The Role of Noncoding RNAs in the Regulation of Anoikis and Anchorage-Independent Growth in Cancer
Abstract
:1. Introduction
2. MiRNAs Positively Regulating Anoikis Resistance and Anchorage-Independent Growth
2.1. MiRNAs Regulating Apoptosis- and Cell Survival-Related Factors
2.1.1. MiR-21-5p and miR-25-3p
2.1.2. MiR-106-5p and miR-141-3p
2.1.3. MiR-145-5p and miR-146-3p
2.1.4. MiR-186-5p, miR-206, and miR-421
2.1.5. MiR-411-5p and miR-520f-5p
2.1.6. MiR-424-5p
2.1.7. MiR-494 and miR-645
2.2. MiRNAs Modulating Cell Cycle Regulators
2.2.1. MiR-27-3p
2.2.2. MiR-141-3p
2.2.3. MiR-182-5p and miR-1303
2.2.4. MiR-186-5p and miR-1288-3p
2.2.5. MiR-362-3p and miR-376c-3p
2.2.6. MiR-527 and miR-760
2.2.7. MiR-582-5p and miR-3607
2.2.8. MiR-766-5p
2.3. MiRNAs Regulating EMT and Stemness
2.3.1. MiR-9-5p
2.3.2. MiR-10b
2.3.3. MiR-139-5p and miR-483-5p
2.3.4. MiR-526b-5p and miR-655-3p
2.3.5. MiR-G-10
2.4. A miRNA Regulating Integrin Expression
MiR-145-5p
3. MiRNAs Suppressing Anoikis Resistance and Anchorage-Independent Growth
3.1. MiRNAs Regulating Apoptosis- and Cell Survival-Related Factors
3.1.1. MiR-10a-5p and miR-22-3p
| miRNA | Type of Cancer | In Vitro Experiment | In Vivo Experiment | Ref. |
|---|---|---|---|---|
| miR-10a-5p | Colorectal cancer | Plating cells on polyHEMA-coated plates + annexin-V/propidium iodide analysis * | Injection of pri-miR-10a-expressing SW620 cells into the spleen | [125] |
| miR-22-3p | Hepatocellular carcinoma | Soft agar assay ** | Orthotopic implantation of galectin-1 silencing MHCC97L cells into the liver | [128] |
| miR-22-3p | Rhabdomyosarcoma | Soft agar assay ** | Subcutaneous injection of RD18 cells conditionally expressing miR-22-3p | [127] |
| miR-23b | Prostate cancer | Soft agar assay ** | Orthotopic injection of PC3-ML cells overexpressing miR-23b and miR-27b | [129] |
| miR-26-5p | Hepatocellular carcinoma | Plating cells on polyHEMA-coated plates + annexin-V/propidium iodide analysis * | Intravenous injection of BEL-7404 cells overexpressing miR-26 for in vivo anoikis assay | [130] |
| miR-27b | Colorectal cancer | Colony formation assay ** | Subcutaneous injection of HCT116 cells overexpressing miR-27-3p | [131] |
| miR-27b | Prostate cancer | Soft agar assay ** | Orthotopic injection of PC3-ML cells overexpressing miR-23b and miR-27b | [129] |
| miR-29-3p | Pancreatic cancer | Soft agar assay ** | - | [132] |
| miR-30-5p | Hepatocellular carcinoma | Plating cells on polyHEMA-coated plates + ethidium homodimer-1 assay * | Examination of malignant ascites for the measure of anoikis. Tail vein injection of HCCLM3 cells overexpressing miR-30-5p for lung metastasis assays | [133] |
| miR-30-5p | Colorectal cancer | Soft agar colony assay ** | - | [134] |
| miR-30-5p | Renal cell carcinoma | Soft agar assay ** | Subcutaneous injection of Caki-1 cells transfected with miR-30-5p | [135] |
| miR-33a | Lung cancer | Soft agar colony formation assay ** | - | [136] |
| miR-124-3p | Colorectal cancer | Plating cells on polyHEMA-coated plates + cell viability assay using 0.4% trypan blue * | Tail vein injection of miR-124-3p-overexpressing Lovo and SW620 cells | [137] |
| miR-133-3p | Esophageal cancer | Plating cells on polyHEMA-coated plates + annexin-V/propidium iodide analysis * | Tail vein injection of KYSE150 and ECa109 cells expressing miR-133-3p | [138] |
| miR-133-3p | Prostate cancer | Spheroid formation assay using ultra-low cluster plates * | Inoculation of PC-3 cells into the left cardiac ventricle + tail vein injection of miR-133-3p inhibitors | [139] |
| miR-137 | Melanoma | Soft agar assay ** | - | [140] |
| miR-137 | Pancreatic cancer | Plating cells on polyHEMA-coated plates + annexin-V/propidium iodide analysis * | Intraperitoneal injection of PANC-1 cells overexpressing miR-137 for in vivo anoikis assay | [141] |
| miR-138 | Ewing’s sarcoma | Annexin-V/propidium iodide analysis following miR-138 transfection * | Tail vein injection of SKES1 cells transfected with miR-138 | [142] |
| miR-141 | Endometrial cancer | Culturing cells on anchorage-resistant plates + calcein AM assay * | - | [143] |
| miR-155-5p | Glioblastoma | Soft agar colony formation assay ** | Injection of miR-155-expressing SNB19 cells into dorsal flank | [144] |
| miR-193b | Pancreatic cancer | Colony-formation assay ** | - | [145] |
| miR-193b | Ewing’s sarcoma | Soft agar assay ** | - | [146] |
| miR-199-5p | Colorectal cancer | Colony-forming assay ** | - | [147] |
| miR-200 | Breast cancer | Soft agar assay ** | - | [148] |
| miR-200 | Bladder cancer | Soft agar assay ** | - | [149] |
| miR-204-5p | Gastric cancer | Plating cells on polyHEMA-coated plates + annexin-V/propidium iodide analysis * | - | [150] |
| miR-204-5p | Medulloblastoma | Soft agar colony formation assay ** | - | [151] |
| miR-335-5p | Gastric cancer | Soft agar colony formation assay ** | - | [152] |
| miR-363-3p | Papillary thyroid cancer | Plating cells on polyHEMA-coated plates + apoptosis analysis by flow cytometry * | - | [153] |
| miR-381 | Rectal cancer | Soft agar assay ** | - | [154] |
| miR-424-5p | Hepatocellular carcinoma | Plating cells on polyHEMA-coated plates + Western blotting for measuring caspase 3 * | Intra-tumoral injection of miR-424-5p expression vectors in a subcutaneous SMMC7721 xenograft model | [155] |
| miR-429 | Soft tissue sarcoma | Soft agar colony formation assay ** | - | [156] |
| miR-450 | Ovarian cancer | Plating cells in ultra-low attachment plates + annexin-V assay * | Intraperitoneal injection of A2780 cells expressing miR-450 | [157] |
| miR-451 | Osteosarcoma | Plating cells on polyHEMA-coated plates + annexin-V/propidium iodide analysis * | Tail vein injection of osteosarcoma cells overexpressing miR-451 | [158] |
| miR-487b-3p | Colorectal cancer | Soft agar assay ** | Subcutaneous injection of CBS cells expressing miR-487b precursor | [159] |
| miR-488-5p | Melanoma | Soft agar assay ** | - | [160] |
| miR-525-5p | Cervical cancer | Plating cells on polyHEMA-coated plates + annexin-V/propidium iodide analysis * | - | [161] |
| miR-592 | Colorectal cancer | Soft agar assay ** | - | [162] |
| miR-630 | Breast cancer | Plating cells on polyHEMA-coated plates + Alamar blue dye assay * | - | [163] |
| miR-1287-5p | Breast cancer | Sphere assay using ultra-low attachment plates ** | Subcutaneous injection of SUM159 cells overexpressing miR-1287-5p | [164] |
| miR-1297 | Colorectal cancer | Colony formation assay ** | - | [165] |
| miR-1827 | Lung cancer | Plating cells on polyHEMA-coated plates + annexin-V/propidium iodide analysis * | - | [166] |
