Targeting the Ubiquitin Signaling Cascade in Tumor Microenvironment for Cancer Therapy
Abstract
1. Introduction
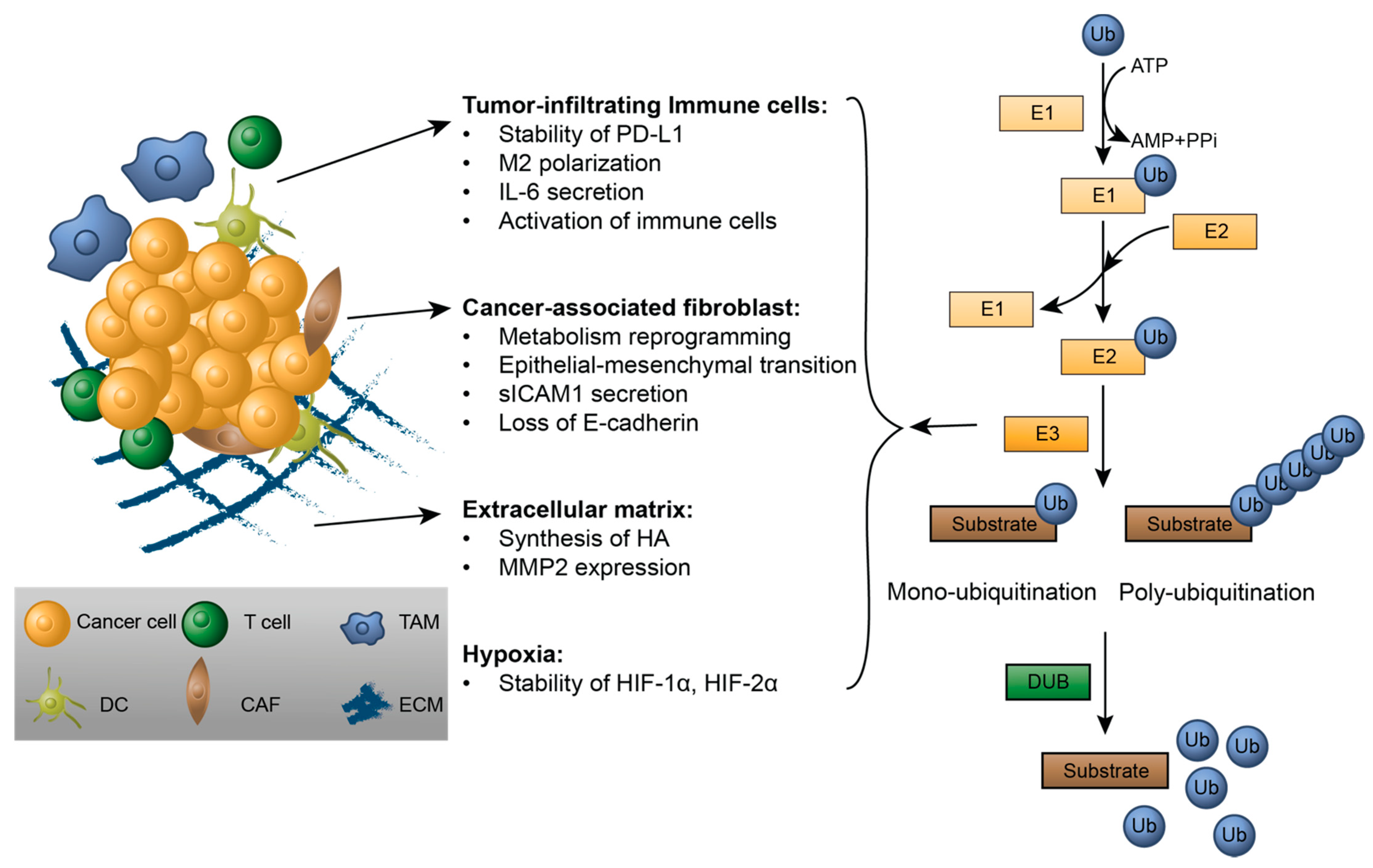
2. Modulating Tumor Microenvironments by Ubiquitination
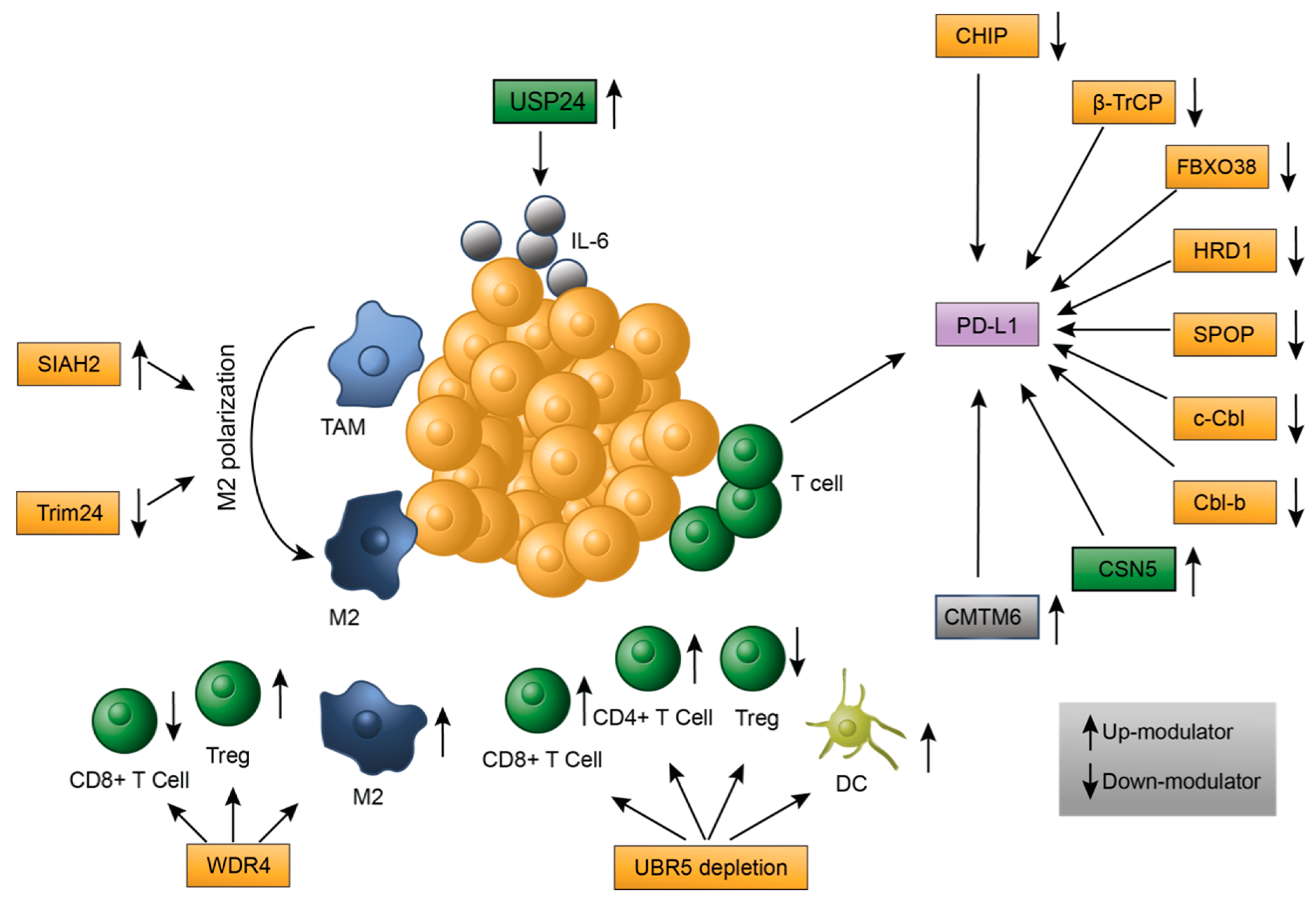
2.1. Tumor-Infiltrating Immune Cells
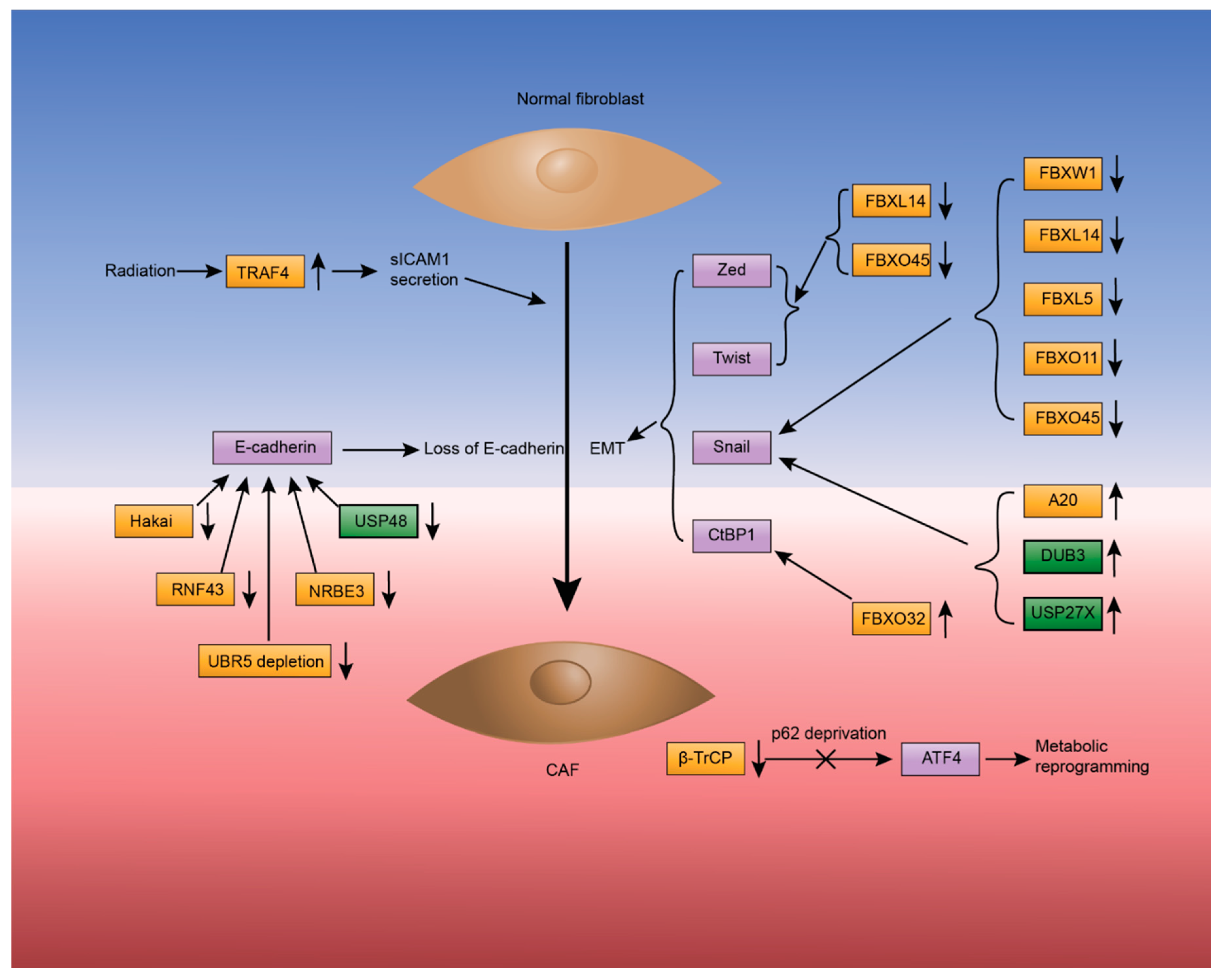
2.2. Cancer-Associated Fibroblasts
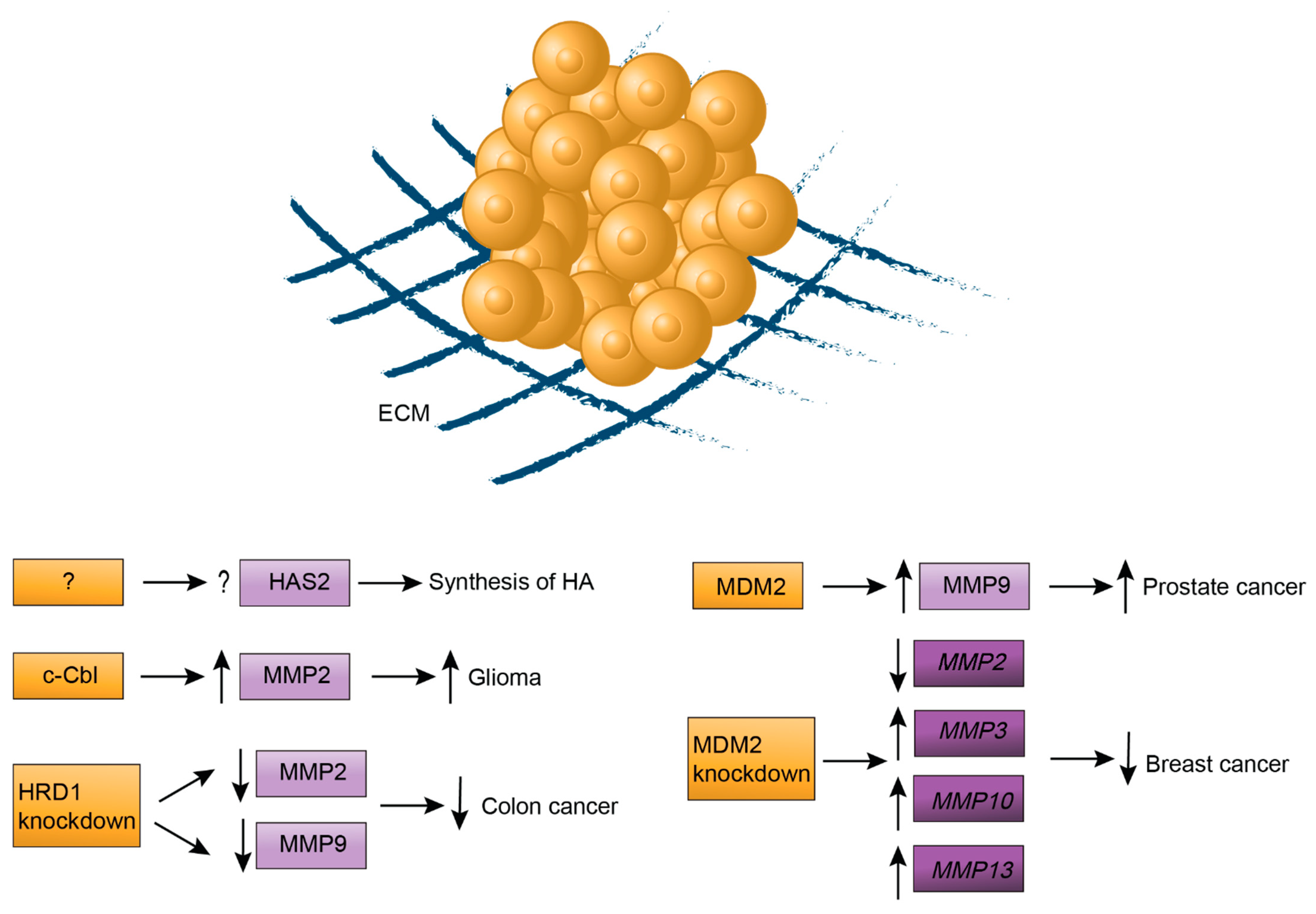
2.3. The Extracellular Matrix
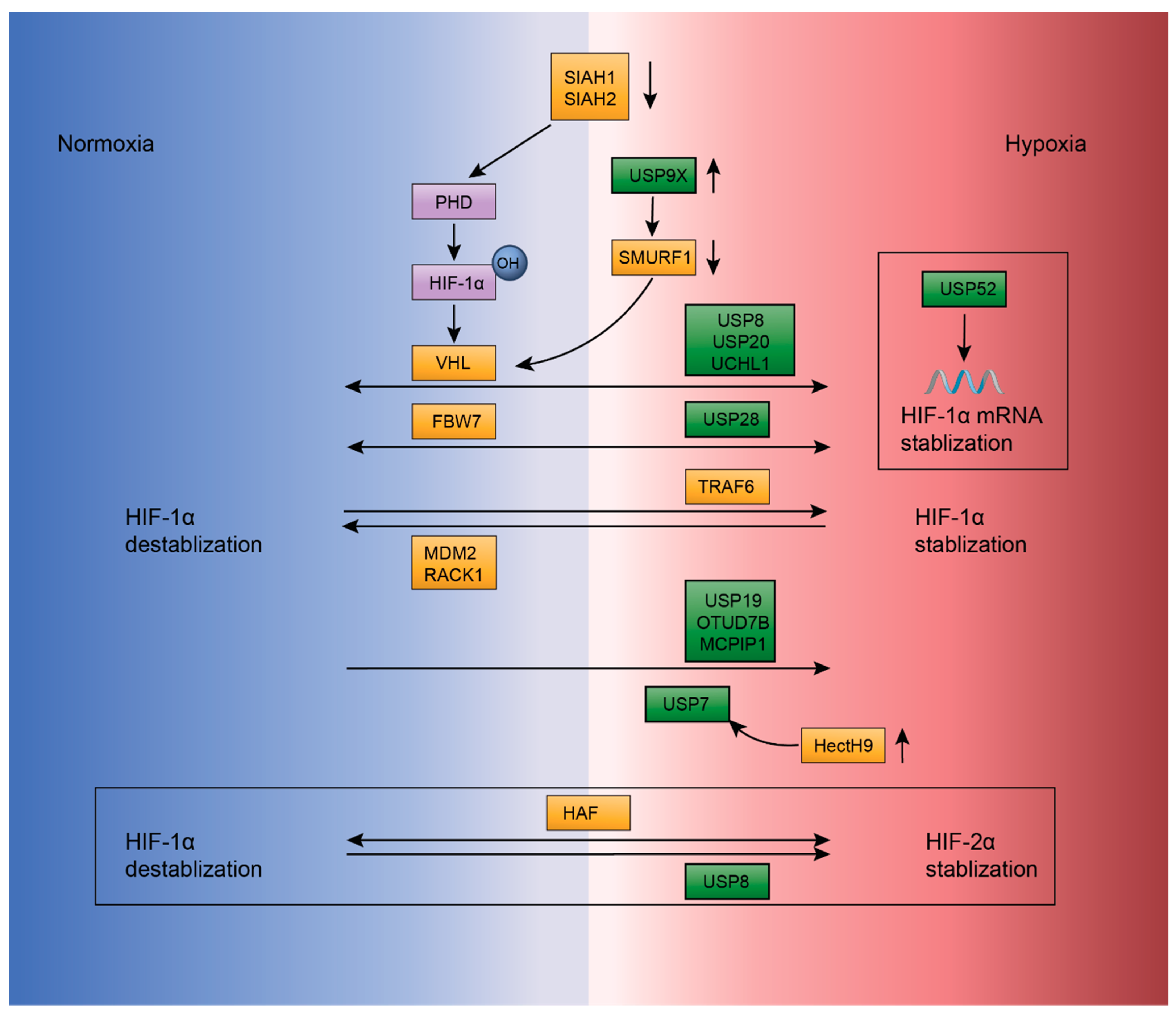
2.4. Ubiquitination Regulation in Hypoxia
3. Modulators Targeting E3s and DUBs in Tumor Microenvironment
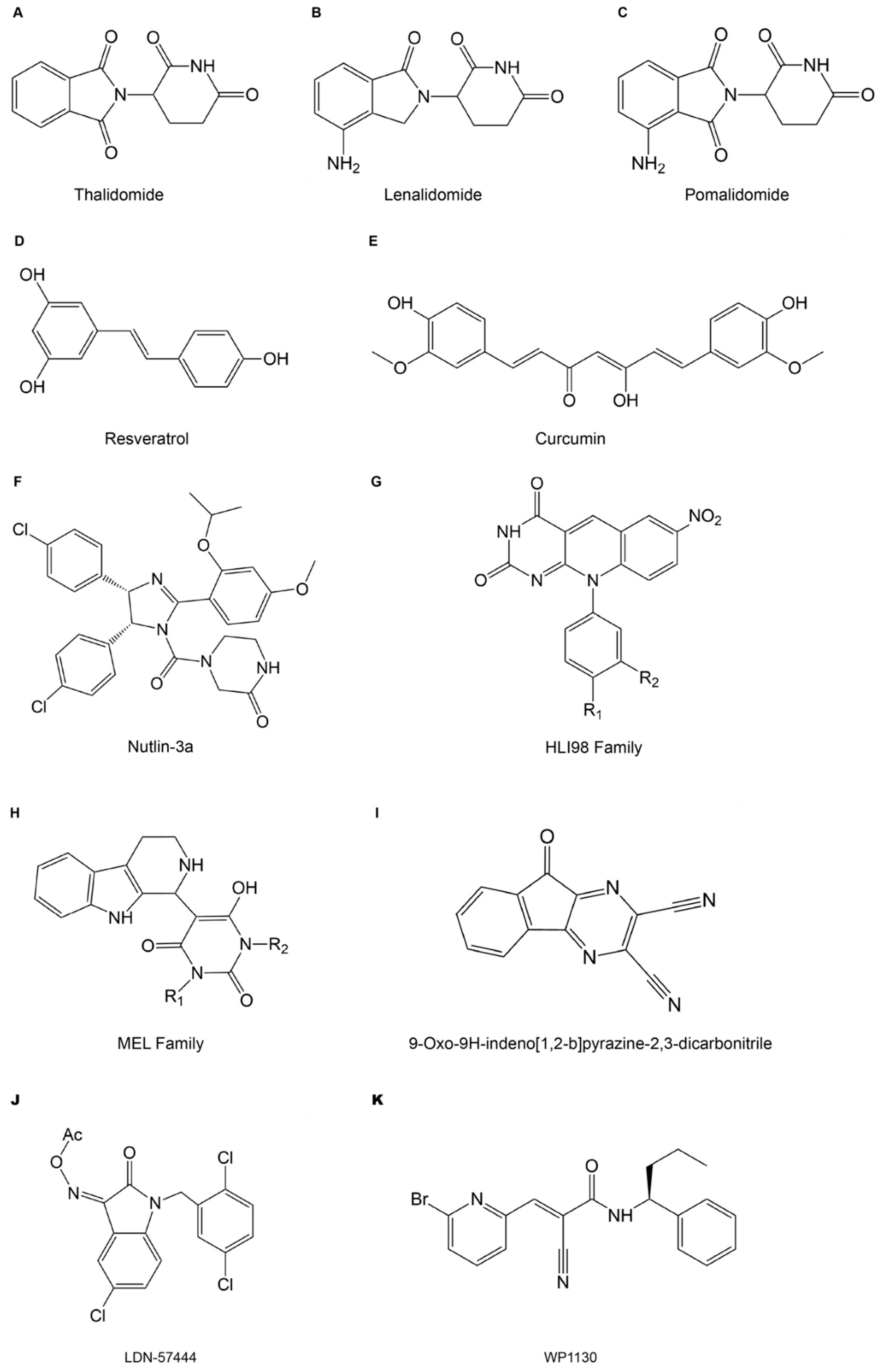
3.1. Small Molecule Inhibitors Targeting E3s and DUBs
3.2. Ubiquitin Variants as Modulators of E3 Ligases and Deubiquitinases
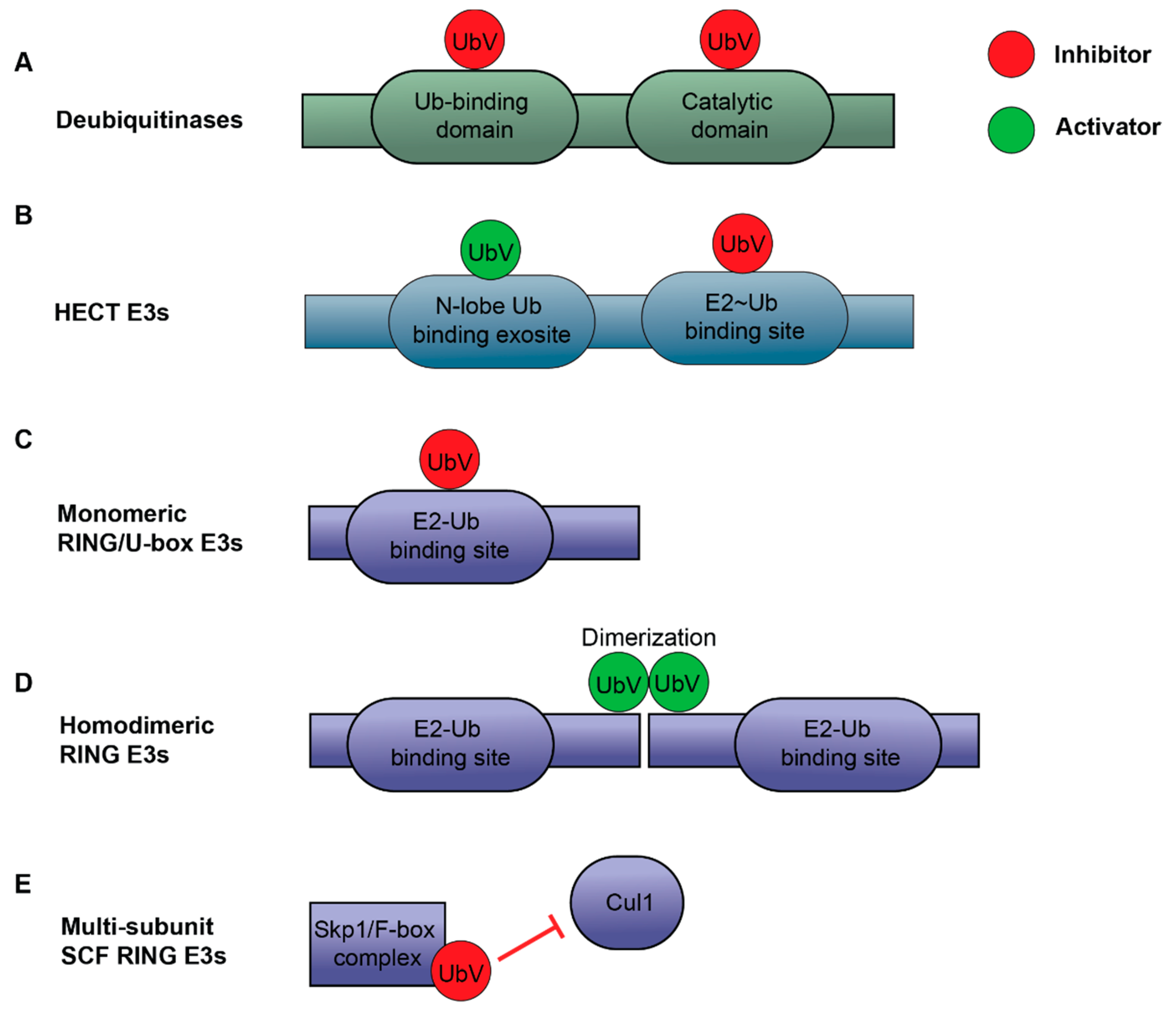
3.2.1. UbV Inhibitors for DUBs
3.2.2. UbV Activators and Inhibitors for E3 Ligases
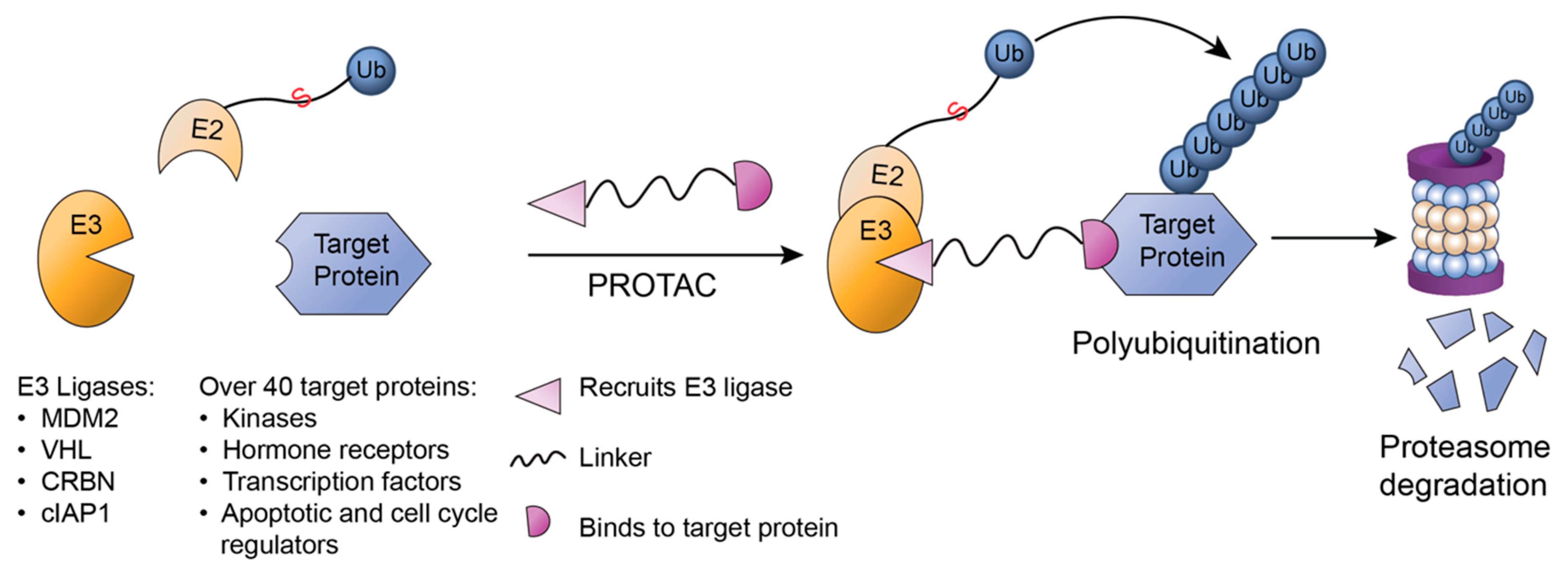
3.3. PROTAC
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HECT | Homologous to the E6AP Carboxyl Terminus |
| RING | Really Interesting New Gene |
| RBR | RING-Between-RING |
| DUB | Deubiquitinase |
| USP | Ubiquitin-Specific Protease |
| UCH | Ubiquitin C-terminal Hydrolase |
| OTU | Ovarian Tumor Protease |
| MJD | Machado-Joseph Disease Protease |
| JAMM | JAB1/MPR/Mov34 Metalloprotease |
| MINDY | Motif Interacting with Ubiquitin-Containing Novel DUB Family |
| TME | Tumor Microenvironment |
| TAM | Tumor-Associated Macrophage |
| MDSC | Myeloid Suppressive Cell |
| DC | Dendritic Cell |
| NK | Natural Killer |
| CD | Cluster of Differentiation |
| Th | T Helper Cell |
| Treg | Regulatory T Cell |
| PD-L1 | Programmed Death-Ligand 1 |
| PD | Programmed Cell Death protein 1 |
| CHIP | Carboxy Terminus of Hsc70 Interacting Protein |
| STUB1 | STIP1 Homology and U-Box Containing Protein 1 |
| β-TrCP | Β-Transducin Repeat-Containing |
| CRL | Cullin-RING E3 Ligase |
| SCF | Skp1-Cullin1-F-Box Protein |
| GSK3β | Glycogen Synthase Kinase 3β |
| FBXO | F-Box Only Protein |
| HRD1 | HMG-CoA Reductase Degradation Protein 1 |
| ERAD | Endoplasmic Reticulum Associated Protein Degradation |
| POZ | Pox Virus and Zinc Finger Protein |
| SPOP | Speckle-Type POZ Protein |
| APC/C | Anaphase-Promoting Complex/Cyclosome |
| CDK | Cyclin Dependent Kinase |
| c-Cbl | Casitas B-Cell Lymphoma |
| Cbl-b | Casitas B-Lineage Lymphoma Proto-Oncogene-B |
| STAT | Signal Transducer and Activator of Transcription Protein |
| ERK | Extracellular Signal Regulated Kinase |
| CSN5 | COP9 Signalosome Complex Subunit 5 |
| NF-κB | Nuclear Factor-κB |
| TNF | Tumor Necrosis Factor |
| CKLF | Chemokine-Like Factor |
| CMTM6 | CKLF-Like Marvel Transmembrane Domain Containing Family Member 6 |
| TAM | Tumor-Associated Macrophage |
| IL | Interleukin |
| PGE2 | Prostaglandin 2 |
| TGF | Transforming Growth Factor |
| SIAH | Seven in Absentia Homologue |
| NRF1 | Nuclear Respiratory Factor 1 |
| Trim24 | Tripartite Motif-Containing Protein 24 |
| CBP | CREB-Binding Protein |
| Stat6 | Signal Transducer and Activator of Transcription 6 |
| IκB | Inhibitor of NF-κB |
| DNMT1 | DNA Methyltransferase 1 |
| WDR4 | WD Repeat 4 |
| PML | Promyelocytic Leukemia |
| UBR5 | Ubiquitin Protein Ligase E3 Component N-Recognin 5 |
| CAF | Cancer-Associated Fibroblasts |
| EndMT | Endothelial-Mesenchymal-Transition |
| EMT | Epithelial-Mesenchymal-Transition |
| MMP | Matrix Metalloproteinase |
| ATF4 | Activating Transcription Factor 4 |
| TRAF | Tumor Necrosis Factor Receptor-Associated Factor |
| sICAM1 | Soluble Intercellular Adhesion Molecule 1 |
| NOX | NADPH Oxidase |
| ROS | Reactive Oxygen Species |
| FBXW | The F-box and WD Repeat Domain Containing Protein |
| FBXL | F-box and Leucine-rich Repeat Protein |
| TNFAIP3 | Tumor Necrosis Factor α-induced Protein 3 |
| CtBP1 | C-Terminal Binding Protein 1 |
| RNF43 | RING Finger Protein 43 |
| NRBE3 | Novel Retinoblastoma E3 Ubiquitin Ligase |
| JNK | c-Jun N-terminal Kinase |
| ECM | Extracellular Matrix |
| HA | Hyaluronan |
| HAS | HA Synthase |
| PSMA | Prostate Specific Membrane Antigen |
| HIF | Hypoxia Inducible Transcription Factor |
| VHL | von Hippel-Lindau E3 ligase |
| ODD | Oxygen Dependent Degradation |
| PHD | Prolyl Hydroxylase |
| FBW | F-box and WD Repeat Domain-containing |
| HAF | Hypoxia Associated Factor |
| MDM2 | Mouse Double Minute 2 Homolog |
| RACK1 | Receptor of Activated Protein C Kinase |
| HSP | Heat Shock Protein |
| UCHL1 | Ubiquitin Carboxyl-terminal Hydrolase L1 |
| PAS | PERN-ARNT-SIM Domain |
| VDU2 | VHL-interacting Deubiquitinating Enzyme 2 |
| HAUSP | Herpesvirus Associated Ubiquitin Specific Protease |
| HectH9 | Homologous to E6AP Carboxyl Terminus Homologous Protein 9 |
