Marker Development for Differentiation of Fusarium oxysporum f. sp. Niveum Race 3 from Races 1 and 2
Abstract
:1. Introduction
2. Results
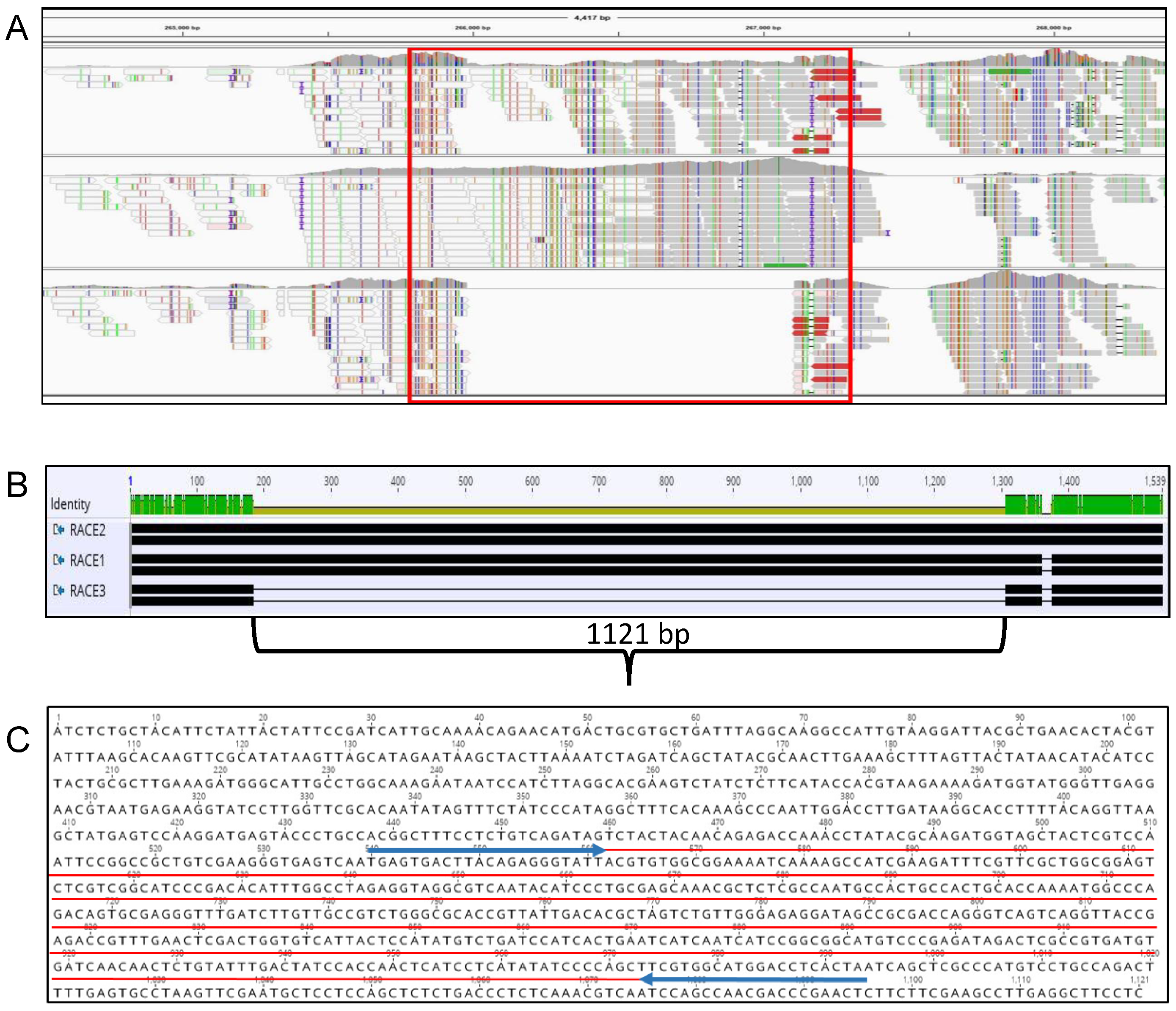
2.1. Sequence Analysis and Primer Design
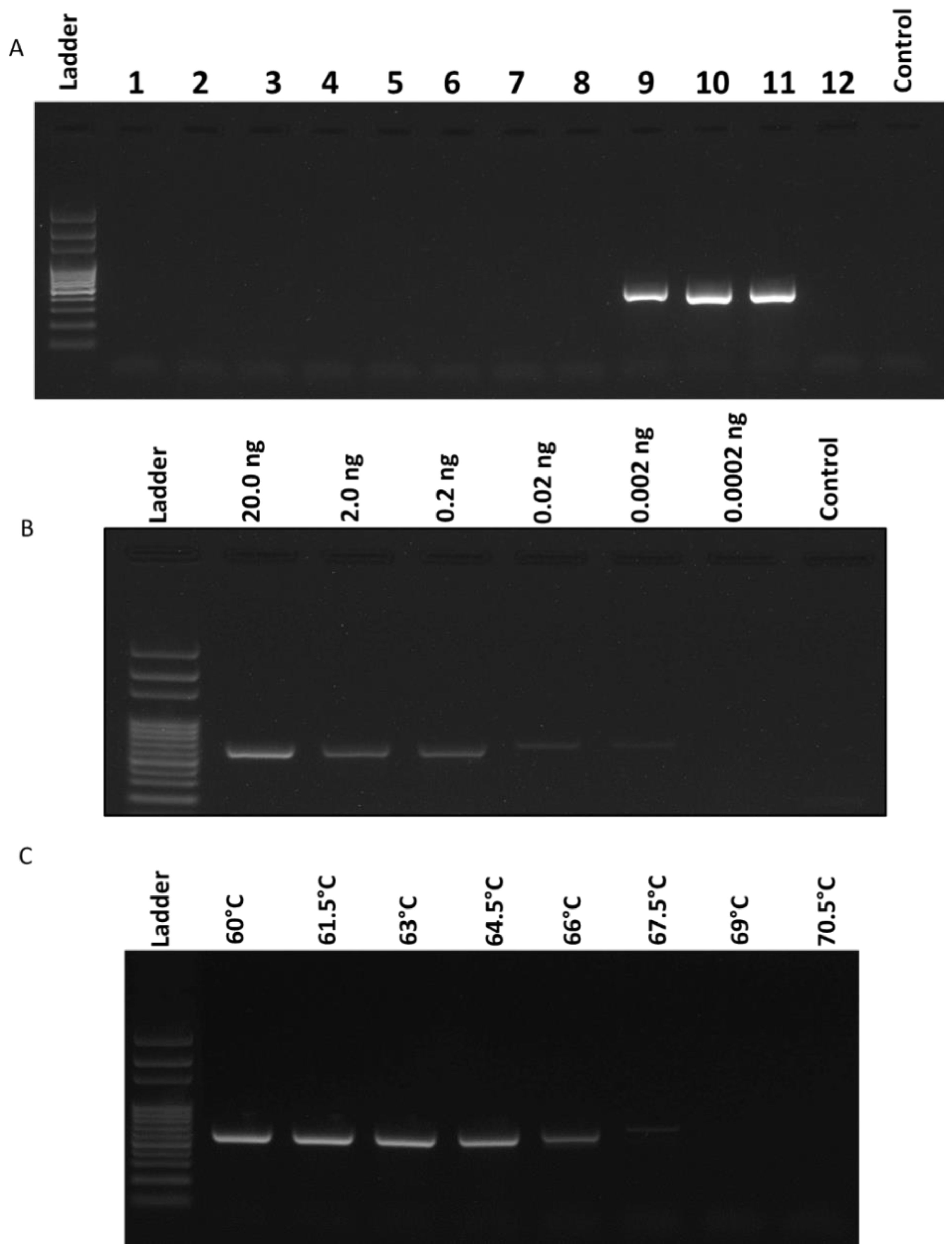
2.2. Specificity and Sensitivity of the Race 3 Marker
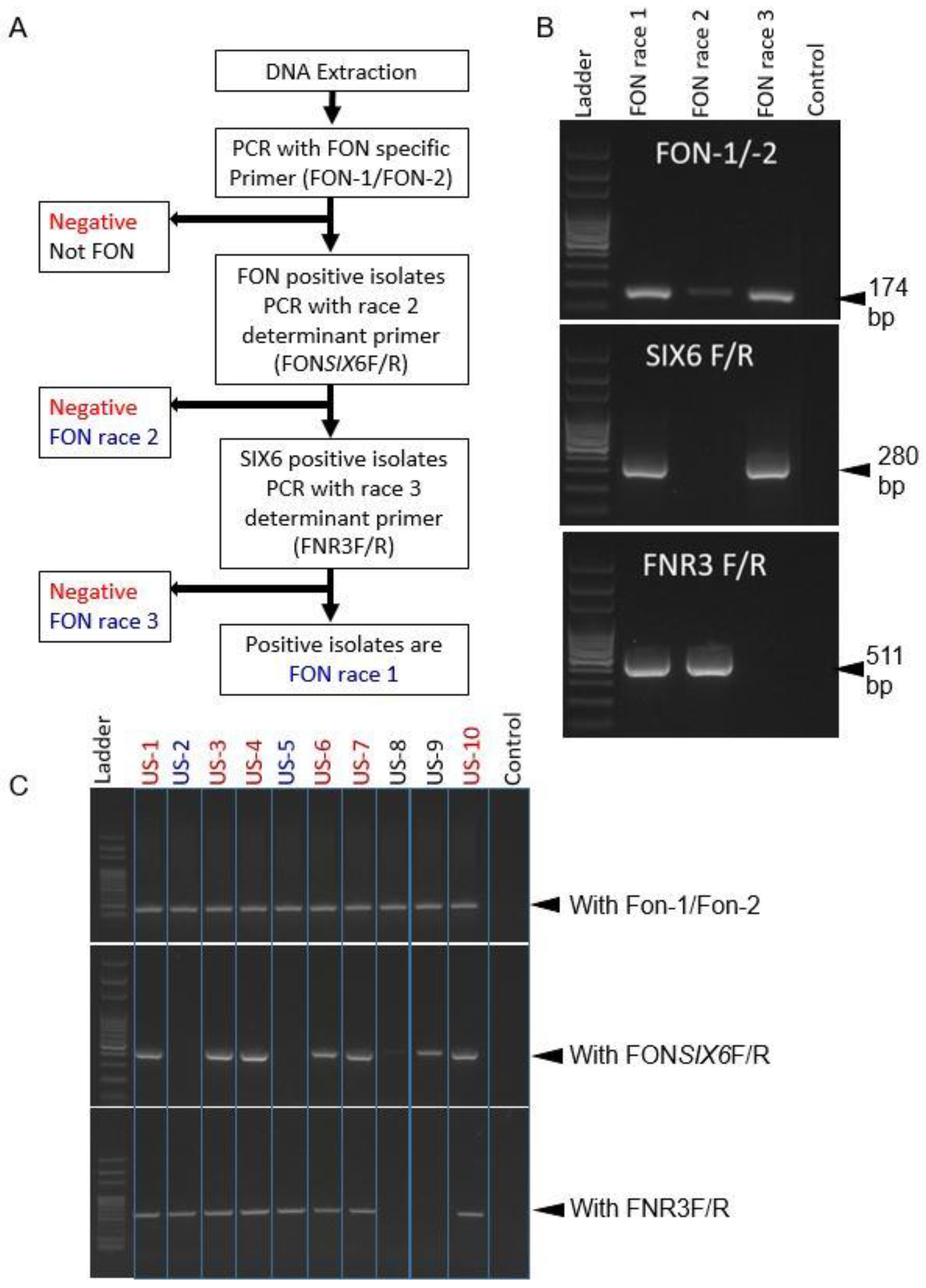
2.3. Development of a Protocol for Race Differentiation
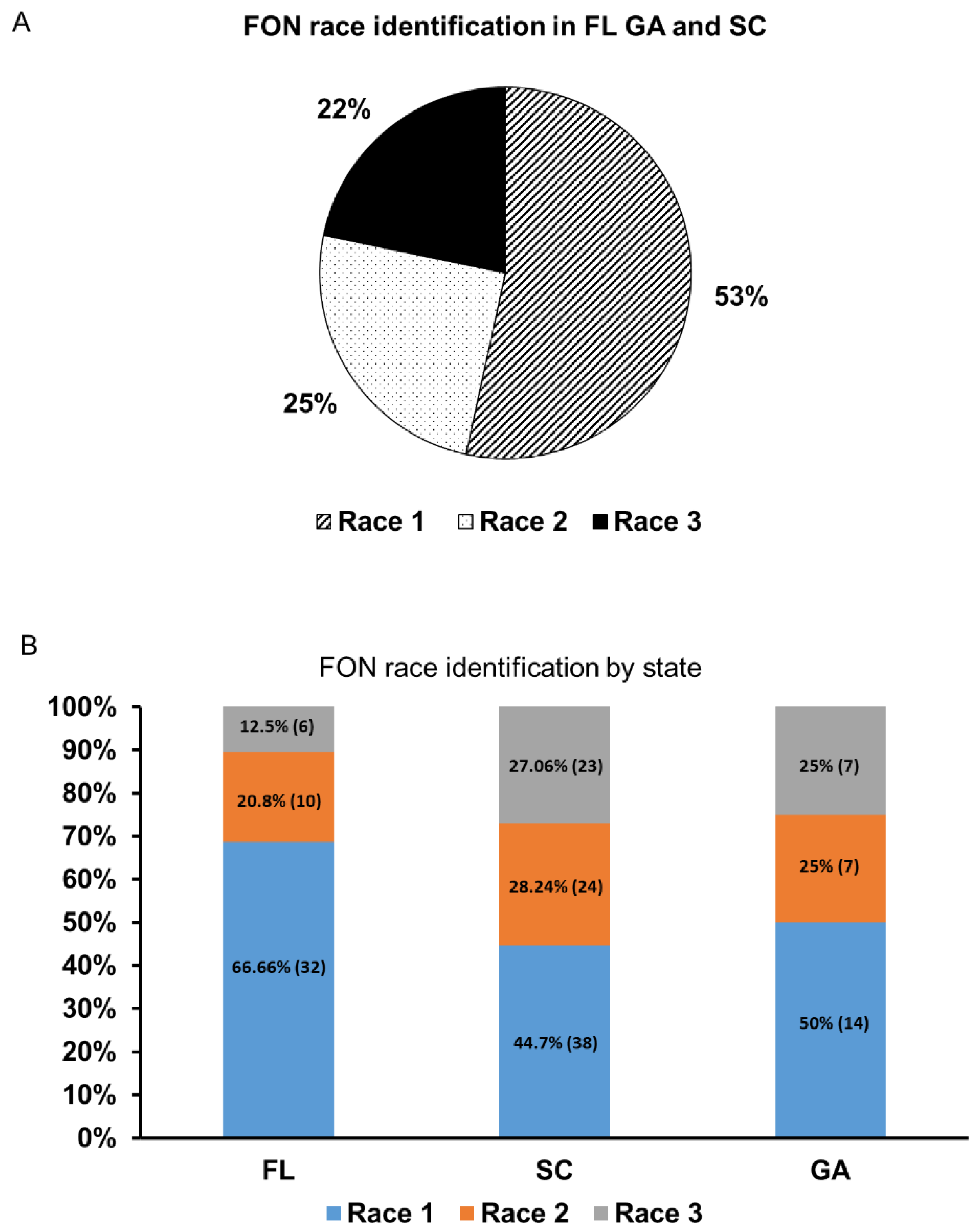
2.4. Race Distribution of Experimental Samples
3. Discussion
4. Materials and Methods
4.1. FON Isolates and DNA Extraction
4.2. Sequence Alignment and Primer Design
4.3. Selection of Race-Specific Diagnostic Marker Primer Set and PCR Conditions
4.4. Optimization of the Developed Marker
4.5. Development and Application of a Protocol for Race Differentiation
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FON | Fusarium oxysporum f. sp. niveum |
| SIX6 | Secreted in Xylem 6 |
| PDA | Potato Dextrose Agar |
| PCR | Polymerase Chain Reaction |
| BLAST | Basic Local Alignment Search Tool |
| FOL | Fusarium oxysporum f. sp. lycopersici |
| IGV | Interactive Genomics Viewer |
| NCBI | National Center for Biotechnology Information |
| HSP | Heat Shock Protein |
| ITS | Internal Transcribed Spacer |
| COX | Cytochrome c Oxidase |
| IGS | Intergenic Spacer |
| AVR | Avirulence Gene |
| PI | Plant Introduction |
References
- Zhou, X.; Everts, K.; Bruton, B. Race 3, a new and highly virulent race of Fusarium oxysporum f. sp. niveum causing Fusarium wilt in watermelon. Plant Dis. 2010, 94, 92–98. [Google Scholar] [CrossRef] [Green Version]
- Netzer, D.; Weintall, C. Inheritance of resistance in watermelon to race 1 of Fusarium oxysporum f. sp. niveum. Plant Dis. 1980, 64, 853–854. [Google Scholar] [CrossRef]
- Niu, X.; Zhao, X.; Ling, K.-S.; Levi, A.; Sun, Y.; Fan, M. The FonSIX6 gene acts as an avirulence effector in the Fusarium oxysporum f. sp. niveum-watermelon pathosystem. Sci. Rep. 2016, 6, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Roberts, P.; Dufault, N.; Hochmuth, R.; Vallad, G.; Paret, M. Fusarium Wilt (Fusarium oxysporum f. sp. niveum) of Watermelon; EDIS, PP352; University of Florida: Gainesville, FL, USA, 2019. [Google Scholar]
- Egel, D.; Martyn, R. Fusarium wilt of watermelon and other cucurbits. Plant Health Instr. 2007, 10, 1094. [Google Scholar]
