Optogenetic Stimulation of Prelimbic Pyramidal Neurons Maintains Fear Memories and Modulates Amygdala Pyramidal Neuron Transcriptome
Abstract
1. Introduction
2. Results
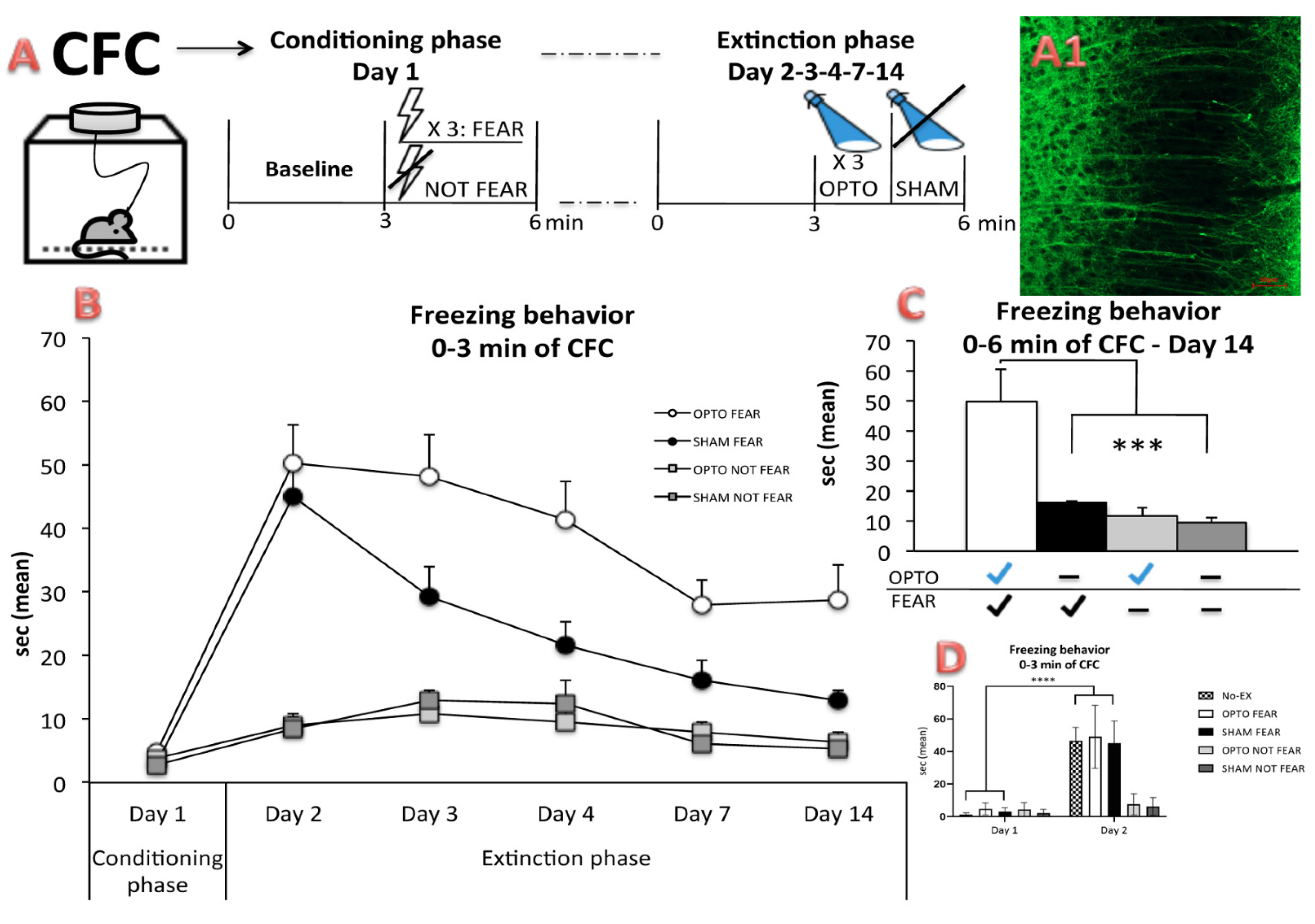
2.1. Behavioral Results: In Vivo Optogenetics of the PrL Pyramidal Neurons during CFC
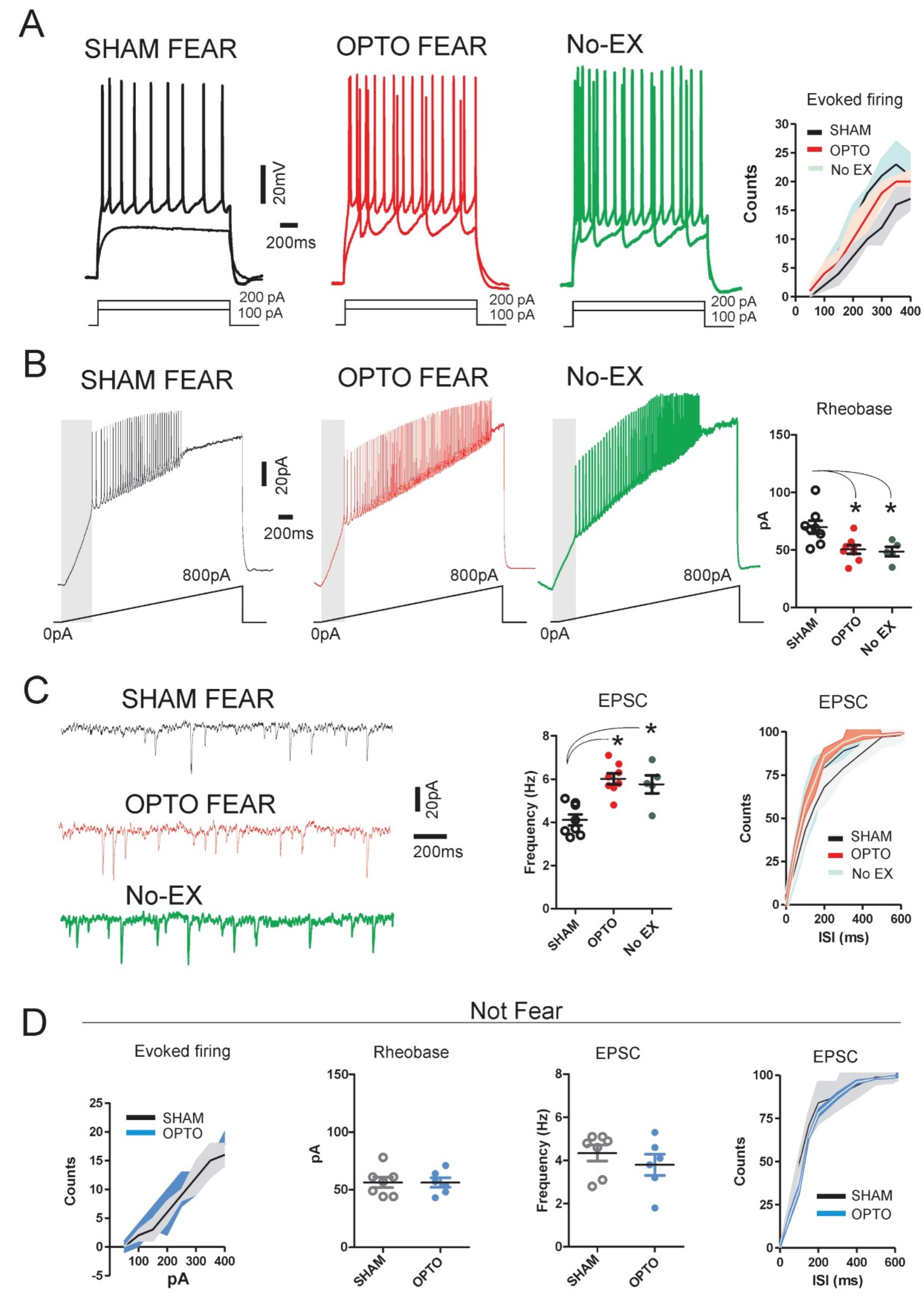
2.2. Electrophysiological Results: Cellular Excitability of PrL Pyramidal Neurons
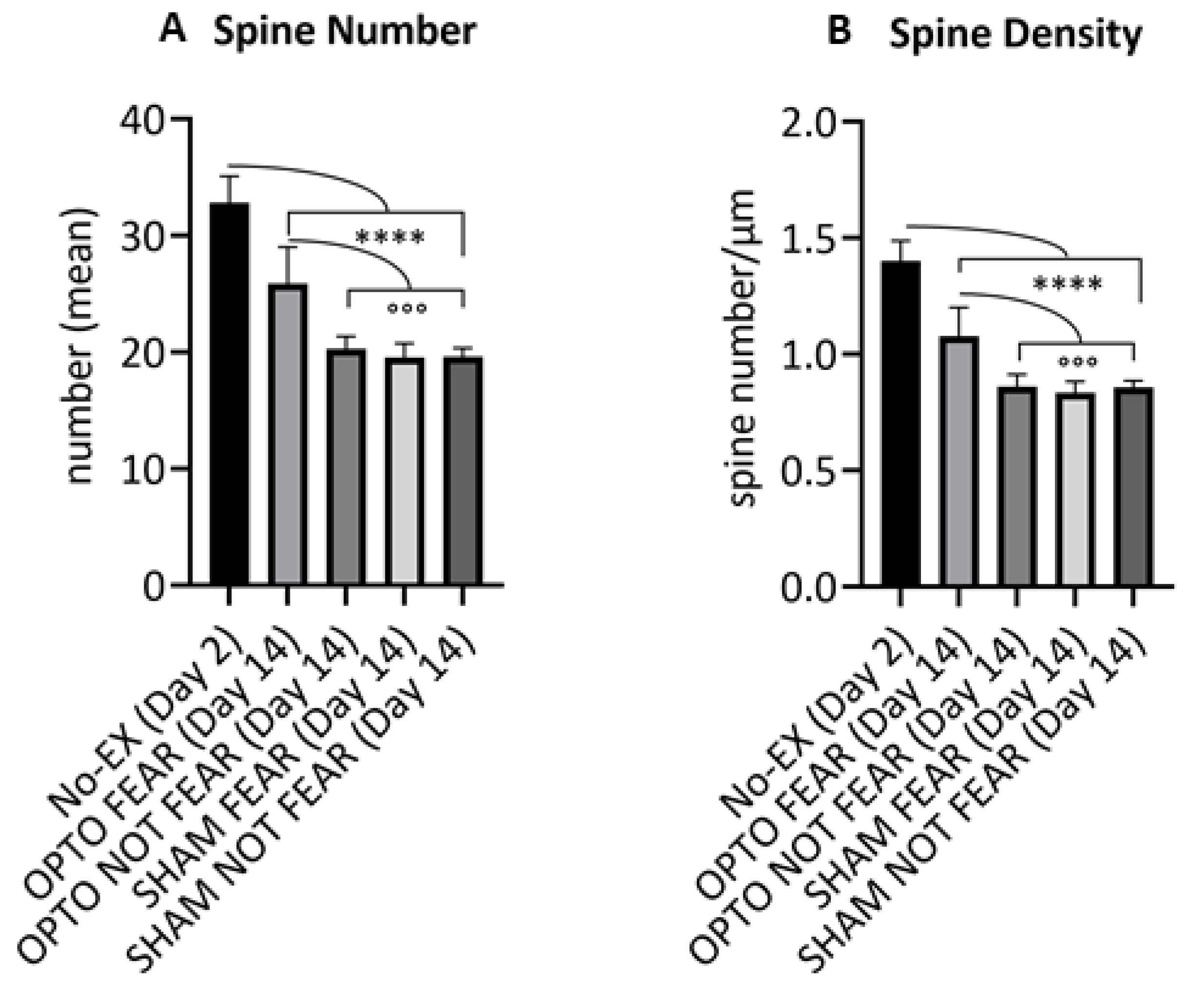
2.3. Morphological Results: Spine Counting of PrL Pyramidal Neurons
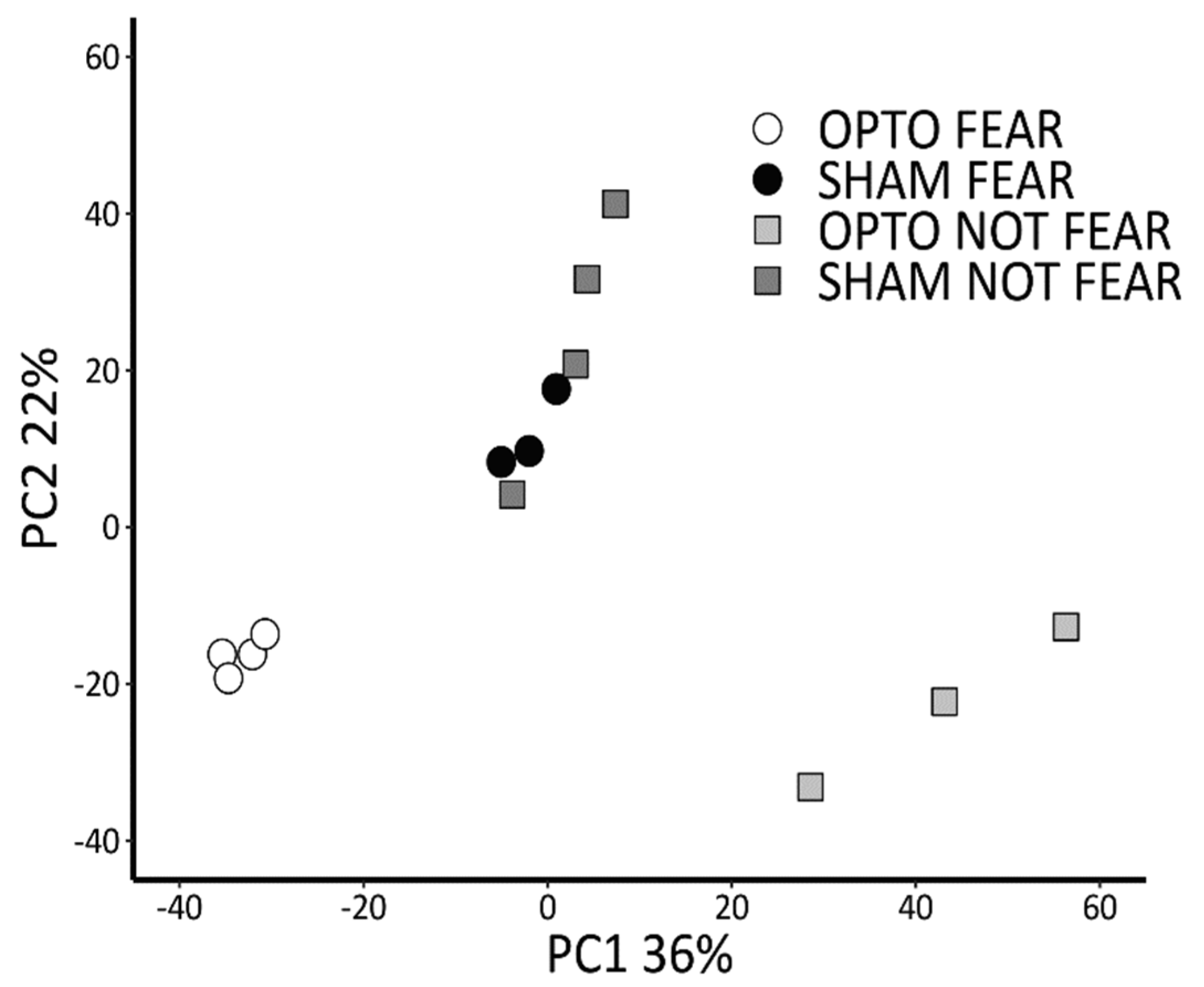
2.4. Transcriptomic Results: Differential Gene Expression Profiling and Functional Enrichment Analysis of RNA Extracted by Sorted Amygdala Pyramidal Neurons
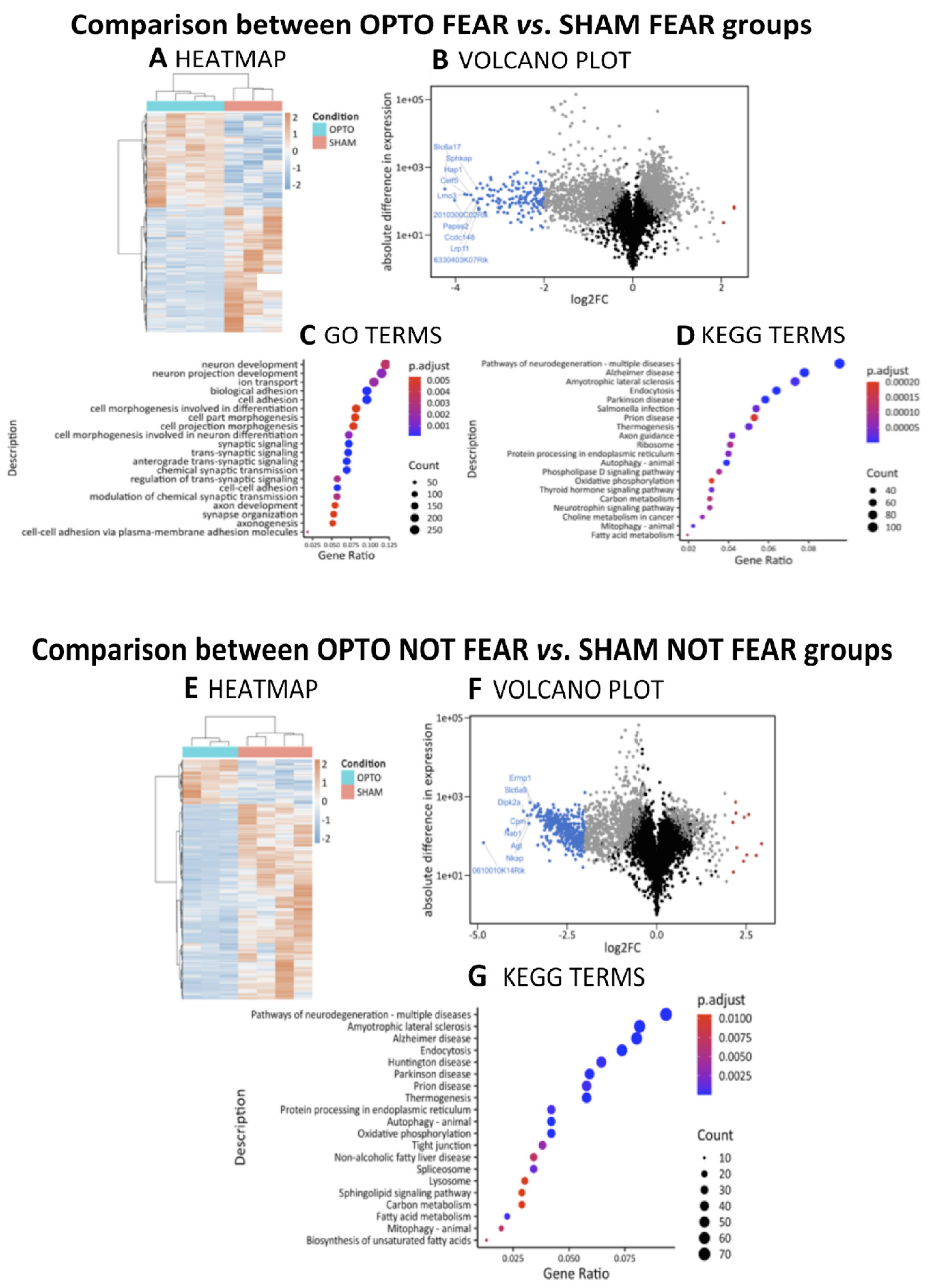
2.4.1. Comparison between OPTO FEAR vs. SHAM FEAR Groups
2.4.2. Comparison between OPTO NOT FEAR vs. SHAM NOT FEAR Groups
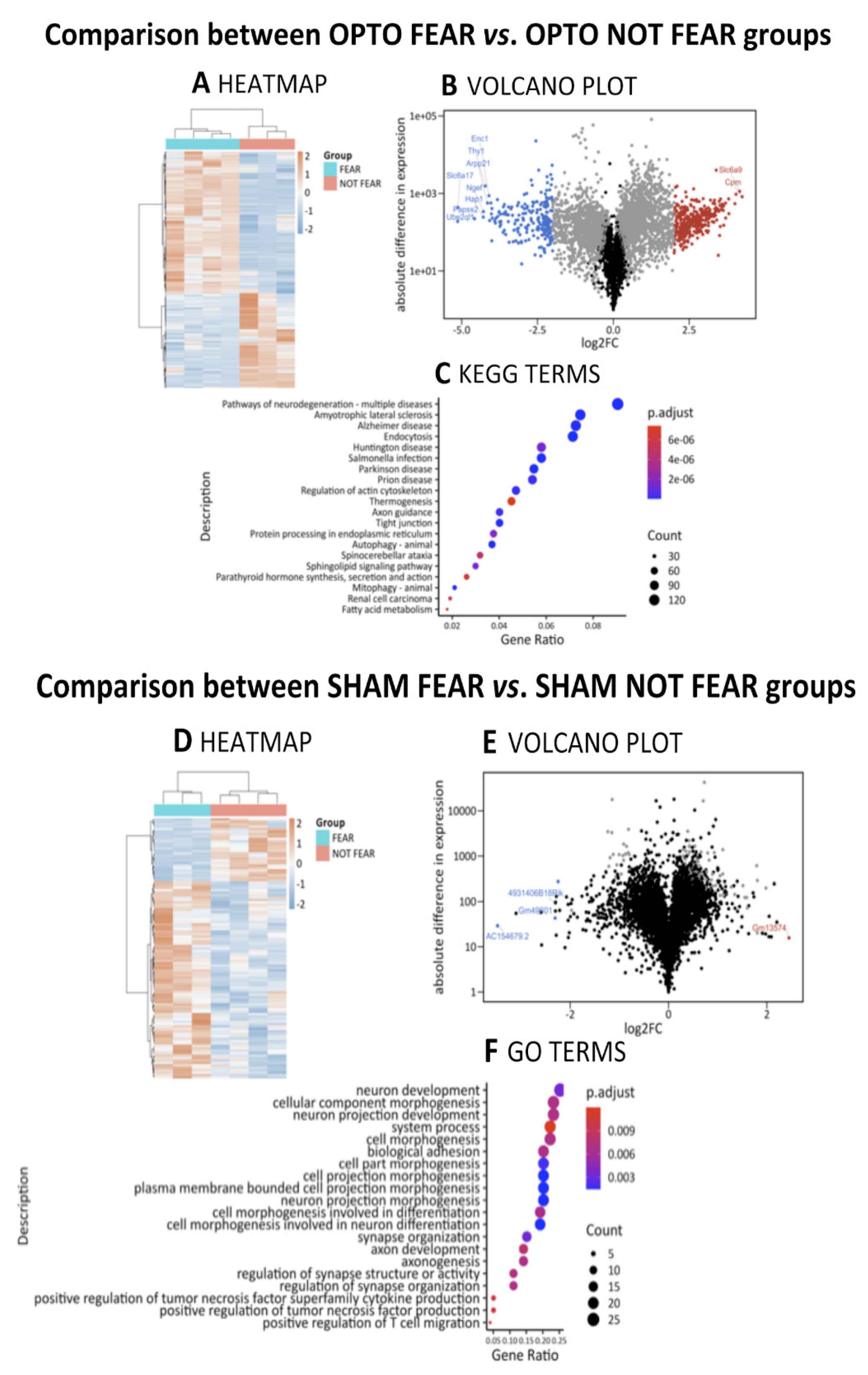
2.4.3. Comparison between OPTO FEAR vs. OPTO NOT FEAR Groups
2.4.4. Comparison between SHAM FEAR vs. SHAM NOT FEAR Groups
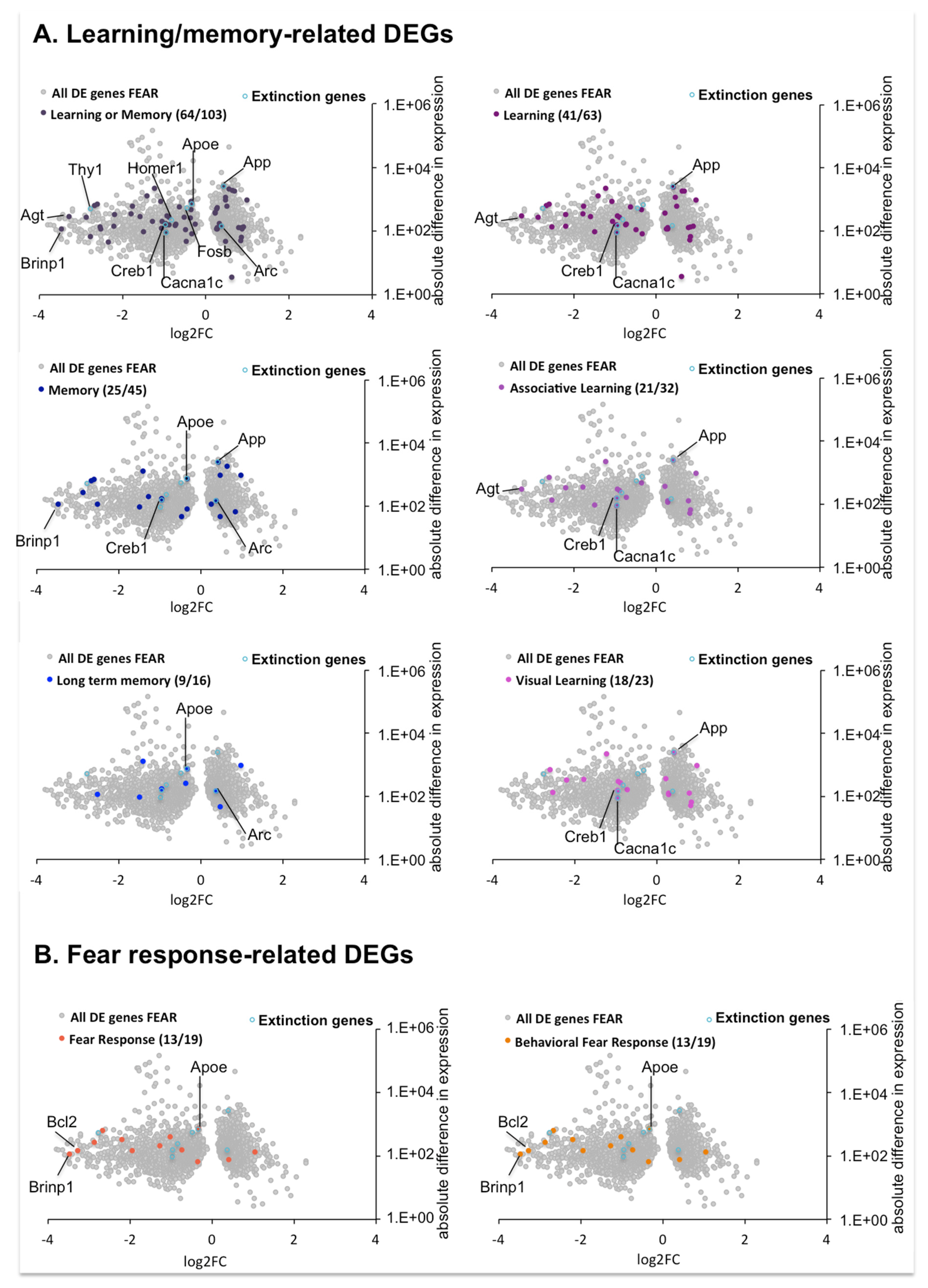
2.4.5. DEGs Involved in Learning/Memory and Fear Response in the Comparison between OPTO FEAR vs. SHAM FEAR Groups
3. Discussion
Limitations
4. Materials and Methods
4.1. Subjects
4.2. Experimental Procedure
4.3. Stereotaxic Surgery and Fiber Optic Implantations
4.4. CFC and In Vivo Optogenetic Stimulations of the PrL Pyramidal Neurons
4.5. Slice Preparation and Electrophysiological Recordings of PrL Pyramidal Neurons
4.6. Spine Counting of PrL Pyramidal Neurons
4.7. Amygdala Pyramidal Neuron-Specific RNA Sequencing
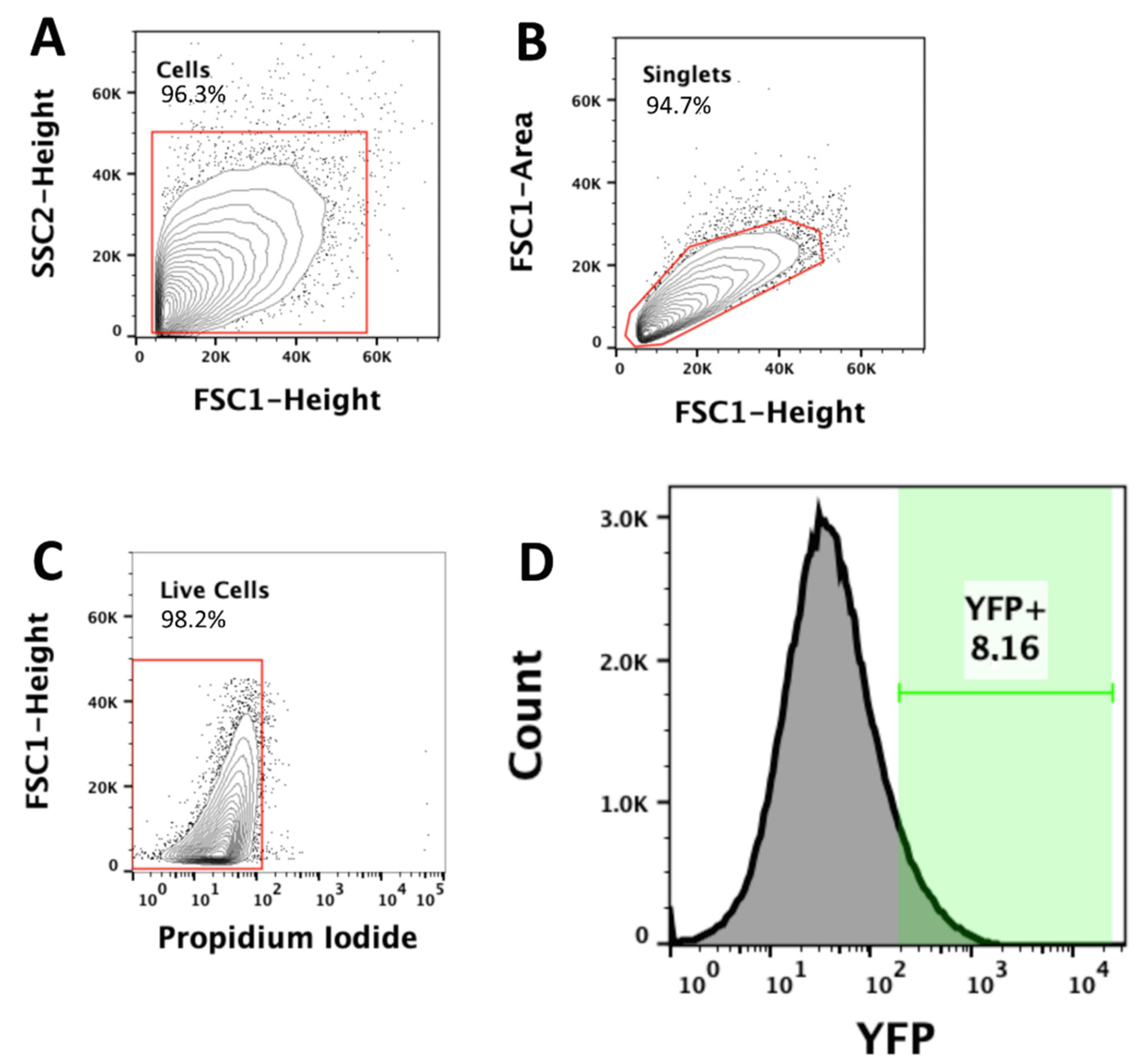
4.7.1. Dissociation of Amygdala Tissue for Fluorescence-Activated Cell Sorting (FACS)
4.7.2. Cell Sorting and Isolation of Purified Pyramidal Neurons
4.7.3. RNA-Seq Library Preparation
4.7.4. Analysis of RNA Sequencing Data
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maren, S.; Fanselow, M.S. The Amygdala and Fear Conditioning: Has the Nut Been Cracked? Neuron 1996, 16, 237–240. [Google Scholar]
- Davis, M. The Role of the Amygdala in Conditioned and Unconditioned Fear and Anxiety. Amygdala 2000, 2, 213–287. [Google Scholar]
- LeDoux, J.E. Emotion Circuits in the Brain. Annu. Rev. Neurosci. 2000, 23, 155–184. [Google Scholar] [PubMed]
- Fanselow, M.S.; Gale, G.D. The Amygdala, Fear, and Memory. Ann. N. Y. Acad. Sci. 2003, 985, 125–134. [Google Scholar] [PubMed]
- Holmes, A.; Singewald, N. Individual Differences in Recovery from Traumatic Fear. Trends Neurosci. 2013, 36, 23–31. [Google Scholar] [PubMed]
- Corcoran, K.A.; Quirk, G.J. Recalling Safety: Cooperative Functions of the Ventromedial Prefrontal Cortex and the Hippocampus in Extinction. CNS Spectr. 2007, 12, 200–206. [Google Scholar]
- Goshen, I.; Brodsky, M.; Prakash, R.; Wallace, J.; Gradinaru, V.; Ramakrishnan, C.; Deisseroth, K. Dynamics of Retrieval Strategies for Remote Memories. Cell 2011, 147, 678–689. [Google Scholar] [PubMed]
- Lacagnina, A.F.; Brockway, E.T.; Crovetti, C.R.; Shue, F.; McCarty, M.J.; Sattler, K.P.; Lim, S.C.; Santos, S.L.; Denny, C.A.; Drew, M.R. Distinct Hippocampal Engrams Control Extinction and Relapse of Fear Memory. Nat. Neurosci. 2019, 22, 753–761. [Google Scholar]
- Tovote, P.; Fadok, J.P.; Lüthi, A. Neuronal Circuits for Fear and Anxiety. Nat. Rev. Neurosci. 2015, 16, 317–331. [Google Scholar]
- Trouche, S.; Sasaki, J.M.; Tu, T.; Reijmers, L.G. Fear Extinction Causes Target-Specific Remodeling of Perisomatic Inhibitory Synapses. Neuron 2013, 80, 1054–1065. [Google Scholar]
