CsCDPK6, a CsSAMS1-Interacting Protein, Affects Polyamine/Ethylene Biosynthesis in Cucumber and Enhances Salt Tolerance by Overexpression in Tobacco
Abstract
:1. Introduction
2. Results
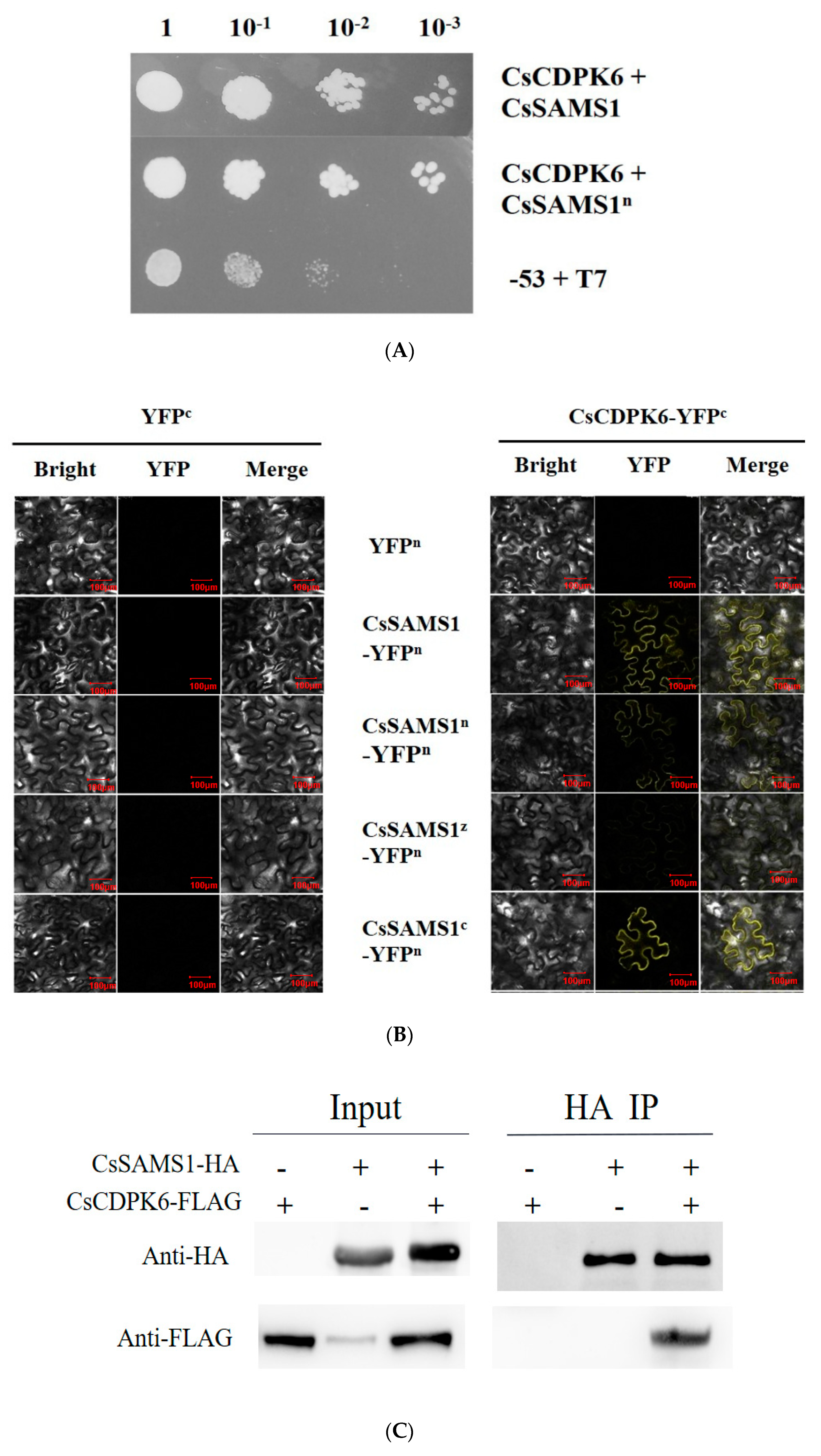
2.1. SAMS1 Interacts with CDPK6
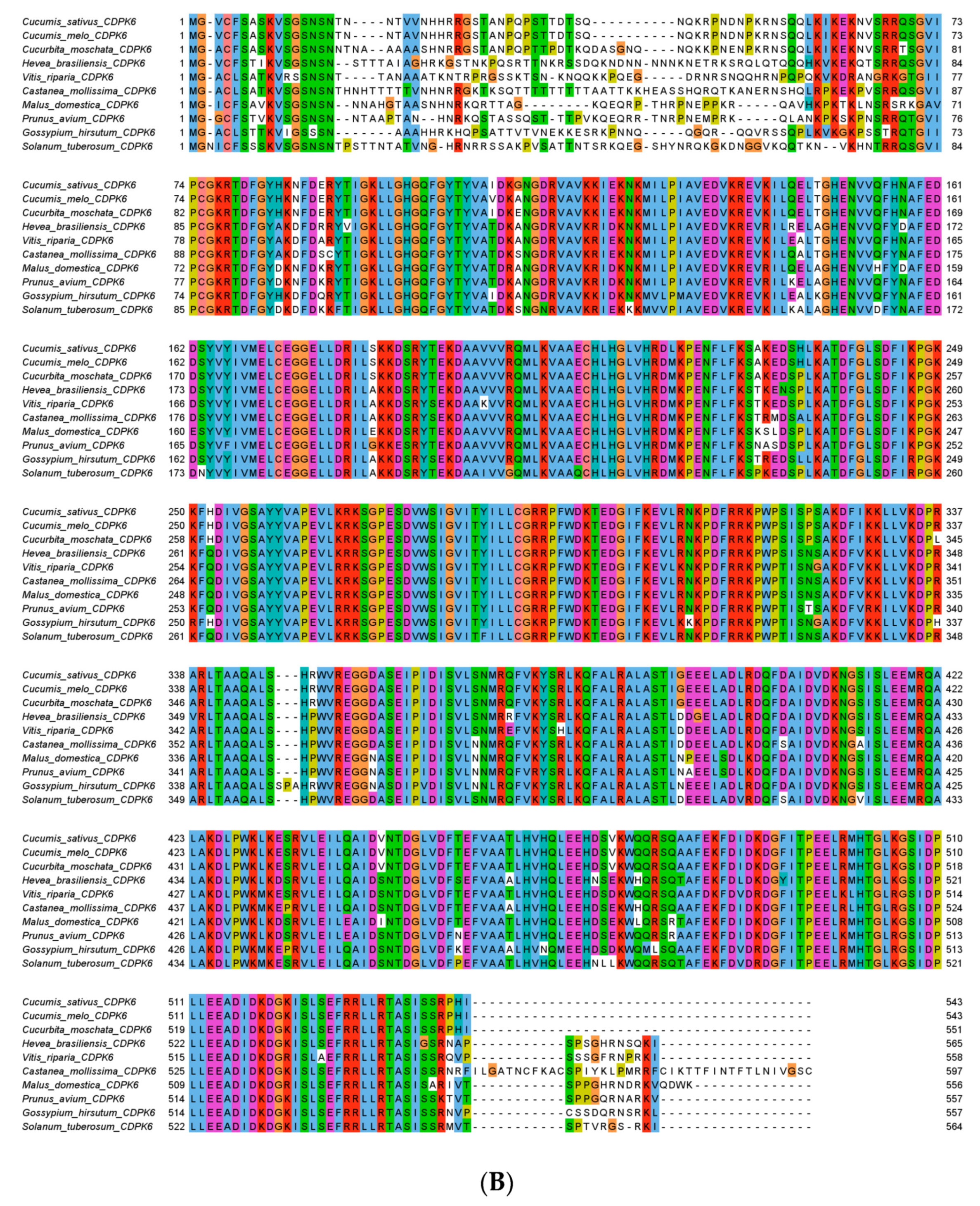
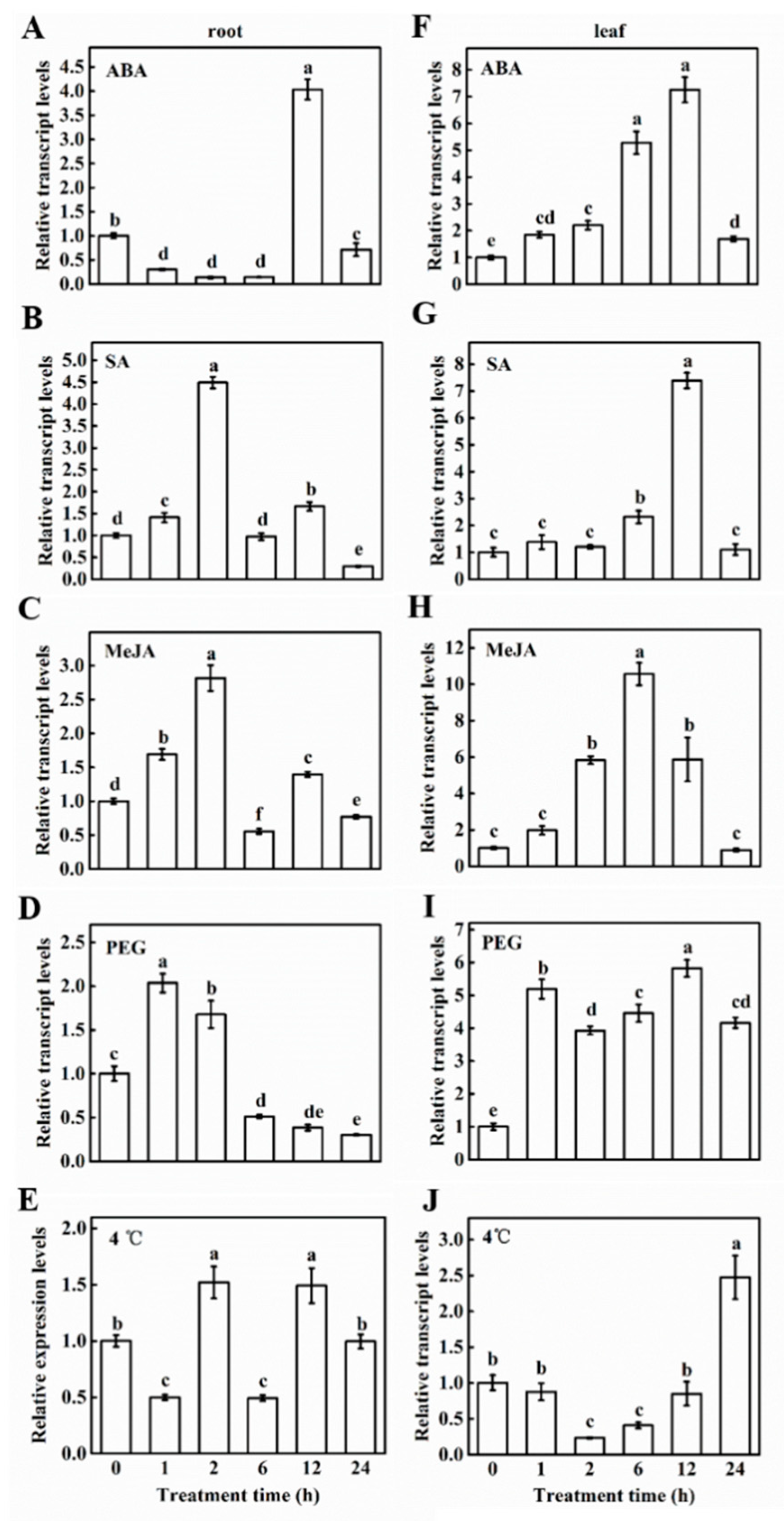
2.2. Phylogenetic and Expression Analysis of CsCDPK6
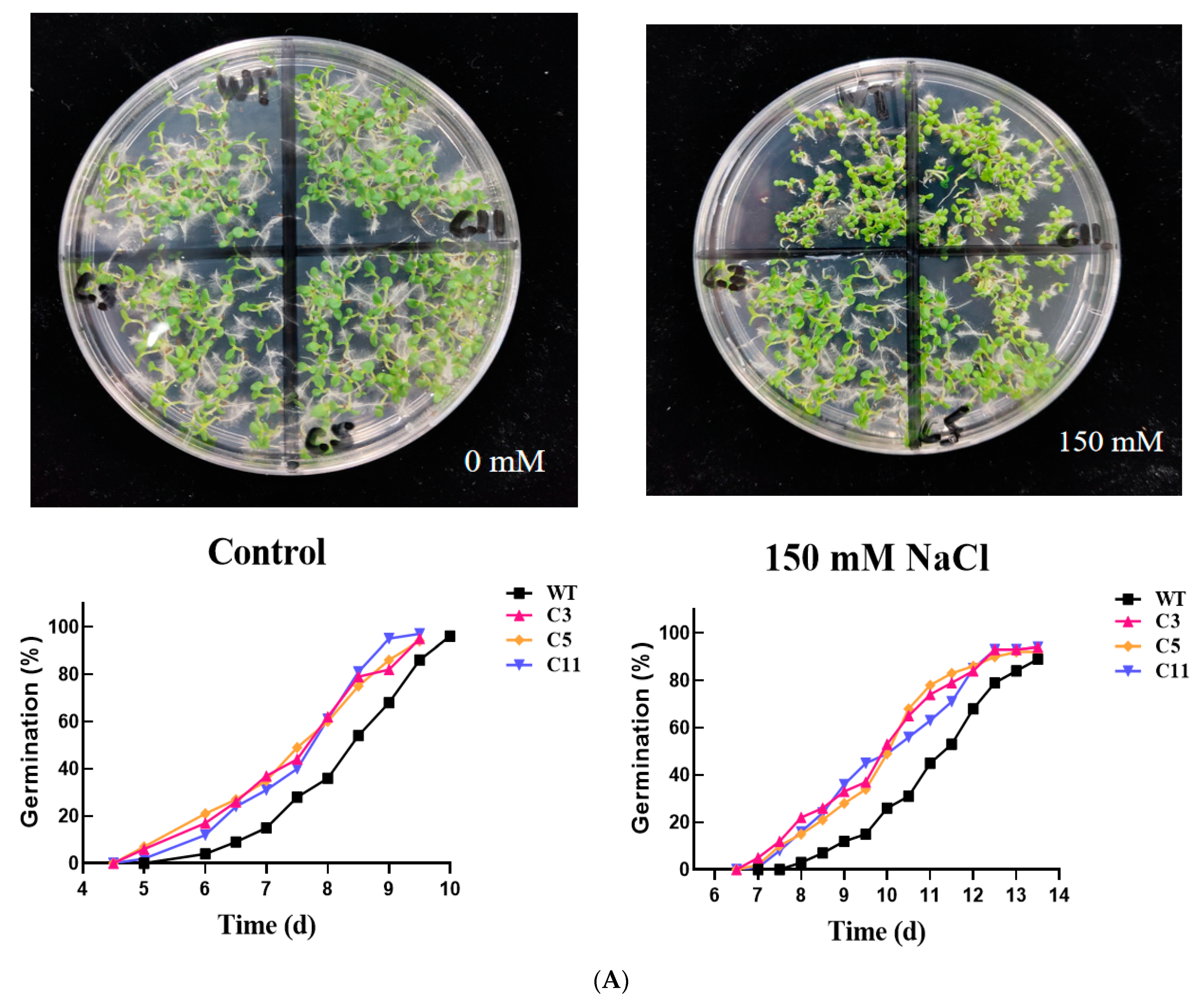
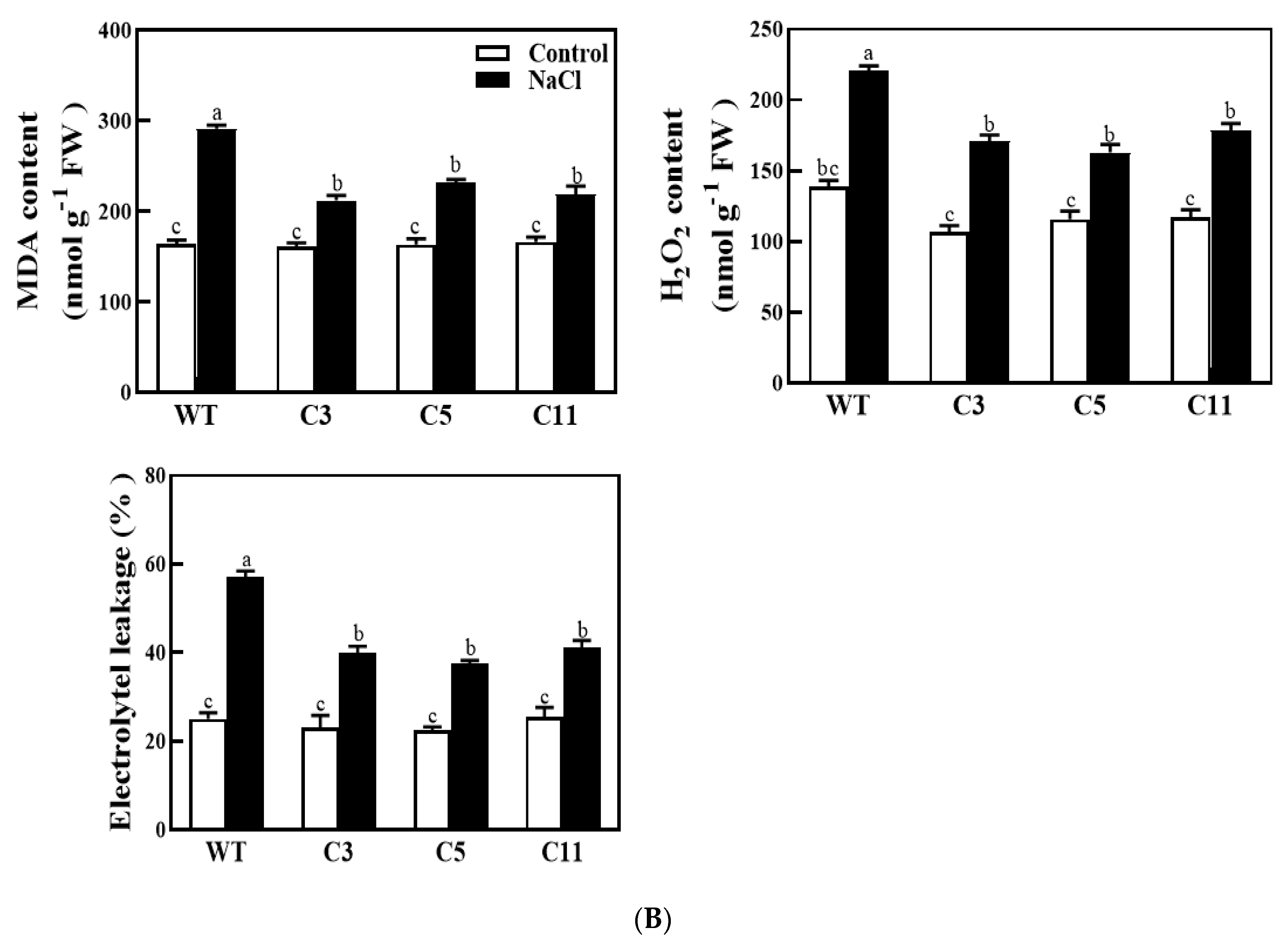
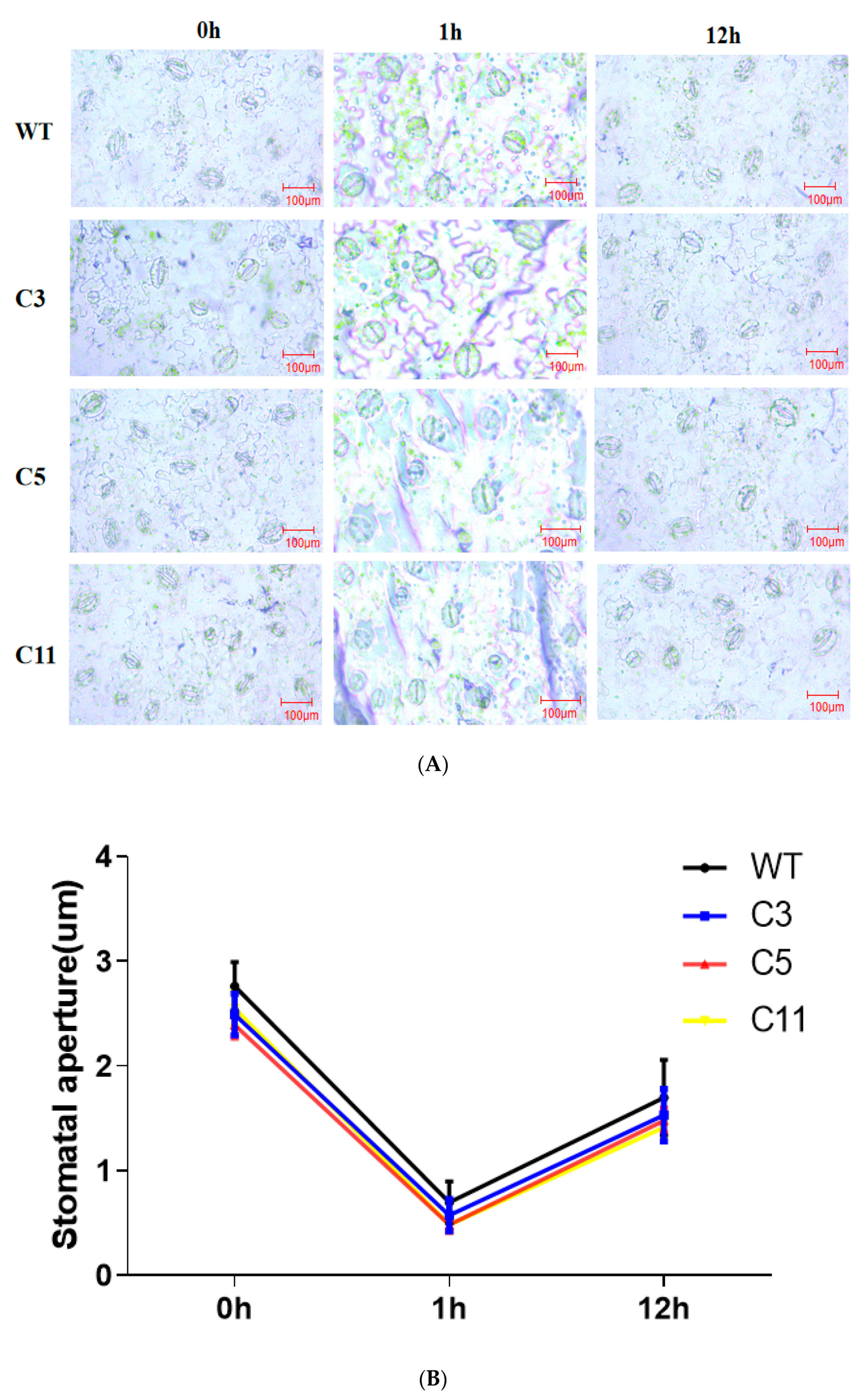
2.3. Overexpression of CsCDPK6 Enhances Tobacco Tolerance to Salt Stress
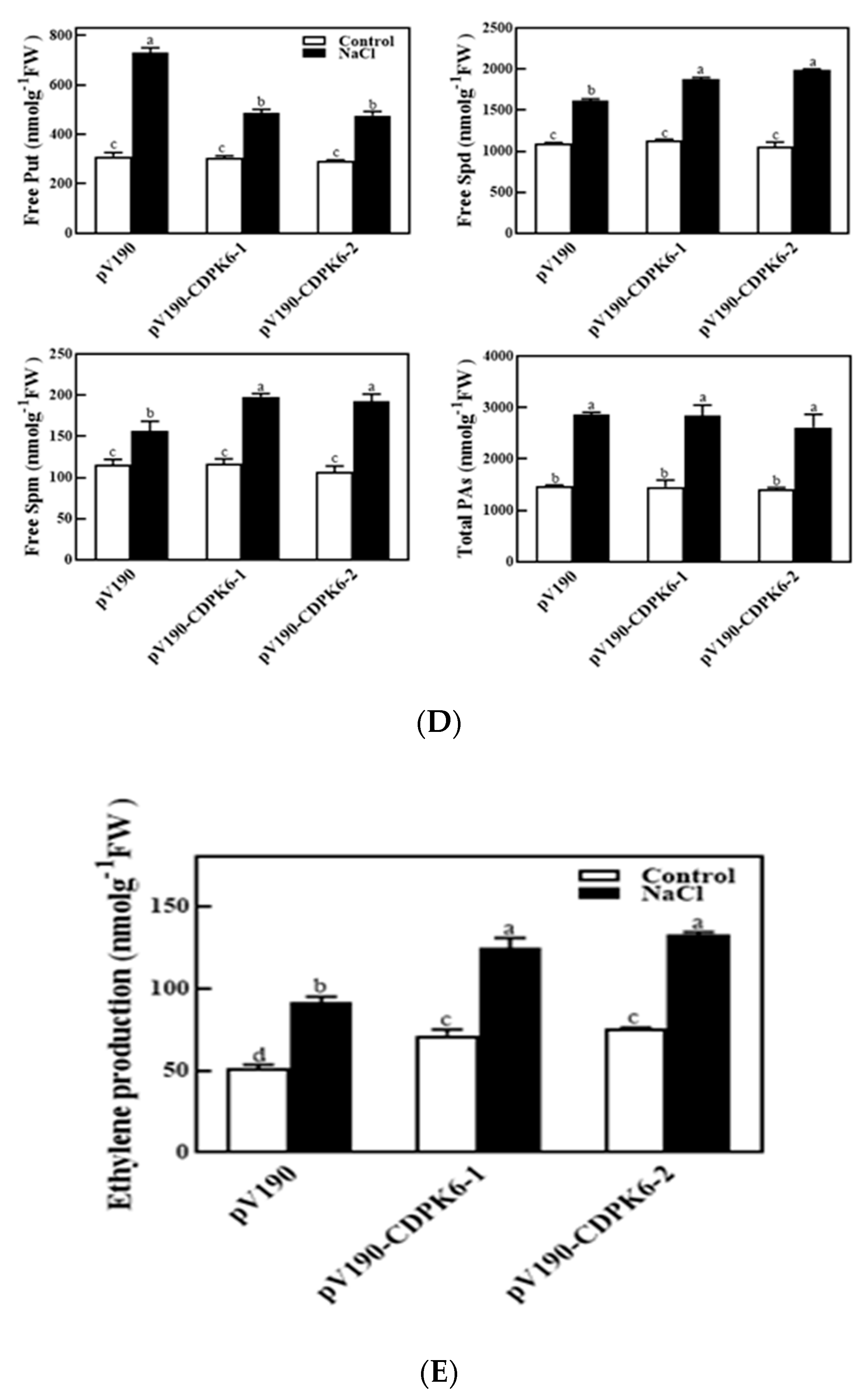
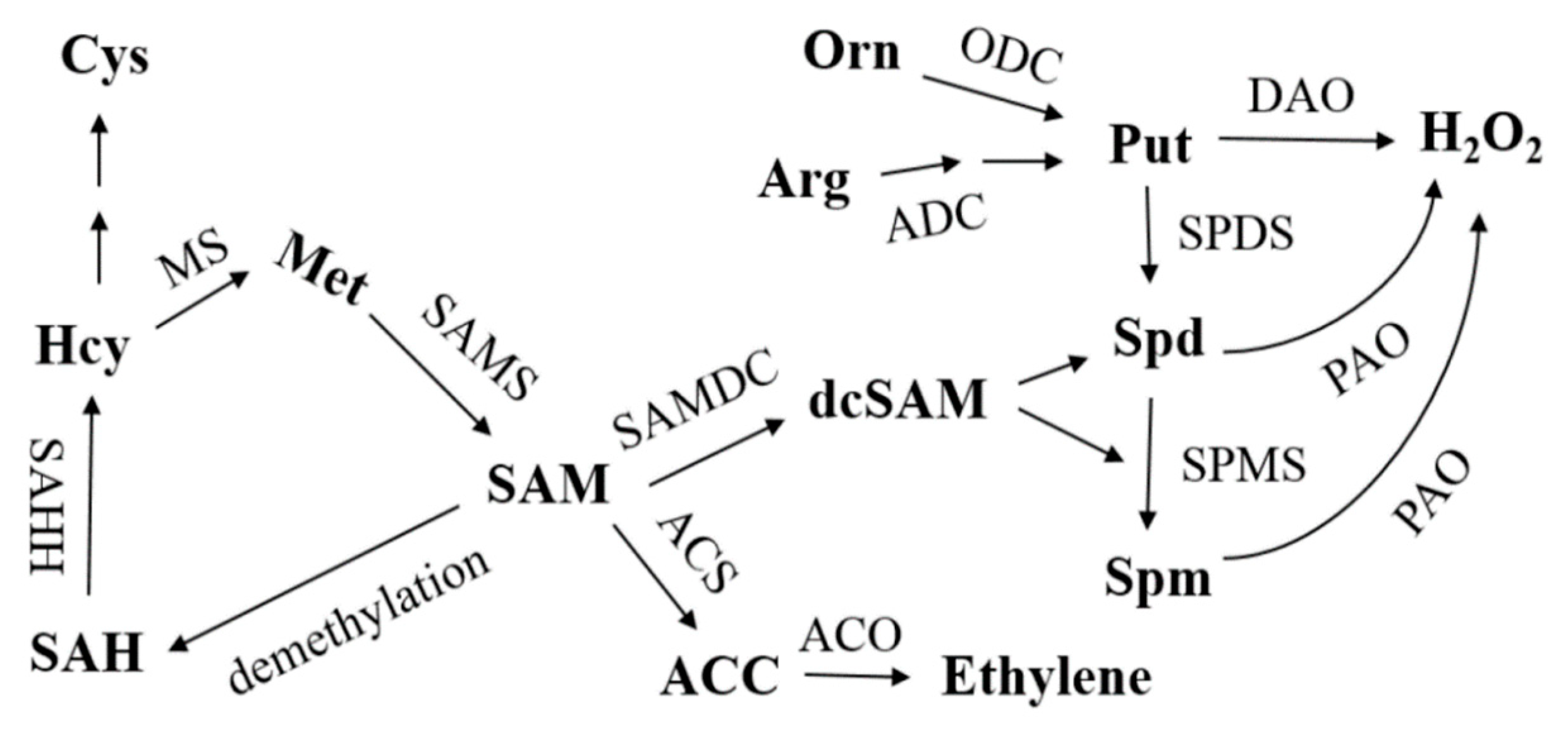
2.4. Silencing of CsCDPK6 Regulates PA and Ethylene Metabolism in Cucumber
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Total RNA Extraction and Gene Expression Analysis
4.3. Y2H Screening and Assays
4.4. BiFC Assays
4.5. Co-IP Assays
4.6. Determination of Plant Germination Rate
4.7. Electrolyte Leakage, Malondialdehyde (MDA) and Hydrogen Peroxide (H2O2) Determination
4.8. Measurement of Stomatal Movement
4.9. SAM Content Determination
4.10. Determination of Free PAs
4.11. Determination of Ethylene Content
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhong, M.; Wang, Y.; Shu, S.; Sun, J.; Guo, S. Ectopic expression of CsTGase enhances salt tolerance by regulating PAs biosynthesis, antioxidant activities and Na(+)/K(+) homeostasis in transgenic tobacco. Plant Sci. 2020, 296, 110492. