A Promiscuous Bacterial P450: The Unparalleled Diversity of BM3 in Pharmaceutical Metabolism
Abstract
1. Context and Introduction to BM3
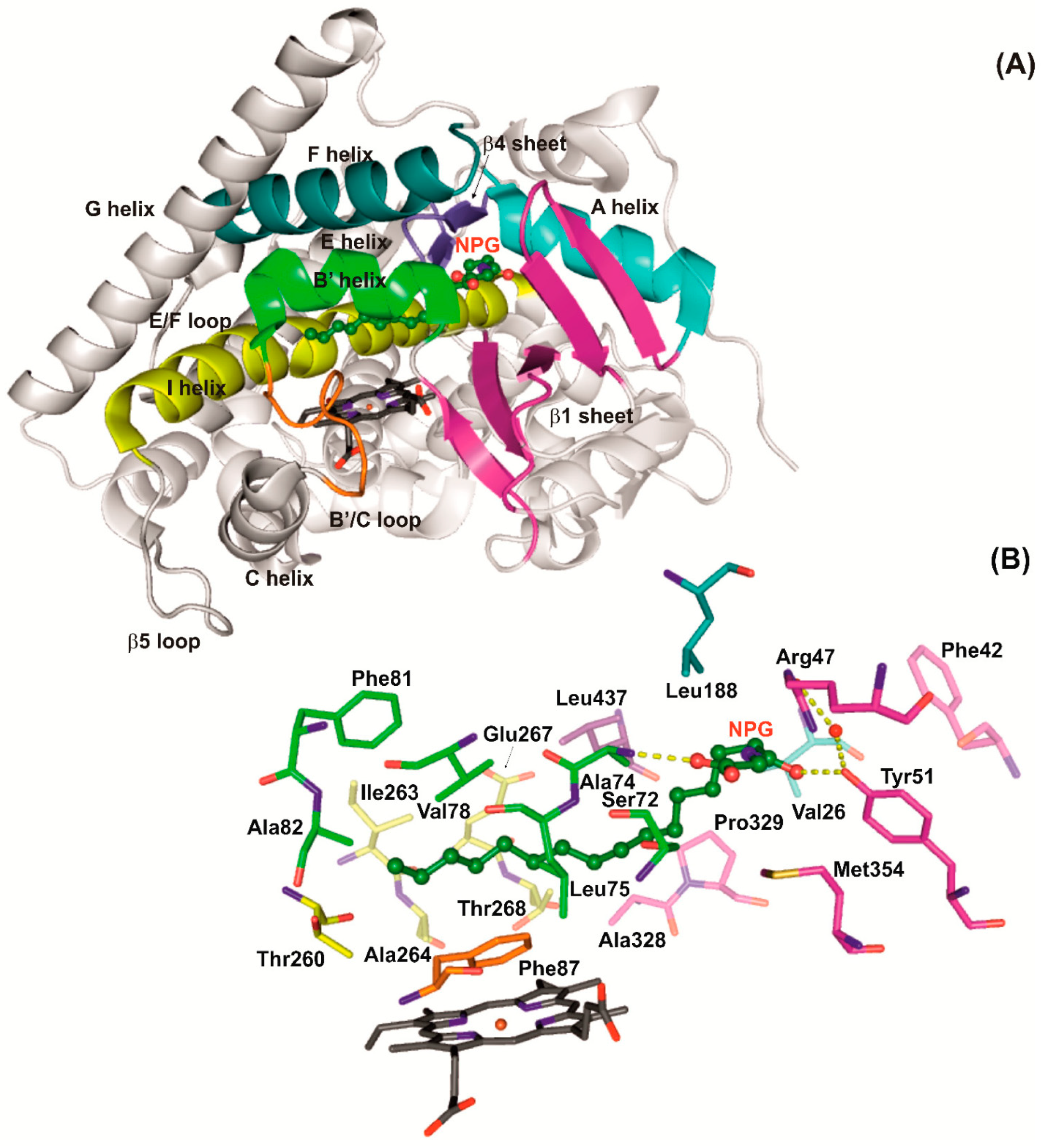
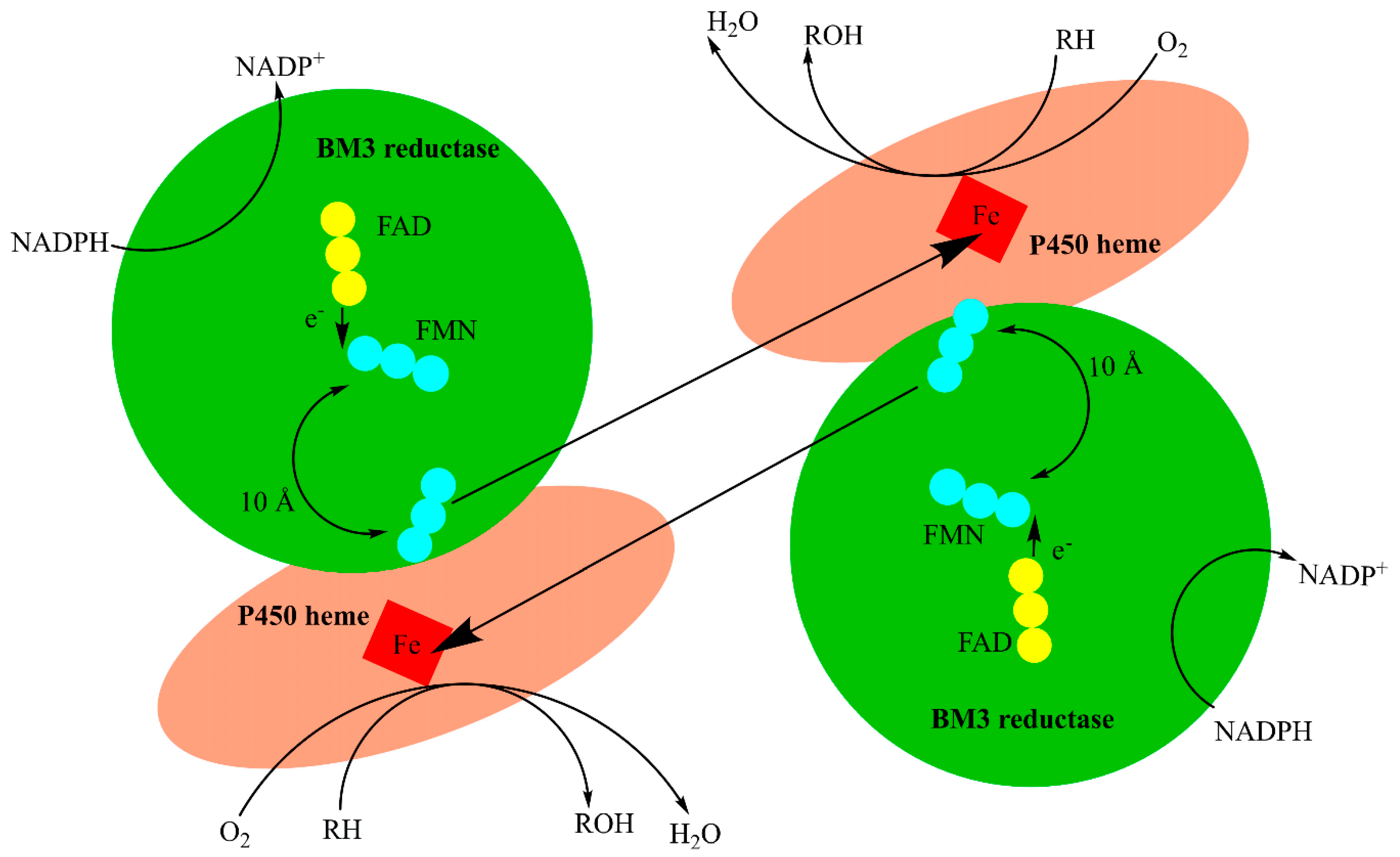
P450 BM3
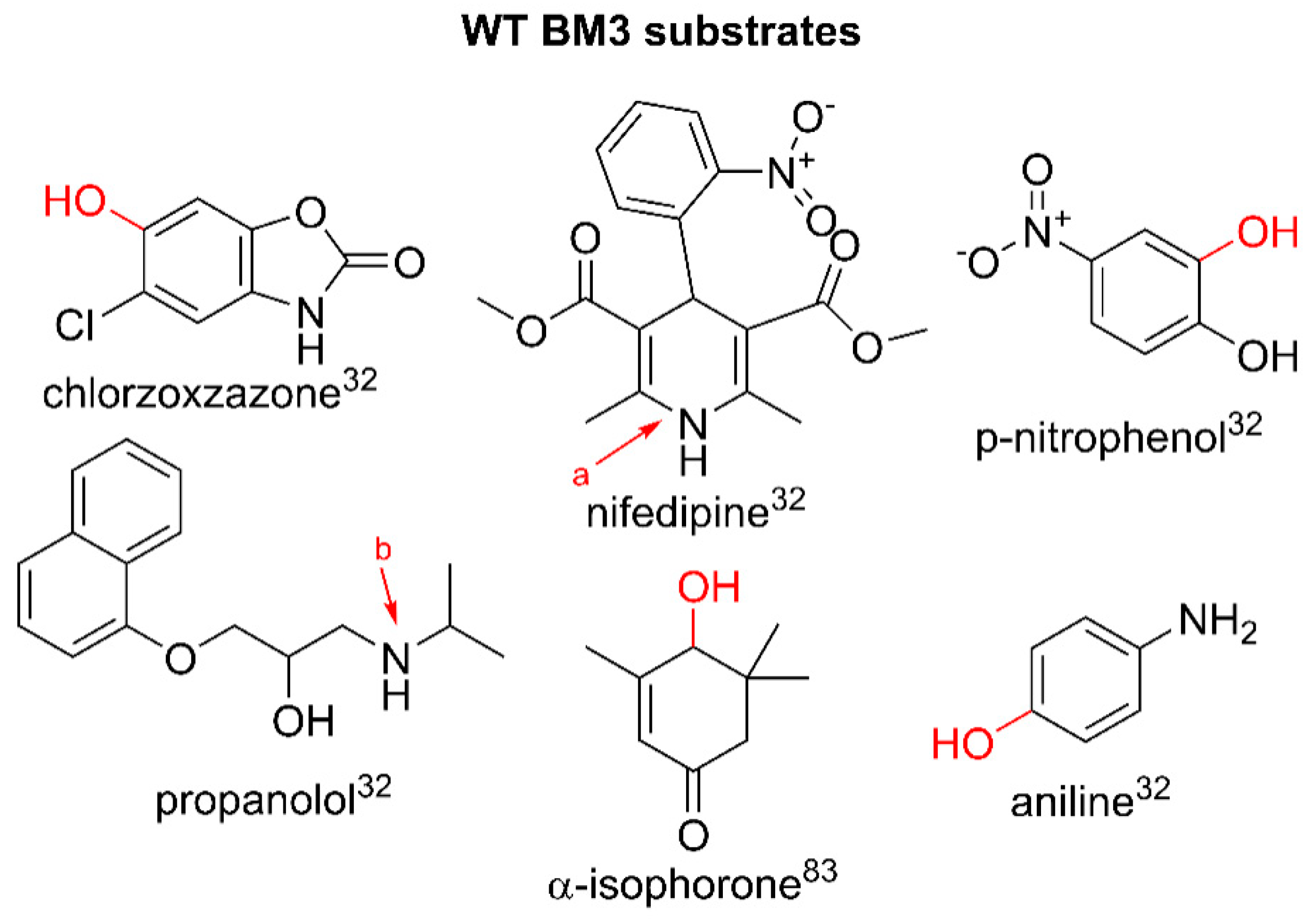
2. Wild Type BM3 and Pharmaceutical Metabolism
3. Engineering BM3 for Drug Metabolite Production
3.1. Key Promiscuous BM3 Variants and Library Screening
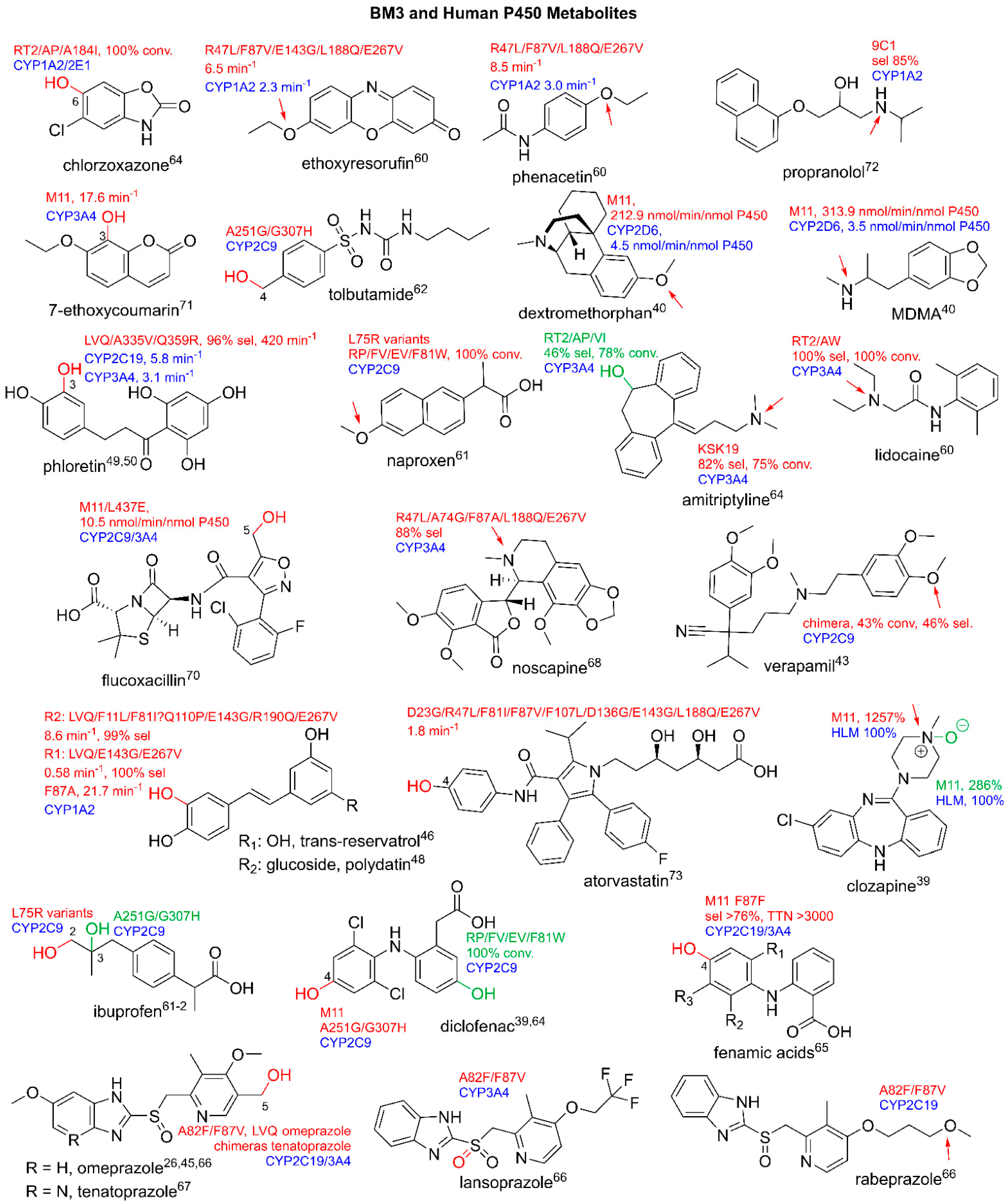
3.2. Production of Human P450 Metabolites
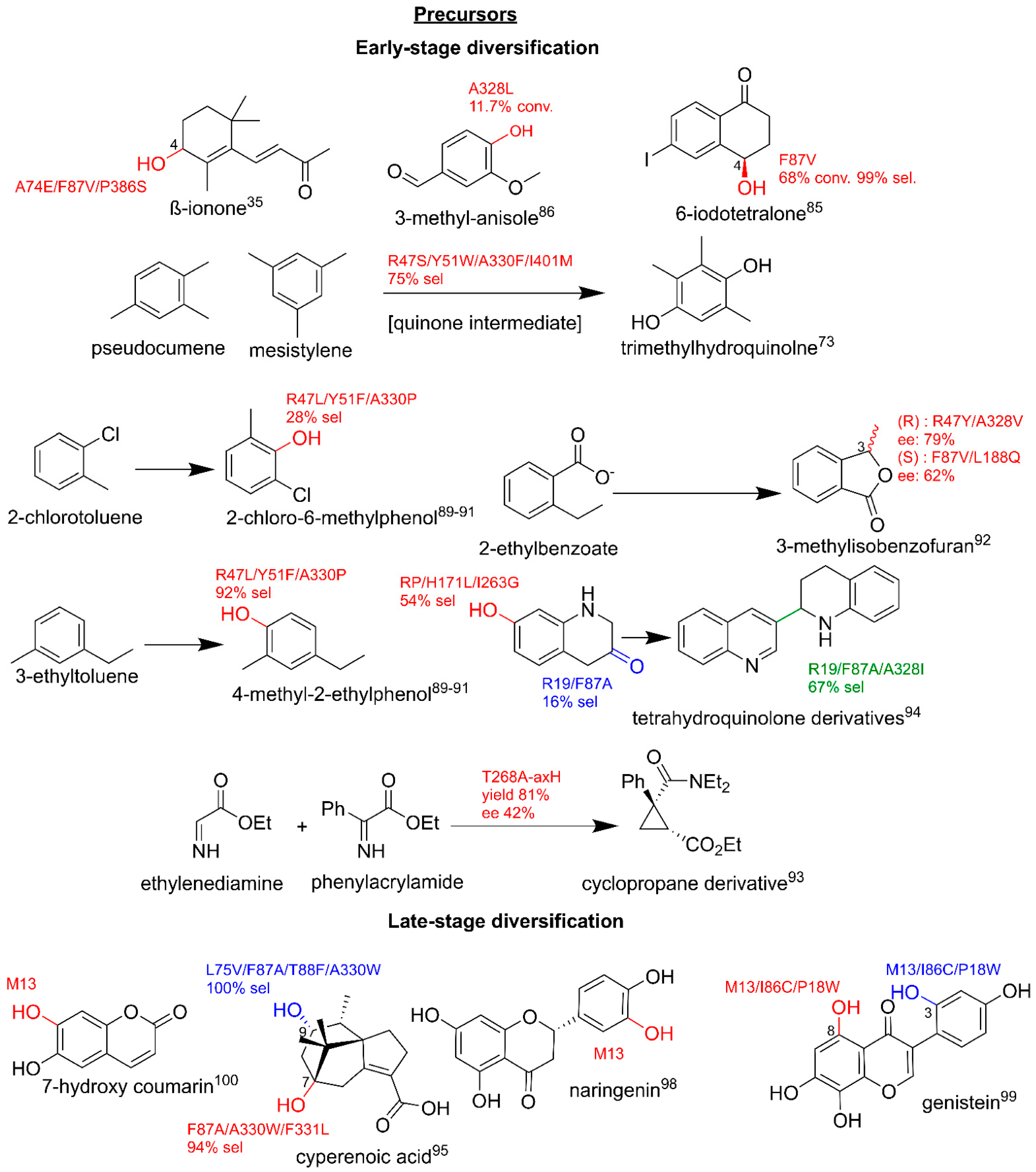
3.3. Production of Clinically Relevant Pharmaceuticals and Related Precursors
3.3.1. Pharmaceutical Intermediate and Final Product Production
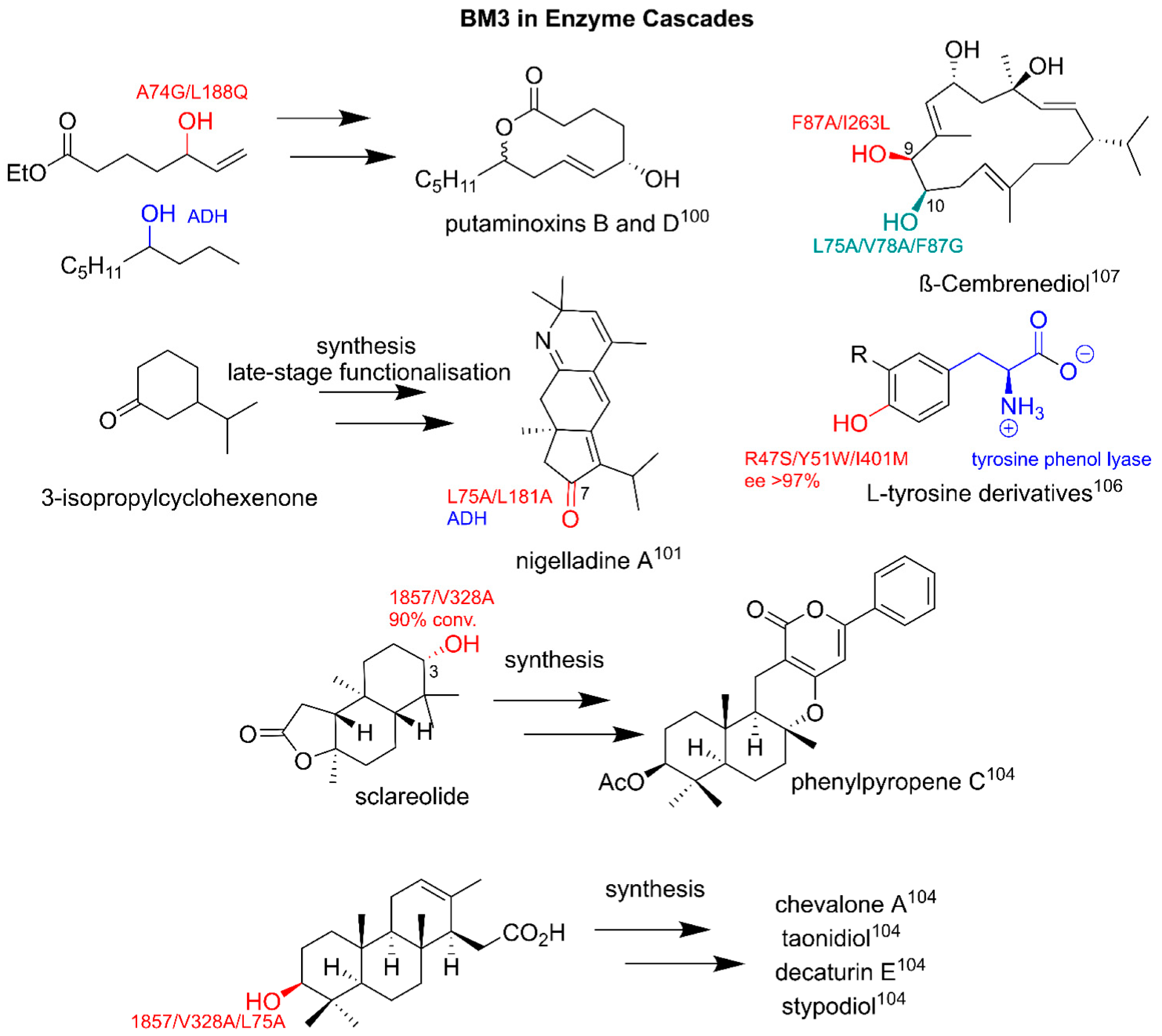
3.3.2. BM3 in Semi-Synthetic Routes and Biosynthetic Cascade Production of Pharmaceuticals
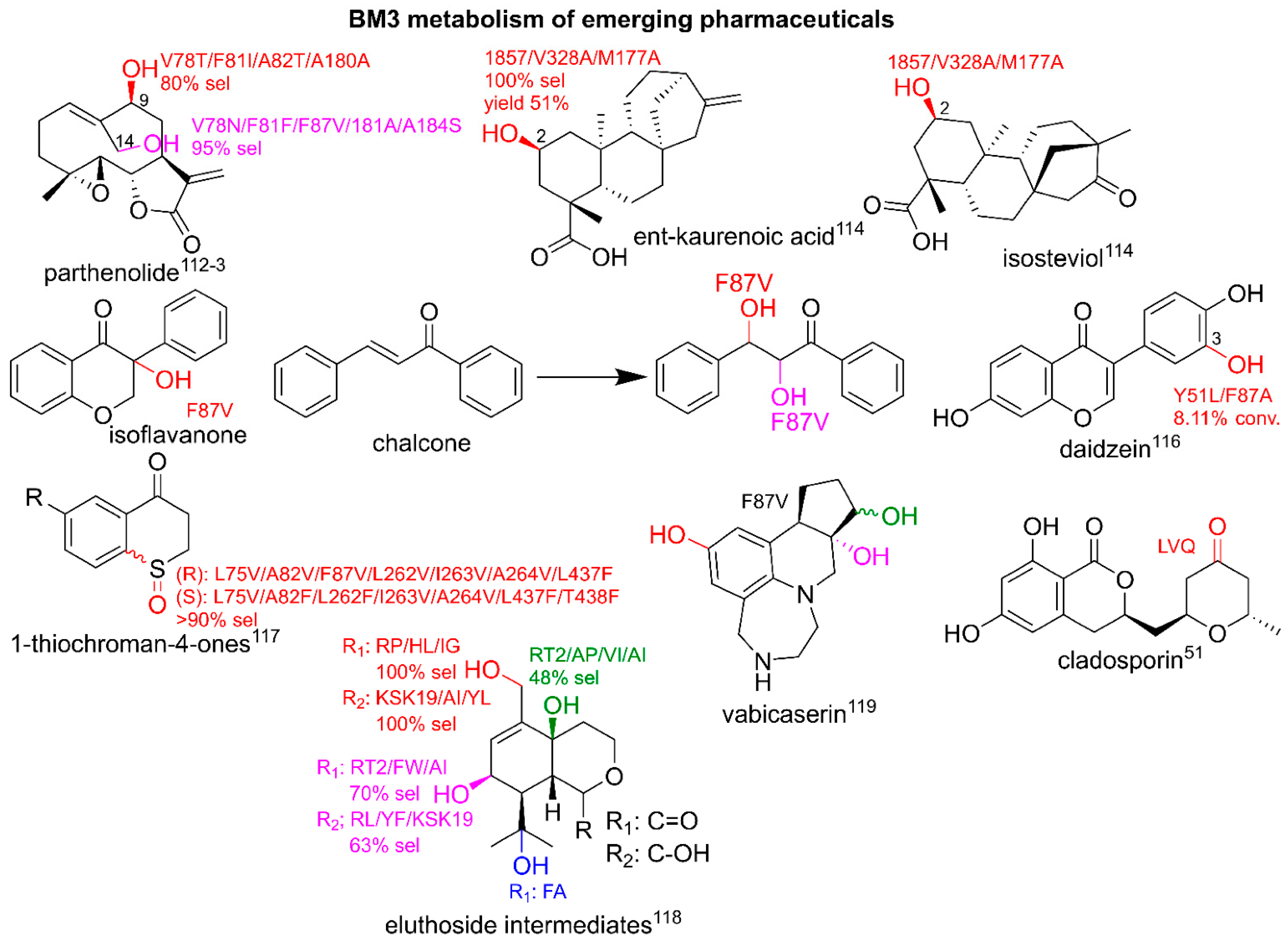
3.4. Utilizing BM3 for Production of Emerging Pharmaceuticals
4. Stretching the Use of BM3
4.1. Decoy Molecules
4.2. Industrial and Preparative Scale-up
4.3. Medicinal Usage of BM3
5. Summary, Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Roberts, A.A.; Ryan, K.S.; Moore, B.S.; Gulder, T.A.M. Total (Bio)synthesis: Strategies of nature and of chemists. Top. Curr. Chem. 2010, 297, 149. [Google Scholar]
- Rupasinghe, S.; Schuler, M.A.; Kagawa, N.; Yuan, H.; Lei, L.; Zhao, B.; Kelly, S.L.; Waterman, M.R.; Lamb, D.C. The cytochrome P450 gene family CYP157 does not contain EXXR in the K-helix reducing the absolute Conserved P450 residues to a single cysteine. FEBS Lett. 2006, 580, 6338–6342. [Google Scholar] [CrossRef]
- Guengerich, F.P.; Macdonald, T.L. Chemical mechanisms of catalysis by cytochromes P-450: A unified view. Acc. Chem. Res. 1984, 17, 9–16. [Google Scholar] [CrossRef]
- Whitehouse, C.J.C.; Bell, S.G.; Wong, L.-L. P450 BM3 (CYP102A1): Connecting the dots. Chem. Soc. Rev. 2012, 41, 1218–1260. [Google Scholar] [CrossRef] [PubMed]
- Iyanagi, T. Molecular mechanism of phase I and phase II drug-metabolizing enzymes: Implications for detoxification. Int. Rev. Cytol. 2007, 260, 35–112. [Google Scholar] [PubMed]
- Miura, Y.; Fulco, A.J. (ω-2) Hydroxylation of fatty acids by a soluble system from Bacillus megaterium. J. Biol. Chem. 1974, 249, 1880–1888. [Google Scholar] [CrossRef]
- Miura, Y.; Fulco, A.J. ω-1, ω-2 and ω-3 hydroxylation of long-chain fatty acids, amides and alcohols by a soluble enzyme system from Bacillus megaterium. Biochim. Biophys. Acta 1975, 388, 305–317. [Google Scholar] [CrossRef]
- Hare, R.S.; Fulco, A.J. Carbon monoxide and hydroxymercuribenzoate sensitivity of a fatty acid (ω-2) hydroxylase from Bacillus megaterium. Biochem. Biophys. Res. Commun. 1975, 65, 665–672. [Google Scholar] [CrossRef]
- Narhi, L.O.; Fulco, A.J. Identification and Characterization of two functional domains in cytochrome P-450BM-3, a catalytically self-sufficient monooxygenase induced by barbiturates in Bacillus megaterium. J. Biol. Chem. 1987, 262, 6683–6690. [Google Scholar] [CrossRef]