| HUWE1 | HECT, UBA and WWE Domain Containing E3 Ubiquitin Protein Ligase 1 |
| MCPIP1 | Monocyte Chemoattractant Protein Induced Protein 1 |
| SMURF | SMAD Specific E3 Ubiquitin Protein Ligase |
| IMiD | Immunomodulatory Drug |
| CRBN | Cereblon |
| CTLA-4 | Cytotoxic T Lymphocyte Associated Protein 4 |
| HLI98 | HDM2(also known as MDM2) ligase inhibitor 98 |
| MEL23/24 | Mdm2 E3 Ligase Inhibitors 23/24 |
| MCL | Myeloid Cell Leukemia |
| T-ALL | T-cell Acute Lymphoblastic Leukemia |
| UbV | Ubiquitin Variant |
| UPS | Ubiquitin Proteasome System |
| BRISC | Brcc36-containing isopeptidase complex |
| DUSP | Domain Present in Ub Specific Proteases |
| MERS-CoV | Middle East Respiratory Syndrome Coronavirus |
| CCHFV | Crimean-Congo Hemorrhagic Fever Virus |
| NEDD4 | Neural Precursor Cells Induced Expression of Developmentally Down-regulated Protein 4 |
| YY1 | Ying-Yang1 |
| UBE4B | Ubiquitination Factor E4B |
| XIAP | X-linked Inhibitor of Apoptosis |
| CDC20 | Cell Division Cycle protein 20 |
| CDH1 | CDC20 Homolog-1 |
| PROTAC | Proteolysis Targeting Chimeras |
| MetAP-2 | Methionine Aminopeptidase-2 |
| cIAP1 | Cellular Inhibitor of Apoptosis Protein 1 |
| IKZF1/3 | Ikaros Family Zinc Finger Protein 1/3 |
| SARM | Steroidal Androgen Receptor Ligand |
| BET | Bromodomain and Extra Terminal Domain |
| BETi | BET inhibitors |
| BETP | BET-Containing Protein |
| BRD4 | Bromodomain-Containing 4 |
| CXCR4 | C-X-C chemokine receptor type 4 |
| AML | Acute Myeloid Leukemia |
References
- Shaid, S.; Brandts, C.H.; Serve, H.; Dikic, I. Ubiquitination and selective autophagy. Cell Death Differ. 2012, 20, 21–30. [Google Scholar] [PubMed]
- Uckelmann, M.; Sixma, T.K. Histone ubiquitination in the DNA damage response. DNA Repair. 2017, 56, 92–101. [Google Scholar] [PubMed]
- Bednash, J.S.; Mallampalli, R.K. Regulation of inflammasomes by ubiquitination. Cell. Mol. Immunol. 2016, 13, 722–728. [Google Scholar] [PubMed]
- Buetow, L.; Huang, D.T. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2016, 17, 626–642. [Google Scholar]
- Metzger, M.B.; Hristova, V.A.; Weissman, A.M. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 2012, 125, 531–537. [Google Scholar]
- Vittal, V.; Stewart, M.D.; Brzovic, P.S.; Klevit, R.E. Regulating the regulators: Recent revelations in the control of E3 ubiquitin ligases. J. Biol. Chem. 2015, 290, 21244–21251. [Google Scholar]
- Poondla, N.; Chandrasekaran, A.P.; Kim, K.S.; Ramakrishna, S. Deubiquitinating enzymes as cancer biomarkers: New therapeutic opportunities? BMB Rep. 2019, 52, 181–189. [Google Scholar]
- Abdul Rehman, S.A.; Kristariyanto, Y.A.; Choi, S.Y.; Nkosi, P.J.; Weidlich, S.; Labib, K.; Hofmann, K.; Kulathu, Y. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol. Cell 2016, 63, 146–155. [Google Scholar]
- Fraile, J.M.; Quesada, V.; Rodríguez, D.; Freije, J.M.P.; López-Otín, C. Deubiquitinases in cancer: New functions and therapeutic options. Oncogene 2012, 31, 2373–2388. [Google Scholar]
- Guo, S.; Deng, C.X. Effect of stromal cells in tumor microenvironment on metastasis initiation. Int. J. Biol. Sci. 2018, 14, 2083–2093. [Google Scholar]
- Najafi, M.; Farhood, B.; Mortezaee, K. Extracellular matrix (ECM) stiffness and degradation as cancer drivers. J. Cell. Biochem. 2019, 120, 2782–2790. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, K. HIF-1 at the crossroads of hypoxia, inflammation, and cancer. Int. J. Cancer 2016, 138, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef] [PubMed]
- Roma-Rodrigues, C.; Mendes, R.; Baptista, P.V.; Fernandes, A.R. Targeting tumor microenvironment for cancer therapy. Int. J. Mol. Sci. 2019, 20, 840. [Google Scholar] [CrossRef]
- Wu, T.; Dai, Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017, 387, 61–68. [Google Scholar] [CrossRef]
- Bhome, R.; Bullock, M.D.; Al Saihati, H.A.; Goh, R.W.; Primrose, J.N.; Sayan, A.E.; Mirnezami, A.H. A top-down view of the tumor microenvironment: Structure, cells and signaling. Front. Cell Dev. Biol. 2015, 3, 33. [Google Scholar] [CrossRef]
- Hui, L.; Chen, Y. Tumor microenvironment: Sanctuary of the devil. Cancer Lett. 2015, 368, 7–13. [Google Scholar] [CrossRef]
- Hsu, J.M.; Li, C.W.; Lai, Y.J.; Hung, M.C. Posttranslational modifications of PD-L1 and their applications in cancer therapy. Cancer Res. 2018, 78, 6349–6353. [Google Scholar] [CrossRef]
- Han, Y.; Liu, D.; Li, L. PD-1/PD-L1 pathway: Current researches in cancer. Am. J. Cancer Res. 2020, 10, 727–742. [Google Scholar]
- Jiang, Y.; Chen, M.; Nie, H.; Yuan, Y. PD-1 and PD-L1 in cancer immunotherapy: Clinical implications and future considerations. Hum. Vaccines Immunother. 2019, 15, 1111–1122. [Google Scholar] [CrossRef]