- Edel-Hermann, V.; Lecomte, C. Current status of Fusarium oxysporum formae speciales and races. Phytopathology 2019, 109, 512–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, J.; Zhan, Y.; Zeng, F.; Long, H.; Pei, Y.; Guo, J. Development of a real-time fluorescence loop-mediated isothermal amplification assay for rapid and quantitative detection of Fusarium oxysporum f. sp. niveum in soil. FEMS Microbiol. Lett. 2013, 349, 127–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhter, A.; Hage-Ahmed, K.; Soja, G.; Steinkellner, S. Potential of Fusarium wilt-inducing chlamydospores, in vitro behaviour in root exudates and physiology of tomato in biochar and compost amended soil. Plant Soil 2016, 406, 425–440. [Google Scholar] [CrossRef] [Green Version]
- McKeen, C.; Wensley, R. Longevity of Fusarium oxysporum in soil tube culture. Science 1961, 134, 1528–1529. [Google Scholar] [CrossRef]
- Kleczewski, N.M.; Egel, D.S. A diagnostic guide for Fusarium wilt of watermelon. Plant Health Prog. 2011. [Google Scholar] [CrossRef] [Green Version]
- Chittarath, K.; Mostert, D.; Crew, K.S.; Viljoen, A.; Kong, G.; Molina, A.; Thomas, J.E. First report of Fusarium oxysporum f. sp. cubense tropical race 4 (VCG 01213/16) associated with Cavendish bananas in Laos. Plant Dis. 2018, 102, 449. [Google Scholar] [CrossRef]
- Manulis, S.; Kogan, N.; Reuven, M.; Ben-Yephet, Y. Use of the RAPD technique for identification of Fusarium oxysporum f. sp. dianthi from carnation. Phytopathology 1994, 84, 98–101. [Google Scholar] [CrossRef]
- Quesada-Ocampo, L. Fusarium Wilt of Watermelon; NC State Extension: Raleigh, NC, USA, 2018. [Google Scholar]
- Martyn, R.D. Fusarium wilt of watermelon: 120 years of research. Hortic. Rev. 2014, 42, 349–442. [Google Scholar]
- Hutson, R.; Smith, I. Phytoalexins and tyloses in tomato cultivars infected with Fusarium oxysporum f. sp. lycopersici or Verticillium albo-atrum. Physiol. Plant Pathol. 1980, 17, 245–257. [Google Scholar] [CrossRef]
- Bruton, B.; Fish, W.; Langston, D. First report of Fusarium wilt caused by Fusarium oxysporum f. sp. niveum race 2 in Georgia watermelons. Plant Dis. 2008, 92, 983. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Xu, J.; Liu, G.; Yao, X.; Li, P.; Yang, X. Characterization of the watermelon seedling infection process by Fusarium oxysporum f. sp. niveum. Plant Pathol. 2015, 64, 1076–1084. [Google Scholar] [CrossRef]