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Wen, W.; Shi, Z.; Gu, Q.; Ahammed, G.J.; Cao, K.; Jahan, M.S.; Shu, S.; Wang, J.; et al. Hydrogen peroxide mediates spermidine-induced autophagy to alleviate salt stress in cucumber. Autophagy 2020, 2020, 2876–2890. [Google Scholar] [CrossRef]
- Malabika, R.; Ra, Y.W. Overexpression of S-adenosylmethionine decarboxylase gene in rice increases polyamine level and enhances sodium chloride stress tolerance. Plant Sci. 2002, 163, 987–992. [Google Scholar]
- Lei, G.; Shen, M.; Li, Z.-G.; Zhang, B.; Duan, K.-X.; Wang, N.; Cao, Y.-R.; Zhang, W.-K.; Ma, B.; Ling, H.-Q.; et al. EIN2 regulates salt stress response and interacts with a MA3 domain-containing protein ECIP1 in Arabidopsis. Plant Cell Environ. 2011, 34, 1678–1692. [Google Scholar] [CrossRef] [PubMed]
- Grillo, M.A.; Colombatto, S. S-adenosylmethionine and its products. Amino Acids 2008, 34, 187–193. [Google Scholar] [CrossRef]
- Markham, G.D.; Pajares, M.A. Structure-function relationships in methionine adenosyltransferases. Cell. Mol. Life Sci. 2009, 66, 636–648. [Google Scholar] [CrossRef]
- Guo, Z.; Tan, J.; Zhuo, C.; Wang, C.; Xiang, B.; Wang, Z. Abscisic acid, H2O2 and nitric oxide interactions mediated cold-induced S-adenosylmethionine synthetase in Medicago sativa subsp. falcata that confers cold tolerance through up-regulating polyamine oxidation. Plant Biotechnol. J. 2014, 12, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Li, D.M.; Staehelin, C.; Zhang, Y.S.; Peng, S.L. Identification of genes differentially expressed in Mikania micrantha during Cuscuta campestris infection by suppression subtractive hybridization. J. Plant Physiol. 2009, 166, 1423–1435. [Google Scholar] [CrossRef]