- Mezzadra, R.; Sun, C.; Jae, L.T.; Gomez-Eerland, R.; De Vries, E.; Wu, W.; Logtenberg, M.E.W.; Slagter, M.; Rozeman, E.A.; Hofland, I.; et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 2017, 549, 106–110. [Google Scholar] [CrossRef]
- Cha, J.H.; Chan, L.C.; Li, C.W.; Hsu, J.L.; Hung, M.C. Mechanisms Controlling PD-L1 Expression in Cancer. Mol. Cell 2019, 76, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Bulatov, E.; Ciulli, A. Targeting Cullin-RING E3 ubiquitin ligases for drug discovery: Structure, assembly and small-molecule modulation. Biochem. J. 2015, 467, 365–386. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Lu, Y.; Zhang, S.; Li, Y.; Huang, J.; Yin, Z.; Ren, J.; Huang, K.; Liu, L.; Yang, K.; et al. β-Trcp ubiquitin ligase and RSK2 kinase-mediated degradation of FOXN2 promotes tumorigenesis and radioresistance in lung cancer. Cell Death Differ. 2018, 25, 1473–1485. [Google Scholar] [CrossRef] [PubMed]
- Li, C.W.; Lim, S.O.; Xia, W.; Lee, H.H.; Chan, L.C.; Kuo, C.W.; Khoo, K.H.; Chang, S.S.; Cha, J.H.; Kim, T.; et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat. Commun. 2016, 7, 12632. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Liu, X.; Guo, X.; Jiang, S.; Chen, T.; Hu, Z.; Liu, H.; Bai, Y.; Xue, M.; Hu, R.; et al. FBXO38 mediates PD-1 ubiquitination and regulates anti-tumour immunity of T cells. Nature 2018, 564, 130–135. [Google Scholar] [CrossRef]
- Cha, J.; Yang, W.; Xia, W.; Wei, Y.; Chan, L.; Lim, S.; Li, C.; Kim, T.; Chang, S.-S.; Lee, H.-H.; et al. Metformin Promotes Antitumor Immunity via Endoplasmic-Reticulum-Associated Degradation of Article Metformin Promotes Antitumor Immunity Degradation of PD-L1. Mol. Cell 2018, 71, 606.e7–620.e7. [Google Scholar] [CrossRef]
- Zhang, J.; Bu, X.; Wang, H.; Zhu, Y.; Geng, Y.; Nihira, N.T.; Tan, Y.; Ci, Y.; Wu, F.; Dai, X.; et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 2018, 553, 91–95. [Google Scholar] [CrossRef]
- Wang, S.; Xu, L.; Che, X.; Li, C.; Xu, L.; Hou, K.; Fan, Y.; Wen, T.; Qu, X.; Liu, Y. E3 ubiquitin ligases Cbl-b and c-Cbl downregulate PD-L1 in EGFR wild-type non-small cell lung cancer. FEBS Lett. 2018, 592, 621–630. [Google Scholar] [CrossRef]
- Lim, S.O.; Li, C.W.; Xia, W.; Cha, J.H.; Chan, L.C.; Wu, Y.; Chang, S.S.; Lin, W.C.; Hsu, J.M.; Hsu, Y.H.; et al. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell 2016, 30, 925–939. [Google Scholar] [CrossRef]
- Petty, A.J.; Yang, Y. Tumor-associated macrophages: Implications in cancer immunotherapy. Immunotherapy 2017, 9, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Tariq, M.; Zhang, J.; Liang, G.; Ding, L.; He, Q.; Yang, B. Macrophage Polarization: Anti-Cancer Strategies to Target Tumor-Associated Macrophage in Breast Cancer. J. Cell. Biochem. 2017, 118, 2484–2501. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Cheng, H.; Mu, C.; Geng, G.; Zhao, T.; Luo, Q.; Ma, K.; Chang, R.; Liu, Q.; Gao, R.; et al. The SIAH2-NRF1 axis spatially regulates tumor microenvironment remodeling for tumor progression. Nat. Commun. 2019, 10, 1034. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Gan, S.; Zhu, Q.; Dai, D.; Li, N.; Wang, H.; Chen, X.; Hou, D.; Wang, Y.; Pan, Q.; et al. Modulation of M2 macrophage polarization by the crosstalk between Stat6 and Trim24. Nat. Commun. 2019, 10, 4353. [Google Scholar] [CrossRef] [PubMed]
- Kumari, N.; Dwarakanath, B.S.; Das, A.; Bhatt, A.N. Role of interleukin-6 in cancer progression and therapeutic resistance. Tumor Biol. 2016, 37, 11553–11572. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Wu, Y.S.; Hung, C.Y.; Wang, S.A.; Young, M.J.; Hsu, T.I.; Hung, J.J. USP24 induces IL-6 in tumor-associated microenvironment by stabilizing p300 and β-TrCP and promotes cancer malignancy. Nat. Commun. 2018, 9, 3996. [Google Scholar] [CrossRef]
- Hadjimichael, C.; Chanoumidou, K.; Nikolaou, C.; Klonizakis, A.; Theodosi, G.I.; Makatounakis, T.; Papamatheakis, J.; Kretsovali, A. Promyelocytic Leukemia Protein Is an Essential Regulator of Stem Cell Pluripotency and Somatic Cell Reprogramming. Stem Cell Rep. 2017, 8, 1366–1378. [Google Scholar] [CrossRef]
- Wang, Y.T.; Chen, R.H. PML degradation fosters an immunosuppressive and pro-metastatic tumor microenvironment. Mol. Cell. Oncol. 2017, 4, e1364212. [Google Scholar] [CrossRef]
- Wang, Y.T.; Chen, J.; Chang, C.W.; Jen, J.; Huang, T.Y.; Chen, C.M.; Shen, R.; Liang, S.Y.; Cheng, I.C.; Yang, S.C.; et al. Ubiquitination of tumor suppressor PML regulates prometastatic and immunosuppressive tumor microenvironment. J. Clin. Investig. 2017, 127, 2982–2997. [Google Scholar] [CrossRef]
- Shearer, R.F.; Iconomou, M.; Watts, C.K.W.; Saunders, D.N. Functional roles of the E3 Ubiquitin Ligase UBR5 in cancer. Mol. Cancer Res. 2015, 13, 1523–1532. [Google Scholar] [CrossRef]