| miR-6744-5p | Breast cancer | Plating cells on low adhesion plates + caspase-3/7 activity assay * | Microinjection of miR-6744-5p transfected MDA-MB-231 cells into the yolk of zebrafish embryos | [167] |
3.1.2. MiR-23b and miR-27b
3.1.3. MiR-30-5p and miR-33a
3.1.4. MiR-133-3p and miR-141
3.1.5. MiR-193b
3.1.6. MiR-335-5p and miR-451
3.1.7. MiR-487b-3p and miR-488-5p
3.1.8. MiR-630 and miR-1287-5p
3.1.9. MiR-1827
3.2. MiRNAs Modulating Cell Cycle Regulators
MiR-592 and miR-1297
3.3. MiRNAs Regulating EMT and Stemness
3.3.1. MiR-22-3p and miR-424-5p
3.3.2. MiR-133-3p, miR-137, and miR-155-5p
3.3.3. MiR-199-5p and miR-200
3.3.4. MiR-204-5p
3.3.5. MiR-381 and miR-525-5p
3.3.6. MiR-429 and miR-450
3.3.7. MiR-6744-5p
3.4. MiRNAs Regulating Autophagy
3.4.1. MiR-29-3p
3.4.2. MiR-30-5p
3.4.3. MiR-204-5p
3.5. MiRNAs Regulating Integrin Levels and FAK Activities
3.5.1. MiR-26-5p, miR-124-3p, and miR-363-3p
3.5.2. MiR-27b, miR-137, and miR-138
4. Long Noncoding RNAs, Anoikis, and Anchorage-Independent Growth
4.1. LncRNA-ANRIL
4.2. LncRNA-FOXD2-AS1
4.3. LncRNA-H19
4.4. LncRNA-HOTAIR
4.5. LncRNA-LEF1-AS1
4.6. LncRNA-MEG3
4.7. LncRNA-NEAT1
4.8. LncRNA-SNHG12
4.9. LncRNA-TINCR
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kim, Y.N.; Koo, K.H.; Sung, J.Y.; Yun, U.J.; Kim, H. Anoikis resistance: An essential prerequisite for tumor metastasis. Int. J. Cell Biol. 2012, 2012, 306879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guadamillas, M.C.; Cerezo, A.; Del Pozo, M.A. Overcoming anoikis—Pathways to anchorage-independent growth in cancer. J. Cell Sci. 2011, 124, 3189–3197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frisch, S.M.; Francis, H. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 1994, 124, 619–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puthalakath, H.; Villunger, A.; O’Reilly, L.A.; Beaumont, J.G.; Coultas, L.; Cheney, R.E.; Huang, D.C.; Strasser, A. Bmf: A proapoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science 2001, 293, 1829–1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mailleux, A.A.; Overholtzer, M.; Schmelzle, T.; Bouillet, P.; Strasser, A.; Brugge, J.S. Bim regulates apoptosis during mammary ductal morphogenesis, and its absence reveals alternative cell death mechanisms. Dev. Cell 2007, 12, 221–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douma, S.; Van Laar, T.; Zevenhoven, J.; Meuwissen, R.; Van Garderen, E.; Peeper, D.S. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature 2004, 430, 1034–1039. [Google Scholar] [CrossRef]
- Gao, Z.; Zhao, G.S.; Lv, Y.; Peng, D.; Tang, X.; Song, H.; Guo, Q.N. Anoikisresistant human osteosarcoma cells display significant angiogenesis by activating the Src kinasemediated MAPK pathway. Oncol. Rep. 2019, 41, 235–245. [Google Scholar] [PubMed] [Green Version]
- Woods, N.T.; Yamaguchi, H.; Lee, F.Y.; Bhalla, K.N.; Wang, H.G. Anoikis, initiated by mcl-1 degradation and bim induction, is deregulated during oncogenesis. Cancer Res. 2007, 67, 10744–10752. [Google Scholar] [CrossRef] [Green Version]
- Gan, L.; Liu, P.; Lu, H.; Chen, S.; Yang, J.; McCarthy, J.B.; Knudsen, K.E.; Huang, H. Cyclin d1 promotes anchorage-independent cell survival by inhibiting foxo-mediated anoikis. Cell Death Differ. 2009, 16, 1408–1417. [Google Scholar] [CrossRef] [Green Version]
- Jolly, M.K.; Ware, K.E.; Xu, S.; Gilja, S.; Shetler, S.; Yang, Y.; Wang, X.; Austin, R.G.; Runyambo, D.; Hish, A.J.; et al. E-cadherin represses anchorage-independent growth in sarcomas through both signaling and mechanical mechanisms. Mol. Cancer Res. 2019, 17, 1391–1402. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Hong, S.H.; Basse, P.H.; Wu, C.; Bartlett, D.L.; Kwon, Y.T.; Lee, Y.J. Cancer stem cells protect non-stem cells from anoikis: Bystander effects. J. Cell. Biochem. 2016, 117, 2289–2301. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.L.; David, J.; Cook-Spaeth, D.; Casey, S.; Cohen, D.; Selvendiran, K.; Bekaii-Saab, T.; Hays, J.L. Autophagy induction results in enhanced anoikis resistance in models of peritoneal disease. Mol. Cancer Res. 2017, 15, 26–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talukdar, S.; Pradhan, A.K.; Bhoopathi, P.; Shen, X.N.; August, L.A.; Windle, J.J.; Sarkar, D.; Furnari, F.B.; Cavenee, W.K.; Das, S.K.; et al. MDA-9/syntenin regulates protective autophagy in anoikis-resistant glioma stem cells. Proc. Natl. Acad. Sci. USA 2018, 115, 5768–5773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alanko, J.; Ivaska, J. Integrin “endoadhesome” signaling suppresses anoikis. Cell Cycle 2016, 15, 605–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stokes, J.B.; Adair, S.J.; Slack-Davis, J.K.; Walters, D.M.; Tilghman, R.W.; Hershey, E.D.; Lowrey, B.; Thomas, K.S.; Bouton, A.H.; Hwang, R.F.; et al. Inhibition of focal adhesion kinase by PF-562,271 inhibits the growth and metastasis of pancreatic cancer concomitant with altering the tumor microenvironment. Mol. Cancer Ther. 2011, 10, 2135–2145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duxbury, M.S.; Ito, H.; Zinner, M.J.; Ashley, S.W.; Whang, E.E. Focal adhesion kinase gene silencing promotes anoikis and suppresses metastasis of human pancreatic adenocarcinoma cells. Surgery 2004, 135, 555–562. [Google Scholar] [CrossRef]
- Yang, Z.; Sun, Q.; Guo, J.; Wang, S.; Song, G.; Liu, W.; Liu, M.; Tang, H. GRSF1-mediated miR-G-1 promotes malignant behavior and nuclear autophagy by directly upregulating TMED5 and LMNB1 in cervical cancer cells. Autophagy 2019, 15, 668–685. [Google Scholar] [CrossRef]
- Lai, X.N.; Li, J.; Tang, L.B.; Chen, W.T.; Zhang, L.; Xiong, L.X. MiRNAs and lncRNAs: Dual roles in TGF-β signaling-regulated metastasis in lung cancer. Int. J. Mol. Sci. 2020, 21, 1193. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef] [Green Version]
- Wei, J.L.; Li, Y.C.; Ma, Z.L.; Jin, Y.X. MiR-181a-5p promotes anoikis by suppressing autophagy during detachment induction in the mammary epithelial cell line MCF10A. Protein Cell 2016, 7, 305–309. [Google Scholar] [CrossRef] [Green Version]
- Seo, H.A.; Moeng, S.; Sim, S.; Kuh, H.J.; Choi, S.Y.; Park, J.K. MicroRNA-based combinatorial cancer therapy: Effects of microRNAs on the efficacy of anti-cancer therapies. Cells 2019, 9, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Son, S.W.; Lee, H.Y.; Moeng, S.; Kuh, H.J.; Choi, S.Y.; Park, J.K. Participation of microRNAs in the treatment of cancer with phytochemicals. Molecules 2020, 25, 4701. [Google Scholar] [CrossRef] [PubMed]
