Identification and Functional Analysis of Tomato TPR Gene Family
Abstract
1. Introduction
2. Results
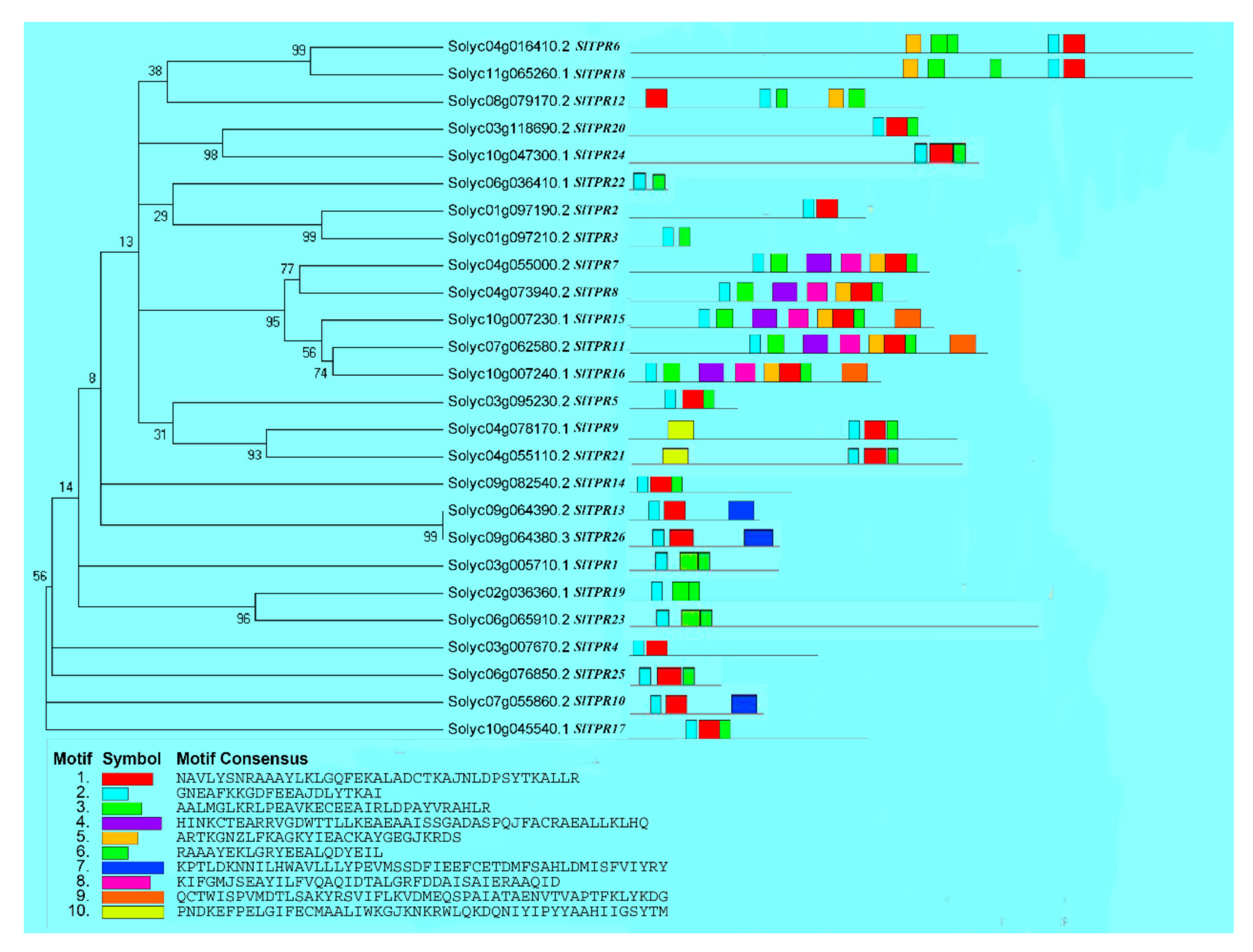
2.1. Identification and Phylogenetic Analysis of Tomato TPR Genes
2.2. Analysis of Tomato TPR Gene Conserved Motif and Gene Structure
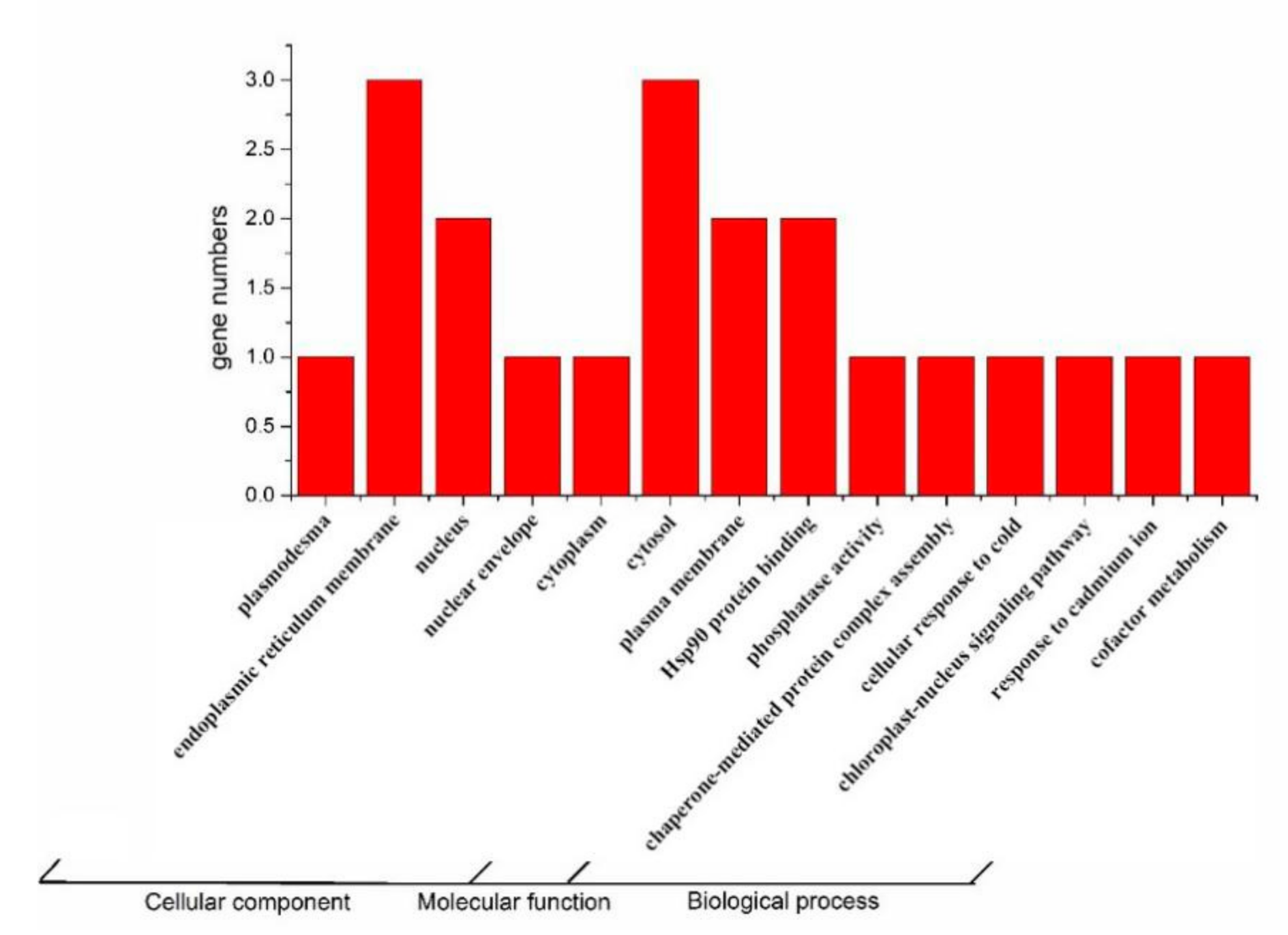
2.3. Expression Analysis and GO Analysis of Tomato TPRs Gene
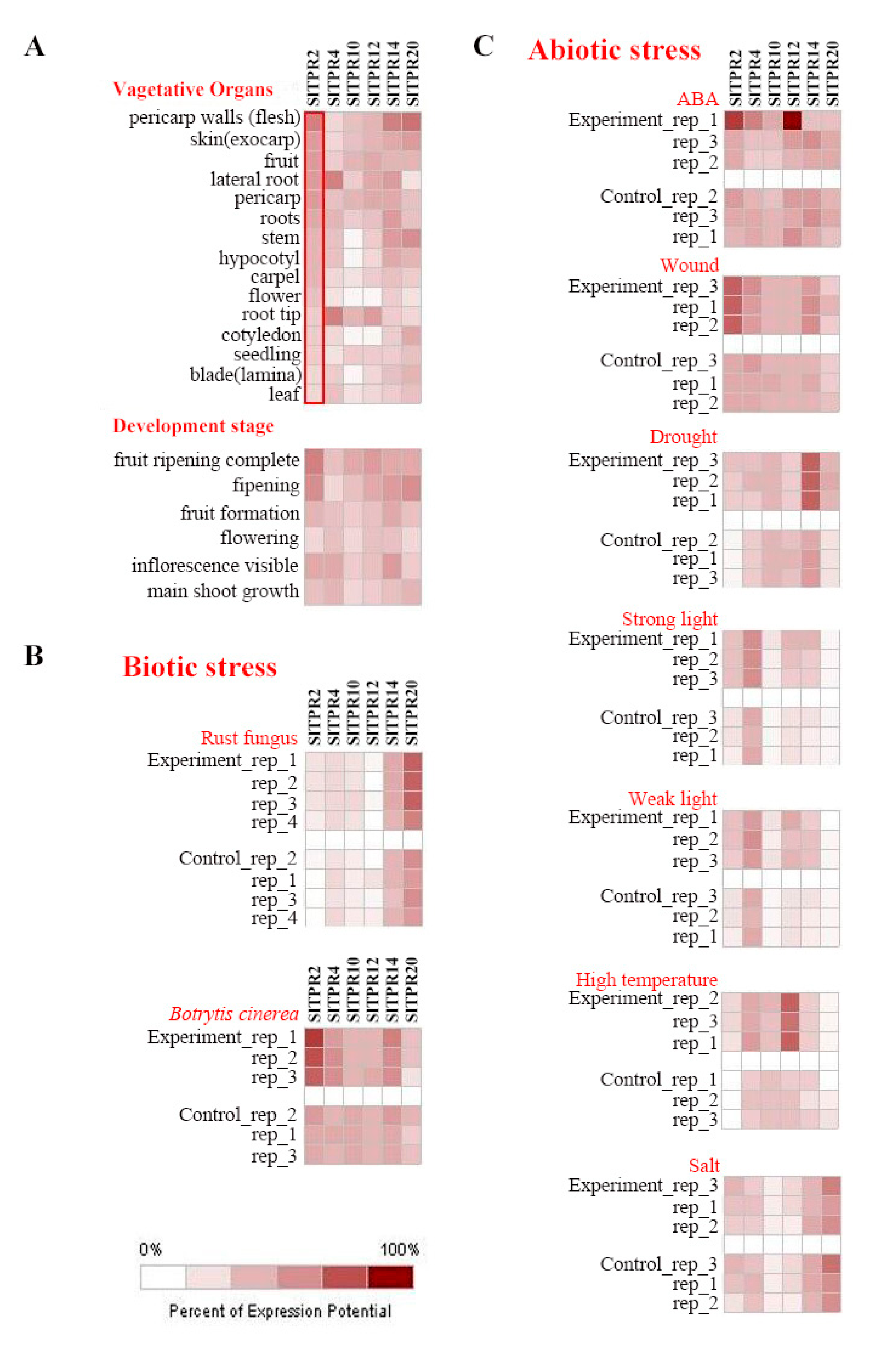
2.4. Tissue Expression Analysis of Tomato TPRs Gene
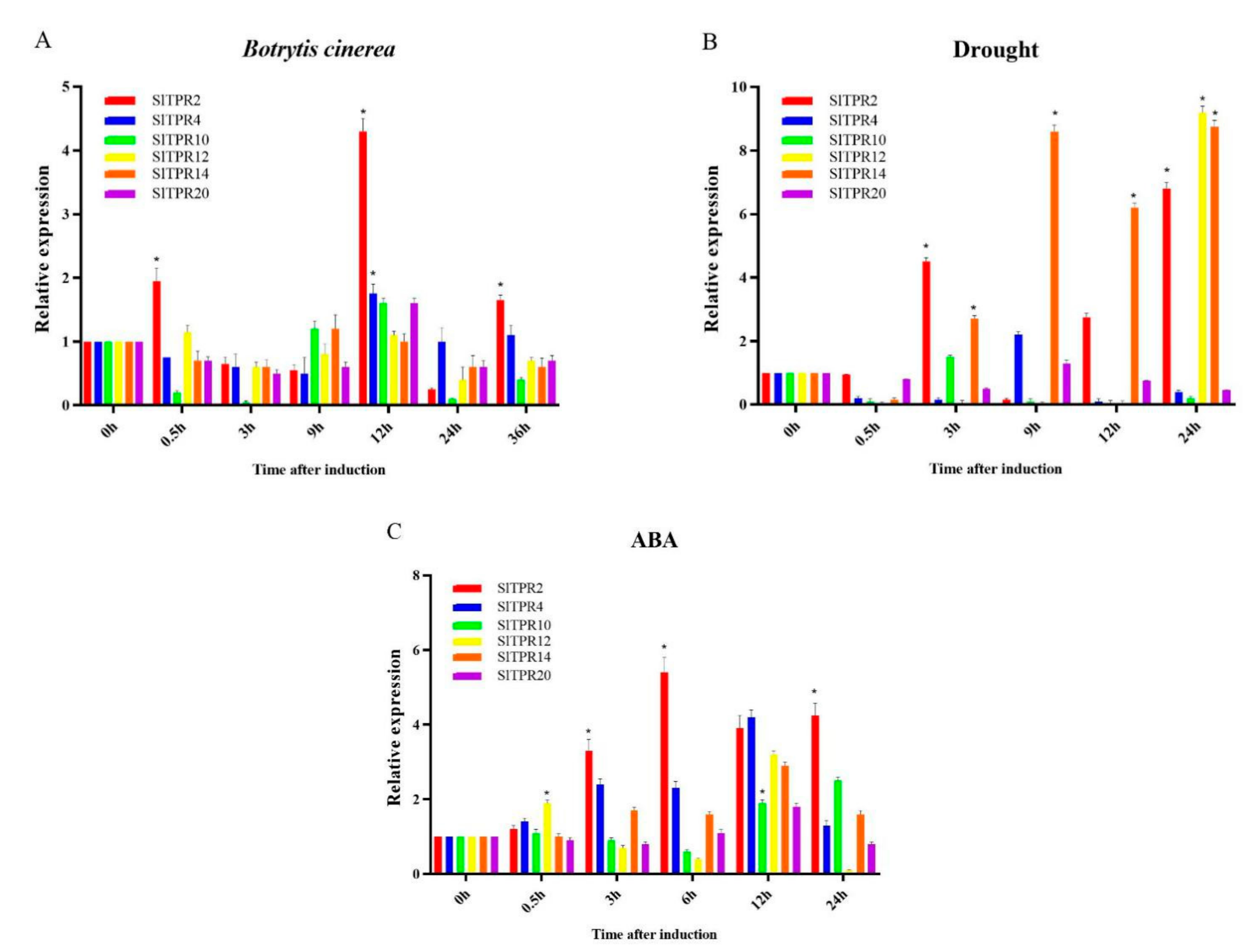
2.5. Expression Analysis of Tomato TPR Gene under Stress
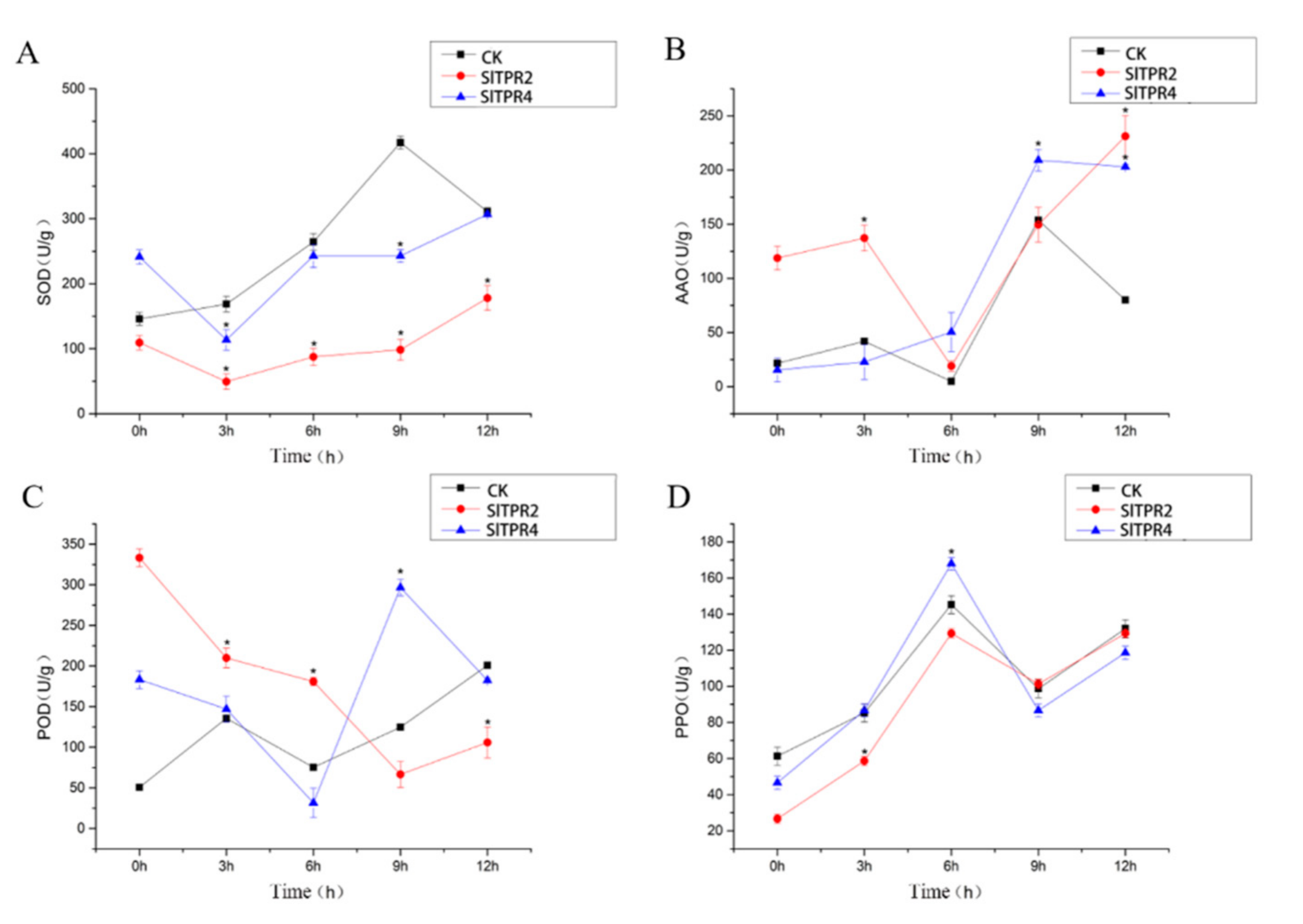
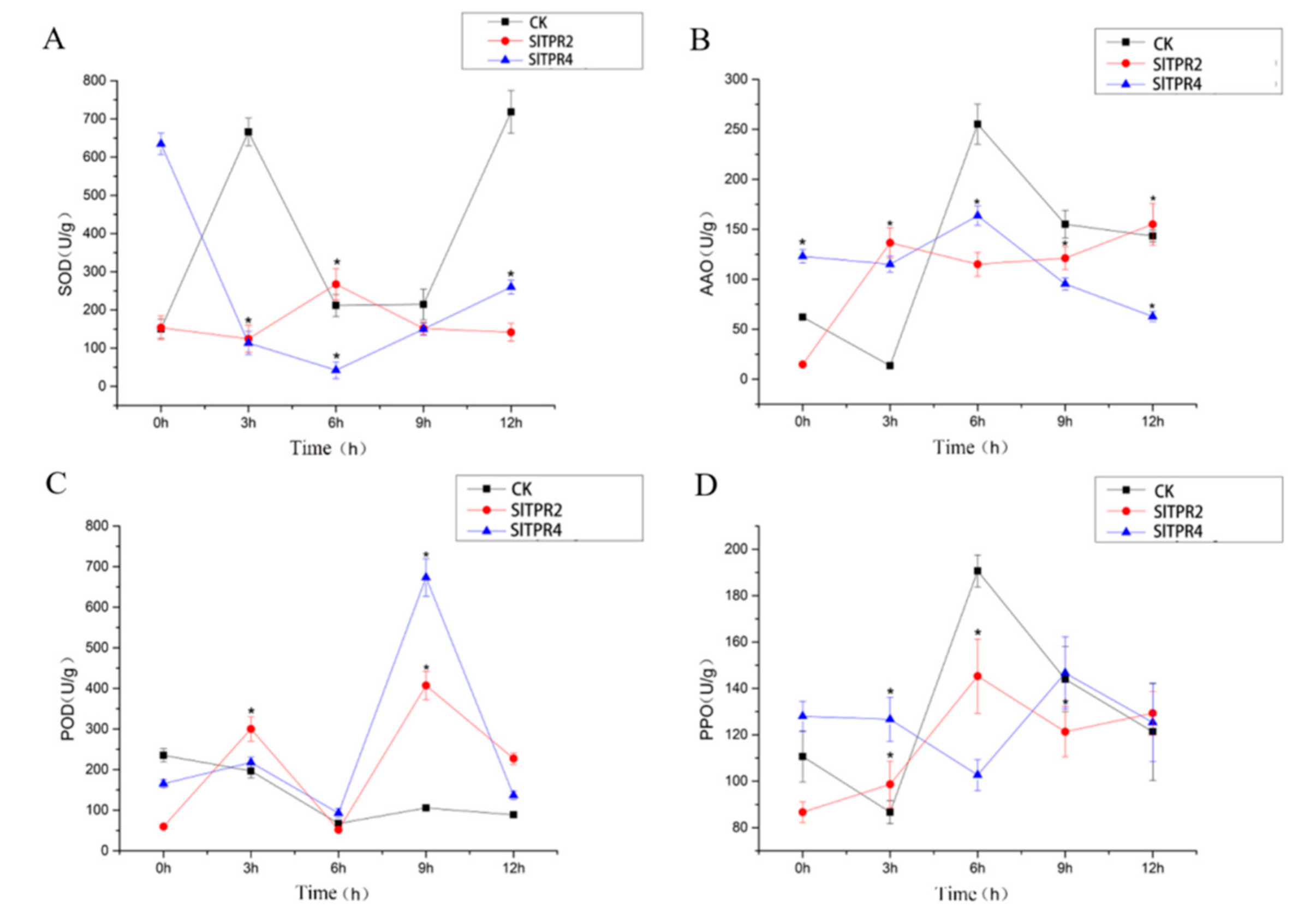
2.6. Functional Analysis of Tomato SlTPR2 and SlTPR4 Silencing in Tomato Stress
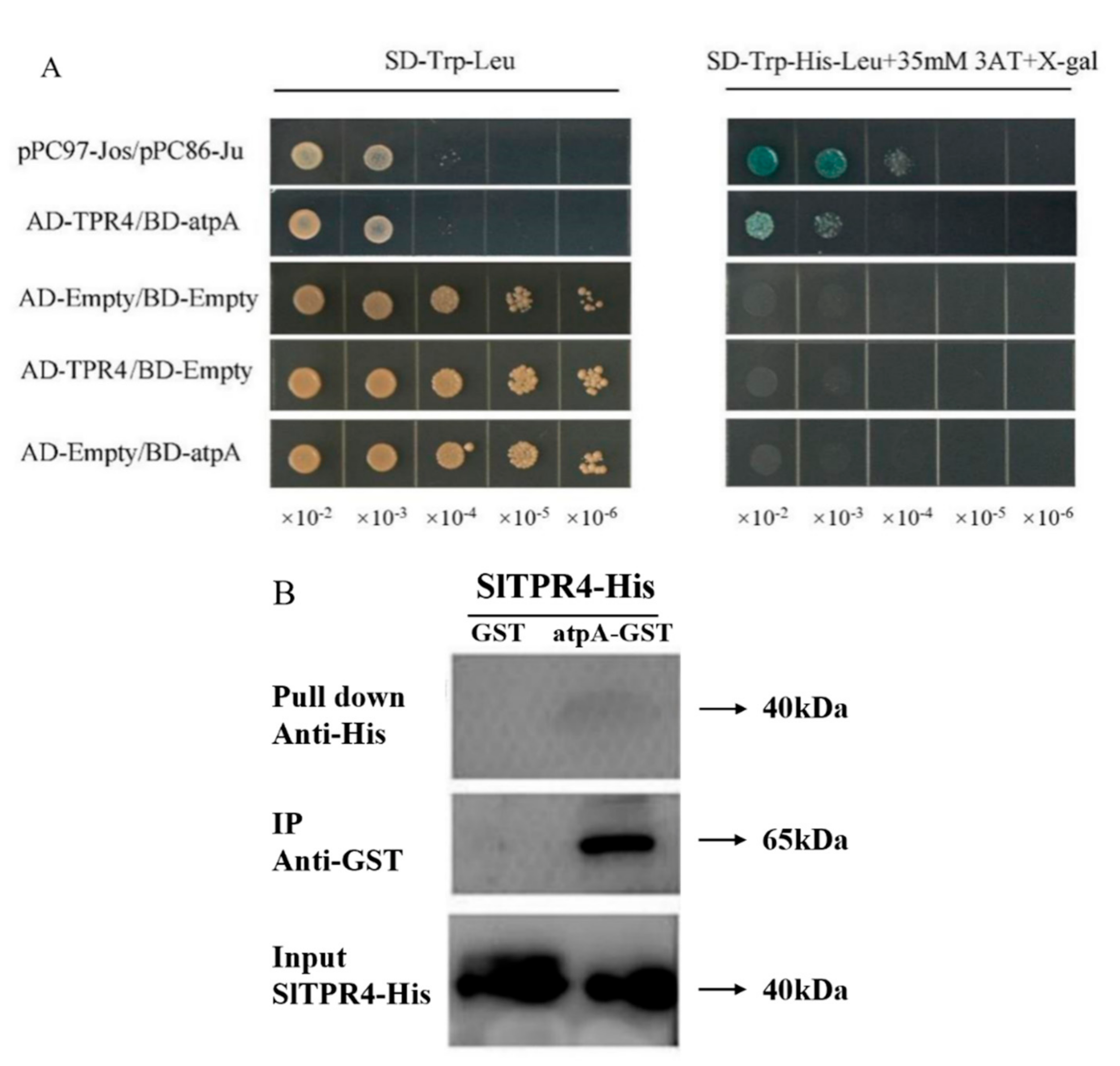
2.7. TPR4 Interacts with atpA
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Materials
5.2. Bioinformatics Analysis of TPR Gene Family
5.3. Quantitative Real Time Polymerase Chain Reaction Analysis
5.4. Virus-Induced Gene Silencing (VIGS)
5.5. Yeast Two-Hybrid Assay
5.6. Pull-Down Assay
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schauer, N.; Semel, Y.; Balbo, I.; Steinfath, M.; Repsilber, D.; Selbig, J.; Pleban, T.; Zamir, D.; Fernie, A.R. Mode of inheritance of primary metabolic traits in tomato. Plant Cell 2008, 20, 509–523. [Google Scholar] [CrossRef]
- Klee, H.J.; Giovannoni, J.J. Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 2011, 45, 41–59. [Google Scholar] [CrossRef]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef]
- Goebl, M.; Yahagida, M. The TPR snap helix: A novel protein repeat motif from mitosis to transcription. Trends Biochem. Sci. 1991, 16, 173. [Google Scholar] [CrossRef]
- Cerveny, L.; Straskova, A.; Dankova, V.; Hartlova, A.; Ceckova, M.; Staud, F.; Stulik, J. Tetratricopeptide repeat motifs in the world of bacterial pathogens: Role in virulence mechanisms. Infect. Immun. 2013, 81, 629–635. [Google Scholar] [CrossRef]