- Liao, L.; Song, M.; Li, X.; Tang, L.; Zhang, T.; Zhang, L.; Pan, Y.; Chouchane, L.; Ma, X. E3 ubiquitin ligase UBR5 drives the growth and metastasis of triple-negative breast cancer. Cancer Res. 2017, 77, 2090–2101. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Wang, C.; Wang, H.; Zhang, T.; Li, J.; Benezra, R.; Chouchane, L.; Sun, Y.H.; Cui, X.G.; Ma, X. Targeting ubiquitin protein ligase E3 component N-recognin 5 in cancer cells induces a CD8+ T cell mediated immune response. Oncoimmunology 2020, 9, 1746148. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Jiang, Y.; Luo, Q.; Li, L.; Jia, L. Neddylation: A novel modulator of the tumor microenvironment. Mol. Cancer 2019, 18, 77. [Google Scholar] [CrossRef] [PubMed]
- Wortel, I.M.N.; van der Meer, L.T.; Kilberg, M.S.; van Leeuwen, F.N. Surviving Stress: Modulation of ATF4-Mediated Stress Responses in Normal and Malignant Cells. Trends Endocrinol. Metab. 2017, 28, 794–806. [Google Scholar] [CrossRef] [PubMed]
- Linares, J.F.; Cordes, T.; Duran, A.; Reina-Campos, M.; Valencia, T.; Ahn, C.S.; Castilla, E.A.; Moscat, J.; Metallo, C.M.; Diaz-Meco, M.T. ATF4-Induced Metabolic Reprograming Is a Synthetic Vulnerability of the p62-Deficient Tumor Stroma. Cell Metab. 2017, 26, 817.e6–829.e6. [Google Scholar] [CrossRef]
- Kim, E.; Kim, W.; Lee, S.; Chun, J.; Kang, J.; Park, G. TRAF4 promotes lung cancer aggressiveness by modulating tumor microenvironment in normal fibroblasts. Sci. Rep. 2017, 7, 8923. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, F.; Vinuesa, A.G.; DeKruijf, E.M.; Mesker, W.E.; Hui, L.; Drabsch, Y.; Li, Y.; Bauer, A.; Rousseau, A.; et al. TRAF4 Promotes TGF-β Receptor Signaling and Drives Breast Cancer Metastasis. Mol. Cell 2013, 51, 559–572. [Google Scholar] [CrossRef]
- Díaz, V.M.; de Herreros, A.G. F-box proteins: Keeping the epithelial-to-mesenchymal transition (EMT) in check. Semin. Cancer Biol. 2016, 36, 71–79. [Google Scholar] [CrossRef]
- Wartz, I.E.; O’Rourke, K.M.; Zhou, H.; Eby, M.; Aravind, L.; Seshagiri, S.; Wu, P.; Wiesmann, C.; Baker, R.; Boone, D.L.; et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 2004, 430, 694–699. [Google Scholar] [CrossRef]
- Lee, J.H.; Jung, S.M.; Yang, K.M.; Bae, E.; Ahn, S.G.; Park, J.S.; Seo, D.; Kim, M.; Ha, J.; Lee, J.; et al. A20 promotes metastasis of aggressive basal-like breast cancers through multi-monoubiquitylation of Snail1. Nat. Cell Biol. 2017, 19, 1260–1273. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Lin, Y.; Liu, Y.; Wang, Y.; Jia, J.; Singh, P.; Chi, Y.I.; Wang, C.; Dong, C.; et al. Dub3 inhibition suppresses breast cancer invasion and metastasis by promoting Snail1 degradation. Nat. Commun. 2017, 8, 14228. [Google Scholar] [CrossRef]
- Liu, T.; Yu, J.; Deng, M.; Yin, Y.; Zhang, H.; Luo, K.; Qin, B.; Li, Y.; Wu, C.; Ren, T.; et al. CDK4/6-dependent activation of DUB3 regulates cancer metastasis through SNAIL1. Nat. Commun. 2017, 8, 13923. [Google Scholar] [CrossRef] [PubMed]
- Lambies, G.; Miceli, M.; Martínez-Guillamon, C.; Olivera-Salguero, R.; Peña, R.; Frías, C.P.; Calderon, I.; Atanassov, B.S.; Dent, S.Y.R.; Arribas, J.; et al. TGFβ-activated USP27X deubiquitinase regulates cell migration and chemoresistance via stabilization of Snail1. Cancer Res. 2019, 79, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Sahu, S.K.; Tiwari, N.; Pataskar, A.; Zhuang, Y.; Borisova, M.; Diken, M.; Strand, S.; Beli, P.; Tiwari, V.K. FBXO32 promotes microenvironment underlying epithelial-mesenchymal transition via CtBP1 during tumour metastasis and brain development. Nat. Commun. 2017, 8, 1523. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Krause, G.; Scheffner, M.; Zechner, D.; Leddy, H.E.M.; Behrens, J.; Sommer, T.; Birchmeier, W. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat. Cell Biol. 2002, 4, 222–231. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, L.; Gao, X.; Guo, A.; Diao, Y.; Zhao, Y. RNF43 ubiquitinates and degrades phosphorylated E-cadherin by c-Src to facilitate epithelial-mesenchymal transition in lung adenocarcinoma. BMC Cancer 2019, 19, 670. [Google Scholar] [CrossRef]
- Zheng, T.; Lu, M.; Wang, T.; Zhang, C.; Du, X. NRBE3 promotes metastasis of breast cancer by down-regulating E-cadherin expression. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 1869–1877. [Google Scholar] [CrossRef]
- Li, S.; Wang, D.; Zhao, J.; Weathington, N.M.; Shang, D.; Zhao, Y. The deubiquitinating enzyme USP48 stabilizes TRAF2 and reduces E-cadherin-mediated adherens junctions. FASEB J. 2018, 32, 230–242. [Google Scholar] [CrossRef]
- Karousou, E.; Misra, S.; Ghatak, S.; Dobra, K. Roles and targeting of the HAS/hyaluronan/CD44 molecular system in cancer. Matrix Biol. 2017, 59, 3–22. [Google Scholar] [CrossRef]