- Keinath, A.P.; DuBose, V.; Katawczik, M.; Wechter, P. Identifying races of Fusarium oxysporum f. sp. niveum in South. Carolina Recovered from Watermelon Seedlings, Plants, and Field Soil. Plant Dis. 2020, 104, 2481–2488. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-H.; Chen, K.-S.; Chang, J.-Y.; Wan, Y.-L.; Hsu, C.-C.; Huang, J.-W.; Chang, P.-F.L. Development of the molecular methods for rapid detection and differentiation of Fusarium oxysporum and F. oxysporum f. sp. niveum in Taiwan. New Biotechnol. 2010, 27, 409–418. [Google Scholar] [CrossRef]
- Sumner, D.R.; Johnson, A.W. Effect of root-knot nematodes on Fusarium wilt of watermelon. Phytopathology 1973, 63, 857–861. [Google Scholar] [CrossRef]
- Hua, G.K.H.; Timper, P.; Ji, P. Meloidogyne incognita intensifies the severity of Fusarium wilt on watermelon caused by Fusarium oxysporum f. sp. niveum. Can. J. Plant Pathol. 2019, 41, 261–269. [Google Scholar] [CrossRef]
- Keinath, A.P.; Coolong, T.W.; Lanier, J.D.; Ji, P.J.P.D. Managing Fusarium wilt of watermelon with delayed transplanting and cultivar resistance. Plant Dis. 2019, 103, 44–50. [Google Scholar] [CrossRef] [Green Version]
- Okungbowa, F.; Shittu, H. Fusarium wilts: An overview. Environ. Res. J. 2012, 6, 83–102. [Google Scholar]
- Kurt, S.; Dervis, S.; Soylu, E.M.; Tok, F.M.; Yetisir, H.; Soylu, S. Pathogenic races and inoculum density of Fusarium oxysporum f. sp. niveum in commercial watermelon fields in southern Turkey. Phytoparasitica 2008, 36, 107–116. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, J.; Wang, Y.; Zheng, X. Molecular detection of Fusarium oxysporum f. sp. niveum and Mycosphaerella melonis in infected plant tissues and soil. FEMS Microbiol. Lett. 2005, 249, 39–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Everts, K.; Hochmuth, M. Field evaluation of triploid cultivars for resistance to Fusarium wilt of watermelon in Delaware, 2010. Plant Dis. Manag. Rep. 2011, 5, V175. [Google Scholar]
- Everts, K.L.; Himmelstein, J.C. Fusarium wilt of watermelon: Towards sustainable management of a re-emerging plant disease. Crop Prot. 2015, 73, 93–99. [Google Scholar] [CrossRef]
- Bruton, B.; Damicone, J. Fusarium wilt of watermelon: Impact of race 2 of Fusarium oxysporum f. sp. niveum on watermelon production in Texas and Oklahoma. Subtrop. Plant Sci. 1999, 51, 4–9. [Google Scholar]