- Zhu, H.; Pleil, K.E.; Urban, D.J.; Moy, S.S.; Kash, T.L.; Roth, B.L. Chemogenetic Inactivation of Ventral Hippocampal Glutamatergic Neurons Disrupts Consolidation of Contextual Fear Memory. Neuropsychopharmacology 2014, 39, 1880–1892. [Google Scholar] [PubMed]
- Johansen, J.P.; Hamanaka, H.; Monfils, M.H.; Behnia, R.; Deisseroth, K.; Blair, H.T.; LeDoux, J.E. Optical Activation of Lateral Amygdala Pyramidal Cells Instructs Associative Fear Learning. Proc. Natl. Acad. Sci. USA 2010, 107, 12692–12697. [Google Scholar]
- Tye, K.M.; Prakash, R.; Kim, S.-Y.; Fenno, L.E.; Grosenick, L.; Zarabi, H.; Thompson, K.R.; Gradinaru, V.; Ramakrishnan, C.; Deisseroth, K. Amygdala Circuitry Mediating Reversible and Bidirectional Control of Anxiety. Nature 2011, 471, 358–362. [Google Scholar]
- Klavir, O.; Prigge, M.; Sarel, A.; Paz, R.; Yizhar, O. Manipulating Fear Associations via Optogenetic Modulation of Amygdala Inputs to Prefrontal Cortex. Nat. Neurosci. 2017, 20, 836–844. [Google Scholar] [PubMed]
- Sierra-Mercado, D.; Padilla-Coreano, N.; Quirk, G.J. Dissociable Roles of Prelimbic and Infralimbic Cortices, Ventral Hippocampus, and Basolateral Amygdala in the Expression and Extinction of Conditioned Fear. Neuropsychopharmacology 2011, 36, 529–538. [Google Scholar] [PubMed]
- Beyeler, A.; Eckhardt, C.A.; Tye, K.M. Deciphering memory function with optogenetics. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2014; Volume 122, pp. 341–390. ISBN 1877-1173. [Google Scholar]
- Kim, H.-S.; Cho, H.-Y.; Augustine, G.J.; Han, J.-H. Selective Control of Fear Expression by Optogenetic Manipulation of Infralimbic Cortex after Extinction. Neuropsychopharmacology 2016, 41, 1261–1273. [Google Scholar] [PubMed]
- Hardt, O.; Nadel, L. Systems Consolidation Revisited, but Not Revised: The Promise and Limits of Optogenetics in the Study of Memory. Neurosci. Lett. 2018, 680, 54–59. [Google Scholar]
- Senn, V.; Wolff, S.B.; Herry, C.; Grenier, F.; Ehrlich, I.; Gründemann, J.; Fadok, J.P.; Müller, C.; Letzkus, J.J.; Lüthi, A. Long-Range Connectivity Defines Behavioral Specificity of Amygdala Neurons. Neuron 2014, 81, 428–437. [Google Scholar]
- Sotres-Bayon, F.; Sierra-Mercado, D.; Pardilla-Delgado, E.; Quirk, G.J. Gating of Fear in Prelimbic Cortex by Hippocampal and Amygdala Inputs. Neuron 2012, 76, 804–812. [Google Scholar]
- Cho, J.-H.; Deisseroth, K.; Bolshakov, V.Y. Synaptic Encoding of Fear Extinction in MPFC-Amygdala Circuits. Neuron 2013, 80, 1491–1507. [Google Scholar]
- Bredy, T.W.; Wu, H.; Crego, C.; Zellhoefer, J.; Sun, Y.E.; Barad, M. Histone Modifications around Individual BDNF Gene Promoters in Prefrontal Cortex Are Associated with Extinction of Conditioned Fear. Learn. Mem. 2007, 14, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Chwang, W.B.; O’Riordan, K.J.; Levenson, J.M.; Sweatt, J.D. ERK/MAPK Regulates Hippocampal Histone Phosphorylation Following Contextual Fear Conditioning. Learn. Mem. 2006, 13, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.A.; Sweatt, J.D. Covalent Modification of DNA Regulates Memory Formation. Neuron 2007, 53, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Itzhak, Y.; Anderson, K.L.; Kelley, J.B.; Petkov, M. Histone Acetylation Rescues Contextual Fear Conditioning in NNOS KO Mice and Accelerates Extinction of Cued Fear Conditioning in Wild Type Mice. Neurobiol. Learn. Mem. 2012, 97, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Monsey, M.S.; Ota, K.T.; Akingbade, I.F.; Hong, E.S.; Schafe, G.E. Epigenetic Alterations Are Critical for Fear Memory Consolidation and Synaptic Plasticity in the Lateral Amygdala. PLoS ONE 2011, 6, e19958. [Google Scholar] [CrossRef]