- Karousou, E.; Kamiryo, M.; Skandalis, S.S.; Ruusala, A.; Asteriou, T.; Passi, A.; Yamashita, H.; Hellman, U.; Heldin, C.H.; Heldin, P. The activity of hyaluronan synthase 2 is regulated by dimerization and ubiquitination. J. Biol. Chem. 2010, 285, 23647–23654. [Google Scholar] [CrossRef]
- Lee, H.; Tsygankov, A.Y. c-Cbl regulates glioma invasion through matrix metalloproteinase 2. J. Cell. Biochem. 2010, 111, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, C.; Vaidya, S.; Wadhwan, V.; Hitesh; Kaur, G.; Pathak, A. Seesaw of matrix metalloproteinases (MMPs). J. Cancer Res. Ther. 2016, 12, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; He, X.; Fan, Z. Upregulation of HRD1 promotes cell migration and invasion in colon cancer. Mol. Cell. Biochem. 2019, 454, 1–9. [Google Scholar] [CrossRef]
- Venkatesan, T.; Alaseem, A.; Chinnaiyan, A.; Dhandayuthapani, S.; Kanagasabai, T.; Alhazzani, K.; Dondapati, P.; Alobid, S.; Natarajan, U.; Schwartz, R.; et al. MDM2 Overexpression Modulates the Angiogenesis-Related Gene Expression Profile of Prostate Cancer Cells. Cells 2018, 7, 41. [Google Scholar] [CrossRef]
- Bradbury, R.; Jiang, W.G.; Cui, Y.X. Mdm2 and psma play inhibitory roles in metastatic breast cancer cells through regulation of matrix metalloproteinases. Anticancer Res. 2016, 36, 1143–1152. [Google Scholar]
- Zhang, J.; Zhang, Q. VHL and hypoxia signaling: Beyond HIF in cancer. Biomedicines 2018, 6, 35. [Google Scholar] [CrossRef]
- Cassavaugh, J.M.; Hale, S.A.; Wellman, T.L.; Howe, A.K.; Wong, C.; Lounsbury, K.M. Negative regulation of HIF-1α by an FBW7-mediated degradation pathway during hypoxia. J. Cell. Biochem. 2011, 112, 3882–3890. [Google Scholar] [CrossRef]
- Nakayama, K.; Ronai, Z. Siah: New players in the cellular response to hypoxia. Cell Cycle 2004, 3, 1345–1347. [Google Scholar] [CrossRef] [PubMed]
- Mennerich, D.; Kubaichuk, K.; Kietzmann, T. DUBs, Hypoxia, and Cancer. Trends Cancer 2019, 5, 632–653. [Google Scholar] [CrossRef]
- Koh, M.Y.; Darnay, B.G.; Powis, G. Hypoxia-Associated Factor, a Novel E3-Ubiquitin Ligase, Binds and Ubiquitinates Hypoxia-Inducible Factor 1, Leading to Its Oxygen-Independent Degradation. Mol. Cell. Biol. 2008, 28, 7081–7095. [Google Scholar] [CrossRef]
- Guan, Z.; Ding, C.; Du, Y.; Zhang, K.; Zhu, J.N.; Zhang, T.; He, D.; Xu, S.; Wang, X.; Fan, J. HAF drives the switch of HIF-1α to HIF-2α by activating the NF-κB pathway, leading to malignant behavior of T24 bladder cancer cells. Int. J. Oncol. 2014, 44, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Li, X.B.; Meng, Y.; Fan, L.; Li, M.; Fang, J. TRAF6 upregulates expression of HIF-1α and promotes tumor angiogenesis. Cancer Res. 2013, 73, 4950–4959. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Singh, A.R.; Durden, D.L. MDM2 regulates hypoxic hypoxia-inducible factor 1α stability in an E3 ligase, proteasome, and PTEN-phosphatidylinositol 3-kinase-AKT-dependent manner. J. Biol. Chem. 2014, 289, 22785–22797. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.V.; Semenza, G.L. RACK1 vs. HSP90: Competition for HIF-1α degradation vs. stabilization. Cell Cycle 2007, 6, 656–659. [Google Scholar] [CrossRef]
- Troilo, A.; Alexander, I.; Muehl, S.; Jaramillo, D.; Knobeloch, K.P.; Krek, W. HIF1α deubiquitination by USP8 is essential for ciliogenesis in normoxia. EMBO Rep. 2014, 15, 77–85. [Google Scholar] [CrossRef]
- Li, Z.; Wang, D.; Messing, E.M.; Wu, G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1α. EMBO Rep. 2005, 6, 373–378. [Google Scholar] [CrossRef]
- Goto, Y.; Zeng, L.; Yeom, C.J.; Zhu, Y.; Morinibu, A.; Shinomiya, K.; Kobayashi, M.; Hirota, K.; Itasaka, S.; Yoshimura, M.; et al. UCHL1 provides diagnostic and antimetastatic strategies due to its deubiquitinating effect on HIF-1α. Nat. Commun. 2015, 6, 6153. [Google Scholar] [CrossRef]
- Li, Z.; Wang, D.; Na, X.; Schoen, S.R.; Messing, E.M.; Wu, G. Identification of a deubiquitinating enzyme subfamily as substrates of the von Hippel-Lindau tumor suppressor. Biochem. Biophys. Res. Commun. 2002, 294, 700–709. [Google Scholar] [CrossRef]
- Flügel, D.; Görlach, A.; Kietzmann, T. GSK-3β regulates cell growth, migration, and angiogenesis via Fbw7 and USP28-dependent degradation of HIF-1α. Blood 2012, 119, 1292–1301. [Google Scholar] [CrossRef]
- Wu, H.T.; Kuo, Y.C.; Hung, J.J.; Huang, C.H.; Chen, W.Y.; Chou, T.Y.; Chen, Y.; Chen, Y.J.; Chen, Y.J.; Cheng, W.C.; et al. K63-polyubiquitinated HAUSP deubiquitinates HIF-1α and dictates H3K56 acetylation promoting hypoxia-induced tumour progression. Nat. Commun. 2016, 7, 13644. [Google Scholar] [CrossRef]