- Hirana, T.; Kinoshita, N.; Morikawa, K. Snap helix with knob and hole: Essential repeats in S. pombe nuclear protein nuc2+. Cell 1990, 60, 319–328. [Google Scholar] [CrossRef]
- Goodarzi, M.O.; Xu, N.; Cui, J.; Guo, X.; Chen, Y.I.; Azziz, R. Small glutamine-rich tetratricopeptide repeat-containing protein alpha (SGTA), a candidate gene for polycystic ovary syndrome. Hum. Reprod. 2008, 23, 1214–1219. [Google Scholar] [CrossRef]
- Groshong, A.M.; Fortune, D.E.; Moore, B.P.; Spencer, H.J.; Skinner, R.A.; Bellamy, W.T.; Blevins, J.S. BB0238, a presumed tetratricopeptide repeat-containing protein, is required during Borrelia burgdorferi mammalian infection. Infect. Immun. 2014, 82, 4292–4306. [Google Scholar] [CrossRef] [PubMed]
- Sohocki, M.M.; Browne, S.J.; Sullivan, L.S.; Blackshaw, S.; Cepko, C.L.; Payne, A.M.; Bhattacharya, S.S.; Khaliq, S.; Mehdi, S.Q.; Birch, D.G.; et al. Mutations in a new photoreceptor-pineal gene on 17p cause Leber congenital amaurosis. Nat. Genet. 2000, 24, 79–83. [Google Scholar] [CrossRef]
- Grizot, S.; Fieschi, F.; Dagher, M.C.; Pebay-Peyroula, E. The active N-terminal region of p67phox. Structure at 1.8 A resolution and biochemical characterizations of the A128V mutant implicated in chronic granulomatous disease. J. Biol. Chem. 2013, 276, 21627–21631. [Google Scholar] [CrossRef]
- Tsukahara, F.; Urakawa, I.; Hattori, M.; Hirai, M.; Ohba, K.; Yoshioka, T.; Sakaki, Y.; Muraki, T. Molecular characterization of the mouse mTPRd gene, a homologue of human TPRD: Unique gene expression suggesting its critical role in the pathophysiology of Down syndrome. J. Biochem. 1998, 123, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Kotaka, M.; Tan, Y.J. Expression, purification and preliminary crystallographic analysis of recombinant human small glutamine-rich tetratricopeptide-repeat protein. Acta Crystallogr. 2008, 64, 602–604. [Google Scholar] [CrossRef] [PubMed]