- Wechter, W.P.; Kousik, C.; McMillan, M.; Levi, A. Identification of resistance to Fusarium oxysporum f. sp. niveum race 2 in Citrullus lanatus var. citroides plant introductions. HortScience 2012, 47, 334–338. [Google Scholar] [CrossRef] [Green Version]
- Keinath, A.P.; Hassell, R.L.; Everts, K.L.; Zhou, X.-G. Cover crops of hybrid common vetch reduce Fusarium wilt of seedless watermelon in the eastern United States. Plant Health Prog. 2010. [Google Scholar] [CrossRef] [Green Version]
- Amaradasa, B.; Beckham, K.; Dufault, N.; Sanchez, T.; Ertek, T.; Iriarte, F.; Paret, M.; Ji, P. First report of Fusarium oxysporum f. sp. niveum race 3 causing wilt of watermelon in Florida, USA. Plant Dis. 2018, 102, 1029. [Google Scholar] [CrossRef]
- Petkar, A.; Harris-Shultz, K.; Wang, H.; Brewer, M.T.; Sumabat, L.; Ji, P. Genetic and phenotypic diversity of Fusarium oxysporum f. sp. niveum populations from watermelon in the southeastern United States. PLoS ONE 2019, 14, e0219821. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Lv, H.; Yang, Y.; Xing, M.M.; Kong, C.C.; Fang, Z.; Yang, L.; Zhang, Y.; Wang, Y.; Ling, J. Genetic diversity, virulence, race profiling and comparative genomic analysis of the Fusarium oxysporum f. sp. conglutinans strains infecting cabbages in China. Front. Microbiol. 2019, 10, 1373. [Google Scholar] [CrossRef] [PubMed]
- Martyn, R. An aggressive race of Fusarium oxysporum f. sp. niveum new to the United States. Plant Dis. 1985, 69, 493–495. [Google Scholar] [CrossRef]
- Martyn, R. An initial survey of the United States for races of Fursarium oxysporum f. sp. niveum. HortScience 1989, 24, 696–698. [Google Scholar]
- Larkin, R.; Hopkins, D.; Martin, F. Vegetative compatibility within Fusarium oxysporum f. sp. niveum and its relationship to virulence, aggressiveness, and race. Can. J. Microbiol. 1990, 36, 352–358. [Google Scholar] [CrossRef]
- Fulton, C.J.; Amaradasa, B.S.; Ertek, S.T.; Iriarte, F.; Sanchez, T.; Ji, P.; Paret, M.; Hudson, O.; Ali, E.; Dufault, S.N. Phylogenetic and phenotypic characterization of Fusarium oxysporum f. sp. niveum isolates from Florida-grown watermelon. PLoS ONE 2020, in press. [Google Scholar]
- Hudson, O.; Hudson, D.; Ji, P.; Ali, M.E. Draft genome sequences of three Fusarium oxysporum f. sp. niveum isolates used in designing markers for race differentiation. Microbiol. Resour. Announc. 2020, 9. [Google Scholar] [CrossRef]