- Altun, M.; Zhao, B.; Velasco, K.; Liu, H.; Hassink, G.; Paschke, J.; Pereira, T.; Lindsten, K. Ubiquitin-specific protease 19 (USP19) regulates hypoxia-inducible factor 1α (HIF-1α) during hypoxia. J. Biol. Chem. 2012, 287, 1962–1969. [Google Scholar] [CrossRef] [PubMed]
- Bremm, A.; Moniz, S.; Mader, J.; Rocha, S.; Komander, D. Cezanne (OTUD 7 B) regulates HIF- 1 a homeostasis in a proteasome-independent manner. EMBO Rep. 2014, 15, 1268–1272. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Lu, Y.X.; Cheng, D.; Zhang, K.; Zheng, J.; Liu, Y.; Wang, X.; Yuan, Y.F.; Tang, Y. Da Monocyte Chemoattractant Protein-Induced Protein 1 Targets Hypoxia-Inducible Factor 1α to Protect Against Hepatic Ischemia/Reperfusion Injury. Hepatology 2018, 68, 2359–2375. [Google Scholar] [CrossRef] [PubMed]
- Bett, J.S.; Ibrahim, A.F.M.; Garg, A.K.; Kelly, V.; Pedrioli, P.; Rocha, S.; Hay, R.T. The P-body component USP52/PAN2 is a novel regulator of HIF1A mRNA stability. Biochem. J. 2013, 451, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Peng, Z.; Zhu, M.; Wang, P.; Du, X.; Li, X.; Liu, Y.; Jin, Y.; McNutt, M.A.; Yin, Y. USP9X destabilizes pVHL and promotes cell proliferation. Oncotarget 2016, 7, 60519–60534. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ito, T.; Handa, H. Cereblon and its downstream substrates as molecular targets of immunomodulatory drugs. Int. J. Hematol. 2016, 104, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Gribben, J.G.; Fowler, N.; Morschhauser, F. Mechanisms of action of lenalidomide in B-cell non-hodgkin lymphoma. J. Clin. Oncol. 2015, 33, 2803–2811. [Google Scholar] [CrossRef] [PubMed]
- Verdura, S.; Cuyàs, E.; Cortada, E.; Brunet, J.; Lopez-Bonet, E.; Martin-Castillo, B.; Bosch-Barrera, J.; Encinar, J.A.; Menendez, J.A. Resveratrol targets PD-L1 glycosylation and dimerization to enhance antitumor T-cell immunity. Aging (Albany NY) 2020, 12, 8–34. [Google Scholar] [CrossRef] [PubMed]
- Seidel, J.A.; Otsuka, A.; Kabashima, K. Anti-PD-1 and anti-CTLA-4 therapies in cancer: Mechanisms of action, efficacy, and limitations. Front. Oncol. 2018, 8, 86. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, S.; Okita, N.; Oku, M.; Nagai, W.; Kobayashi, M.; Higami, Y. An Mdm2 antagonist, Nutlin-3a, induces p53-dependent and proteasome-mediated poly(ADP-ribose) polymerase1 degradation in mouse fibroblasts. Biochem. Biophys. Res. Commun. 2011, 407, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ludwig, R.L.; Jensen, J.P.; Pierre, S.A.; Medaglia, M.V.; Davydov, I.V.; Safiran, Y.J.; Oberoi, P.; Kenten, J.H.; Phillips, A.C.; et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell 2005, 7, 547–559. [Google Scholar] [CrossRef] [PubMed]
- Herman, A.G.; Hayano, M.; Poyurovsky, M.V.; Shimada, K.; Skouta, R.; Prives, C.; Stockwell, B.R. Discovery of Mdm2-MdmX E3 ligase inhibitors using a cell-based ubiquitination assay. Cancer Discov. 2011, 1, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Patterson, D.M.; Gao, D.; Trahan, D.N.; Johnson, B.A.; Ludwig, A.; Barbieri, E.; Chen, Z.; Diaz-Miron, J.; Vassilev, L.; Shohet, J.M.; et al. Effect of MDM2 and vascular endothelial growth factor inhibition on tumor angiogenesis and metastasis in neuroblastoma. Angiogenesis 2011, 14, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; An, S.; Choi, Y.M.; Jung, J.H.; Li, L.; Meng, H.; Dong, Y.; Ahn, K.J.; An, I.S.; Bae, S. TRIAD1 Is a Novel Transcriptional Target of p53 and Regulates Nutlin-3a-Induced Cell Death. J. Cell. Biochem. 2017, 118, 1733–1740. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Vallese, S.; Peretto, I.; Jacq, X.; Rain, J.C.; Colland, F.; Guedat, P. Synthesis and biological evaluation of 9-oxo-9hindeno[1,2-b]pyrazine-2,3- dicarbonitrile analogues as potential inhibitors of deubiquitinating enzymes. ChemMedChem 2010, 5, 552–558. [Google Scholar] [CrossRef]
- Bi, H.L.; Zhang, Y.L.; Yang, J.; Shu, Q.; Yang, X.L.; Yan, X.; Chen, C.; Li, Z.; Li, H.H. Inhibition of UCHL1 by LDN-57444 attenuates Ang II–Induced atrial fibrillation in mice. Hypertens. Res. 2019, 43, 168–177. [Google Scholar] [CrossRef]
- Kapuria, V.; Peterson, L.F.; Fang, D.; Bornmann, W.G.; Talpaz, M.; Donato, N.J. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010, 70, 9265–9276. [Google Scholar] [CrossRef]
- Fu, P.; Du, F.; Liu, Y.; Yao, M.; Zhang, S.; Zheng, X.; Zheng, S. WP1130 increases cisplatin sensitivity through inhibition of usp9x in estrogen receptor-negative breast cancer cells. Am. J. Transl. Res. 2017, 9, 1783–1791. [Google Scholar]
- Liu, H.; Chen, W.; Liang, C.; Chen, B.W.; Zhi, X.; Zhang, S.; Zheng, X.; Bai, X.; Liang, T. WP1130 increases doxorubicin sensitivity in hepatocellular carcinoma cells through usp9x-dependent p53 degradation. Cancer Lett. 2015, 361, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Jing, B.; Xia, Y.; Zhang, Y.; Hu, M.; Cai, H.; Tong, Y.; Zhou, L.; Yang, L.; Yang, J.; et al. WP1130 reveals USP24 as a novel target in T-cell acute lymphoblastic leukemia. Cancer Cell Int. 2019, 19, 56. [Google Scholar] [CrossRef]