- Jang, W.H.; Jeong, Y.J.; Choi, S.H.; Kim, S.J.; Seog, D.H. Dynamin-1-like protein (Dnm1L) interaction with kinesin light chain 1 (KLC1) through the tetratricopeptide repeat (TPR) domains. Biosci. Biotechnol. Biochem. 2014, 78, 2069–2072. [Google Scholar] [CrossRef]
- Li, J.; Liu, J.; Wang, G.; Cha, J.Y.; Li, G.; Chen, S.; Li, Z.; Guo, J.; Zhang, C.; Yang, Y.; et al. A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 2015, 27, 908–925. [Google Scholar] [CrossRef]
- Greenboim-Wainberg, Y.; Maymon, I.; Borochov, R.; Alvarez, J.; Olszewski, N.; Ori, N.; Eshed, Y.; Weiss, D. Cross talk between gibberellin and cytokinin: The Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 2005, 17, 92–102. [Google Scholar] [CrossRef]
- Rosado, A.; Schapire, A.L.; Bressan, R.A.; Antoine, L.; Harfouche, A.L.; Hasegawa, P.M.; Botella, M.A. The Arabidopsis tetratricopeptide repeat-containing protein TTL1 is required for osmotic stress responses and abscisic acid sensitivity. Plant Physiol. 2006, 142, 1113–1126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, F.; Fang, Y.; Chen, X.M.; Chen, Y.H.; Zhang, W.X.; Dai, H.E.; Liu, L. The Non-canonical Tetratricopeptide Repeat (TPR) Domain of Fluorescent (FLU) Mediates Complex Formation with Glutamyl-tRNA Reductase. J. Biol. Chem. 2015, 290, 17559–17565. [Google Scholar] [CrossRef]
- Tseng, T.S.; Salom, P.A.; Mcclung, C.R. SPINDLY and GIGANTEA Interact and Act in Arabidopsis thaliana Pathways Involved in Light Responses, Flowering and Rhythms in Cotyledon Movements. Plant Cell 2004, 16, 1550. [Google Scholar] [CrossRef]
- Schweiger, R.; Mller, N.C.; Schmitt, M.J.; Schwenkert, S. AtTPR7 is a chaperone-docking protein of the Sec translocon in Arabidopsis. J. Cell Sci. 2012, 125, 5196–5207. [Google Scholar] [CrossRef]