- Stafford, J.M.; Raybuck, J.D.; Ryabinin, A.E.; Lattal, K.M. Increasing Histone Acetylation in the Hippocampus-Infralimbic Network Enhances Fear Extinction. Biol. Psychiatry 2012, 72, 25–33. [Google Scholar] [CrossRef]
- Duke, C.G.; Kennedy, A.J.; Gavin, C.F.; Day, J.J.; Sweatt, J.D. Experience-Dependent Epigenomic Reorganization in the Hippocampus. Learn. Mem. 2017, 24, 278–288. [Google Scholar] [CrossRef]
- Gelernter, J.; Sun, N.; Polimanti, R.; Pietrzak, R.; Levey, D.F.; Bryois, J.; Lu, Q.; Hu, Y.; Li, B.; Radhakrishnan, K. Genome-Wide Association Study of Post-Traumatic Stress Disorder Reexperiencing Symptoms in >165,000 US Veterans. Nat. Neurosci. 2019, 22, 1394–1401. [Google Scholar] [CrossRef]
- Halder, R.; Hennion, M.; Vidal, R.O.; Shomroni, O.; Rahman, R.-U.; Rajput, A.; Centeno, T.P.; Van Bebber, F.; Capece, V.; Vizcaino, J.C.G. DNA Methylation Changes in Plasticity Genes Accompany the Formation and Maintenance of Memory. Nat. Neurosci. 2016, 19, 102–110. [Google Scholar] [CrossRef]
- Ryan, T.J.; Roy, D.S.; Pignatelli, M.; Arons, A.; Tonegawa, S. Engram Cells Retain Memory under Retrograde Amnesia. Science 2015, 348, 1007–1013. [Google Scholar] [CrossRef]
- Yehuda, R.; LeDoux, J. Response Variation Following Trauma: A Translational Neuroscience Approach to Understanding PTSD. Neuron 2007, 56, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Binder, E.B.; Bradley, R.G.; Liu, W.; Epstein, M.P.; Deveau, T.C.; Mercer, K.B.; Tang, Y.; Gillespie, C.F.; Heim, C.M.; Nemeroff, C.B. Association of FKBP5 Polymorphisms and Childhood Abuse with Risk of Posttraumatic Stress Disorder Symptoms in Adults. JAMA 2008, 299, 1291–1305. [Google Scholar] [CrossRef] [PubMed]
- Champagne, F.A. Epigenetic Mechanisms and the Transgenerational Effects of Maternal Care. Front. Neuroendocrinol. 2008, 29, 386–397. [Google Scholar] [CrossRef]
- Franklin, T.B.; Russig, H.; Weiss, I.C.; Gräff, J.; Linder, N.; Michalon, A.; Vizi, S.; Mansuy, I.M. Epigenetic Transmission of the Impact of Early Stress across Generations. Biol. Psychiatry 2010, 68, 408–415. [Google Scholar] [CrossRef]
- Koenen, K.C.; Uddin, M. FKBP5 Polymorphisms Modify the Effects of Childhood Trauma. Neuropsychopharmacology 2010, 35, 1623–1624. [Google Scholar] [CrossRef]
- Yehuda, R.; Bierer, L.M. The Relevance of Epigenetics to PTSD: Implications for the DSM-V. J. Trauma. Stress 2009, 22, 427–434. [Google Scholar] [CrossRef]
- Timmons, J.A.; Szkop, K.J.; Gallagher, I.J. Multiple Sources of Bias Confound Functional Enrichment Analysis of Global-Omics Data. Genome Biol. 2015, 16, 186. [Google Scholar] [CrossRef]
- Alberini, C.M.; LeDoux, J.E. Memory Reconsolidation. Curr. Biol. 2013, 23, R746–R750. [Google Scholar] [CrossRef]
- Vaverková, Z.; Milton, A.L.; Merlo, E. Retrieval-Dependent Mechanisms Affecting Emotional Memory Persistence: Reconsolidation, Extinction, and the Space in Between. Front. Behav. Neurosci. 2020, 14, 574358. [Google Scholar] [CrossRef]
- McCullough, K.M.; Chatzinakos, C.; Hartmann, J.; Missig, G.; Neve, R.L.; Fenster, R.J.; Carlezon, W.A.; Daskalakis, N.P.; Ressler, K.J. Genome-Wide Translational Profiling of Amygdala Crh-Expressing Neurons Reveals Role for CREB in Fear Extinction Learning. Nat. Commun. 2020, 11, 5180. [Google Scholar] [CrossRef]
- Shen, H.; Sabaliauskas, N.; Sherpa, A.; Fenton, A.A.; Stelzer, A.; Aoki, C.; Smith, S.S. A Critical Role for A4βδ GABAA Receptors in Shaping Learning Deficits at Puberty in Mice. Science 2010, 327, 1515–1518. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.; Forsythe, I.D.; Kopp-Scheinpflug, C. SYMPOSIUM REVIEW: Going Native: Voltage-gated Potassium Channels Controlling Neuronal Excitability. J. Physiol. 2010, 588, 3187–3200. [Google Scholar] [CrossRef] [PubMed]
