amini, B.; Andersen, E.; Andersson, A.-C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A Human Protein Atlas for Normal and Cancer Tissues Based on Antibody Proteomics. Mol. Cell. Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef]
- Xu, H.; Dembski, M.; Yang, Q.; Yang, D.; Moriarty, A.; Tayber, O.; Chen, H.; Kapeller, R.; Tartaglia, L.A. Dual Specificity Mitogen-Activated Protein (MAP) Kinase Phosphatase-4 Plays a Potential Role in Insulin Resistance. J. Biol. Chem. 2003, 278, 30187–30192. [Google Scholar] [CrossRef] [PubMed]
- Jiapaer, Z.; Li, G.; Ye, D.; Bai, M.; Li, J.; Guo, X.; Du, Y.; Su, D.; Jia, W.; Chen, W.; et al. LincU Preserves Naive Pluripotency by Restricting ERK Activity in Embryonic Stem Cells. Stem Cell Rep. 2018, 11, 395–409. [Google Scholar] [CrossRef]
- Imajo, M.; Kondoh, K.; Yamamoto, T.; Nakayama, K.; Nakajima-Koyama, M.; Nishida, E. Antagonistic Interactions between Extracellular Signal-Regulated Kinase Mitogen-Activated Protein Kinase and Retinoic Acid Receptor Signaling in Colorectal Cancer Cells. Mol. Cell. Biol. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthi, V.P.; Ratri, A.; Masumi, S.; Borosha, S.; Ghosh, S.; Christenson, L.K.; Roby, K.F.; Wolfe, M.W.; Rumi, M.A.K. Granulosa Cell Genes That Regulate Ovarian Follicle Development beyond the Antral Stage: The Role of Estrogen Receptor β. Mol. Cell. Endocrinol. 2021, 528, 111212. [Google Scholar] [CrossRef]
- Trezeguet, V.; Grosset, C.F. Les MicroARN et Leur Potentiel Thérapeutique En Cancérologie: Le Point En 2020. J. Biol. Méd. 2020, 32, 271–277. [Google Scholar]
- Cartier, F.; Indersie, E.; Lesjean, S.; Charpentier, J.; Hooks, K.B.; Ghousein, A.; Desplat, A.; Dugot-Senant, N.; Trézéguet, V.; Sagliocco, F.; et al. New Tumor Suppressor MicroRNAs Target Glypican-3 in Human Liver Cancer. Oncotarget 2017, 8, 41211–41226. [Google Scholar] [CrossRef]
- Indersie, E.; Lesjean, S.; Hooks, K.B.; Sagliocco, F.; Ernault, T.; Cairo, S.; Merched-Sauvage, M.; Rullier, A.; Le Bail, B.; Taque, S.; et al. MicroRNA Therapy Inhibits Hepatoblastoma Growth in Vivo by Targeting Beta-Catenin and Wnt Signaling. Hepatol. Commun. 2017, 1, 168–183. [Google Scholar] [CrossRef] [PubMed]
- Maurel, M.; Jalvy, S.; Ladeiro, Y.; Combe, C.; Vachet, L.; Sagliocco, F.; Bioulac-Sage, P.; Pitard, V.; Jacquemin-Sablon, H.; Zucman-Rossi, J.; et al. A Functional Screening Identifies Five Micrornas Controlling Glypican-3: Role of Mir-1271 down-Regulation in Hepatocellular Carcinoma. Hepatology 2013, 57, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Xiao, X.; Dong, Y.; Zhou, X.H. Fermented Barley Extracts with Lactobacillus Plantarum Dy-1 Rich in Vanillic Acid Modulate Glucose Consumption in Human HepG2 Cells. Biomed. Environ. Sci. 2018, 31, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Liang, N.; Huang, Q.; Sun, T.; Xue, H.; Xie, T.; Wang, X.; Wang, Q. Downregulation of DUSP9 Promotes Tumor Progression and Contributes to Poor Prognosis in Human Colorectal Cancer. Front. Oncol. 2020, 10, 547011. [Google Scholar] [CrossRef]
- Chang, J.; Huang, L.; Cao, Q.; Liu, F. Identification of Colorectal Cancer-Restricted MicroRNAs and Their Target Genes Based on High-Throughput Sequencing Data. OncoTargets Ther. 2016, 9, 1787–1794. [Google Scholar] [CrossRef]
- Choi, J.; Clement, K.; Huebner, A.J.; Webster, J.; Rose, C.M.; Brumbaugh, J.; Walsh, R.M.; Lee, S.; Savol, A.; Etchegaray, J.P.; et al. DUSP9 Modulates DNA Hypomethylation in Female Mouse Pluripotent Stem Cells. Cell Stem Cell 2017, 20, 706–719.e7. [Google Scholar] [CrossRef]