- Kwon, S.I.; Kim, S.H.; Bhattacharjee, S.; Noh, J.J.; Gassmann, W. SRFR1, a suppressor of effector-triggered immunity, encodes a conserved tetratricopeptide repeat protein with similarity to transcriptional repressors. Plant J. 2009, 57, 109–119. [Google Scholar] [CrossRef]
- Nicolet, C.M.; Craig, E.A. Isolation and characterization of STI1, a stress inducible gene from Saccharomyces cerevisiae. Mol. Cell. Biol. 1989, 9, 3638–3646. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.F.; Sullivan, W.P.; Marion, T.N.; Zaitsu, K.; Madden, B.; McCormick, D.J.; Toft, D.O. Identification of a 60-kilodalton stress-related protein, p60, which interacts with hsp90 and hsp70. Mol. Cell. Biol. 1993, 13, 869–876. [Google Scholar] [CrossRef]
- Chang, H.-C.J.; Lindquist, S. Conservation of Hsp90 macromolecular complexes in Saccharomyces cerevisiae. J. Biol. Chem. 1994, 269, 24983–24988. [Google Scholar] [PubMed]
- Zhu, X.; Xiao, K.; Cui, H.; Hu, J. Overexpression of the Prunus sogdiana NBS-LRR subgroup gene PsoRPM2 promotes resistance to the root-knot nematode Meloidogyne incognita in tobacco. Front. Microbiol. 2017, 8, 2113. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Kud, J.; Zhao, Z.; Du, X.; Liu, Y.; Zhao, Y.; Xiao, F. SGT1 interacts with the Prf resistance protein and is required for Prf accumulation and Prf-mediated defense signaling. Biochem. Biophys. Res. Commun. 2013, 431, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, C.; Betsuyaku, S.; Peart, J.; Takahashi, A.; Noël, L.; Sadanandom, A. Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 2006, 25, 2007–2016. [Google Scholar] [CrossRef] [PubMed]
- Leone, V.; Faraldo-Gomez, J.D. Structure and mechanism of the ATP synthase membrane motor inferred from quantitative integrative modeling. J. Gen. Physiol. 2016, 148, 441–457. [Google Scholar] [CrossRef]
- Cherian, S.; Figueroa, C.R.; Nair, H. ‘Movers and shakers’ in the regulation of fruit ripening: A cross-dissection of climacteric versus non-climacteric fruit. J. Exp. Bot. 2014, 65, 4705–4722. [Google Scholar] [CrossRef]