- Miyake, A.; Mochizuki, S.; Yokoi, H.; Kohda, M.; Furuichi, K. New Ether-à-Go-Go K(+) Channel Family Members Localized in Human Telencephalon. J. Biol. Chem. 1999, 274, 25018–25025. [Google Scholar] [CrossRef] [PubMed]
- Kraus, D.M.; Elliott, G.S.; Chute, H.; Horan, T.; Pfenninger, K.H.; Sanford, S.D.; Foster, S.; Scully, S.; Welcher, A.A.; Holers, V.M. CSMD1 Is a Novel Multiple Domain Complement-Regulatory Protein Highly Expressed in the Central Nervous System and Epithelial Tissues. J. Immunol. 2006, 176, 4419–4430. [Google Scholar] [CrossRef] [PubMed]
- Sentman, C.L.; Shutter, J.R.; Hockenbery, D.; Kanagawa, O.; Korsmeyer, S.J. Bcl-2 Inhibits Multiple Forms of Apoptosis but Not Negative Selection in Thymocytes. Cell 1991, 67, 879–888. [Google Scholar] [CrossRef]
- Williams, G.T. Programmed Cell Death: Apoptosis and Oncogenesis. Cell 1991, 65, 1097–1098. [Google Scholar] [CrossRef]
- Farlie, P.G.; Dringen, R.; Rees, S.M.; Kannourakis, G.; Bernard, O. Bcl-2 Transgene Expression Can Protect Neurons against Developmental and Induced Cell Death. Proc. Natl. Acad. Sci. USA 1995, 92, 4397–4401. [Google Scholar] [CrossRef]
- Clifton, N.E.; Cameron, D.; Trent, S.; Sykes, L.H.; Thomas, K.L.; Hall, J. Hippocampal Regulation of Postsynaptic Density Homer1 by Associative Learning. Neural Plast. 2017, 2017, 5959182. [Google Scholar] [CrossRef]
- Olsen, R.H.; Agam, M.; Davis, M.J.; Raber, J. ApoE Isoform-dependent Deficits in Extinction of Contextual Fear Conditioning. Genes Brain Behav. 2012, 11, 806–812. [Google Scholar] [CrossRef]
- Pardon, M.-C.; Sarmad, S.; Rattray, I.; Bates, T.E.; Scullion, G.A.; Marsden, C.A.; Barrett, D.A.; Lowe, J.; Kendall, D.A. Repeated Novel Cage Exposure-Induced Improvement of Early Alzheimer’s-like Cognitive and Amyloid Changes in TASTPM Mice Is Unrelated to Changes in Brain Endocannabinoids Levels. Neurobiol. Aging 2009, 30, 1099–1113. [Google Scholar] [CrossRef]
- Rattray, I.; Scullion, G.A.; Soulby, A.; Kendall, D.A.; Pardon, M.-C. The Occurrence of a Deficit in Contextual Fear Extinction in Adult Amyloid-over-Expressing TASTPM Mice Is Independent of the Strength of Conditioning but Can Be Prevented by Mild Novel Cage Stress. Behav. Brain Res. 2009, 200, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Temme, S.J.; Murphy, G.G. The L-Type Voltage-Gated Calcium Channel CaV1.2 Mediates Fear Extinction and Modulates Synaptic Tone in the Lateral Amygdala. Learn. Mem. 2017, 24, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Trent, S.; Barnes, P.; Hall, J.; Thomas, K.L. AMPA Receptors Control Fear Extinction through an Arc-Dependent Mechanism. Learn. Mem. 2017, 24, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Shamah, S.M.; Lin, M.Z.; Goldberg, J.L.; Estrach, S.; Sahin, M.; Hu, L.; Bazalakova, M.; Neve, R.L.; Corfas, G.; Debant, A.; et al. EphA Receptors Regulate Growth Cone Dynamics through the Novel Guanine Nucleotide Exchange Factor Ephexin. Cell 2001, 105, 233–244. [Google Scholar] [CrossRef]
- Fu, W.-Y.; Chen, Y.; Sahin, M.; Zhao, X.-S.; Shi, L.; Bikoff, J.B.; Lai, K.-O.; Yung, W.-H.; Fu, A.K.; Greenberg, M.E. Cdk5 Regulates EphA4-Mediated Dendritic Spine Retraction through an Ephexin1-Dependent Mechanism. Nat. Neurosci. 2007, 10, 67–76. [Google Scholar] [CrossRef]
- Frank, C.A.; Pielage, J.; Davis, G.W. A Presynaptic Homeostatic Signaling System Composed of the Eph Receptor, Ephexin, Cdc42, and CaV2.1 Calcium Channels. Neuron 2009, 61, 556–569. [Google Scholar] [CrossRef]