- Song, J.; Janiszewski, A.; De Geest, N.; Vanheer, L.; Talon, I.; El Bakkali, M.; Oh, T.; Pasque, V. X-Chromosome Dosage Modulates Multiple Molecular and Cellular Properties of Mouse Pluripotent Stem Cells Independently of Global DNA Methylation Levels. Stem Cell Rep. 2019, 12, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Genolet, O.; Monaco, A.A.; Dunkel, I.; Boettcher, M.; Schulz, E.G. Identification of X-Chromosomal Genes That Drive Sex Differences in Embryonic Stem Cells through a Hierarchical CRISPR Screening Approach. Genome Biol. 2021, 22, 110. [Google Scholar] [CrossRef] [PubMed]
- Yaribeygi, H.; Sathyapalan, T.; Atkin, S.L.; Sahebkar, A. Molecular Mechanisms Linking Oxidative Stress and Diabetes Mellitus. Oxid. Med. Cell. Longev. 2020, 2020, 8609213. [Google Scholar] [CrossRef]
- Jung, H.J.; Suh, Y. Regulation of IGF -1 Signaling by MicroRNAs. Front. Genet. 2015, 5. [Google Scholar] [CrossRef]
- Mukherjee, B.; Hossain, C.M.; Mondal, L.; Paul, P.; Ghosh, M.K. Obesity and Insulin Resistance: An Abridged Molecular Correlation. Lipid Insights 2013, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ruderman, N.B.; Carling, D.; Prentki, M.; Cacicedo, J.M. AMPK, Insulin Resistance, and the Metabolic Syndrome. J. Clin. Investig. 2013, 123, 2764–2772. [Google Scholar] [CrossRef]
- Christie, G.R.; Williams, D.J.; Macisaac, F.; Dickinson, R.J.; Rosewell, I.; Keyse, S.M. The Dual-Specificity Protein Phosphatase DUSP9/MKP-4 Is Essential for Placental Function but Is Not Required for Normal Embryonic Development. Mol. Cell. Biol. 2005, 25, 8323–8333. [Google Scholar] [CrossRef] [PubMed]
- Bazuine, M.; Carlotti, F.; Tafrechi, R.S.; Hoeben, R.C.; Maassen, J.A. Mitogen-Activated Protein Kinase (MAPK) Phosphatase-1 and -4 Attenuate P38 MAPK during Dexamethasone-Induced Insulin Resistance in 3T3-L1 Adipocytes. Mol. Endocrinol. 2004, 18, 1697–1707. [Google Scholar] [CrossRef] [PubMed]
- Emanuelli, B.; Eberle, D.; Suzuki, R.; Kahn, C.R. Overexpression of the Dual-Specificity Phosphatase MKP-4/DUSP-9 Protects against Stress-Induced Insulin Resistance. Proc. Natl. Acad. Sci. USA 2008, 105, 3545–3550. [Google Scholar] [CrossRef]
- Petrochilos, D.; Shojaie, A.; Gennari, J.; Abernethy, N. Using Random Walks to Identify Cancer-Associated Modules in Expression Data. BioData Min. 2013, 6, 17. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Luo, X.; Liu, X.; Fang, Z.; Xu, J.; Li, L. DUSP9 Suppresses Proliferation and Migration of Clear Cell Renal Cell Carcinoma via the MTOR Pathway. OncoTargets Ther. 2020, 13, 1321–1330. [Google Scholar] [CrossRef]
- Wu, F.; Lv, T.; Chen, G.; Ye, H.; Wu, W.; Li, G.; Zhi, F.-C. Epigenetic Silencing of DUSP9 Induces the Proliferation of Human Gastric Cancer by Activating JNK Signaling. Oncol. Rep. 2015, 34, 121–128. [Google Scholar] [CrossRef]
- Liu, Y.; Lagowski, J.; Sundholm, A.; Sundberg, A.; Kulesz-Martin, M. Microtubule Disruption and Tumor Suppression by Mitogen-Activated Protein Kinase Phosphatase 4. Cancer Res. 2007, 67, 10711–10719. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Wang, H.; Xiao, H.; Lan, B.; Liu, J.; Yang, Z. EEF1A2 and ERN2 Could Potentially Discriminate Metastatic Status of Mediastinal Lymph Node in Lung Adenocarcinomas Harboring EGFR 19Del/L858R Mutations. Thorac. Cancer 2020, 11, 2755–2766. [Google Scholar] [CrossRef]
- Cairo, S.; Armengol, C.; De Reyniès, A.; Wei, Y.; Thomas, E.; Renard, C.-A.; Goga, A.; Balakrishnan, A.; Semeraro, M.; Gresh, L.; et al. Hepatic Stem-like Phenotype and Interplay of Wnt/β-Catenin and Myc Signaling in Aggressive Childhood Liver Cancer. Cancer Cell 2008, 14, 471–484. [Google Scholar] [CrossRef]