- Narhi, L.O.; Fulco, A.J. Characterization of a catalytically self-sufficient 119,000-dalton Cytochrome P-450 monooxygenase induced by barbiturates in Bacillus megaterium. J. Biol. Chem. 1986, 261, 7160–7169. [Google Scholar] [CrossRef]
- Porter, T.D. An unusual yet strongly conserved flavoprotein reductase in bacteria and mammals. Trends Biochem. Sci. 1991, 16, 154–158. [Google Scholar] [CrossRef]
- Lewis, D.F.V.; Watson, E.; Lake, B.G. Evolution of the cytochrome P450 superfamily: Sequence alignments and pharmacogenetics. Mutat. Res. 1998, 410, 245–270. [Google Scholar] [CrossRef]
- Fulco, A.J. Chain elongation, 20hydroxylation, and decarboxylation of long chain fatty acids by yeast. J. Biol. Chem. 1967, 242, 3608–3613. [Google Scholar] [CrossRef]
- English, N.; Hughes, V.; Wolf, C.R. Common pathways of cytochrome P450 gene regulation by peroxisome proliferators and barbiturates in Bacillus megaterium ATCC14581. J. Biol. Chem. 1994, 269, 26836–26841. [Google Scholar] [CrossRef]
- Ravichandran, K.G.; Boddupalli, S.S.; Hasemann, C.A.; Peterson, J.A.; Deisenhofer, J. Crystal structure of hemoprotein domain of P450BM-3, a prototype for microsomal P450′s. Science 1993, 261, 731–736. [Google Scholar] [CrossRef]
- Hasemann, C.A.; Kurumbail, R.G.; Boddupalli, S.S.; Peterson, J.A.; Deisenhofer, J. Structure and function of cytochromes P450: A comparative analysis of three crystal structures. Structure 1995, 3, 41–62. [Google Scholar] [CrossRef]
- Anzenbacher, P.; Hudeček, J. Differences in flexibility of active sites of cytochromes P450 probed by resonance raman and UV-Vis absorption spectroscopy. J. Inorg. Biochem. 2001, 87, 209–213. [Google Scholar] [CrossRef]
- Li, H.; Poulos, T.L. Modeling protein–substrate interactions in the heme domain of cytochrome P450BM−3. Acta Crystallogr. Sect. D Biol. Crystallogr. 1995, 51, 21–32. [Google Scholar] [CrossRef]
- Joyce, M.G.; Girvan, H.M.; Munro, A.W.; Leys, D. A single mutation in cytochrome P450 BM3 induces the conformational rearrangement seen upon substrate binding in the wild-type Enzyme. J. Biol. Chem. 2004, 279, 23287–23293. [Google Scholar] [CrossRef]
- Daff, S.N.; Chapman, S.K.; Turner, K.L.; Holt, R.A.; Govindaraj, S.; Poulos, T.L.; Munro, A.W. Redox control of the catalytic cycle of flavocytochrome P-450 BM3. Biochemistry 1997, 36, 13816–13823. [Google Scholar] [CrossRef] [PubMed]
- Gonvindaraj, S.; Li, H.; Poulos, T.L. Flavin supported fatty acid oxidation by the heme domain of Bacillus megaterium cytochrome P450BM-3. Biochem. Biophys. Res. Commun. 1994, 203, 1745–1749. [Google Scholar] [CrossRef] [PubMed]
- Sevrioukova, I.F.; Peterson, J.A. Reaction of carbon-monoxide and molecular-Oxygen with P450terp (CYP108) and P450BM-3 (CYP102). Arch. Biochem. Biophys. 1995, 317, 397–404. [Google Scholar] [CrossRef]
- Neeli, R.; Girvan, H.M.; Lawrence, A.; Warren, M.J.; Leys, D.; Scrutton, N.S.; Munro, A.W. The dimeric form of flavocytochrome P450 BM3 is catalytically functional as a fatty acid hydroxylase. FEBS Lett. 2005, 579, 5582–5588. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Poulos, T.L. The structure of the cytochrome P450BM-3 haem domain complexed with the fatty acid substrate, palmitoleic acid. Nat. Struct. Biol. 1997, 4, 140–146. [Google Scholar] [CrossRef]