- Park, J.K.; Doseff, A.I.; Schmittgen, T.D. MicroRNAs targeting caspase-3 and -7 in PANC-1 cells. Int. J. Mol. Sci. 2018, 19, 1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, F.; Wang, C.; Li, T.; Cai, W.; Sun, J. Role of miR-21 in the growth and metastasis of human salivary adenoid cystic carcinoma. Mol. Med. Rep. 2018, 17, 4237–4244. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Tan, Z.; Hu, H.; Liu, H.; Wu, T.; Zheng, C.; Wang, X.; Luo, Z.; Wang, J.; Liu, S.; et al. MicroRNA-21 promotes breast cancer proliferation and metastasis by targeting LZTFL1. BMC Cancer 2019, 19, 738. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.L.; Wang, H.; Liu, J.; Wang, Z.X. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol. Cell. Biochem. 2013, 372, 35–45. [Google Scholar] [CrossRef]
- Zhao, M.Y.; Wang, L.M.; Liu, J.; Huang, X.; Liu, J.; Zhang, Y.F. MiR-21 suppresses anoikis through targeting PDCD4 and PTEN in human esophageal adenocarcinoma. Curr. Med. Sci. 2018, 38, 245–251. [Google Scholar] [CrossRef]
- Wan, W.; Wan, W.; Long, Y.; Li, Q.; Jin, X.; Wan, G.; Zhang, F.; Lv, Y.; Zheng, G.; Li, Z.; et al. MiR-25-3p promotes malignant phenotypes of retinoblastoma by regulating PTEN/Akt pathway. Biomed. Pharmacother. 2019, 118, 109111. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, H.; Yang, L.; Wu, Q.; Liu, W.; Fu, Q.; Zhang, W.; Zhang, H.; Xu, J.; Gu, J. Loss of N-acetylgalactosaminyltransferase-4 orchestrates oncogenic microRNA-9 in hepatocellular carcinoma. J. Biol. Chem. 2017, 292, 3186–3200. [Google Scholar] [CrossRef] [Green Version]
- Datar, I.; Kalpana, G.; Choi, J.; Basuroy, T.; Trumbly, R.; Chaitanya Arudra, S.K.; McPhee, M.D.; de la Serna, I.; Yeung, K.C. Critical role of miR-10b in B-RafV600E dependent anchorage independent growth and invasion of melanoma cells. PLoS ONE 2019, 14, e0204387. [Google Scholar] [CrossRef] [Green Version]
- Ye, P.; Ke, X.; Zang, X.; Sun, H.; Dong, Z.; Lin, J.; Wang, L.; Liu, W.; Miao, G.; Tan, Y.; et al. Up-regulated miR-27-3p promotes the g1-s phase transition by targeting inhibitor of growth family member 5 in osteosarcoma. Biomed. Pharmacother. 2018, 101, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Zhang, Y.; Zhang, Z.; Yang, Y.; Song, T. Effect of miR-106b on invasiveness of pituitary adenoma via PTEN-PI3K/Akt. Med. Sci. Monit. 2017, 23, 1277–1285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agosta, C.; Laugier, J.; Guyon, L.; Denis, J.; Bertherat, J.; Libe, R.; Boisson, B.; Sturm, N.; Feige, J.J.; Chabre, O.; et al. MiR-483-5p and miR-139-5p promote aggressiveness by targeting n-myc downstream-regulated gene family members in adrenocortical cancer. Int. J. Cancer 2018, 143, 944–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mak, C.S.; Yung, M.M.; Hui, L.M.; Leung, L.L.; Liang, R.; Chen, K.; Liu, S.S.; Qin, Y.; Leung, T.H.; Lee, K.F.; et al. MicroRNA-141 enhances anoikis resistance in metastatic progression of ovarian cancer through targeting klf12/sp1/survivin axis. Mol. Cancer 2017, 16, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Tian, Z.; Jin, H.; Xu, J.; Hua, X.; Yan, H.; Liufu, H.; Wang, J.; Li, J.; Zhu, J.; et al. Decreased c-myc mRNA stability via the microRNA 141-3p/auf1 axis is crucial for p63alpha inhibition of cyclin d1 gene transcription and bladder cancer cell tumorigenicity. Mol. Cell. Biol. 2018, 38, e00273-18. [Google Scholar] [CrossRef] [Green Version]
- Derouet, M.F.; Dakpo, E.; Wu, L.; Zehong, G.; Conner, J.; Keshavjee, S.; de Perrot, M.; Waddell, T.; Elimova, E.; Yeung, J.; et al. MiR-145 expression enhances integrin expression in sk-gt-4 cell line by down-regulating c-myc expression. Oncotarget 2018, 9, 15198–15207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, G.; Huang, C.; Li, J.; Huang, H.; Jin, H.; Zhu, J.; Wu, X.R.; Huang, C. Role of stat3 and foxo1 in the divergent therapeutic responses of non-metastatic and metastatic bladder cancer cells to miR-145. Mol. Cancer Ther. 2017, 16, 924–935. [Google Scholar] [CrossRef] [Green Version]
- Yao, S.; Xu, J.; Zhao, K.; Song, P.; Yan, Q.; Fan, W.; Li, W.; Lu, C. Down-regulation of hpgd by miR-146b-3p promotes cervical cancer cell proliferation, migration and anchorage-independent growth through activation of stat3 and akt pathways. Cell Death Dis. 2018, 9, 1055. [Google Scholar] [CrossRef] [Green Version]
- Jiang, G.; Huang, C.; Li, J.; Huang, H.; Wang, J.; Li, Y.; Xie, F.; Jin, H.; Zhu, J.; Huang, C. Transcriptional and post-transcriptional upregulation of p27 mediates growth inhibition of isorhapontigenin (iso) on human bladder cancer cells. Carcinogenesis 2018, 39, 482–492. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.Z.; Schmidt, M.L.; Suman, S.; Hobbing, K.R.; Barve, S.S.; Gobejishvili, L.; Brock, G.; Klinge, C.M.; Rai, S.N.; Park, J.; et al. Micro-RNA-186-5p inhibition attenuates proliferation, anchorage independent growth and invasion in metastatic prostate cancer cells. BMC Cancer 2018, 18, 421. [Google Scholar] [CrossRef] [Green Version]
- Qiu, H.; Yuan, S.; Lu, X. MiR-186 suppressed cyld expression and promoted cell proliferation in human melanoma. Oncol. Lett. 2016, 12, 2301–2306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.C.; Sharma, S.B.; Farrugia, M.K.; McLaughlin, S.L.; Ice, R.J.; Loskutov, Y.V.; Pugacheva, E.N.; Brundage, K.M.; Chen, D.; Ruppert, J.M. Kruppel-like factor 4 signals through microRNA-206 to promote tumor initiation and cell survival. Oncogenesis 2015, 4, e155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, H.; Li, W.; Tian, Y.; Xu, P.; Wang, H.; Zhang, J.; Li, Y. Upregulation of miR-362-3p modulates proliferation and anchorage-independent growth by directly targeting tob2 in hepatocellular carcinoma. J. Cell. Biochem. 2015, 116, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Hung, P.S.; Chen, C.Y.; Chen, W.T.; Kuo, C.Y.; Fang, W.L.; Huang, K.H.; Chiu, P.C.; Lo, S.S. MiR-376c promotes carcinogenesis and serves as a plasma marker for gastric carcinoma. PLoS ONE 2017, 12, e0177346. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Zhang, Y.; Cao, S.; Wu, Y.; Guo, W.; Yuan, W.; Zhang, S. MiR-411 regulated itch expression and promoted cell proliferation in human hepatocellular carcinoma cells. Biomed. Pharmacother. 2015, 70, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cui, J.; Li, Z.; Fu, X.; Li, J.; Li, H.; Wang, S.; Zhang, M. Orp8 induces apoptosis by releasing cytochrome c from mitochondria in nonsmall cell lung cancer. Oncol. Rep. 2020, 43, 1516–1524. [Google Scholar] [PubMed]