- Sun, T.; Wu, W.; Wu, H.; Rou, W.; Zhou, Y.; Zhuo, T.; Fan, X.; Hu, X.; Zou, H. Ralstonia solanacearum elicitor RipX induces defense reaction by suppressing the mitochondrial atpA gene in host plant. Int. J. Mol. Sci. 2020, 21, 2000. [Google Scholar] [CrossRef]
- Xu, X.M.; Rose, A.; Muthuswamy, S.; Jeong, S.Y.; Venkatakrishnan, S.; Zhao, Q.; Meier, I. NUCLEAR-PORE ANCHOR, the Arabidopsis Homolog of TPR/MIp1/MIp2/Megator, is Involved in mRNA Export, SUMO Homeostasis and Affects Diverse Aspects of Plant Development. Plant Cell 2007, 19, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, A.; Tanaka, H.; Kato, C.; Iwasaki, Y.; Asahi, T. Molecular Characterization of the ZKT Gene Encoding a Protein with PDZ, K-Box and TPR Motifs in Arabidopsis. J. Agric. Chem. Soc. Jpn. 2005, 69, 972–978. [Google Scholar]
- Panigrahi, R.; Adina-Zada, A.; Whelan, J.; Vrielink, A. Ligand Recognition by the TPR Domain of the Import Factor Toc64 from Arabidopsis thaliana. PLoS ONE 2013, 8, e83461. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Han, P. Comparative functional genomics of the TPR gene family in Arabidopsis, rice and maize. Mol. Breed. 2017, 37, 152. [Google Scholar] [CrossRef]
- An, J.; Lee, J.; Hur, Y. Study of Gene (AtTPR) Coding TPR (Tetratricopeptide Repeat) Motif Protein in Arabidopsis. Energies 2007, 6, 5597–5608. [Google Scholar]
- Song, W.; Wang, B.; Li, X.; Wei, J.; Chen, L.; Zhang, D.; Zhang, W.; Li, R. Identification of Immune Related LRR-Containing Genes in Maize (Zea mays L.) by Genome-Wide Sequence Analysis. Int. J. Genom. 2015, 231358. [Google Scholar] [CrossRef]
- Inz, D. CORRECTION: A Repressor Protein Complex Regulates Leaf Growth in Arabidopsis. Plant Cell 2016, 40, 49–56. [Google Scholar]