- Noble, M.A.; Miles, C.S.; Chapman, S.K.; Lysek, D.A.; Mackay, A.C.; Reid, G.A.; Hanzlik, R.P.; Munro, A.W. Roles of key active-site residues in flavocytochrome P450 BM3. Biochem. J. 1999, 339, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Butler, C.F.; Peet, C.; Mason, A.E.; Voice, M.W.; Leys, D.; Munro, A.W. Key mutations alter the cytochrome P450 BM3 conformational landscape and remove inherent substrate bias. J. Biol. Chem. 2013, 288, 25387–25399. [Google Scholar] [CrossRef] [PubMed]
- Volz, T.J.; Rock, D.A.; Jones, J.P. Evidence for two different active oxygen species in cytochrome P450 BM3 mediated sulfoxidation and N-Dealkylation Reactions. J. Am. Chem. Soc. 2002, 124, 9724–9725. [Google Scholar] [CrossRef] [PubMed]
- Cryle, M.J.; De Voss, J.J. Is the ferric hydroperoxy species responsible for sulfur oxidation in cytochrome P450s? Angew. Chemie Int. Ed. 2006, 45, 8221–8223. [Google Scholar] [CrossRef]
- Haines, D.C.; Tomchick, D.R.; Machius, M.; Peterson, J.A. Pivotal role of water in the mechanism of P450BM-3. Biochemistry 2001, 40, 13456–13465. [Google Scholar] [CrossRef] [PubMed]
- Ost, T.W.B.; Clark, J.; Mowat, C.G.; Miles, C.S.; Walkinshaw, M.D.; Reid, G.A.; Chapman, S.K.; Daff, S. Oxygen activation and electron transfer in flavocytochrome P450 BM3. J. Am. Chem. Soc. 2003, 125, 15010–15020. [Google Scholar] [CrossRef]
- Ost, T.W.B.; Miles, C.S.; Munro, A.W.; Murdoch, J.; Reid, G.A.; Chapman, S.K. Phenylalanine 393 exerts thermodynamic control over the heme of flavocytochrome P450 BM3. Biochemistry 2001, 40, 13421–13429. [Google Scholar] [CrossRef]
- Di Nardo, G.; Fantuzzi, A.; Sideri, A.; Panicco, P.; Sassone, C.; Giunta, C.; Gilardi, G. Wild-type CYP102A1 as a biocatalyst: Turnover of drugs usually metabolised by human liver enzymes. J. Biol. Inorg. Chem. 2007, 12, 313–323. [Google Scholar] [CrossRef]
- Whitehouse, C.J.C.; Yang, W.; Yorke, J.A.; Tufton, H.G.; Ogilvie, L.C.I.; Bell, S.G.; Zhou, W.; Bartlam, M.; Rao, Z.; Wong, L.L. Structure, electronic properties and catalytic behaviour of an activity-Enhancing CYP102A1 (P450 BM3) Variant. Dalt. Trans. 2011, 40, 10383–10396. [Google Scholar] [CrossRef]
- Li, Q.S.; Schwaneberg, U.; Fischer, M.; Schmitt, J.; Pleiss, J.; Lutz-Wahl, S.; Schmid, R.D. Rational evolution of a medium chain-specific cytochrome P-450 BM-3 variant. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2001, 1545, 114–121. [Google Scholar] [CrossRef]
- Urlacher, V.B.; Makhsumkhanov, A.; Schmid, R.D. Biotransformation of β-ionone by engineered cytochrome P450 BM-3. Appl. Microbiol. Biotechnol. 2006, 70, 53–59. [Google Scholar] [CrossRef]
- Peters, M.W.; Meinhold, P.; Glieder, A.; Arnold, F.H. Regio- and enantioselective alkane hydroxylation with engineered cytochromes P450 BM-3. J. Am. Chem. Soc. 2003, 125, 13442–13450. [Google Scholar] [CrossRef]
- Lewis, J.C.; Mantovani, S.M.; Fu, Y.; Snow, C.D.; Komor, R.S.; Wong, C.-H.; Arnold, F.H. Combinatorial alanine substitution enables rapid optimization of cytochrome P450BM3 for selective hydroxylation of large substrates. ChemBioChem 2010, 11, 2502–2505. [Google Scholar] [CrossRef]