- Kemp, M. Recent Advances in the Discovery of Deubiquitinating Enzyme Inhibitors. Prog. Med. Chem. 2016, 55, 149–192. [Google Scholar] [PubMed]
- Ernst, A.; Avvakumov, G.; Tong, J.; Fan, Y.; Zhao, Y.; Alberts, P.; Persaud, A.; Walker, J.R.; Neculai, A.M.; Neculai, D.; et al. A strategy for modulation of enzymes in the ubiquitin system. Science 2013, 339, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Veggiani, G.; Gerpe, M.C.R.; Sidhu, S.S.; Zhang, W. Emerging drug development technologies targeting ubiquitination for cancer therapeutics. Pharmacol. Ther. 2019, 199, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wu, K.P.; Sartori, M.A.; Kamadurai, H.B.; Ordureau, A.; Jiang, C.; Mercredi, P.Y.; Murchie, R.; Hu, J.; Persaud, A.; et al. System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes. Mol. Cell 2016, 62, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, L.; Rouge, L.; Phillips, A.H.; Lam, C.; Liu, P.; Sandoval, W.; Helgason, E.; Murray, J.M.; Wertz, I.E.; et al. Conformational stabilization of ubiquitin yields potent and selective inhibitors of USP7. Nat. Chem. Biol. 2013, 9, 51–58. [Google Scholar] [CrossRef]
- Phillips, A.H.; Zhang, Y.; Cunningham, C.N.; Zhou, L.; Forrest, W.F.; Liu, P.S.; Steffek, M.; Lee, J.; Tam, C.; Helgason, E.; et al. Conformational dynamics control ubiquitin-deubiquitinase interactions and influence in vivo signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 11379–11384. [Google Scholar] [CrossRef]
- Teyra, J.; Singer, A.U.; Schmitges, F.W.; Jaynes, P.; Kit Leng Lui, S.; Polyak, M.J.; Fodil, N.; Krieger, J.R.; Tong, J.; Schwerdtfeger, C.; et al. Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15. Structure 2019, 27, 590.e5–605.e5. [Google Scholar] [CrossRef]
- Zhang, W.; Sartori, M.A.; Makhnevych, T.; Federowicz, K.E.; Dong, X.; Liu, L.; Nim, S.; Dong, A.; Yang, J.; Li, Y.; et al. Generation and Validation of Intracellular Ubiquitin Variant Inhibitors for USP7 and USP10. J. Mol. Biol. 2017, 429, 3546–3560. [Google Scholar] [CrossRef]
- Zhang, W.; Bailey-Elkin, B.A.; Knaap, R.C.M.; Khare, B.; Dalebout, T.J.; Johnson, G.G.; van Kasteren, P.B.; McLeish, N.J.; Gu, J.; He, W.; et al. Potent and selective inhibition of pathogenic viruses by engineered ubiquitin variants. PLOS Pathog. 2017, 13, e1006372. [Google Scholar] [CrossRef]
- Gabrielsen, M.; Buetow, L.; Nakasone, M.A.; Ahmed, S.F.; Sibbet, G.J.; Smith, B.O.; Zhang, W.; Sidhu, S.S.; Huang, D.T. A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants. Mol. Cell 2017, 68, 456.e10–470.e10. [Google Scholar] [CrossRef]
- Gorelik, M.; Orlicky, S.; Sartori, M.A.; Tang, X.; Marcon, E.; Kurinov, I.; Greenblatt, J.F.; Tyers, M.; Moffat, J.; Sicheri, F.; et al. Inhibition of SCF ubiquitin ligases by engineered ubiquitin variants that target the Cul1 binding site on the Skp1-F-box interface. Proc. Natl. Acad. Sci. USA 2016, 113, 3527–3532. [Google Scholar] [CrossRef] [PubMed]
- Watson, E.R.; Grace, C.R.R.; Zhang, W.; Miller, D.J.; Davidson, I.F.; Prabu, J.R.; Yu, S.; Bolhuis, D.L.; Kulko, E.T.; Vollrath, R.; et al. Protein engineering of a ubiquitin-variant inhibitor of APC/C identifies a cryptic K48 ubiquitin chain binding site. Proc. Natl. Acad. Sci. USA 2019, 116, 17280–17289. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.G.; VanderLinden, R.; Watson, E.R.; Weissmann, F.; Ordureau, A.; Wu, K.P.; Zhang, W.; Yu, S.; Mercredi, P.Y.; Harrison, J.S.; et al. Dual RING E3 architectures regulate multiubiquitination and ubiquitin chain elongation by APC/C. Cell 2016, 165, 1440–1453. [Google Scholar] [CrossRef] [PubMed]
- Leung, I.; Dekel, A.; Shifman, J.M.; Sidhu, S.S. Saturation scanning of ubiquitin variants reveals a common hot spot for binding to USP2 and USP21. Proc. Natl. Acad. Sci. USA 2016, 113, 8705–8710. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.C.; Steffen, A.M.; Oldham, M.L.; Chen, J.; Huibregtse, J.M. Structure and function of a HECT domain ubiquitin-binding site. EMBO Rep. 2011, 12, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.-F.; Zhang, Z.; Yang, J.; McLaughlin, S.H.; Barford, D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature 2014, 513, 388–393. [Google Scholar] [CrossRef]
- Ottis, P.; Crews, C.M. Proteolysis-Targeting Chimeras: Induced Protein Degradation as a Therapeutic Strategy. ACS Chem. Biol. 2017, 12, 892–898. [Google Scholar] [CrossRef]
- Salami, J.; Crews, C.M. Waste disposal—An attractive strategy for cancer therapy. Science 2017, 355, 1163–1167. [Google Scholar] [CrossRef]