- Du, C.-X.; Fan, H.-F.; Guo, S.-R.; Tezuka, T.; Li, J. Proteomic analysis of cucumber seedling roots subjected to salt stress. Phytochemistry 2010, 71, 1450–1459. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; He, L.; Guo, S.; Li, J.; Yang, Y.; Yan, B.; Sun, J.; Li, J. Proteomics reveal cucumber Spd-responses under normal condition and salt stress. Plant Physiol. Biochem. 2013, 67, 7–14. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, Y.; Luo, W.; Li, W.; Chen, N.; Zhang, D.; Chong, K. The F-Box Protein OsFBK12 Targets OsSAMS1 for Degradation and Affects Pleiotropic Phenotypes, Including Leaf Senescence, in Rice. Plant Physiol. 2013, 163, 1673–1685. [Google Scholar] [CrossRef]
- Mao, D.; Yu, F.; Li, J.; Van de Poel, B.; Tan, D.; Li, J.; Liu, Y.; Li, X.; Dong, M.; Chen, L.; et al. FERONIA receptor kinase interacts withS-adenosylmethionine synthetase and suppressesS-adenosylmethionine production and ethylene biosynthesis inArabidopsis. Plant Cell Environ. 2015, 38, 2566–2574. [Google Scholar] [CrossRef]
- Jia, M.; Du, P.; Ding, N.; Zhang, Q.; Xing, S.; Wei, L.; Zhao, Y.; Mao, W.; Li, J.; Li, B.; et al. Two FERONIA-Like Receptor Kinases Regulate Apple Fruit Ripening by Modulating Ethylene Production. Front. Plant Sci. 2017, 8, 1406. [Google Scholar] [CrossRef]