- Hooks, K.B.; Audoux, J.; Fazli, H.; Lesjean, S.; Ernault, T.; Dugot-Senant, N.; Leste-Lasserre, T.; Hagedorn, M.; Rousseau, B.; Danet, C.; et al. New Insights into Diagnosis and Therapeutic Options for Proliferative Hepatoblastoma. Hepatology 2018, 68, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Bartholomeusz, C.; Gonzalez-Angulo, A.M.; Liu, P.; Hayashi, N.; Lluch, A.; Ferrer-Lozano, J.; Hortobágyi, G.N. High ERK Protein Expression Levels Correlate with Shorter Survival in Triple-Negative Breast Cancer Patients. Oncologist 2012, 17, 766–774. [Google Scholar] [CrossRef]
- Sansom, O.J.; Meniel, V.; Wilkins, J.A.; Cole, A.M.; Oien, K.A.; Marsh, V.; Jamieson, T.J.; Guerra, C.; Ashton, G.H.; Barbacid, M.; et al. Loss of Apc Allows Phenotypic Manifestation of the Transforming Properties of an Endogenous K-Ras Oncogene in Vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 14122–14127. [Google Scholar] [CrossRef] [PubMed]
- Jenner, S.; Wiedorn, K.H.; Techel, D. Development of a DUSP9 Methylation Screening Assay. Pathol. Oncol. Res. 2015, 21, 123–130. [Google Scholar] [CrossRef]
- Llovet, J.M.; Villanueva, A.; Lachenmayer, A.; Finn, R.S. Advances in Targeted Therapies for Hepatocellular Carcinoma in the Genomic Era. Nat. Rev. Clin. Oncol. 2015, 12, 408–424. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ni, W.; Xiao, M.; Jiang, F.; Ni, R. Decreased Expression and Prognostic Role of Mitogen-Activated Protein Kinase Phosphatase 4 in Hepatocellular Carcinoma. J. Gastrointest. Surg. 2013, 17, 756–765. [Google Scholar] [CrossRef] [PubMed]
- López-Terrada, D.; Cheung, S.W.; Finegold, M.J.; Knowles, B.B. Hep G2 Is a Hepatoblastoma-Derived Cell Line. Hum. Pathol. 2009, 40, 1512–1515. [Google Scholar] [CrossRef]
- Sanders, J.A.; Brilliant, K.E.; Clift, D.; Patel, A.; Cerretti, B.; Claro, P.; Mills, D.R.; Hixson, D.C.; Gruppuso, P.A. The Inhibitory Effect of Rapamycin on the Oval Cell Response and Development of Preneoplastic Foci in the Rat. Exp. Mol. Pathol. 2012, 93, 40–49. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Y.; Zang, R.K.; Du, Y.N. HSA_CIRC_0004050 on Proliferation and Apoptosis of A549 Cells through ERK/JNK Signaling Pathway. J. Biol. Regul. Homeost. Agents 2020, 34, 2037–2047. [Google Scholar] [CrossRef] [PubMed]
- Laczmanska, I.; Laczmanski, L.; Sasiadek, M.M. Expression Analysis of Tyrosine Phosphatase Genes at Different Stages of Renal Cell Carcinoma. Anticancer Res. 2020, 40, 5667–5671. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Chen, J.; Li, Z.; Li, X.; Hu, X.; Huang, Y.; Zhao, X.; Liang, C.; Wang, Y.; Sun, L.; et al. Integrated Profiling of MicroRNAs and MRNAs: MicroRNAs Located on Xq27.3 Associate with Clear Cell Renal Cell Carcinoma. PLoS ONE 2010, 5, e15224. [Google Scholar] [CrossRef] [PubMed]
- Blaskovich, M.A.T. Drug Discovery and Protein Tyrosine Phosphatases. Curr. Med. Chem. 2009, 16, 2095–2176. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Z.-Y. PTP1B as a Drug Target: Recent Developments in PTP1B Inhibitor Discovery. Drug Discov. Today 2007, 12, 373–381. [Google Scholar] [CrossRef]
- Yoon, S.Y.; Lee, J.H.; Kwon, S.J.; Kang, H.J.; Chung, S.J. Ginkgolic Acid as a Dual-Targeting Inhibitor for Protein Tyrosine Phosphatases Relevant to Insulin Resistance. Bioorg. Chem. 2018, 81, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Na, N.; Sheng, H.; Feng, B.; Wang, H.; Zhu, P.; Zhang, W.; Zhang, M.; Deng, Z. Ginkgolic Acid Inhibits the Growth of Renal Cell Carcinoma Cells via Inactivation of the EGFR Signaling Pathway. Exp. Ther. Med. 2020, 19, 2949–2956. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.-R.; Yang, H. Ginkgolic Acid (GA) Suppresses Gastric Cancer Growth by Inducing Apoptosis and Suppressing STAT3/JAK2 Signaling Regulated by ROS. Biomed. Pharmacother. 2020, 125, 109585. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).