- Alapin, J.M.; Dines, M.; Vassiliev, M.; Tamir, T.; Ram, A.; Locke, C.; Yu, J.; Lamprecht, R. Activation of EphB2 Forward Signaling Enhances Memory Consolidation. Cell Rep. 2018, 23, 2014–2025. [Google Scholar] [CrossRef]
- Kim, T.A.; Lim, J.; Ota, S.; Raja, S.; Rogers, R.; Rivnay, B.; Avraham, H.; Avraham, S. NRP/B, a Novel Nuclear Matrix Protein, Associates with P110(RB) and Is Involved in Neuronal Differentiation. J. Cell Biol. 1998, 141, 553–566. [Google Scholar] [CrossRef]
- Cooper, E.C.; Aldape, K.D.; Abosch, A.; Barbaro, N.M.; Berger, M.S.; Peacock, W.S.; Jan, Y.N.; Jan, L.Y. Colocalization and Coassembly of Two Human Brain M-Type Potassium Channel Subunits That Are Mutated in Epilepsy. Proc. Natl. Acad. Sci. USA 2000, 97, 4914–4919. [Google Scholar] [CrossRef]
- Jiang, L.; Kundu, S.; Lederman, J.D.; López-Hernández, G.Y.; Ballinger, E.C.; Wang, S.; Talmage, D.A.; Role, L.W. Cholinergic Signaling Controls Conditioned Fear Behaviors and Enhances Plasticity of Cortical-Amygdala Circuits. Neuron 2016, 90, 1057–1070. [Google Scholar] [CrossRef]
- Kowalewski, B.; Lamanna, W.C.; Lawrence, R.; Damme, M.; Stroobants, S.; Padva, M.; Kalus, I.; Frese, M.-A.; Lübke, T.; Lüllmann-Rauch, R.; et al. Arylsulfatase G Inactivation Causes Loss of Heparan Sulfate 3-O-Sulfatase Activity and Mucopolysaccharidosis in Mice. Proc. Natl. Acad. Sci. USA 2012, 109, 10310–10315. [Google Scholar] [CrossRef] [PubMed]
- Burgute, B.D.; Peche, V.S.; Steckelberg, A.-L.; Glöckner, G.; Gaßen, B.; Gehring, N.H.; Noegel, A.A. NKAP Is a Novel RS-Related Protein That Interacts with RNA and RNA Binding Proteins. Nucleic Acids Res. 2014, 42, 3177–3193. [Google Scholar] [CrossRef] [PubMed]
- Lingawi, N.W.; Holmes, N.M.; Westbrook, R.F.; Laurent, V. The Infralimbic Cortex Encodes Inhibition Irrespective of Motivational Significance. Neurobiol. Learn. Mem. 2018, 150, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Rothenhoefer, K.M.; Stauffer, W.R. Dopamine Signals Learn New Tricks. Neuron 2020, 106, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Bocchio, M.; Nabavi, S.; Capogna, M. Synaptic Plasticity, Engrams, and Network Oscillations in Amygdala Circuits for Storage and Retrieval of Emotional Memories. Neuron 2017, 94, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Iakoubova, O.A.; Shiffman, D.; Devlin, J.J.; Forrester, J.S.; Superko, H.R. KIF6 Polymorphism as a Predictor of Risk of Coronary Events and of Clinical Event Reduction by Statin Therapy. Am. J. Cardiol. 2010, 106, 994–998. [Google Scholar] [CrossRef] [PubMed]
- Svendsen, J.M.; Smogorzewska, A.; Sowa, M.E.; O’Connell, B.C.; Gygi, S.P.; Elledge, S.J.; Harper, J.W. Mammalian BTBD12/SLX4 Assembles a Holliday Junction Resolvase and Is Required for DNA Repair. Cell 2009, 138, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Min, J.-N.; Tian, Y.; Xiao, Y.; Wu, L.; Li, L.; Chang, S. The MINO80 Chromatin Remodeling Complex Is Required for Efficient Telomere Replication and Maintenance of Genome Stability. Cell Res. 2013, 23, 1396–1413. [Google Scholar] [CrossRef]
- Ressler, K.J.; Mercer, K.B.; Bradley, B.; Jovanovic, T.; Mahan, A.; Kerley, K.; Norrholm, S.D.; Kilaru, V.; Smith, A.K.; Myers, A.J.; et al. Post-Traumatic Stress Disorder Is Associated with PACAP and the PAC1 Receptor. Nature 2011, 470, 492–497. [Google Scholar] [CrossRef]
- Sananbenesi, F.; Fischer, A.; Wang, X.; Schrick, C.; Neve, R.; Radulovic, J.; Tsai, L.-H. A Hippocampal Cdk5 Pathway Regulates Extinction of Contextual Fear. Nat. Neurosci. 2007, 10, 1012–1019. [Google Scholar] [CrossRef]