- Uhrig, R.G.; Moorhead, G. AtSLP2 is an intronless protein phosphatase that co-expresses with intronless mitochondrial pentatricopeptide repeat (PPR) and tetratricopeptide (TPR) protein encoding genes. Plant Signal. Behav. 2017, 12, 443–450. [Google Scholar] [CrossRef]
- Tör, M.; Gordon, P.; Cuzick, A.; Eulgem, T.; Sinapidou, E.; Mert-Türk, F.; Can, C.; Dangl, J.L.; Holub, E.B. Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell 2002, 14, 993–1003. [Google Scholar] [CrossRef]
- Gray, W.M.; Muskett, P.R.; Chuang, H.W.; Parker, J.E. Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 2003, 15, 1310–1319. [Google Scholar] [CrossRef]
- Bhattarai, K.K.; Li, Q.; Liu, Y.; Dinesh-Kumar, S.P.; Kaloshian, I. The MI-1-mediated pest resistance requires Hsp90 and Sgt1. Plant Physiol. 2007, 144, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Shirasu, K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 2009, 60, 139–164. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S. Low Temperature-Induced Cytoplasmic Acidosis in Cultured Mung Bean (Vigna radiata [L.] Wilczek) Cells. Plant Physiol. 1994, 104, 1131–1138. [Google Scholar] [CrossRef] [PubMed]
- Sugimota, K.; Matsui, K.; Ozawa, R.; Kuramitsu, Y.; Kley, J.; David, A.; Mück, A.; Nakamura, K.; Boland, W.; Takabayashi, J. Induced defence in lima bean plants exposed to the volatiles from two-spotted spider mite-infested conspecifics is independent of the major protein expression. J. Plant Interact. 2013, 8, 219–224. [Google Scholar] [CrossRef]
- Takken, F.L.W.; Tameling, W.I.L. To nibble at plant resistance proteins. Science 2009, 324, 744–746. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, X.; Yue, J.X.; Tian, D.; Chen, J.Q. Recent duplications dominate nbs-encoding gene expansion in two woody species. Mol. Genet. Genom. 2008, 280, 187. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhang, X.; Yang, S. A novel chloroplast-localized protein EMB1303 is required for chloroplast development in Arabidopsis. Cell Res. 2009, 19, 1205–1216. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, X.; Liu, S.; Yu, A.; Yang, C.; Chen, X.; Liu, J.; Wang, A. Identification and Functional Analysis of Tomato CIPK Gene Family. Int. J. Mol. Sci. 2020, 21, 110. [Google Scholar] [CrossRef]
- Wang, Z.; Meng, P.; Zhang, X.; Ren, D.; Yang, S. BON1 interacts with the protein kinases BIR1 and BAK1 in modulation of temperature dependent plant growth and cell death in Arabidopsis. Plant J. 2011, 67, 1081–1093. [Google Scholar] [CrossRef]

| Gene Name | Sequence Accession | No. Amino Acid | Chromosome Location | pI | Protein M.W (kDa) | Subcellular Localization |
|---|---|---|---|---|---|---|
| SlTPR1 | solyc03g005710.1 | 378 | 502006–505012 | 4.4 | 42,542 | 1 |
| SlTPR2 | solyc01g097190.2 | 464 | 88125140–8813834 | 5.11 | 49,913 | 1 |
| SlTPR3 | solyc01g097210.2 | 120 | 88147803–88150286 | 8.34 | 13,365 | 1 |
| SlTPR4 | solyc03g007670.2 | 370 | 2200919–2205536 | 5.11 | 41,245 | 1 |
| SlTPR5 | solyc03g095230.2 | 211 | 56209836–56215404 | 9.23 | 23,613 | 1 |
| SlTPR6 | solyc04g016410.1 | 1420 | 7200714–7218394 | 8.47 | 156,043 | 2 |
| SlTPR7 | solyc04g055000.2 | 587 | 53530227–53533608 | 9.26 | 64,012 | 3,4 |
| SlTPR8 | solyc04g073940.2 | 543 | 59949691..59951735 | 9.5 | 61,374 | 2 |
| SlTPR9 | solyc04g078170.1 | 644 | 62975642..62978282 | 8.92 | 73,681 | 1 |
| SlTPR10 | solyc07g055860.2 | 532 | 63773890..63783760 | 5.89 | 59,541 | 1 |
| SlTPR11 | solyc07g062580.2 | 701 | 65259956..65263961 | 9.05 | 76,013 | 2 |
| SlTPR12 | solyc08g079170.2 | 579 | 62804339..62810456 | 5.99 | 65,166 | 3 |
| SlTPR13 | solyc09g064390.2 | 256 | 61561701..61566057 | 6.34 | 28,958 | 1 |
| SlTPR14 | solyc09g082540.2 | 318 | 68259584..68265551 | 8.76 | 36,210 | 3 |
| SlTPR15 | solyc10g007230.2 | 596 | 1650247..1653367 | 8.72 | 65,786 | 2,1 |
| SlTPR16 | solyc10g007240.2 | 594 | 1660427..1662715 | 8.6 | 65,206 | 2,1 |