- Neklesa, T.; Jin, M.; Crew, A.P.; Rossi, A.; Willard, R.; Dong, H.Q.; Siu, K.; Wang, J.; Gordon, D.; Chen, X.; et al. ARV-330: Androgen receptor PROTAC degrader for prostate cancer. J. Clin. Oncol. 2016, 34. [Google Scholar] [CrossRef]
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559. [Google Scholar] [CrossRef]
- Schneekloth, A.R.; Pucheault, M.; Tae, H.S.; Crews, C.M. Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorg. Med. Chem. Lett. 2008, 18, 5904–5908. [Google Scholar] [CrossRef] [PubMed]
- Bondeson, D.P.; Mares, A.; Smith, I.E.D.; Ko, E.; Campos, S.; Miah, A.H.; Mulholland, K.E.; Routly, N.; Buckley, D.L.; Gustafson, J.L.; et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 2015, 11, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Buckley, D.L.; Gustafson, J.L.; Van Molle, I.; Roth, A.G.; Tae, H.S.; Gareiss, P.C.; Jorgensen, W.L.; Ciulli, A.; Crews, C.M. Small-molecule inhibitors of the interaction between the E3 ligase VHL and HIF1α. Angew. Chem. Int. Ed. Engl. 2012, 51, 11463–11467. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Ishikawa, M.; Naito, M.; Hashimoto, Y. Protein Knockdown Using Methyl Bestatin−Ligand Hybrid Molecules: Design and Synthesis of Inducers of Ubiquitination-Mediated Degradation of Cellular Retinoic Acid-Binding Proteins. J. Am. Chem. Soc. 2010, 132, 5820–5826. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Ando, H.; Suzuki, T.; Ogura, T.; Hotta, K.; Imamura, Y.; Yamaguchi, Y.; Handa, H. Identification of a Primary Target of Thalidomide Teratogenicity. Science 2010, 327, 1345–1350. [Google Scholar] [CrossRef]
- Lopez-Girona, A.; Mendy, D.; Ito, T.; Miller, K.; Gandhi, A.K.; Kang, J.; Karasawa, S.; Carmel, G.; Jackson, P.; Abbasian, M.; et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia 2012, 26, 2326–2335. [Google Scholar] [CrossRef]
- Sun, X.; Gao, H.; Yang, Y.; He, M.; Wu, Y.; Song, Y.; Tong, Y.; Rao, Y. PROTACs: Great opportunities for academia and industry. Signal Transduct. Target. Ther. 2019, 4, 64. [Google Scholar] [CrossRef]
- Chamberlain, P.P.; Lopez-Girona, A.; Miller, K.; Carmel, G.; Pagarigan, B.; Chie-Leon, B.; Rychak, E.; Corral, L.G.; Ren, Y.J.; Wang, M.; et al. Structure of the human Cereblon–DDB1–lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat. Struct. Mol. Biol. 2014, 21, 803–809. [Google Scholar] [CrossRef]
- Fischer, E.S.; Böhm, K.; Lydeard, J.R.; Yang, H.; Stadler, M.B.; Cavadini, S.; Nagel, J.; Serluca, F.; Acker, V.; Lingaraju, G.M.; et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 2014, 512, 49–53. [Google Scholar] [CrossRef]
- Pettersson, M.; Crews, C.M. PROteolysis TArgeting Chimeras (PROTACs)—Past, present and future. Drug Discov. Today. Technol. 2019, 31, 15–27. [Google Scholar] [CrossRef]
- Saenz, D.T.; Fiskus, W.; Qian, Y.; Manshouri, T.; Rajapakshe, K.; Raina, K.; Coleman, K.G.; Crew, A.P.; Shen, A.; Mill, C.P.; et al. Novel BET protein proteolysis-targeting chimera exerts superior lethal activity than bromodomain inhibitor (BETi) against post-myeloproliferative neoplasm secondary (s) AML cells. Leukemia 2017, 31, 1951–1961. [Google Scholar] [CrossRef] [PubMed]
- Piya, S.; Mu, H.; Bhattacharya, S.; Lorenzi, P.L.; Davis, R.E.; McQueen, T.; Ruvolo, V.; Baran, N.; Wang, Z.; Qian, Y.; et al. BETP degradation simultaneously targets acute myelogenous leukemia stem cells and the microenvironment. J. Clin. Investig. 2019, 129, 1878–1894. [Google Scholar] [CrossRef]
- LeBlanc, N.; Mallette, E.; Zhang, W. Targeted modulation of E3 ligases using engineered ubiquitin variants. FEBS J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Kanner, S.A.; Shuja, Z.; Choudhury, P.; Jain, A.; Colecraft, H.M. Targeted deubiquitination rescues distinct trafficking-deficient ion channelopathies. Nat. Methods 2020, 17, 1245–1253. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Aminu, B.; Roscow, O.; Zhang, W. Targeting the Ubiquitin Signaling Cascade in Tumor Microenvironment for Cancer Therapy. Int. J. Mol. Sci. 2021, 22, 791. https://doi.org/10.3390/ijms22020791
Liu Q, Aminu B, Roscow O, Zhang W. Targeting the Ubiquitin Signaling Cascade in Tumor Microenvironment for Cancer Therapy. International Journal of Molecular Sciences. 2021; 22(2):791. https://doi.org/10.3390/ijms22020791
Chicago/Turabian StyleLiu, Qi, Bayonle Aminu, Olivia Roscow, and Wei Zhang. 2021. "Targeting the Ubiquitin Signaling Cascade in Tumor Microenvironment for Cancer Therapy" International Journal of Molecular Sciences 22, no. 2: 791. https://doi.org/10.3390/ijms22020791
APA StyleLiu, Q., Aminu, B., Roscow, O., & Zhang, W. (2021). Targeting the Ubiquitin Signaling Cascade in Tumor Microenvironment for Cancer Therapy. International Journal of Molecular Sciences, 22(2), 791. https://doi.org/10.3390/ijms22020791