- Jin, Y.; Ye, N.; Zhu, F.; Li, H.; Wang, J.; Jiang, L.; Zhang, J. Calcium-dependent protein kinase CPK28 targets the methionine adenosyltransferases for degradation by the 26S proteasome and affects ethylene biosynthesis and lignin deposition in Arabidopsis. Plant J. 2017, 90, 304–318. [Google Scholar] [CrossRef]
- Roje, S. S-adenosyl-L-methionine: Beyond the universal methyl group donor. Phytochemistry 2006, 67, 1686–1698. [Google Scholar] [CrossRef]
- Fujiwara, T.; Mitsuya, S.; Miyake, H.; Hattori, T.; Takabe, T. Characterization of a novel glycinebetaine/proline transporter gene expressed in the mestome sheath and lateral root cap cells in barley. Planta 2010, 232, 133–143. [Google Scholar] [CrossRef]
- Harper, J.F.; Sussman, M.R.; Schaller, G.E.; Putnam-Evans, C.; Charbonneau, H.; Harmon, A.C. A calcium-dependent protein kinase with a regulatory domain similar to calmodulin. Science 1991, 252, 951–954. [Google Scholar] [CrossRef]
- Harper, J.F.; Breton, G.; Harmon, A. Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 2004, 55, 263–288. [Google Scholar] [CrossRef]
- Klimecka, M.; Grazyna, M. Structure and functions of plant calcium-dependent protein kinases. Acta Biochim. Pol. 2007, 54, 219–233. [Google Scholar] [CrossRef]
- Liese, A.; Romeis, T. Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim. Biophys. Acta 2013, 1833, 1582–1589. [Google Scholar] [CrossRef]
- Zou, J.-J.; Wei, F.-J.; Wang, C.; Wu, J.-J.; Ratnasekera, D.; Liu, W.-X.; Wu, W.-H. Arabidopsis Calcium-Dependent Protein Kinase CPK10 Functions in Abscisic Acid- and Ca2+-Mediated Stomatal Regulation in Response to Drought Stress. Plant Physiol. 2010, 154, 1232–1243. [Google Scholar] [CrossRef]