- Glieder, A.; Farinas, E.T.; Arnold, F.H. Laboratory evolution of a soluble, self-sufficient, highly active alkane hydroxylase. Nat. Biotechnol. 2002, 20, 1135–1139. [Google Scholar] [CrossRef]
- Damsten, M.C.; van Vugt-Lussenburg, B.M.A.; Zeldenthuis, T.; de Vlieger, J.S.B.; Commandeur, J.N.M.; Vermeulen, N.P.E. Application of drug metabolising mutants of cytochrome P450 BM3 (CYP102A1) as biocatalysts for the generation of reactive metabolites. Chem. Biol. Interact. 2008, 171, 96–107. [Google Scholar] [CrossRef]
- Van Vugt-Lussenburg, B.M.A.; Stjernschantz, E.; Lastdrager, J.; Oostenbrink, C.; Vermeulen, N.P.E.; Commandeur, J.N.M. Identification of critical residues in novel drug metabolizing mutants of Cytochrome P450 BM3 using random mutagenesis. J. Med. Chem. 2007, 50, 455–461. [Google Scholar] [CrossRef]
- Budde, M.; Morr, M.; Schmid, R.D.; Urlacher, V.B. Selective Hydroxylation of highly branched fatty acids and their derivatives by CYP102A1 from Bacillus megaterium. ChemBioChem 2006, 7, 789–794. [Google Scholar] [CrossRef]
- Vugt-Lussenburg, B.M.A.; Damsten, M.C.; Maasdijk, D.M.; Vermeulen, N.P.E.; Commandeur, J.N.M. Heterotropic and homotropic cooperativity by a drug-metabolising mutant of cytochrome P450 BM3. Biochem. Biophys. Res. Commun. 2006, 346, 810–818. [Google Scholar] [CrossRef]
- Sawayama, A.M.; Chen, M.M.Y.; Kulanthaivel, P.; Kuo, M.-S.; Hemmerle, H.; Arnold, F.H. A panel of cytochrome P450 BM3 variants to produce drug metabolites and diversify lead compounds. Chem. A Eur. J. 2009, 15, 11723–11729. [Google Scholar] [CrossRef]
- Reinen, J.; Ferman, S.; Vottero, E.; Vermeulen, N.P.E.; Commandeur, J.N.M. Application of a fluorescence-based continuous-flow bioassay to screen for diversity of cytochrome P450 BM3 mutant libraries. J. Biomol. Screen. 2011, 16, 239–250. [Google Scholar] [CrossRef]
- Ryu, S.H.; Park, B.Y.; Kim, S.Y.; Park, S.H.; Jung, H.J.; Park, M.; Park, K.D.; Ahn, T.; Kang, H.S.; Yun, C.H. Regioselective hydroxylation of omeprazole enantiomers by bacterial CYP102A1 mutants. Drug Metab. Dispos. 2014, 42, 1493–1497. [Google Scholar] [CrossRef]
- Kim, D.H.; Ahn, T.; Jung, H.C.; Pan, J.G.; Yun, C.H. Generation of the human metabolite piceatannol from the anticancer-preventive agent resveratrol by bacterial cytochrome P450 BM3. Drug Metab. Dispos. 2009, 37, 932–936. [Google Scholar] [CrossRef]
- Jang, H.H.; Ryu, S.H.; Le, T.K.; Doan, T.T.M.; Nguyen, T.H.H.; Park, K.D.; Yim, D.E.; Kim, D.H.; Kang, C.K.; Ahn, T.; et al. Regioselective C-H hydroxylation of omeprazole Sulfide by Bacillus megaterium CYP102A1 to Produce a Human Metabolite. Biotechnol. Lett. 2017, 39, 105–112. [Google Scholar] [CrossRef]
- Le, T.K.; Jang, H.H.; Nguyen, H.T.H.; Doan, T.T.M.; Lee, G.Y.; Park, K.D.; Ahn, T.; Joung, Y.H.; Kang, H.S.; Yun, C.H. Highly Regioselective hydroxylation of polydatin, a resveratrol glucoside, for one-step synthesis of astringin, a piceatannol glucoside, by P450 BM3. Enzyme Microb. Technol. 2017, 97, 34–42. [Google Scholar] [CrossRef]