- Larkin, R.; Hopkins, D.; Martin, F. A chromosome-scale genome assembly for the Fusarium oxysporum strain Fo5176 to establish a model Arabidopsis-fungal pathosystem. Genes Genomes Genet. 2020, 10, 3549–3555. [Google Scholar]
- Ma, L.-J.; Van Der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.-J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef]
- Inami, K.; Yoshioka-Akiyama, C.; Morita, Y.; Yamasaki, M.; Teraoka, T.; Arie, T. A genetic mechanism for emergence of races in Fusarium oxysporum f. sp. lycopersici: Inactivation of avirulence gene AVR1 by transposon insertion. PLoS ONE 2012, 7, e44101. [Google Scholar] [CrossRef] [Green Version]
- Kistler, H. Genetic diversity in the plant-pathogenic fungus Fusarium oxysporum. Phytopathology 1997, 87, 474–479. [Google Scholar] [CrossRef] [Green Version]
- Mehmood, S.; Sajid, M.; Husnain, S.K.; Zhao, J.; Huang, L.; Kang, Z. Study of inheritance and linkage of virulence genes in a selfing population of a Pakistani dominant race of Puccinia striiformis f. sp. tritici. Int. J. Mol. Sci. 2020, 21, 1685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porter, L.D.; Pasche, J.S.; Chen, W.; Harveson, R.M. Isolation, identification, storage, pathogenicity tests, hosts, and geographic range of Fusarium solani f. sp. pisi causing fusarium root rot of pea. Plant Health Prog. 2015, 16, 136–145. [Google Scholar] [CrossRef] [Green Version]
- Windels, C.E.; Burnes, P.M.; Kommedahl, T. Five-year preservation of Fusarium species on silica gel and soil. Phytopathology 1988, 78, 107–109. [Google Scholar] [CrossRef]
- Webb, K.; Hill, A.; Laufman, J.; Hanson, L.; Panella, L. Long-term preservation of a collection of Rhizoctonia solani using cryogenic storage. Ann. Appl. Biol. 2011, 158, 297–304. [Google Scholar] [CrossRef]
- Zhou, X.; Everts, K. First report of the occurrence of Fusarium oxysporum f. sp. niveum race 2 in commercial watermelon production areas of Maryland and Delaware. Plant Dis. 2001, 85, 1291. [Google Scholar] [CrossRef]
- Dutta, B.; Coolong, T. Fusarium Wilt of Watermelon in Georgia; UGA Extension: Tifton, GA, USA, 2017. [Google Scholar]