- Wang, L.; Yu, C.; Xu, S.; Zhu, Y.; Huang, W. OsDi19-4 acts downstream of OsCDPK14 to positively regulate ABA response in rice. Plant Cell Environ. 2016, 39, 2740–2753. [Google Scholar] [CrossRef]
- Asano, T.; Hayashi, N.; Kobayashi, M.; Aoki, N.; Miyao, A.; Mitsuhara, I.; Ichikawa, H.; Komatsu, S.; Hirochika, H.; Kikuchi, S.; et al. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 2012, 69, 26–36. [Google Scholar] [CrossRef]
- He, M.-W.; Wang, Y.; Wu, J.-Q.; Shu, S.; Sun, J.; Guo, S.-R. Isolation and characterization of S-Adenosylmethionine synthase gene from cucumber and responsive to abiotic stress. Plant Physiol. Biochem. 2019, 141, 431–445. [Google Scholar] [CrossRef]
- Xu, X.; Liu, M.; Lu, L.; He, M.; Qu, W.; Xu, Q.; Qi, X.; Chen, X. Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Mol. Genet. Genom. 2015, 290, 1403–1414. [Google Scholar] [CrossRef]
- Alcazar, R.; Marco, F.; Cuevas, J.C.; Patron, M.; Ferrando, A.; Carrasco, P.; Tiburcio, A.F.; Altabella, T. Involvement of polyamines in plant response to abiotic stress. Biotechnol. Lett. 2006, 28, 1867–1876. [Google Scholar] [CrossRef]
- Duan, J.; Li, J.; Guo, S.; Kang, Y. Exogenous spermidine affects polyamine metabolism in salinity-stressed Cucumis sativus roots and enhances short-term salinity tolerance. J. Plant Physiol. 2008, 165, 1620–1635. [Google Scholar] [CrossRef]
- Shu, S.; Tang, Y.; Zhou, X.; Jahan, M.S.; Sun, J.; Wang, Y.; Guo, S. Physiological mechanism of transglutaminase-mediated improvement in salt tolerance of cucumber seedlings. Plant Physiol. Biochem. 2020, 156, 333–344. [Google Scholar] [CrossRef]