- Liu, X.; Fu, Y.; Zhang, G.; Zhang, D.; Liang, N.; Li, F.; Li, C.; Sui, C.; Jiang, J.; Lu, H.; et al. MiR-424-5p promotes anoikis resistance and lung metastasis by inactivating hippo signaling in thyroid cancer. Mol. Ther. Oncolytics 2019, 15, 248–260. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Xu, J.; Gao, G.; Li, J.; Huang, H.; Jin, H.; Zhu, J.; Che, X.; Huang, C. Tumor-suppressor nfkappab2 p100 interacts with erk2 and stabilizes pten mRNA via inhibition of miR-494. Oncogene 2016, 35, 4080–4090. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.X.; An, Q.; Chen, L.M.; Guo, L. MiR-520f regulated itch expression and promoted cell proliferation in human melanoma cells. Dose Response 2020, 18, 1559325820918450. [Google Scholar] [CrossRef]
- Tordjman, J.; Majumder, M.; Amiri, M.; Hasan, A.; Hess, D.; Lala, P.K. Tumor suppressor role of cytoplasmic polyadenylation element binding protein 2 (cpeb2) in human mammary epithelial cells. BMC Cancer 2019, 19, 561. [Google Scholar] [CrossRef] [Green Version]
- Xian, Q.; Zhao, R.; Fu, J. MicroRNA-527 induces proliferation and cell cycle in esophageal squamous cell carcinoma cells by repressing ph domain leucine-rich-repeats protein phosphatase 2. Dose Response 2020, 18, 1559325820928687. [Google Scholar] [CrossRef] [PubMed]
- Shu, Z.; Chen, L.; Ding, D. MiR-582-5p induces colorectal cancer cell proliferation by targeting adenomatous polyposis coli. World J. Surg. Oncol. 2016, 14, 239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, S.T.; Guo, X.Y.; Wang, J.; Wang, C.Y.; Yang, R.H.; Wang, F.H.; Li, X.Y.; Hondermarck, H.; Thorne, R.F.; Wang, Y.F.; et al. MicroRNA-645 is an oncogenic regulator in colon cancer. Oncogenesis 2017, 6, e335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Deng, Y.; Liu, J.; Ye, Z.; You, Z.; Yao, S.; He, S. MiR-760 overexpression promotes proliferation in ovarian cancer by downregulation of phlpp2 expression. Gynecol. Oncol. 2016, 143, 655–663. [Google Scholar] [CrossRef]
- Li, Y.C.; Li, C.F.; Chen, L.B.; Li, D.D.; Yang, L.; Jin, J.P.; Zhang, B. MicroRNA-766 targeting regulation of sox6 expression promoted cell proliferation of human colorectal cancer. OncoTargets Ther. 2015, 8, 2981–2988. [Google Scholar] [CrossRef] [Green Version]
- Yin, J.; Weng, C.; Ma, J.; Chen, F.; Huang, Y.; Feng, M. MicroRNA1288 promotes cell proliferation of human glioblastoma cells by repressing ubiquitin carboxylterminal hydrolase cyld expression. Mol. Med. Rep. 2017, 16, 6764–6770. [Google Scholar] [CrossRef]
- Li, Z.; Xu, Z.; Xie, Q.; Gao, W.; Xie, J.; Zhou, L. MiR-1303 promotes the proliferation of neuroblastoma cell sh-sy5y by targeting gsk3beta and sfrp1. Biomed. Pharmacother. 2016, 83, 508–513. [Google Scholar] [CrossRef]
- Lin, Y.; Gu, Q.; Sun, Z.; Sheng, B.; Qi, C.; Liu, B.; Fu, T.; Liu, C.; Zhang, Y. Upregulation of miR-3607 promotes lung adenocarcinoma proliferation by suppressing apc expression. Biomed. Pharmacother. 2017, 95, 497–503. [Google Scholar] [CrossRef]
- Sun, Q.; Yang, Z.; Li, P.; Wang, X.; Sun, L.; Wang, S.; Liu, M.; Tang, H. A novel miRNA identified in grsf1 complex drives the metastasis via the pik3r3/akt/nf-kappab and timp3/mmp9 pathways in cervical cancer cells. Cell Death Dis. 2019, 10, 636. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Li, T.E.; Chen, M.; Pan, J.J.; Shen, K.W. MiR-106b-5p contributes to the lung metastasis of breast cancer via targeting cnn1 and regulating rho/rock1 pathway. Aging (Albany NY) 2020, 12, 1867–1887. [Google Scholar] [CrossRef]
- Ni, S.; Weng, W.; Xu, M.; Wang, Q.; Tan, C.; Sun, H.; Wang, L.; Huang, D.; Du, X.; Sheng, W. MiR-106b-5p inhibits the invasion and metastasis of colorectal cancer by targeting ctsa. OncoTargets Ther. 2018, 11, 3835–3845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Y.; Li, S.L.; Ma, Y.Y.; Diao, Y.J.; Yang, L.; Su, M.Q.; Li, Z.; Ji, Y.; Wang, J.; Lei, L.; et al. Exosomal miR-141-3p regulates osteoblast activity to promote the osteoblastic metastasis of prostate cancer. Oncotarget 2017, 8, 94834–94849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meltzer, S.; Bjornetro, T.; Lyckander, L.G.; Flatmark, K.; Dueland, S.; Samiappan, R.; Johansen, C.; Kalanxhi, E.; Ree, A.H.; Redalen, K.R. Circulating exosomal miR-141-3p and miR-375 in metastatic progression of rectal cancer. Transl. Oncol. 2019, 12, 1038–1044. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Rerko, R.M.; Platzer, P.; Dawson, D.; Willis, J.; Tong, M.; Lawrence, E.; Lutterbaugh, J.; Lu, S.; Willson, J.K.; et al. 15-hydroxyprostaglandin dehydrogenase, a cox-2 oncogene antagonist, is a tgf-beta-induced suppressor of human gastrointestinal cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 17468–17473. [Google Scholar] [CrossRef] [Green Version]
- He, P.; Li, K.; Li, S.B.; Hu, T.T.; Guan, M.; Sun, F.Y.; Liu, W.W. Upregulation of akap12 with hdac3 depletion suppresses the progression and migration of colorectal cancer. Int. J. Oncol. 2018, 52, 1305–1316. [Google Scholar] [CrossRef]
- Li, S.; Huang, L.; Gu, J.; Wu, J.; Ou, W.; Feng, J.; Liu, B.; Xu, X.; Zhou, Y. Restoration of klf4 inhibits invasion and metastases of lung adenocarcinoma through suppressing mmp2. J. Cancer 2017, 8, 3480–3489. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Yu, Z.; Wang, J.; Zhou, L.; Zhang, J.; Yao, B.; Dou, J.; Qiu, Z.; Huang, C. Kruppel-like factor 4 inhibits pancreatic cancer epithelial-to-mesenchymal transition and metastasis by down-regulating caveolin-1 expression. Cell. Physiol. Biochem. 2018, 46, 238–252. [Google Scholar] [CrossRef]
- Qi, X.T.; Li, Y.L.; Zhang, Y.Q.; Xu, T.; Lu, B.; Fang, L.; Gao, J.Q.; Yu, L.S.; Zhu, D.F.; Yang, B.; et al. Klf4 functions as an oncogene in promoting cancer stem cell-like characteristics in osteosarcoma cells. Acta Pharmacol. Sin. 2019, 40, 546–555. [Google Scholar] [CrossRef]
- Zhong, W.; Qin, S.; Zhu, B.; Pu, M.; Liu, F.; Wang, L.; Ye, G.; Yi, Q.; Yan, D. Oxysterol-binding protein-related protein 8 (orp8) increases sensitivity of hepatocellular carcinoma cells to fas-mediated apoptosis. J. Biol. Chem. 2015, 290, 8876–8887. [Google Scholar] [CrossRef] [Green Version]