- Nguyen, N.A.; Jang, J.; Le, T.K.; Nguyen, T.H.H.; Woo, S.M.; Yoo, S.K.; Lee, Y.J.; Park, K.D.; Yeom, S.J.; Kim, G.J.; et al. Biocatalytic production of a potent inhibitor of adipocyte differentiation from phloretin using engineered CYP102A1. J. Agric. Food Chem. 2020, 68, 6683–6691. [Google Scholar] [CrossRef]
- Nguyen, N.A.; Cao, N.T.; Nguyen, T.H.H.; Le, T.K.; Cha, G.S.; Choi, S.K.; Pan, J.G.; Yeom, S.J.; Kang, H.S.; Yun, C.H. Regioselective hydroxylation of phloretin, a bioactive compound from apples, by human Cytochrome P450 Enzymes. Pharmaceuticals 2020, 13, 330. [Google Scholar] [CrossRef]
- Fredenhagen, A.; Schroer, K.; Schröder, H.; Hoepfner, D.; Ligibel, M.; Porchet Zemp, L.; Radoch, C.; Freund, E.; Meishammer, A. Cladosporin derivatives obtained by biotransformation provide guidance for the focused derivatization of this antimalarial lead compound. ChemBioChem 2019, 20, 650–654. [Google Scholar] [CrossRef]
- Venkataraman, H.; de Beer, S.B.A.; van Bergen, L.A.H.; van Essen, N.; Geerke, D.P.; Vermeulen, N.P.E.; Commandeur, J.N.M. A Single Active Site Mutation Inverts Stereoselectivity of 16-hydroxylation of testosterone catalyzed by engineered cytochrome P450BM3. ChemBioChem 2012, 13, 520–523. [Google Scholar] [CrossRef]
- Reinen, J.; Vredenburg, G.; Klaering, K.; Vermeulen, N.P.E.; Commandeur, J.N.M.; Honing, M.; Vos, J.C. Selective whole-cell biosynthesis of the designer drug metabolites 15- or 16-betahydroxynorethisterone by engineered cytochrome P450 BM3 Mutants. J. Mol. Catal. B Enzym. 2015, 121, 64–74. [Google Scholar] [CrossRef]
- Venkataraman, H.; te Poele, E.M.; Rosłoniec, K.Z.; Vermeulen, N.; Commandeur, J.N.M.; van der Geize, R.; Dijkhuizen, L. Biosynthesis of a steroid metabolite by an engineered Rhodococcus Erythropolis strain expressing a mutant cytochrome P450 BM3 enzyme. Appl. Microbiol. Biotechnol. 2015, 99, 4713–4721. [Google Scholar] [CrossRef]
- Rea, V.; Kolkman, A.J.; Vottero, E.; Stronks, E.J.; Ampt, K.A.M.; Honing, M.; Vermeulen, N.P.E.; Wijmenga, S.S.; Commandeur, J.N.M. Active site substitution A82W improves the regioselectivity of steroid hydroxylation by cytochrome P450 BM3 mutants as rationalized by spin relaxation nuclear magnetic resonance studies. Biochemistry 2012, 51, 750–760. [Google Scholar] [CrossRef]
- Cha, G.S.; Ryu, S.H.; Ahn, T.; Yun, C.H. Regioselective hydroxylation of 17β-estradiol by mutants of CYP102A1 from Bacillus megaterium. Biotechnol. Lett. 2014, 36, 2501–2506. [Google Scholar] [CrossRef]
- Liu, X.; Kong, J.-Q. Steroids hydroxylation catalyzed by the monooxygenase mutant 139-3 from Bacillus megaterium BM3. Acta Pharm. Sin. B 2017, 7, 510–516. [Google Scholar] [CrossRef]
- Marchetti, S.; Schellens, J.H.M. The impact of FDA and EMEA guidelines on drug development in relation to phase 0 trials. Br. J. Cancer 2007, 97, 577–581. [Google Scholar] [CrossRef]
- Food and Drug Administration Center for Drug Evaluation and Research. Safety Testing of Drug Metabolites: Guidance for Industry; FDA: Silver Spring, MD, USA, 2016.