- Hu, D.-G.; Sun, C.-H.; Sun, M.-H.; Hao, Y.-J. MdSOS2L1 phosphorylates MdVHA-B1 to modulate malate accumulation in response to salinity in apple. Plant Cell Rep. 2016, 35, 705–718. [Google Scholar] [CrossRef]
- Zhu, Y.; Yan, J.; Liu, W.; Liu, L.; Sheng, Y.; Sun, Y.; Li, Y.; Scheller, H.V.; Jiang, M.; Hou, X. Phosphorylation of a NAC transcription factor by ZmCaMK regulates abscisic acid-induced antioxidant defense in maize. Plant Physiol. 2016, 171, 1651–1664. [Google Scholar] [CrossRef]
- Zou, J.-J.; Li, X.-D.; Ratnasekera, D.; Wang, C.; Liu, W.-X.; Song, L.-F.; Zhang, W.-Z.; Wu, W.-H. Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 2015, 27, 1445–1460. [Google Scholar] [CrossRef]
- Ma, S.Y.; Wu, W.H. AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol. Biol. 2007, 65, 511–518. [Google Scholar] [CrossRef]
- Xu, J.; Tian, Y.-S.; Peng, R.-H.; Xiong, A.-S.; Zhu, B.; Jin, X.-F.; Gao, F.; Fu, X.-Y.; Hou, X.-L.; Yao, Q.-H. AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta 2010, 231, 1251–1260. [Google Scholar] [CrossRef]
- Huang, K.; Peng, L.; Liu, Y.; Yao, R.; Liu, Z.; Li, X.; Yang, Y.; Wang, J. Arabidopsis calcium-dependent protein kinase AtCPK1 plays a positive role in salt/drought-stress response. Biochem. Biophys. Res. Commun. 2018, 498, 92–98. [Google Scholar] [CrossRef]