| SlTPR17 | solyc10g045540.2 | 470 | 34548304..34563332 | 8.83 | 52,399 | 2 |
| SlTPR18 | solyc11g065260.1 | 1261 | 50631990..50650877 | 6 | 138,554 | 4 |
| SlTPR19 | solyc02g036360.1 | 761 | 30576501..30578786 | 5.32 | 84,464 | 1,3 |
| SlTPR20 | solyc03g118690.2 | 590 | 67543799..67551645 | 8.94 | 64,110 | 3 |
| SlTPR21 | solyc04g055110.2 | 627 | 53710473..53724633 | 5.83 | 70,253 | 1 |
| SlTPR22 | solyc06g036410.2 | 661 | 25974062..25974262 | 4.95 | 7,364 | 1 |
| SlTPR23 | solyc06g065910.2 | 685 | 41307528..41309760 | 5.47 | 78,976 | 1 |
| SlTPR24 | solyc10g047300.1 | 598 | 40482669..40501656 | 9.2 | 65,597 | 2,3 |
| SlTPR25 | solyc06g076850.2 | 551 | 47742135..47745217 | 5.22 | 62,095 | 2 |
| SlTPR26 | solyc09g064380.3 | 128 | 61558744..61562593 | 6.34 | 28,958 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Zheng, Y.; Cai, Z.; Wang, X.; Liu, Y.; Yu, A.; Chen, X.; Liu, J.; Zhang, Y.; Wang, A. Identification and Functional Analysis of Tomato TPR Gene Family. Int. J. Mol. Sci. 2021, 22, 758. https://doi.org/10.3390/ijms22020758
Zhou X, Zheng Y, Cai Z, Wang X, Liu Y, Yu A, Chen X, Liu J, Zhang Y, Wang A. Identification and Functional Analysis of Tomato TPR Gene Family. International Journal of Molecular Sciences. 2021; 22(2):758. https://doi.org/10.3390/ijms22020758
Chicago/Turabian StyleZhou, Xi’nan, Yangyang Zheng, Zhibo Cai, Xingyuan Wang, Yang Liu, Anzhou Yu, Xiuling Chen, Jiayin Liu, Yao Zhang, and Aoxue Wang. 2021. "Identification and Functional Analysis of Tomato TPR Gene Family" International Journal of Molecular Sciences 22, no. 2: 758. https://doi.org/10.3390/ijms22020758
APA StyleZhou, X., Zheng, Y., Cai, Z., Wang, X., Liu, Y., Yu, A., Chen, X., Liu, J., Zhang, Y., & Wang, A. (2021). Identification and Functional Analysis of Tomato TPR Gene Family. International Journal of Molecular Sciences, 22(2), 758. https://doi.org/10.3390/ijms22020758