- Kim, D.H.; Kim, K.H.; Kim, D.; Jung, H.C.; Pan, J.G.; Chi, Y.T.; Ahn, T.; Yun, C.H. Oxidation of human cytochrome P450 1A2 substrates by Bacillus megaterium cytochrome P450 BM3. J. Mol. Catal. B Enzym. 2010, 63, 179–187. [Google Scholar] [CrossRef]
- Rentmeister, A.; Brown, T.R.; Snow, C.D.; Carbone, M.N.; Arnold, F.H. Engineered Bacterial Mimics of Human Drug Metabolizing Enzyme CYP2C9. ChemCatChem 2011, 3, 1065–1071. [Google Scholar] [CrossRef]
- Tsotsou, G.E.; Sideri, A.; Goyal, A.; Di Nardo, G.; Gilardi, G. Identification of mutant Asp251Gly/Gln307His of cytochrome P 450 BM3 for the generation of metabolites of diclofenac, ibuprofen and tolbutamide. Chem. A Eur. J. 2012, 18, 3582–3588. [Google Scholar] [CrossRef]
- Di Nardo, G.; Dell’Angelo, V.; Catucci, G.; Sadeghi, S.J.; Gilardi, G. Subtle structural changes in the Asp251Gly/Gln307His P450 BM3 mutant responsible for new activity toward diclofenac, tolbutamide and ibuprofen. Arch. Biochem. Biophys. 2016, 602, 106–115. [Google Scholar] [CrossRef][Green Version]
- Ren, X.; Yorke, J.A.; Taylor, E.; Zhang, T.; Zhou, W.; Wong, L.L. Drug oxidation by cytochrome P450BM3: Metabolite synthesis and discovering new P450 reaction types. Chem. A Eur. J. 2015, 21, 15039–15047. [Google Scholar] [CrossRef]
- Venkataraman, H.; Verkade-Vreeker, M.C.A.; Capoferri, L.; Geerke, D.P.; Vermeulen, N.P.E.; Commandeur, J.N.M. Application of engineered cytochrome P450 mutants as biocatalysts for the synthesis of benzylic and aromatic metabolites of fenamic acid NSAIDs. Bioorg. Med. Chem. 2014, 22, 5613–5620. [Google Scholar] [CrossRef]
- Butler, C.F.; Peet, C.; McLean, K.J.; Baynham, M.T.; Blankley, R.T.; Fisher, K.; Rigby, S.E.J.; Leys, D.; Voice, M.W.; Munro, A.W. Human P450-like oxidation of diverse proton pump inhibitor drugs by “gatekeeper” mutants of flavocytochrome P450 BM3. Biochem. J. 2014, 460, 247–259. [Google Scholar] [CrossRef]
- Le, T.K.; Cha, G.S.; Jang, H.H.; Nguyen, T.H.H.; Doan, T.T.M.; Lee, Y.J.; Park, K.D.; Shin, Y.; Kim, D.H.; Yun, C.H. Regioselective hydroxylation pathway of tenatoprazole to produce human metabolites by Bacillus megaterium CYP102A1. Process Biochem. 2019, 87, 95–104. [Google Scholar] [CrossRef]
- Richards, L.; Lutz, A.; Chalmers, D.K.; Jarrold, A.; Bowser, T.; Stevens, G.W.; Gras, S.L. Production of metabolites of the anti-cancer drug noscapine Using a P450BM3 mutant library. Biotechnol. Rep. 2019, 24, e00372. [Google Scholar] [CrossRef]
- Koyani, R.D.; Vazquez-Duhalt, R. Enzymatic activation of the emerging drug resveratrol. Appl. Biochem. Biotechnol. 2018, 185, 248–256. [Google Scholar] [CrossRef]
- Luirink, R.A.; Dekker, S.J.; Capoferri, L.; Janssen, L.F.H.; Kuiper, C.L.; Ari, M.E.; Vermeulen, N.P.E.; Vos, J.C.; Commandeur, J.N.M.; Geerke, D.P. A combined computational and experimental study on selective flucloxacillin hydroxylation by cytochrome P450 BM3 variants. J. Inorg. Biochem. 2018, 184, 115–122. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, K.H.; Kim, D.H.; Liu, K.H.; Jung, H.C.; Pan, J.G.; Yun, C.H. Generation of human metabolites of 7-ethoxycoumarin by bacterial cytochrome P450 BM3. Drug Metab. Dispos. 2008, 36, 2166–2170. [Google Scholar] [CrossRef]
- Otey, C.R.; Bandara, G.; Lalonde, J.; Takahashi, K.; Arnold, F.H. Preparation of human metabolites of propranolol using laboratory-evolved bacterial cytochromes P450. Biotechnol. Bioeng. 2006, 93, 494–499. [Google Scholar] [CrossRef]
- Nguyen, T.; Yeom, S.-J.; Yun, C.-H. Production of a human metabolite of atorvastatin by bacterial CYP102A1 peroxygenase. Appl. Sci. 2021, 11, 603. [Google Scholar] [CrossRef]
- Fairhead, M.; Giannini, S.; Gillam, E.M.J.; Gilardi, G. Functional characterisation of an engineered multidomain human P450 2E1 by molecular lego. J. Biol. Inorg. Chem. 2005, 10, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Dodhia, V.R.; Fantuzzi, A.; Gilardi, G. Engineering Human Cytochrome P450 enzymes into catalytically self-sufficient chimeras using molecular lego. J. Biol. Inorg. Chem. 2006, 11, 903–916. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, S.J.; Di Nardo, G.; Gilardi, G. Chimeric cytochrome P450 3A4 used for in Vitro prediction of food–drug interactions. Biotechnol. Appl. Biochem. 2020, 67, 541–548. [Google Scholar] [CrossRef]
- Castrignanò, S.; D’Avino, S.; Di Nardo, G.; Catucci, G.; Sadeghi, S.J.; Gilardi, G. Modulation of the interaction between human P450 3A4 and B. megaterium reductase via engineered loops. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Degregorio, D.; D’Avino, S.; Castrignanò, S.; di Nardo, G.; Sadeghi, S.J.; Catucci, G.; Gilardi, G. Human cytochrome P450 3A4 as a biocatalyst: Effects of the engineered linker in modulation of coupling efficiency in 3A4-BMR chimeras. Front. Pharmacol. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zuo, R.; Zhang, Y.; Jiang, C.; Hackett, J.C.; Loria, R.; Bruner, S.D.; Ding, Y. Engineered P450 biocatalysts show improved activity and regio-promiscuity in aromatic nitration. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, G.; Nandekar, P.P.; Wade, R.C. Electron transfer from cytochrome P450 reductase to cytochrome p450: Towards a structural and dynamic understanding. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lechner, A.; Brunk, E.; Keasling, J.D. The need for integrated approaches in metabolic engineering. Cold Spring Harb. Perspect. Biol. 2016, 8, a023903. [Google Scholar] [CrossRef]
- Urlacher, V.B.; Girhard, M. Cytochrome P450 monooxygenases in biotechnology and synthetic biology. Trends Biotechnol. 2019, 37, 882–897. [Google Scholar] [CrossRef]
- Dezvarei, S.; Lee, J.H.Z.; Bell, S.G. Stereoselective hydroxylation of isophorone by variants of the cytochromes P450 CYP102A1 and CYP101A1. Enzyme Microb. Technol. 2018, 111, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Kaluzna, I.; Schmitges, T.; Straatman, H.; Van Tegelen, D.; Mü, M.; Schü, M.; Mink, D. Enabling selective and sustainable p450 oxygenation technology. Production of 4-hydroxy-α-isophorone on kilogram scale. Org. Process Res. Dev. 2016, 20, 814–819. [Google Scholar] [CrossRef]
- Ilie, A.; Harms, K.; Reetz, M.T. P450-catalyzed regio- and stereoselective oxidative hydroxylation of 6-Iodotetralone: Preparative-scale synthesis of a key intermediate for Pd-catalyzed transformations. J. Org. Chem. 2018, 83, 7504–7508. [Google Scholar] [CrossRef] [PubMed]
- Klaus, T.; Seifert, A.; Häbe, T.; Nestl, B.M.; Hauer, B. An Enzyme cascade synthesis of vanillin. Catalysts 2019, 9, 252. [Google Scholar] [CrossRef]
- Dennig, A.; Weingartner, A.M.; Kardashliev, T.; Müller, C.A.; Tassano, E.; Schürmann, M.; Ruff, A.J.; Schwaneberg, U. An enzymatic route to α-tocopherol synthons: Aromatic hydroxylation of pseudocumene and mesitylene with P450 BM3. Chem. A Eur. J. 2017, 23, 17981–17991. [Google Scholar] [CrossRef]
- Weingartner, A.M.; Sauer, D.F.; Dhoke, G.V.; Davari, M.D.; Ruff, A.J.; Schwaneberg, U. A hydroquinone-specific screening system for directed P450 evolution. Appl. Microbiol. Biotechnol. 2018, 102, 9657–9667. [Google Scholar] [CrossRef] [PubMed]
- Munday, S.D.; Dezvarei, S.; Lau, I.C.K.; Bell, S.G. Examination of Selectivity in the Oxidation of Ortho- and Meta-Disubstituted Benzenes by CYP102A1 (P450 Bm3) Variants. ChemCatChem 2017, 9, 2512–2522. [Google Scholar] [CrossRef]