- Laricchiuta, D.; Centonze, D.; Petrosini, L. Effects of Endocannabinoid and Endovanilloid Systems on Aversive Memory Extinction. Behav. Brain Res. 2013, 256, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Laricchiuta, D.; Saba, L.; De Bartolo, P.; Caioli, S.; Zona, C.; Petrosini, L. Maintenance of Aversive Memories Shown by Fear Extinction-Impaired Phenotypes Is Associated with Increased Activity in the Amygdaloid-Prefrontal Circuit. Sci. Rep. 2016, 6, 21205. [Google Scholar] [CrossRef] [PubMed]
- Jasnow, A.M.; Ehrlich, D.E.; Choi, D.C.; Dabrowska, J.; Bowers, M.E.; McCullough, K.M.; Rainnie, D.G.; Ressler, K.J. Thy1-Expressing Neurons in the Basolateral Amygdala May Mediate Fear Inhibition. J. Neurosci. 2013, 33, 10396–10404. [Google Scholar] [CrossRef]
- Franklin, K.B.J.; Paxinos, G. The Mouse Brain in Stereotaxic Coordinates; Academic Press: Cambridge, MA, USA, 1997; p. 186. [Google Scholar]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential Analyses for RNA-Seq: Transcript-Level Estimates Improve Gene-Level Inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J. Bioconductor: Open Software Development for Computational Biology and Bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef]
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Pietro, A.D.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data Quality Aware Analysis of Differential Expression in RNA-Seq with NOISeq R/Bioc Package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A Scaling Normalization Method for Differential Expression Analysis of RNA-Seq Data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- Nueda, M.J.; Ferrer, A.; Conesa, A. ARSyN: A Method for the Identification and Removal of Systematic Noise in Multifactorial Time Course Microarray Experiments. Biostatistics 2012, 13, 553–566. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Merico, D.; Isserlin, R.; Stueker, O.; Emili, A.; Bader, G.D. Enrichment Map: A Network-Based Method for Gene-Set Enrichment Visualization and Interpretation. PLoS ONE 2010, 5, e13984. [Google Scholar] [CrossRef]
| Gene Symbol | Gene Name | Fold Change (log2 Scale) | OPTO FEAR adj.mean | SHAM FEAR adj.mean |
|---|---|---|---|---|
| Slc6a17 | solute carrier family 6 (neurotransmitter transporter), member 17 | −4.24 | 12.86 | 242.95 |
| 2010300C02Rik | RIKEN cDNA 2010300C02 gene | −4.02 | 6.95 | 113.03 |
| Lmo3 | LIM domain only 3 | −3.80 | 12.43 | 172.75 |
| Celf5 | CUGBP, Elav-like family member 5 | −3.74 | 12.58 | 168.18 |
| Hap1 | huntingtin-associated protein 1 | −3.65 | 13.22 | 166.13 |
| Papss2 | 3′-phosphoadenosine 5′-phosphosulfate synthase 2 | −3.60 | 9.79 | 118.38 |
| Sphkap | SPHK1 interactor, AKAP domain containing | −3.52 | 23.12 | 265.56 |
| Ccdc148 | coiled-coil domain containing 148 | −3.51 | 9.18 | 104.53 |
| Lrp11 | low density lipoprotein receptor-related protein 11 | −3.50 | 8.59 | 97.11 |
| 6330403K07Rik | RIKEN cDNA 6330403K07 gene | −3.48 | 6.08 | 67.76 |
| Ube2ql1 | ubiquitin-conjugating enzyme E2Q family-like 1 | −3.47 | 5.54 | 61.52 |
| Brinp1 | bone morphogenic protein/retinoic acid inducible neural specific 1 | −3.47 | 11.69 | 129.49 |
| Arpp21 | cyclic AMP-regulated phosphoprotein, 21 | −3.44 | 33.80 | 366.38 |
| Gabra4 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 | −3.41 | 12.24 | 130.15 |
| Csmd1 | CUB and Sushi multiple domains 1 | −3.39 | 25.79 | 269.68 |
| Kcnh3 | potassium voltage-gated channel, subfamily H (eag-related), member 3 | −3.34 | 5.02 | 50.67 |
| Agt | angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | −3.28 | 33.63 | 327.00 |
| St6gal2 | beta galactoside alpha 2,6 sialyltransferase 2 | −3.28 | 9.03 | 87.47 |
| Bcl2 | B cell leukemia/lymphoma 2 | −3.27 | 17.48 | 169.00 |
| Cbarp | calcium channel, voltage-dependent, beta subunit associated regulatory protein | −3.23 | 16.69 | 156.53 |