- Ronzier, E.; Corratgé-Faillie, C.; Sanchez, F.; Prado, K.; Brière, C.; Leonhardt, N.; Thibaud, J.; Xiong, T.C. CPK13, a Noncanonical Ca2+-Dependent Protein Kinase, Specifically Inhibits KAT2 and KAT1 Shaker K+ Channels and Reduces Stomatal Opening. Plant Physiol. 2014, 166, 314–326. [Google Scholar] [CrossRef]
- Zhong, M.; Yuan, Y.; Shu, S.; Sun, J.; Guo, S.; Yuan, R.; Tang, Y. Effects of exogenous putrescine on glycolysis and Krebs cycle metabolism in cucumber leaves subjected to salt stress. Plant Growth Regul. 2016, 79, 319–330. [Google Scholar] [CrossRef]
- Saha, J.; Brauer, E.K.; Sengupta, A.; Popescu, S.C.; Gupta, K.; Gupta, B. Polyamines as redox homeostasis regulators during salt stress in plants. Front. Environ. Sci. 2015, 3, 21. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wang, N.; Xu, H.; Jiang, S.; Zhang, Z.; Lu, N.; Qiu, H.; Qu, C.; Wang, Y.; Wu, S.; Chen, X. MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 2017, 90, 276–292. [Google Scholar] [CrossRef]
- Dhindsa, R.S.; Plumb-Dhindsa, P.; Thorpe, T.A. Leaf senescence: Correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J. Exp. Bot. 1981, 32, 93–101. [Google Scholar] [CrossRef]
- Kramer, G.F.; Norman, H.A.; Krizek, D.T.; Mirecki, R.M. influence of UV-B radiation on polyamines, lipid peroxidation and membrane lipids in cucumber. Phytochemistry 1991, 30, 2101–2108. [Google Scholar] [CrossRef]
- Alexieva, V.; Sergiev, I.; Mapelli, S.; Karanov, E. The effect of drought and ultraviolet radiation on growth and stress markers in pea and wheat. Plant Cell Environ. 2001, 24, 1337–1344. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhong, M.; Shu, S.; Du, N.; Sun, J.; Guo, S. Proteomic and Physiological Analyses Reveal Putrescine Responses in Roots of Cucumber Stressed by NaCl. Front. Plant Sci. 2016, 7, 1035. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, H.; He, M.; Jahan, M.S.; Wu, J.; Gu, Q.; Shu, S.; Sun, J.; Guo, S. CsCDPK6, a CsSAMS1-Interacting Protein, Affects Polyamine/Ethylene Biosynthesis in Cucumber and Enhances Salt Tolerance by Overexpression in Tobacco. Int. J. Mol. Sci. 2021, 22, 11133. https://doi.org/10.3390/ijms222011133
Zhu H, He M, Jahan MS, Wu J, Gu Q, Shu S, Sun J, Guo S. CsCDPK6, a CsSAMS1-Interacting Protein, Affects Polyamine/Ethylene Biosynthesis in Cucumber and Enhances Salt Tolerance by Overexpression in Tobacco. International Journal of Molecular Sciences. 2021; 22(20):11133. https://doi.org/10.3390/ijms222011133
Chicago/Turabian StyleZhu, Heyuan, Meiwen He, Mohammad Shah Jahan, Jianqiang Wu, Qinsheng Gu, Sheng Shu, Jin Sun, and Shirong Guo. 2021. "CsCDPK6, a CsSAMS1-Interacting Protein, Affects Polyamine/Ethylene Biosynthesis in Cucumber and Enhances Salt Tolerance by Overexpression in Tobacco" International Journal of Molecular Sciences 22, no. 20: 11133. https://doi.org/10.3390/ijms222011133
APA StyleZhu, H., He, M., Jahan, M. S., Wu, J., Gu, Q., Shu, S., Sun, J., & Guo, S. (2021). CsCDPK6, a CsSAMS1-Interacting Protein, Affects Polyamine/Ethylene Biosynthesis in Cucumber and Enhances Salt Tolerance by Overexpression in Tobacco. International Journal of Molecular Sciences, 22(20), 11133. https://doi.org/10.3390/ijms222011133