- Carmichael, A.B.; Wong, L.L. Protein engineering of Bacillus megaterium CYP102A1. The oxidation of polycyclic aromatic hydrocarbons. Eur. J. Biochem. 2001, 268, 3117–3125. [Google Scholar] [CrossRef] [PubMed]
- Whitehouse, C.J.C.; Bell, S.G.; Tufton, H.G.; Kenny, R.J.P.; Ogilvie, L.C.I.; Wong, L.L. Evolved CYP102A1 (P450BM3) variants oxidise a range of non-natural substrates and offer new selectivity options. Chem. Commun. 2008, 8, 966–968. [Google Scholar] [CrossRef]
- Holec, C.; Hartrampf, U.; Neufeld, K.; Pietruszka, J. P450 BM3-catalyzed regio- and stereoselective hydroxylation aiming at the synthesis of phthalides and isocoumarins. ChemBioChem 2017, 18, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.J.; Renata, H.; Peck, N.E.; Farwell, C.C.; Coelho, P.S.; Arnold, F.H. Improved Cyclopropanation activity of histidine-ligated Cytochrome P450 enables the enantioselective formal synthesis of levomilnacipran. Angew. Chemie Int. Ed. 2014, 53, 6810–6813. [Google Scholar] [CrossRef]
- Li, Y.; Wong, L.L. Multi-functional oxidase activity of CYP102A1 (P450BM3) in the oxidation of quinolines and tetrahydroquinolines. Angew. Chemie Int. Ed. 2019, 58, 9551–9555. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qin, B.; Li, X.; Tang, J.; Chen, Y.; Zhou, L.; You, S. Selective oxidations of cyperenoic acid by slightly reshaping the binding pocket of cytochrome P450 BM3. ChemCatChem 2018, 10, 559–565. [Google Scholar] [CrossRef]
- Acevedo-Rocha, C.G.; Gamble, C.G.; Lonsdale, R.; Li, A.; Nett, N.; Hoebenreich, S.; Lingnau, J.B.; Wirtz, C.; Fares, C.; Hinrichs, H.; et al. P450-catalyzed regio-and diastereoselective steroid hydroxylation: Efficient directed evolution enabled by mutability Landscaping. ACS Catal. 2018, 8, 3395–3410. [Google Scholar] [CrossRef]
- Chu, L.L.; Pandey, R.P.; Lim, H.N.; Jung, H.J.; Thuan, N.H.; Kim, T.S.; Sohng, J.K. Synthesis of umbelliferone derivatives in Escherichia Coli and their biological activities. J. Biol. Eng. 2017, 11, 1–11. [Google Scholar] [CrossRef]
- Chu, L.L.; Pandey, R.P.; Jung, N.; Jung, H.J.; Kim, E.H.; Sohng, J.K. Hydroxylation of Diverse Flavonoids by CYP450 BM3 variants: Biosynthesis of eriodictyol from naringenin in whole cells and its biological activities. Microb. Cell Fact. 2016, 15, 135. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.L.; Kong, J.Q. Altering the Regioselectivity of Cytochrome P450 BM3 variant M13 toward genistein through protein engineering and variation of reaction conditions. ACS Omega 2020, 5, 32059–32066. [Google Scholar] [CrossRef]
- Bisterfeld, C.; Holec, C.; Böse, D.; Marx, P.; Pietruszka, J. Chemoenzymatic total synthesis of the proposed structures of putaminoxins B and D. J. Nat. Prod. 2017, 80, 1563–1574. [Google Scholar] [CrossRef]
- Loskot, S.A.; Romney, D.K.; Arnold, F.H.; Stoltz, B.M. Enantioselective total synthesis of Nigelladine A via Late-Stage C-H Oxidation Enabled by an Engineered P450 Enzyme. J. Am. Chem. Soc. 2017, 139, 10196–10199. [Google Scholar] [CrossRef]
- Falck, J.R.; Reddy, Y.K.; Haines, D.C.; Reddy, K.M.; Krishna, U.M.; Graham, S.; Murry, B.; Peterson, J.A. Practical, Enantiospecific Syntheses of 14,15-EET and Leukotoxin B (Vernolic Acid). Tetrahedron Lett. 2001, 42, 4131–4133. [Google Scholar] [CrossRef]
- Zhang, K.; El Damaty, S.; Fasan, R. P450 fingerprinting method for rapid discovery of terpene hydroxylating P450 catalysts with diversified regioselectivity. J. Am. Chem. Soc. 2011, 133, 3242–3245. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, F.; King-Smith, E.; Renata, H. Merging chemoenzymatic and radical-based retrosynthetic logic for rapid and modular synthesis of oxidized meroterpenoids. Nat. Chem. 2020, 12, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Dennig, A.; Marienhagen, J.; Ruff, A.J.; Guddat, L.; Schwaneberg, U. Directed evolution of P450 BM3 into a P-xylene hydroxylase. ChemCatChem 2012, 4, 771–773. [Google Scholar] [CrossRef]
- Dennig, A.; Busto, E.; Kroutil, W.; Faber, K. Biocatalytic one-pot synthesis of l-tyrosine derivatives from monosubstituted benzenes, pyruvate, and Ammonia. ACS Catal. 2015, 5, 7503–7506. [Google Scholar] [CrossRef]
- Le-Huu, P.; Petrović, D.; Strodel, B.; Urlacher, V.B. One-pot, two-step hydroxylation of the macrocyclic diterpenoid β-cembrenediol catalyzed by P450 BM3 Mutants. ChemCatChem 2016, 8, 3755–3761. [Google Scholar] [CrossRef]
- Wu, S.; Snajdrova, R.; Moore, J.C.; Baldenius, K.; Bornscheuer, U.T. Biocatalysis: Enzymatic synthesis for industrial applications. Angew. Chemie Int. Ed. 2021, 60, 88–119. [Google Scholar] [CrossRef]
- Chen, W.; Fisher, M.J.; Leung, A.; Cao, Y.; Wong, L.L. Oxidative diversification of steroids by nature-inspired scanning glycine Mutagenesis of P450BM3 (CYP102A1). ACS Catal. 2020, 10, 8334–8343. [Google Scholar] [CrossRef]
- Li, A.; Acevedo-Rocha, C.G.; D’Amore, L.; Chen, J.; Peng, Y.; Garcia-Borràs, M.; Gao, C.; Zhu, J.; Rickerby, H.; Osuna, S.; et al. Regio- and stereoselective steroid hydroxylation at C7 by cytochrome P450 monooxygenase mutants. Angew. Chemie Int. Ed. 2020, 59, 12499–12505. [Google Scholar] [CrossRef]
- Kolev, J.N.; O’Dwyer, K.M.; Jordan, C.T.; Fasan, R. Discovery of potent parthenolide-based antileukemic agents enabled by late-stage P450-mediated C—H functionalization. ACS Chem. Biol. 2014, 9, 164–173. [Google Scholar] [CrossRef]
- Alwaseem, H.; Frisch, B.J.; Fasan, R. Anticancer activity Profiling of Parthenolide Analogs Generated via P450-mediated chemoenzymatic synthesis. Bioorg. Med. Chem. 2018, 26, 1365–1373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; King-Smith, E.; Dong, L.B.; Yang, L.C.; Rudolf, J.D.; Shen, B.; Renata, H. Divergent synthesis of complex diterpenes through a hybrid oxidative approach. Science 2020, 369, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, E.; Otomatsu, T.; Maeda, C.; Aoki, Y.; Ota, C.; Misawa, N.; Shindo, K. Production of hydroxlated flavonoids with cytochrome P450 BM3 variant F87V and their antioxidative activities. Biosci. Biotechnol. Biochem. 2013, 77, 1340–1343. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.; Yang, Y.H.; Choi, K.Y.; Kim, B.G. Rational design and directed evolution of CYP102A1 (BM3) for regio-specific hydroxylation of isoflavone. Biotechnol. Bioprocess Eng. 2015, 20, 225–233. [Google Scholar] [CrossRef]
- Wang, J.B.; Ilie, A.; Reetz, M.T. Chemo- and stereoselective cytochrome P450-BM3-catalyzed sulfoxidation of 1-Thiochroman-4-Ones enabled by directed evolution. Adv. Synth. Catal. 2017, 359, 2056–2060. [Google Scholar] [CrossRef]
- Syntrivanis, L.-D.; Wong, L.L.; Robertson, J. Hydroxylation of eleuthoside synthetic Intermediates by P450 BM3 (CYP102A1). Eur. J. Org. Chem. 2018, 2018, 6369–6378. [Google Scholar] [CrossRef]
- Vickers, C.; Backfisch, G.; Oellien, F.; Piel, I.; Lange, U.E.W. Enzymatic Late-stage oxidation of lead compounds with solubilizing biomimetic docking/protecting groups. Chem. A Eur. J. 2018, 24, 17936–17947. [Google Scholar] [CrossRef]
- Shoji, O.; Yanagisawa, S.; Stanfield, J.K.; Suzuki, K.; Cong, Z.; Sugimoto, H.; Shiro, Y.; Watanabe, Y. Direct hydroxylation of benzene to phenol by cytochrome P450BM3 triggered by amino acid derivatives. Angew. Chemie Int. Ed. 2017, 56, 10324–10329. [Google Scholar] [CrossRef]