- van Dam, P.; Fokkens, L.; Ayukawa, Y.; van der Gragt, M.; Ter Horst, A.; Brankovics, B.; Houterman, P.M.; Arie, T.; Rep, M. A mobile pathogenicity chromosome in Fusarium oxysporum for infection of multiple cucurbit species. Sci. Rep. 2017, 7, 9042. [Google Scholar] [CrossRef]
- Vlaardingerbroek, I.; Beerens, B.; Rose, L.; Fokkens, L.; Cornelissen, B.J.; Rep, M. Exchange of core chromosomes and horizontal transfer of lineage-specific chromosomes in Fusarium oxysporum. Environ. Microbiol. 2016, 18, 3702–3713. [Google Scholar] [CrossRef]
- Mehrabi, R.; Bahkali, A.H.; Abd-Elsalam, K.A.; Moslem, M.; Ben M’Barek, S.; Gohari, A.M.; Jashni, M.K.; Stergiopoulos, I.; Kema, G.H.; de Wit, P.J. Horizontal gene and chromosome transfer in plant pathogenic fungi affecting host range. FEMS Microbiol. Rev. 2011, 35, 542–554. [Google Scholar] [CrossRef] [Green Version]
- Sakr, N. Pathogenic, morphological and genetic diversity in Plasmopara halstedii, the causal agent of sunflower downy mildew. Acta Scientiarum. Agronomy 2013, 35, 9–19. [Google Scholar]
- Martín-Sanz, A.; Rueda, S.; García-Carneros, A.B.; González-Fernández, S.; Miranda-Fuentes, P.; Castuera-Santacruz, S.; Molinero-Ruiz, L. Genetics, host range, and molecular and pathogenic characterization of Verticillium dahliae from sunflower reveal two differentiated groups in Europe. Front. Plant Sci. 2018, 9, 288. [Google Scholar] [CrossRef] [Green Version]
- Henrique, F.H.; Carbonell, S.A.M.; Ito, M.F.; Gonçalves, J.G.R.; Sasseron, G.R.; Chiorato, A.F. Classification of physiological races of Fusarium oxysporum f. sp. phaseoli in common bean. Bragantia 2015, 74, 84–92. [Google Scholar] [CrossRef]
- Liu, S.; Wu, B.; Shen, Z.; Li, R.; Ganjun, Y.; Li, C.; Guo, X. Genetic diversity in FUB genes of Fusarium oxysporum f. sp. cubense suggest horizontal gene transfer. Front. Plant Sci. 2019, 10, 1069. [Google Scholar] [CrossRef] [PubMed]
- Laurence, M.; Summerell, B.; Liew, E. Fusarium oxysporum f. sp. canariensis: Evidence for horizontal gene transfer of putative pathogenicity genes. Plant Pathol. 2015, 64, 1068–1075. [Google Scholar]
- Czislowski, E.; Fraser-Smith, S.; Zander, M.; O’Neill, W.T.; Meldrum, R.A.; Tran-Nguyen, L.T.; Batley, J.; Aitken, E.A. Investigation of the diversity of effector genes in the banana pathogen, Fusarium oxysporum f. sp. cubense, reveals evidence of horizontal gene transfer. Mol. Plant Pathol. 2018, 19, 1155–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pal, S.; Rao, E.S.; Thontadarya, R.; Chandran, N.K.; Sriram, S. First report on the occurrence of races 1 and 2 of Fusarium oxysporum f. sp. niveum infecting watermelon in India. Indian Phytopathol. 2020, 73, 793–796. [Google Scholar] [CrossRef]