- Le Clorennec, C.; Lazrek, Y.; Dubreuil, O.; Sampaio, C.; Larbouret, C.; Lanotte, R.; Poul, M.A.; Barret, J.M.; Prost, J.F.; Pelegrin, A.; et al. Itch-dependent proteasomal degradation of c-flip induced by the anti-her3 antibody 9f7-f11 promotes dr5/caspase 8-mediated apoptosis of tumor cells. Cell Commun. Signal. 2019, 17, 106. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.M.; Pan, Y.; Wei, Y.; Cheng, X.; Zhou, B.P.; Tan, M.; Zhou, X.; Xia, W.; Hortobagyi, G.N.; Yu, D.; et al. Upregulation of cxcr4 is essential for her2-mediated tumor metastasis. Cancer Cell 2004, 6, 459–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.X.; Guan, K.L. The hippo pathway: Regulators and regulations. Genes Dev. 2013, 27, 355–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, S.J.; Ni, L.; Osinski, A.; Tomchick, D.R.; Brautigam, C.A.; Luo, X. Sav1 promotes hippo kinase activation through antagonizing the pp2a phosphatase stripak. Elife 2017, 6, e30278. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Feldmann, G.; Huang, J.; Wu, S.; Zhang, N.; Comerford, S.A.; Gayyed, M.F.; Anders, R.A.; Maitra, A.; Pan, D. Elucidation of a universal size-control mechanism in drosophila and mammals. Cell 2007, 130, 1120–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, L.; Chen, Y.; Ji, M.; Dong, J. Kibra regulates hippo signaling activity via interactions with large tumor suppressor kinases. J. Biol. Chem. 2011, 286, 7788–7796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, B.; Li, L.; Wang, L.; Wang, C.Y.; Yu, J.; Guan, K.L. Cell detachment activates the hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012, 26, 54–68. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Guo, L.; Li, Y.; Feng, G.H.; Teng, F.; Li, W.; Zhou, Q. MicroRNA-494 promotes cancer progression and targets adenomatous polyposis coli in colorectal cancer. Mol. Cancer 2018, 17, 1. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Liu, W.; Jiang, X.; Shi, X.; Yin, L.; Ao, L.; Cui, Z.; Li, Y.; Huang, C.; Cao, J.; et al. Sox30, a novel epigenetic silenced tumor suppressor, promotes tumor cell apoptosis by transcriptional activating p53 in lung cancer. Oncogene 2015, 34, 4391–4402. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Hou, X.; Wu, C.; Han, L.; Li, Q.; Wang, J.; Luo, S. MiR-645 promotes invasiveness, metastasis and tumor growth in colorectal cancer by targeting efna5. Biomed. Pharmacother. 2020, 125, 109889. [Google Scholar] [CrossRef]
- Rao, X.; Wan, L.; Jie, Z.; Zhu, X.; Yin, J.; Cao, H. Upregulated miR-27a-3p indicates a poor prognosis in pancreatic carcinoma patients and promotes the angiogenesis and migration by epigenetic silencing of gata6 and activating vegfa/vegfr2 signaling pathway. OncoTargets Ther. 2019, 12, 11241–11254. [Google Scholar] [CrossRef] [Green Version]
- Zhao, N.; Sun, H.; Sun, B.; Zhu, D.; Zhao, X.; Wang, Y.; Gu, Q.; Dong, X.; Liu, F.; Zhang, Y.; et al. MiR-27a-3p suppresses tumor metastasis and vm by down-regulating ve-cadherin expression and inhibiting emt: An essential role for twist-1 in hcc. Sci. Rep. 2016, 6, 23091. [Google Scholar] [CrossRef] [PubMed]
- Al-Khalaf, H.H.; Colak, D.; Al-Saif, M.; Al-Bakheet, A.; Hendrayani, S.F.; Al-Yousef, N.; Kaya, N.; Khabar, K.S.; Aboussekhra, A. P16(ink4a) positively regulates cyclin d1 and e2f1 through negative control of auf1. PLoS ONE 2011, 6, e21111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melino, G. P63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011, 18, 1487–1499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, X.; Chakravarti, D.; Cho, M.S.; Liu, L.; Gi, Y.J.; Lin, Y.L.; Leung, M.L.; El-Naggar, A.; Creighton, C.J.; Suraokar, M.B.; et al. Tap63 suppresses metastasis through coordinate regulation of dicer and miRNAs. Nature 2010, 467, 986–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirata, H.; Ueno, K.; Shahryari, V.; Tanaka, Y.; Tabatabai, Z.L.; Hinoda, Y.; Dahiya, R. Oncogenic miRNA-182-5p targets smad4 and reck in human bladder cancer. PLoS ONE 2012, 7, e51056. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Zeng, Q.; Yu, G.; Li, S.; Wang, C.Y. Wnt/beta-catenin signaling inhibits death receptor-mediated apoptosis and promotes invasive growth of hnscc. Cell Signal. 2006, 18, 679–687. [Google Scholar] [CrossRef]
- Kawada, M.; Uehara, Y.; Mizuno, S.; Yamori, T.; Tsuruo, T. Up-regulation of p27kip1 correlates inversely with anchorage-independent growth of human cancer cell lines. Jpn. J. Cancer Res. 1998, 89, 110–115. [Google Scholar] [CrossRef]
- Zheng, J.Y.; Wang, W.Z.; Li, K.Z.; Guan, W.X.; Yan, W. Effect of p27(kip1) on cell cycle and apoptosis in gastric cancer cells. World J. Gastroenterol. 2005, 11, 7072–7077. [Google Scholar] [CrossRef]
- Shi, H.; Li, H.; Yuan, R.; Guan, W.; Zhang, X.; Zhang, S.; Zhang, W.; Tong, F.; Li, L.; Song, Z.; et al. Pcbp1 depletion promotes tumorigenesis through attenuation of p27(kip1) mRNA stability and translation. J. Exp. Clin. Cancer Res. 2018, 37, 187. [Google Scholar] [CrossRef]
- Collins, N.L.; Reginato, M.J.; Paulus, J.K.; Sgroi, D.C.; Labaer, J.; Brugge, J.S. G1/s cell cycle arrest provides anoikis resistance through erk-mediated bim suppression. Mol. Cell. Biol. 2005, 25, 5282–5291. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.C.; Wu, Y.P.; Ye, B.; Lin, D.C.; Feng, Y.B.; Zhang, Z.Q.; Xu, X.; Han, Y.L.; Cai, Y.; Dong, J.T.; et al. Suppression of anoikis by skp2 amplification and overexpression promotes metastasis of esophageal squamous cell carcinoma. Mol. Cancer Res. 2009, 7, 12–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Zhou, W.; Jiang, H.; Xiang, Z.; Wang, L. MiR-1303 promotes the proliferation, migration and invasion of prostate cancer cells through regulating the wnt/beta-catenin pathway by targeting dkk3. Exp. Ther. Med. 2019, 18, 4747–4757. [Google Scholar]
- Massoumi, R.; Chmielarska, K.; Hennecke, K.; Pfeifer, A.; Fassler, R. Cyld inhibits tumor cell proliferation by blocking bcl-3-dependent nf-kappab signaling. Cell 2006, 125, 665–677. [Google Scholar] [CrossRef] [Green Version]
- Ikematsu, N.; Yoshida, Y.; Kawamura-Tsuzuku, J.; Ohsugi, M.; Onda, M.; Hirai, M.; Fujimoto, J.; Yamamoto, T. Tob2, a novel anti-proliferative tob/btg1 family member, associates with a component of the ccr4 transcriptional regulatory complex capable of binding cyclin-dependent kinases. Oncogene 1999, 18, 7432–7441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, M.; Li, Z.; Aau, M.; Wong, C.H.; Yang, X.; Yu, Q. Myc/miR-378/tob2/cyclin d1 functional module regulates oncogenic transformation. Oncogene 2011, 30, 2242–2251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Z.; DeGregori, J.; Shohet, R.; Leone, G.; Stillman, B.; Nevins, J.R.; Williams, R.S. Cdc6 is regulated by e2f and is essential for DNA replication in mammalian cells. Proc. Natl. Acad. Sci. USA 1998, 95, 3603–3608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollern, D.P.; Honeysett, J.; Cardiff, R.D.; Andrechek, E.R. The e2f transcription factors regulate tumor development and metastasis in a mouse model of metastatic breast cancer. Mol. Cell. Biol. 2014, 34, 3229–3243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, A.; Marcellus, R.C.; Corbeil, H.B.; Branton, P.E. Rbp1 induces growth arrest by repression of e2f-dependent transcription. Oncogene 1999, 18, 2091–2100. [Google Scholar] [CrossRef] [Green Version]