| Gene Symbol | Gene Name | Fold Change (log2 Scale) | OPTO NOT FEAR adj.mean | SHAM NOT FEAR adj.mean |
|---|---|---|---|---|
| 0610010K14Rik | RIKEN cDNA 0610010K14 gene | −4.83 | 2.44 | 69.50 |
| Gm10163 | predicted pseudogene 10163 | −4.15 | 8.60 | 153.17 |
| Dipk2a | divergent protein kinase domain 2A | −3.72 | 34.80 | 457.61 |
| Nab1 | Ngfi-A binding protein 1 | −3.61 | 30.00 | 366.69 |
| Nkap | NFKB activating protein | −3.56 | 19.30 | 227.98 |
| Slc6a9 | solute carrier family 6 (neurotransmitter transporter, glycine), member 9 | −3.53 | 67.13 | 778.02 |
| Agt | angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | −3.51 | 33.15 | 378.72 |
| Cpm | carboxypeptidase M | −3.47 | 45.96 | 509.16 |
| Ermp1 | endoplasmic reticulum metallopeptidase 1 | −3.40 | 61.64 | 652.31 |
| Gm15500 | predicted pseudogene 15500 | −3.40 | 54.43 | 573.54 |
| Dazap2 | DAZ associated protein 2 | −3.36 | 59.16 | 608.47 |
| Rtl8b | retrotransposon Gag like 8B | −3.35 | 33.48 | 340.80 |
| Polm | polymerase (DNA directed), mu | −3.32 | 6.50 | 64.73 |
| Gpx4 | glutathione peroxidase 4 | −3.31 | 39.34 | 391.42 |
| Sos1 | SOS Ras/Rac guanine nucleotide exchange factor 1 | −3.31 | 73.41 | 727.62 |
| Ctr9 | CTR9 homolog, Paf1/RNA polymerase II complex component | −3.30 | 67.94 | 667.74 |
| Tmem184c | transmembrane protein 184C | −3.29 | 47.58 | 464.32 |
| Dzank1 | double zinc ribbon and ankyrin repeat domains 1 | −3.27 | 36.48 | 352.91 |
| D430019H16Rik | RIKEN cDNA D430019H16 gene | −3.27 | 35.99 | 346.81 |
| Klhl9 | kelch-like 9 | −3.24 | 57.35 | 543.23 |
| Gene Symbol | Gene Name | Fold Change (log2 Scale) | OPTO FEAR adj.mean | OPTO NOT FEAR adj.mean |
|---|---|---|---|---|
| Ube2ql1 | ubiquitin-conjugating enzyme E2Q family-like 1 | −5.13 | 5.54 | 194.27 |
| Slc6a17 | solute carrier family 6 (neurotransmitter transporter), member 17 | −5.12 | 12.86 | 448.21 |
| Papss2 | 3′-phosphoadenosine 5′-phosphosulfate synthase 2 | −4.57 | 9.79 | 232.51 |
| Hap1 | huntingtin-associated protein 1 | −4.29 | 13.22 | 258.18 |
| Ngef | neuronal guanine nucleotide exchange factor | −4.28 | 18.41 | 358.22 |
| Cpm | carboxypeptidase M | 4.24 | 871.36 | 45.96 |
| Thy1 | thymus cell antigen 1, theta | −4.22 | 88.57 | 1649.90 |
| Slc6a9 | solute carrier family 6 (neurotransmitter transporter, glycine), member 9 | 4.16 | 1202.20 | 67.13 |
| Arpp21 | cyclic AMP-regulated phosphoprotein, 21 | −4.15 | 33.80 | 599.42 |
| Enc1 | ectodermal-neural cortex 1 | −4.10 | 54.26 | 928.57 |
| Celf5 | CUGBP, Elav-like family member 5 | −4.08 | 12.58 | 212.32 |
| Ermp1 | endoplasmic reticulum metallopeptidase 1 | 4.04 | 1016.54 | 61.64 |
| Ptprg | protein tyrosine phosphatase, receptor type, G | −4.04 | 36.77 | 603.55 |
| Cxcl14 | chemokine (C-X-C motif) ligand 14 | −4.02 | 19.49 | 315.59 |
| Chrna4 | cholinergic receptor, nicotinic, alpha polypeptide 4 | −4.01 | 3.93 | 63.10 |
| Arsg | arylsulfatase G | 4.01 | 580.93 | 36.15 |
| Kcnq2 | potassium voltage-gated channel, subfamily Q, member 2 | −4.00 | 27.21 | 436.00 |
| Sfxn1 | sideroflexin 1 | −3.99 | 15.50 | 246.04 |
| Thrb | thyroid hormone receptor beta | −3.91 | 13.85 | 208.27 |
| Lmo3 | LIM domain only 3 | −3.89 | 12.43 | 183.70 |
| Gene Symbol | Gene Name | Fold Change (log2 Scale) | SHAM FEAR adj.mean | SHAM NOT FEAR adj.mean |
|---|---|---|---|---|
| Gm13574 | predicted gene 13574 | −3.48 | 2.78 | 30.95 |
| Gm49601 | predicted gene, 49601 | 2.45 | 17.99 | 3.30 |
| 4931406B18Rik | RIKEN cDNA 4931406B18 gene | −2.31 | 10.68 | 52.82 |
| D430019H16Rik | RIKEN cDNA D430019H16 gene | −2.30 | 23.45 | 115.51 |