- Boughalleb, N.; El-Mahjoub, M. Detection of races 0, 1 and 2 of Fusarium oxysporum f. sp. niveum and their distribution in the watermelon-growing regions of Tunisia. EPPO Bull. 2005, 35, 253–260. [Google Scholar] [CrossRef]
- Boughalleb-M’Hamdi, N.; Salem, I.B.; BenFradj, N.; Abad-Campus, P. Genetic diversity of Fusarium oxysporum f. sp. niveum responsible of watermelon Fusarium wilt in Tunisia and Spain. J. Phytopathol. Pest Manag. 2020, 7, 54–63. [Google Scholar]
- Rep, M.; Kistler, H.C. The genomic organization of plant pathogenicity in Fusarium species. Curr. Opin. Plant Biol. 2010, 13, 420–426. [Google Scholar] [CrossRef]
- Ma, L.-J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O’Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef] [Green Version]
- Taylor, A.; Vágány, V.; Jackson, A.C.; Harrison, R.J.; Rainoni, A.; Clarkson, J.P. Identification of pathogenicity-related genes in Fusarium oxysporum f. sp. cepae. Mol. Plant Pathol. 2016, 17, 1032–1047. [Google Scholar] [CrossRef] [Green Version]
- Rep, M.; Meijer, M.; Houterman, P.; Van Der Does, H.; Cornelissen, B. Fusarium oxysporum evades I-3-mediated resistance without altering the matching avirulence gene. Mol. Plant Microbe Interact. 2005, 18, 15–23. [Google Scholar] [CrossRef] [Green Version]
- van Dam, P.; de Sain, M.; Ter Horst, A.; van der Gragt, M.; Rep, M. Use of comparative genomics-based markers for discrimination of host specificity in Fusarium oxysporum. Appl. Environ. Microbiol. 2018, 84, e01868–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gawehns, F.; Houterman, P.; Ichou, F.A.; Michielse, C.; Hijdra, M.; Cornelissen, B.; Rep, M.; Takken, F. The Fusarium oxysporum effector Six6 contributes to virulence and suppresses I-2-mediated cell death. Mol. Plant Microbe Interact. 2014, 27, 336–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oelke, L.M.; Bosland, P.W.; Steiner, R. Differentiation of race specific resistance to Phytophthora root rot and foliar blight in Capsicum annuum. J. Am. Soc. Hortic. Sci. 2003, 128, 213–218. [Google Scholar] [CrossRef] [Green Version]