- Heinen, C.D.; Goss, K.H.; Cornelius, J.R.; Babcock, G.F.; Knudsen, E.S.; Kowalik, T.; Groden, J. The apc tumor suppressor controls entry into s-phase through its ability to regulate the cyclin d/rb pathway. Gastroenterology 2002, 123, 751–763. [Google Scholar] [CrossRef]
- Shtutman, M.; Zhurinsky, J.; Simcha, I.; Albanese, C.; D’Amico, M.; Pestell, R.; Ben-Ze’ev, A. The cyclin d1 gene is a target of the beta-catenin/lef-1 pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 5522–5527. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Azuma, Y.; Moore, D.; Osheroff, N.; Neufeld, K.L. Interaction between tumor suppressor adenomatous polyposis coli and topoisomerase iialpha: Implication for the g2/m transition. Mol. Biol. Cell 2008, 19, 4076–4085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orford, K.; Orford, C.C.; Byers, S.W. Exogenous expression of beta-catenin regulates contact inhibition, anchorage-independent growth, anoikis, and radiation-induced cell cycle arrest. J. Cell Biol. 1999, 146, 855–868. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Yuan, Q.; Jiang, Y.; Huang, L.; Chen, C.; Hu, G.; Wan, R.; Wang, X.; Yang, L. Identification of sox6 as a regulator of pancreatic cancer development. J. Cell. Mol. Med. 2018, 22, 1864–1872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Y.R.; Tang, H.; Xie, F.; Liu, H.; Zhu, Y.; Ai, J.; Chen, L.; Li, Y.; Kwong, D.L.; Fu, L.; et al. Characterization of tumor-suppressive function of sox6 in human esophageal squamous cell carcinoma. Clin. Cancer Res. 2011, 17, 46–55. [Google Scholar] [CrossRef] [Green Version]
- Jia, B.; Xia, L.; Cao, F. The role of miR-766-5p in cell migration and invasion in colorectal cancer. Exp. Ther. Med. 2018, 15, 2569–2574. [Google Scholar] [CrossRef]
- Chen, W.; Cai, G.; Liao, Z.; Lin, K.; Li, G.; Li, Y. MiRNA-766 induces apoptosis of human colon cancer cells through the p53/bax signaling pathway by mdm4. Exp. Ther. Med. 2019, 17, 4100–4108. [Google Scholar] [CrossRef] [Green Version]
- Lauzier, A.; Normandeau-Guimond, J.; Vaillancourt-Lavigueur, V.; Boivin, V.; Charbonneau, M.; Rivard, N.; Scott, M.S.; Dubois, C.M.; Jean, S. Colorectal cancer cells respond differentially to autophagy inhibition in vivo. Sci. Rep. 2019, 9, 11316. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.Q.; Jiao, X.L.; Zhang, S.Y.; Xu, Y.; Li, S.; Kong, B.H. MiR-9-5p could promote angiogenesis and radiosensitivity in cervical cancer by targeting socs5. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7314–7326. [Google Scholar]
- Li, G.; Wu, F.; Yang, H.; Deng, X.; Yuan, Y. MiR-9-5p promotes cell growth and metastasis in non-small cell lung cancer through the repression of tgfbr2. Biomed. Pharmacother. 2017, 96, 1170–1178. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, J.; Wang, B.; Ren, J.C.; Su, W.; Zhang, T. MicroRNA-10b triggers the epithelial-mesenchymal transition (emt) of laryngeal carcinoma hep-2 cells by directly targeting the e-cadherin. Appl. Biochem. Biotechnol. 2015, 176, 33–44. [Google Scholar] [CrossRef]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Lopez-Beltran, A.; Massari, F.; MacLennan, G.T.; Montironi, R. Molecular testing for braf mutations to inform melanoma treatment decisions: A move toward precision medicine. Mod. Pathol. 2018, 31, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Kim, H.S.; Lee, S.H.; Yang, Y.; Lee, M.S.; Lim, J.S. Ndrg2 controls cox-2/pge(2)-mediated breast cancer cell migration and invasion. Mol. Cells 2014, 37, 759–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jandrey, E.H.F.; Moura, R.P.; Andrade, L.N.S.; Machado, C.L.; Campesato, L.F.; Leite, K.R.M.; Inoue, L.T.; Asprino, P.F.; da Silva, A.P.M.; de Barros, A.; et al. Ndrg4 promoter hypermethylation is a mechanistic biomarker associated with metastatic progression in breast cancer patients. NPJ Breast Cancer 2019, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Yao, L.; Chau, L.; Ng, S.S.; Peng, Y.; Liu, X.; Au, W.S.; Wang, J.; Li, F.; Ji, S.; et al. N-myc downstream-regulated gene 2 (ndrg2) inhibits glioblastoma cell proliferation. Int. J. Cancer 2003, 106, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Lai, X.; Zhao, Y.; Zhang, Y.; Li, M.; Li, D.; Kong, J.; Zhang, Y.; Jing, P.; Li, H.; et al. Loss of ndrg2 in liver microenvironment inhibits cancer liver metastasis by regulating tumor associate macrophages polarization. Cell Death Dis. 2018, 9, 248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLigio, J.T.; Lin, G.; Chalfant, C.E.; Park, M.A. Splice variants of cytosolic polyadenylation element-binding protein 2 (cpeb2) differentially regulate pathways linked to cancer metastasis. J. Biol. Chem. 2017, 292, 17909–17918. [Google Scholar] [CrossRef] [Green Version]
- Johnson, R.M.; Vu, N.T.; Griffin, B.P.; Gentry, A.E.; Archer, K.J.; Chalfant, C.E.; Park, M.A. The alternative splicing of cytoplasmic polyadenylation element binding protein 2 drives anoikis resistance and the metastasis of triple negative breast cancer. J. Biol. Chem. 2015, 290, 25717–25727. [Google Scholar] [CrossRef] [Green Version]
- Yin, S.; Cheryan, V.T.; Xu, L.; Rishi, A.K.; Reddy, K.B. Myc mediates cancer stem-like cells and emt changes in triple negative breast cancers cells. PLoS ONE 2017, 12, e0183578. [Google Scholar] [CrossRef] [Green Version]
- Carabet, L.A.; Rennie, P.S.; Cherkasov, A. Therapeutic inhibition of myc in cancer. Structural bases and computer-aided drug discovery approaches. Int. J. Mol. Sci. 2019, 20, 120. [Google Scholar] [CrossRef] [Green Version]
- Elbadawy, M.; Usui, T.; Yamawaki, H.; Sasaki, K. Emerging roles of c-myc in cancer stem cell-related signaling and resistance to cancer chemotherapy: A potential therapeutic target against colorectal cancer. Int. J. Mol. Sci. 2019, 20, 2340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.Y.; Cha, J.; Kim, S.K.; Park, J.H.; Song, K.H.; Kim, P.; Kim, M.Y. C-myc drives breast cancer metastasis to the brain, but promotes synthetic lethality with trail. Mol. Cancer Res. 2019, 17, 544–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Radisky, D.C.; Yang, D.; Xu, R.; Radisky, E.S.; Bissell, M.J.; Bishop, J.M. Myc suppresses cancer metastasis by direct transcriptional silencing of alphav and beta3 integrin subunits. Nat. Cell Biol. 2012, 14, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, H.; Ivaska, J. Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhang, Y.; Wu, H.; Li, Y.; Zhang, Y.; Liu, M.; Li, X.; Tang, H. MiR-10a suppresses colorectal cancer metastasis by modulating the epithelial-to-mesenchymal transition and anoikis. Cell Death Dis. 2017, 8, e2739. [Google Scholar] [CrossRef]