| Sphkap | SPHK1 interactor, AKAP domain containing | −2.25 | 73.14 | 346.81 |
| Slx4ip | SLX4 interacting protein | 1.98 | 265.56 | 67.29 |
| Smcr8 | Smith-Magenis syndrome chromosome region, candidate 8 homolog (human) | −1.97 | 21.56 | 84.55 |
| Olfr1233 | olfactory receptor 1233 | 1.81 | 403.22 | 115.33 |
| Hcn2 | hyperpolarization-activated, cyclic nucleotide-gated K+ 2 | 1.80 | 27.04 | 7.74 |
| Sumf1 | sulfatase modifying factor 1 | 1.79 | 859.93 | 248.23 |
| Cd99l2 | CD99 antigen-like 2 | 1.60 | 106.02 | 34.94 |
| Kif6 | kinesin family member 6 | 1.59 | 182.58 | 60.68 |
| Plaa | phospholipase A2, activating protein | 1.55 | 185.44 | 63.29 |
| Clstn3 | calsyntenin 3 | 1.51 | 344.56 | 120.72 |
| Adcyap1r1 | adenylate cyclase activating polypeptide 1 receptor 1 | 1.35 | 235.84 | 92.81 |
| Ino80 | INO80 complex subunit | −1.28 | 326.26 | 789.84 |
| Cpe | carboxypeptidase E | 1.27 | 359.63 | 148.70 |
| Uty | ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome | −1.23 | 2559.64 | 6021.69 |
| Vmn2r114 | vomeronasal 2, receptor 114 | −1.21 | 448.82 | 1040.79 |
| Gm13574 | predicted gene 13,574 | −1.19 | 720.62 | 1641.20 |
| Conditioning | PrL Stimulation | Sample Name | RNA (ng) | Library (ng) | Reads (M) |
|---|---|---|---|---|---|
| FEAR | OPTO | ID_FO1 | 0.540 | 0.270 | 33.954 |
| FEAR | OPTO | ID_FO2 | 0.846 | 0.423 | 38.971 |
| FEAR | OPTO | ID_FO3 | 0.225 | 0.112 | 40.455 |
| FEAR | OPTO | ID_FO4 | 0.410 | 0.205 | 34.162 |
| FEAR | SHAM | ID_FS1 | 0.140 | 0.070 | 34.804 |
| FEAR | SHAM | ID_FS2 | 2.760 | 1.380 | 37.340 |
| FEAR | SHAM | ID_FS3 | 0.640 | 0.320 | 30.547 |
| NOT FEAR | OPTO | ID_NF01 | 1.770 | 0.885 | 30.483 |
| NOT FEAR | OPTO | ID_NF02 | 0.240 | 0.120 | 32.432 |
| NOT FEAR | OPTO | ID_NF03 | 0.510 | 0.255 | 34.756 |
| NOT FEAR | SHAM | ID_NFS1 | 0.360 | 0.170 | 31.432 |
| NOT FEAR | SHAM | ID_NFS2 | 0.520 | 0.260 | 30.438 |
| NOT FEAR | SHAM | ID_NFS3 | 0.110 | 0.055 | 36.824 |
| NOT FEAR | SHAM | ID_NFS4 | 1.060 | 0.530 | 54.741 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laricchiuta, D.; Sciamanna, G.; Gimenez, J.; Termine, A.; Fabrizio, C.; Caioli, S.; Balsamo, F.; Panuccio, A.; De Bardi, M.; Saba, L.; et al. Optogenetic Stimulation of Prelimbic Pyramidal Neurons Maintains Fear Memories and Modulates Amygdala Pyramidal Neuron Transcriptome. Int. J. Mol. Sci. 2021, 22, 810. https://doi.org/10.3390/ijms22020810
Laricchiuta D, Sciamanna G, Gimenez J, Termine A, Fabrizio C, Caioli S, Balsamo F, Panuccio A, De Bardi M, Saba L, et al. Optogenetic Stimulation of Prelimbic Pyramidal Neurons Maintains Fear Memories and Modulates Amygdala Pyramidal Neuron Transcriptome. International Journal of Molecular Sciences. 2021; 22(2):810. https://doi.org/10.3390/ijms22020810
Chicago/Turabian StyleLaricchiuta, Daniela, Giuseppe Sciamanna, Juliette Gimenez, Andrea Termine, Carlo Fabrizio, Silvia Caioli, Francesca Balsamo, Anna Panuccio, Marco De Bardi, Luana Saba, and et al. 2021. "Optogenetic Stimulation of Prelimbic Pyramidal Neurons Maintains Fear Memories and Modulates Amygdala Pyramidal Neuron Transcriptome" International Journal of Molecular Sciences 22, no. 2: 810. https://doi.org/10.3390/ijms22020810
APA StyleLaricchiuta, D., Sciamanna, G., Gimenez, J., Termine, A., Fabrizio, C., Caioli, S., Balsamo, F., Panuccio, A., De Bardi, M., Saba, L., Passarello, N., Cutuli, D., Mattioni, A., Zona, C., Orlando, V., & Petrosini, L. (2021). Optogenetic Stimulation of Prelimbic Pyramidal Neurons Maintains Fear Memories and Modulates Amygdala Pyramidal Neuron Transcriptome. International Journal of Molecular Sciences, 22(2), 810. https://doi.org/10.3390/ijms22020810