- Kashiwa, T.; Suzuki, T.; Sato, A.; Akai, K.; Teraoka, T.; Komatsu, K.; Arie, T. A new biotype of Fusarium oxysporum f. sp. lycopersici race 2 emerged by a transposon-driven mutation of avirulence gene AVR1. FEMS Microbiol. Lett. 2016, 363, fnw132. [Google Scholar] [CrossRef] [Green Version]
- Lievens, B.; Houterman, P.M.; Rep, M. Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbiol. Lett. 2009, 300, 201–215. [Google Scholar] [CrossRef] [Green Version]
| FON Race | Number of Isolates Tested Using Both Methods | Percentage (%) of Molecular Results Agree with the Bioassay Race Results |
|---|---|---|
| Race 1 | 29 | 89.65% |
| Race 2 | 41 | 80.49% |
| Race 3 | 23 | 60.87% |
| Assay | Primers | Sequence (5′–3′) | Product Size (bp) | Source |
|---|---|---|---|---|
| FON specific primer | Fon-1 | CGATTAGCGAAGACATTCACAAGACT | 174 | [19] |
| Fon-2 | ACGGTCAAGAAGATGCAGGGTAAAGGT | |||
| Race 2 differentiating primer | FONSIX6F | CGCTCTTATCGCATCAATCT | 453 | [3] |
| FONSIX6R | GGGTTGACTGAGGTCGTGGT | |||
| Race 3 differentiating primer | FNR3F | CGGCTTTCCTCTGTCAGATAGT | 511 | This study |
| FNR3R | TAGTGAGGTCCATGCCACGAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hudson, O.; Waliullah, S.; Fulton, J.C.; Ji, P.; Dufault, N.S.; Keinath, A.; Ali, M.E. Marker Development for Differentiation of Fusarium oxysporum f. sp. Niveum Race 3 from Races 1 and 2. Int. J. Mol. Sci. 2021, 22, 822. https://doi.org/10.3390/ijms22020822
Hudson O, Waliullah S, Fulton JC, Ji P, Dufault NS, Keinath A, Ali ME. Marker Development for Differentiation of Fusarium oxysporum f. sp. Niveum Race 3 from Races 1 and 2. International Journal of Molecular Sciences. 2021; 22(2):822. https://doi.org/10.3390/ijms22020822
Chicago/Turabian StyleHudson, Owen, Sumyya Waliullah, James C. Fulton, Pingsheng Ji, Nicholas S. Dufault, Anthony Keinath, and Md Emran Ali. 2021. "Marker Development for Differentiation of Fusarium oxysporum f. sp. Niveum Race 3 from Races 1 and 2" International Journal of Molecular Sciences 22, no. 2: 822. https://doi.org/10.3390/ijms22020822
APA StyleHudson, O., Waliullah, S., Fulton, J. C., Ji, P., Dufault, N. S., Keinath, A., & Ali, M. E. (2021). Marker Development for Differentiation of Fusarium oxysporum f. sp. Niveum Race 3 from Races 1 and 2. International Journal of Molecular Sciences, 22(2), 822. https://doi.org/10.3390/ijms22020822