- Cully, M.; Shiu, J.; Piekorz, R.P.; Muller, W.J.; Done, S.J.; Mak, T.W. Transforming acidic coiled coil 1 promotes transformation and mammary tumorigenesis. Cancer Res. 2005, 65, 10363–10370. [Google Scholar] [CrossRef] [Green Version]
- Bersani, F.; Lingua, M.F.; Morena, D.; Foglizzo, V.; Miretti, S.; Lanzetti, L.; Carra, G.; Morotti, A.; Ala, U.; Provero, P.; et al. Deep sequencing reveals a novel miR-22 regulatory network with therapeutic potential in rhabdomyosarcoma. Cancer Res. 2016, 76, 6095–6106. [Google Scholar] [CrossRef] [Green Version]
- Leung, Z.; Ko, F.C.F.; Tey, S.K.; Kwong, E.M.L.; Mao, X.; Liu, B.H.M.; Ma, A.P.Y.; Fung, Y.M.E.; Che, C.M.; Wong, D.K.H.; et al. Galectin-1 promotes hepatocellular carcinoma and the combined therapeutic effect of otx008 galectin-1 inhibitor and sorafenib in tumor cells. J. Exp. Clin. Cancer Res. 2019, 38, 423. [Google Scholar] [CrossRef]
- Rice, M.A.; Ishteiwy, R.A.; Magani, F.; Udayakumar, T.; Reiner, T.; Yates, T.J.; Miller, P.; Perez-Stable, C.; Rai, P.; Verdun, R.; et al. The microRNA-23b/-27b cluster suppresses prostate cancer metastasis via huntingtin-interacting protein 1-related. Oncogene 2016, 35, 4752–4761. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Cheng, S.L.; Bian, K.; Wang, L.; Zhang, X.; Yan, B.; Jia, L.T.; Zhao, J.; Gammoh, N.; Yang, A.G.; et al. MicroRNA-26a promotes anoikis in human hepatocellular carcinoma cells by targeting alpha5 integrin. Oncotarget 2015, 6, 2277–2289. [Google Scholar] [CrossRef] [Green Version]
- Matsuyama, R.; Okuzaki, D.; Okada, M.; Oneyama, C. MicroRNA-27b suppresses tumor progression by regulating arfgef1 and focal adhesion signaling. Cancer Sci. 2016, 107, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.J.; Willy, J.A.; Quirin, K.A.; Wek, R.C.; Korc, M.; Yin, X.M.; Kota, J. Novel role of miR-29a in pancreatic cancer autophagy and its therapeutic potential. Oncotarget 2016, 7, 71635–71650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, X.T.; Shi, Y.H.; Zhou, J.; Peng, Y.F.; Liu, W.R.; Shi, G.M.; Gao, Q.; Wang, X.Y.; Song, K.; Fan, J.; et al. MicroRNA-30a suppresses autophagy-mediated anoikis resistance and metastasis in hepatocellular carcinoma. Cancer Lett. 2018, 412, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Xing, C.; Cui, K.; Li, Y.; Zhang, J.; Du, R.; Zhang, X.; Li, Y. MicroRNA-30a attenuates mutant kras-driven colorectal tumorigenesis via direct suppression of me1. Cell Death Differ. 2017, 24, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Song, E.; Shen, G.; Zhu, T.; Jiang, T.; Shen, H.; Niu, L.; Wang, B.; Lu, Z.; Qian, J. Expression of microRNA-30c via lentivirus vector inhibits the proliferation and enhances the sensitivity of highly aggressive ccrcc caki-1 cells to anticancer agents. OncoTargets Ther. 2017, 10, 579–590. [Google Scholar] [CrossRef] [Green Version]
- Du, M.; Zhang, Y.; Mao, Y.; Mou, J.; Zhao, J.; Xue, Q.; Wang, D.; Huang, J.; Gao, S.; Gao, Y. MiR-33a suppresses proliferation of nsclc cells via targeting mettl3 mRNA. Biochem. Biophys. Res. Commun. 2017, 482, 582–589. [Google Scholar] [CrossRef]
- Sa, K.D.; Zhang, X.; Li, X.F.; Gu, Z.P.; Yang, A.G.; Zhang, R.; Li, J.P.; Sun, J.Y. A miR-124/itga3 axis contributes to colorectal cancer metastasis by regulating anoikis susceptibility. Biochem. Biophys. Res. Commun. 2018, 501, 758–764. [Google Scholar] [CrossRef]
- Zhu, J.F.; Liu, Y.; Huang, H.; Shan, L.; Han, Z.G.; Liu, J.Y.; Li, Y.L.; Dong, X.; Zeng, W. MicroRNA-133b/egfr axis regulates esophageal squamous cell carcinoma metastases by suppressing anoikis resistance and anchorage-independent growth. Cancer Cell Int. 2018, 18, 193. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Pan, J.; Huang, S.; Peng, X.; Zou, X.; Luo, Y.; Ren, D.; Zhang, X.; Li, R.; He, P.; et al. Downregulation of miR-133a-3p promotes prostate cancer bone metastasis via activating pi3k/akt signaling. J. Exp. Clin. Cancer Res. 2018, 37, 160. [Google Scholar] [CrossRef]
- Peres, J.; Kwesi-Maliepaard, E.M.; Rambow, F.; Larue, L.; Prince, S. The tumour suppressor, miR-137, inhibits malignant melanoma migration by targetting the tbx3 transcription factor. Cancer Lett. 2017, 405, 111–119. [Google Scholar] [CrossRef]
- Li, L.; He, Z.; Zhu, C.; Chen, S.; Yang, Z.; Xu, J.; Bi, N.; Yu, C.; Sun, C. MiR-137 promotes anoikis through modulating the akt signaling pathways in pancreatic cancer. J. Cancer 2020, 11, 6277–6285. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Kawano, M.; Itonaga, I.; Iwasaki, T.; Miyazaki, M.; Ikeda, S.; Tsumura, H. Tumor suppressive microRNA-138 inhibits metastatic potential via the targeting of focal adhesion kinase in ewing’s sarcoma cells. Int. J. Oncol. 2016, 48, 1135–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozak, J.; Wdowiak, P.; Maciejewski, R.; Torres, A. Interactions between microRNA-200 family and sestrin proteins in endometrial cancer cell lines and their significance to anoikis. Mol. Cell. Biochem. 2019, 459, 21–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Srivastava, N.; Yadav, A.; Ateeq, B. Targeting agtr1/nf-kappab/cxcr4 axis by miR-155 attenuates oncogenesis in glioblastoma. Neoplasia 2020, 22, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Sun, Y.; Yang, H.; Li, J.; Yu, S.; Chang, X.; Lu, Z.; Chen, J. Deregulation of the miR-193b-kras axis contributes to impaired cell growth in pancreatic cancer. PLoS ONE 2015, 10, e0125515. [Google Scholar] [CrossRef]
- Moore, C.; Parrish, J.K.; Jedlicka, P. MiR-193b, downregulated in ewing sarcoma, targets the erbb4 oncogene to inhibit anchorage-independent growth. PLoS ONE 2017, 12, e0178028. [Google Scholar] [CrossRef]
- Cristobal, I.; Torrejon, B.; Rubio, J.; Santos, A.; Pedregal, M.; Carames, C.; Zazo, S.; Luque, M.; Sanz-Alvarez, M.; Madoz-Gurpide, J.; et al. Deregulation of set is associated with tumor progression and predicts adverse outcome in patients with early-stage colorectal cancer. J. Clin. Med. 2019, 8, 346. [Google Scholar] [CrossRef] [Green Version]