- Karasawa, M.; Stanfield, J.K.; Yanagisawa, S.; Shoji, O.; Watanabe, Y. Whole-cell biotransformation of benzene to phenol catalysed by intracellular Cytochrome P450BM3 activated by external additives. Angew. Chemie Int. Ed. 2018, 57, 12264–12269. [Google Scholar] [CrossRef]
- Shoji, O.; Watanabe, Y. Monooxygenation of nonnative substrates catalyzed by bacterial cytochrome P450s Facilitated by decoy molecules. Chem. Lett. 2017, 46, 278–288. [Google Scholar] [CrossRef]
- Dubey, K.K.; Haque, S.; Jawed, A.; Singh, B.P.; Behera, B.K. Construction of recombinant Escherichia coli for enhanced bioconversion of colchicine into 3-demethylated colchicine at 70l Bioreactor Level. Process Biochem. 2010, 45, 1036–1042. [Google Scholar] [CrossRef]
- Yari, M.; Ghoshoon, M.B.; Vakili, B.; Ghasemi, Y. Therapeutic enzymes: Applications and Approaches to Pharmacological Improvement. Curr. Pharm. Biotechnol. 2017, 18, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, M.G.; Westmeyer, G.G.; Romero, P.A.; Szablowski, J.O.; Küster, B.; Shah, A.; Otey, C.R.; Langer, R.; Arnold, F.H.; Jasanoff, A. Directed evolution of a magnetic resonance imaging contrast agent for noninvasive imaging of Dopamine. Nat. Biotechnol. 2010, 28, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Brustad, E.M.; Lelyveld, V.S.; Snow, C.D.; Crook, N.; Jung, S.T.; Martinez, F.M.; Scholl, T.J.; Jasanoff, A.; Arnold, F.H. Structure-Guided Directed evolution of highly selective P450-based magnetic resonance imaging sensors for dopamine and serotonin. J. Mol. Biol. 2012, 422, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Vredenburg, G.; den Braver-Sewradj, S.; van Vugt-Lussenburg, B.M.A.; Vermeulen, N.P.E.; Commandeur, J.N.M.; Vos, J.C. Activation of the anticancer drugs cyclophosphamide and ifosfamide by Cytochrome P450 BM3 Mutants. Toxicol. Lett. 2015, 232, 182–192. [Google Scholar] [CrossRef] [PubMed]
- González-Davis, O.; Chauhan, K.; Zapian-Merino, S.J.; Vazquez-Duhalt, R. Bi-enzymatic virus-like bionanoreactors for the transformation of endocrine disruptor compounds. Int. J. Biol. Macromol. 2020, 146, 415–421. [Google Scholar] [CrossRef]
- Cirino, P.C.; Arnold, F.H. A self-sufficient peroxide-driven hydroxylation biocatalyst. Angew. Chemie Int. Ed. 2003, 42, 3299–3301. [Google Scholar] [CrossRef]
- Chauhan, K.; Sengar, P.; Juarez-Moreno, K.; Hirata, G.A.; Vazquez-Duhalt, R. Camouflaged, activatable and therapeutic tandem bionanoreactors for breast cancer theranosis. J. Colloid Interface Sci. 2020, 580, 365–376. [Google Scholar] [CrossRef]
| BM3 Variant | Point Mutations | Substrates |
|---|---|---|
| LVQ [39] | R47L/F87V/L188Q | clozapine [39], diclofenac [39], acetaminophen [39], omeprazole [45], reservatrol * [46], polydatin [47,48], phloretin * [49,50], cladosporin [51] |
| M01 [39] | LVQ/E267V/G415S/G1049E | clozapine [39], diclofenac [39], acetaminophen [39], testosterone * [52], noresthisterone * [53], methoxyresorufin [44], ethoxyresorufin [44] |
| M02 [39] | LVQ/L86I/N319T/A964V | clozapine [39], diclofenac [39], acetaminophen [39], norandrostenedione [54], methoxyresorufin [44], ethoxyresorufin [44], buspirone [44], amitriptyline [44], aripiprazole [44] |
| M05 [39] | LVQ/F81I/E267V/G415S | clozapine [39], diclofenac [39], acetaminophen [39], methoxyresorufin [44], ethoxyresorufin [44] |
| M11 [39] | LVQ/E64G/F81I/E143G/E267V/G415S | clozapine [39], diclofenac [39], acetaminophen [39], Testosterone * [52,55], norethisterone * [53,55], dextromethorphan [40], nifedipine [40], midazolam [40], 3,4-methylenedioxymethylamphetamine [40], estradiol [14,56], methoxyresorufin [44], ethoxyresorufin [44], buspirone [44], duloxetine [44], thioridazine [44], ondansetron [44], imatinib [44], omeprazole [44], rosiglitazone [44], paroxetine [44], norethisterone [44], resveratrol [44], dihydrobenzophenone [44], 17α-ethinylestradiol [44], bisphenol A [44], diethylstilbestrol [44], hexestrol [44], estriol [44], benzophenone-3 [44], aldrin [44], testosterone [44], tramadol [44], imidazole [44], ketoconazole [44] |
| 9-10A [36] | R47C/V78A/K94I/P142S/T175I/A184V/F205C /S226R/H236Q/E252G/R255S/A290V/L353V | verapamil [43], astemizole [43], LY294002 [43] |
| 139-3 [38] | V78A/H138Y/T175I/V178I/A184V/ H236Q/ E252G/R255S/A290V/A295T/L353V | androstenedione [57] |
| GVQ [41] | A74G/F87V/L188Q | testosterone [42], amodiaquine [42], dextromethorphan [42], acetaminophen [42], 3,4-methylenedioxymethylamphetamine [42] |
| Steroid | Structure | OH- Position | Variant | Parameters |
|---|---|---|---|---|
| Testosterone | 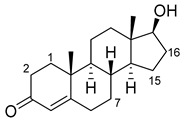 | 2β * | RLYF/KSK19 * [64] | 100% conv, 61% sel |
| 1β | H171L/Q307H/N319Y/F87A/T260G/P329G/A330W [109] | 71% sel, 76% conv | ||
| 7β | R47W/S72W/F77Y/V78L/F81I/A82L/T88S/M177T/M185Q/L188Q/I209T [110] | 90% sel | ||
| 15β * | KSK19/FV/QP [64] | 96% sel, 83% conv | ||
| 16α * | M11/V87I/S72I [52] M01/A82W/S72I [52] | 90% ee 95% ee | ||
| 16β * | M01/A82W [52] M11/V87I [52] | 100% ee 100% ee | ||
| Estradiol | 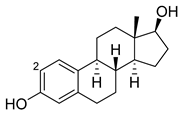 | 2 | M11 [56] | 47 min−1 |
| Norandrostenedione |  | 16β * | M02 [54] | 95% sel |
| Nandrolone | 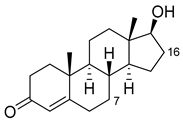 | 7β | R47W/S72W/F77Y/V78L/F81I/A82L/T88S/M177T/M185Q/L188Q/I209T [110] | 75% sel |
| 16α | R47L/S72I/A82F/F87I/L188C/A330W [96] | 98% sel | ||
| 16β | R47W/A82W/F87V/L181Q [96] | 90% sel | ||
| Norethindrone | 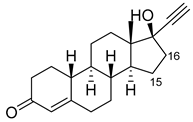 | 15β * | M11/A82W/V87A [53] | 100% sel |
| 16β * | M01/A82W [53] | 96% sel | ||
| Boldenone | 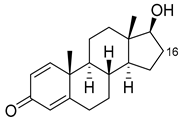 | 16α | R47L/Y51W/S72I/A82W/F87I/L181C [96] | 97% sel |
| 16β | R47W/A82W/F87V/L181Q [96] | 72% sel | ||
| Androstenedione | 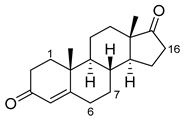 | 1α | 139-3 [57] | |
| 6β | H171L/Q307H/N319Y/F87V/I263G [109] | 78% sel, 29% conv | ||
| 7β | R47W/S72W/F77Y/V78L/F81I/A82L/T88S/M177T/M185Q/L188Q/I209T [110] R47L/Y51F/H171L/Q307H/N319Y/ F87A/A184I/T260G/A328G [109] | 90% sel 81% sel, 94% conv | ||
| 16α | R47W/Y51W/S72I/A82F/F87I/L181C [96] | 95% sel | ||
| 16β | R47W/Y51H/A82W/F87V/L181Q [96] | 100% sel |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thistlethwaite, S.; Jeffreys, L.N.; Girvan, H.M.; McLean, K.J.; Munro, A.W. A Promiscuous Bacterial P450: The Unparalleled Diversity of BM3 in Pharmaceutical Metabolism. Int. J. Mol. Sci. 2021, 22, 11380. https://doi.org/10.3390/ijms222111380
Thistlethwaite S, Jeffreys LN, Girvan HM, McLean KJ, Munro AW. A Promiscuous Bacterial P450: The Unparalleled Diversity of BM3 in Pharmaceutical Metabolism. International Journal of Molecular Sciences. 2021; 22(21):11380. https://doi.org/10.3390/ijms222111380
Chicago/Turabian StyleThistlethwaite, Sian, Laura N. Jeffreys, Hazel M. Girvan, Kirsty J. McLean, and Andrew W. Munro. 2021. "A Promiscuous Bacterial P450: The Unparalleled Diversity of BM3 in Pharmaceutical Metabolism" International Journal of Molecular Sciences 22, no. 21: 11380. https://doi.org/10.3390/ijms222111380
APA StyleThistlethwaite, S., Jeffreys, L. N., Girvan, H. M., McLean, K. J., & Munro, A. W. (2021). A Promiscuous Bacterial P450: The Unparalleled Diversity of BM3 in Pharmaceutical Metabolism. International Journal of Molecular Sciences, 22(21), 11380. https://doi.org/10.3390/ijms222111380