- Rogers, T.J.; Christenson, J.L.; Greene, L.I.; O’Neill, K.I.; Williams, M.M.; Gordon, M.A.; Nemkov, T.; D’Alessandro, A.; Degala, G.D.; Shin, J.; et al. Reversal of triple-negative breast cancer emt by miR-200c decreases tryptophan catabolism and a program of immunosuppression. Mol. Cancer Res. 2019, 17, 30–41. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Zeng, X.; Jiang, G.; Liao, X.; Liu, C.; Li, J.; Jin, H.; Zhu, J.; Sun, H.; Wu, X.R.; et al. Xiap bir domain suppresses miR-200a expression and subsequently promotes egfr protein translation and anchorage-independent growth of bladder cancer cell. J. Hematol. Oncol. 2017, 10, 6. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wang, X.; Chen, P. MiR-204 down regulates sirt1 and reverts sirt1-induced epithelial-mesenchymal transition, anoikis resistance and invasion in gastric cancer cells. BMC Cancer 2013, 13, 290. [Google Scholar] [CrossRef] [Green Version]
- Bharambe, H.S.; Paul, R.; Panwalkar, P.; Jalali, R.; Sridhar, E.; Gupta, T.; Moiyadi, A.; Shetty, P.; Kazi, S.; Deogharkar, A.; et al. Downregulation of miR-204 expression defines a highly aggressive subset of group 3/group 4 medulloblastomas. Acta Neuropathol. Commun. 2019, 7, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandoval-Borquez, A.; Polakovicova, I.; Carrasco-Veliz, N.; Lobos-Gonzalez, L.; Riquelme, I.; Carrasco-Avino, G.; Bizama, C.; Norero, E.; Owen, G.I.; Roa, J.C.; et al. MicroRNA-335-5p is a potential suppressor of metastasis and invasion in gastric cancer. Clin. Epigenet. 2017, 9, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Y.; Zhu, X.; Wang, K.; Chen, Y. MicroRNA-363-3p suppresses anoikis resistance in human papillary thyroid carcinoma via targeting integrin alpha 6. Acta Biochim. Biophys. Sin. 2019, 51, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tian, S.; Li, X.; Ji, Y.; Wang, Z.; Liu, C. Ube2c promotes rectal carcinoma via miR-381. Cancer Biol. Ther. 2018, 19, 230–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Li, T.; Guo, P.; Kang, J.; Wei, Q.; Jia, X.; Zhao, W.; Huai, W.; Qiu, Y.; Sun, L.; et al. MiR-424-5p reversed epithelial-mesenchymal transition of anchorage-independent hcc cells by directly targeting icat and suppressed hcc progression. Sci. Rep. 2014, 4, 6248. [Google Scholar] [CrossRef] [PubMed]
- Samantarrai, D.; Mallick, B. MiR-429 inhibits metastasis by targeting kiaa0101 in soft tissue sarcoma. Exp. Cell Res. 2017, 357, 33–39. [Google Scholar] [CrossRef]
- Muys, B.R.; Sousa, J.F.; Placa, J.R.; de Araujo, L.F.; Sarshad, A.A.; Anastasakis, D.G.; Wang, X.; Li, X.L.; de Molfetta, G.A.; Ramao, A.; et al. MiR-450a acts as a tumor suppressor in ovarian cancer by regulating energy metabolism. Cancer Res. 2019, 79, 3294–3305. [Google Scholar] [CrossRef]
- Zhai, Y.; Liu, M.; Zheng, Y. MicroRNA-451 dictates the anoikis resistance of osteosarcoma by targeting rab14. Int. J. Clin. Exp. Pathol. 2017, 10, 10989–10997. [Google Scholar]
- Yi, H.; Geng, L.; Black, A.; Talmon, G.; Berim, L.; Wang, J. The miR-487b-3p/grm3/tgfbeta signaling axis is an important regulator of colon cancer tumorigenesis. Oncogene 2017, 36, 3477–3489. [Google Scholar] [CrossRef] [Green Version]
- Arnold, J.; Engelmann, J.C.; Schneider, N.; Bosserhoff, A.K.; Kuphal, S. MiR-488-5p and its role in melanoma. Exp. Mol. Pathol. 2020, 112, 104348. [Google Scholar] [CrossRef]
- Chen, M.; Liu, L.X. MiR-525-5p repressed metastasis and anoikis resistance in cervical cancer via blocking ube2c/zeb1/2 signal axis. Dig. Dis. Sci. 2020, 65, 2442–2451. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wu, R.; Li, G.; Sun, P.; Xu, Q.; Liu, Z. MiR-592 inhibited cell proliferation of human colorectal cancer cells by suppressing of ccnd3 expression. Int. J. Clin. Exp. Med. 2015, 8, 3490–3497. [Google Scholar] [PubMed]
- Corcoran, C.; Rani, S.; Breslin, S.; Gogarty, M.; Ghobrial, I.M.; Crown, J.; O’Driscoll, L. MiR-630 targets igf1r to regulate response to her-targeting drugs and overall cancer cell progression in her2 over-expressing breast cancer. Mol. Cancer 2014, 13, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarzenbacher, D.; Klec, C.; Pasculli, B.; Cerk, S.; Rinner, B.; Karbiener, M.; Ivan, C.; Barbano, R.; Ling, H.; Wulf-Goldenberg, A.; et al. MiR-1287-5p inhibits triple negative breast cancer growth by interaction with phosphoinositide 3-kinase cb, thereby sensitizing cells for pi3kinase inhibitors. Breast Cancer Res. 2019, 21, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Xue, J.; Kuang, H.; Zhou, X.; Liao, L.; Yin, F. MicroRNA-1297 inhibits the growth and metastasis of colorectal cancer by suppressing cyclin d2 expression. DNA Cell Biol. 2017, 36, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, Z.; Sun, Q.; Sun, C.; Hua, H.; Huang, Q. The inhibitory effect of microRNA-1827 on anoikis resistance in lung adenocarcinoma a549 cells via targeting caveolin-1. Acta Biochim. Biophys. Sin. 2020, 52, 1148–1155. [Google Scholar] [CrossRef]
- Malagobadan, S.; Ho, C.S.; Nagoor, N.H. MicroRNA-6744-5p promotes anoikis in breast cancer and directly targets nat1 enzyme. Cancer Biol. Med. 2020, 17, 101–111. [Google Scholar]
- Hyun, T.S.; Rao, D.S.; Saint-Dic, D.; Michael, L.E.; Kumar, P.D.; Bradley, S.V.; Mizukami, I.F.; Oravecz-Wilson, K.I.; Ross, T.S. Hip1 and hip1r stabilize receptor tyrosine kinases and bind 3-phosphoinositides via epsin n-terminal homology domains. J. Biol. Chem. 2004, 279, 14294–14306. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Yao, H.; Li, C.; Shi, H.; Lan, J.; Li, Z.; Zhang, Y.; Liang, L.; Fang, J.Y.; Xu, J. Hip1r targets pd-l1 to lysosomal degradation to alter t cell-mediated cytotoxicity. Nat. Chem. Biol. 2019, 15, 42–50. [Google Scholar] [CrossRef]
- Wang, H.; Xu, B.; Shi, J. N6-methyladenosine mettl3 promotes the breast cancer progression via targeting bcl-2. Gene 2020, 722, 144076. [Google Scholar] [CrossRef]
- Mahoney, M.G.; Simpson, A.; Jost, M.; Noe, M.; Kari, C.; Pepe, D.; Choi, Y.W.; Uitto, J.; Rodeck, U. Metastasis-associated protein (mta)1 enhances migration, invasion, and anchorage-independent survival of immortalized human keratinocytes. Oncogene 2002, 21, 2161–2170. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Wu, Y.; Li, Q.; Liang, J.; He, Q.; Zhao, L.; Chen, J.; Cheng, M.; Huang, Z.; Ren, H.; et al. Mettl3 promotes tumorigenesis and metastasis through bmi1 m(6)a methylation in oral squamous cell carcinoma. Mol. Ther. 2020, 28, 2177–2190. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Dhar, S.; Campanelli, G.; Butt, N.A.; Schallheim, J.M.; Gomez, C.R.; Levenson, A.S. Mta1 drives malignant progression and bone metastasis in prostate cancer. Mol. Oncol. 2018, 12, 1596–1607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, N.; Gui, B.; Kumar, R. Role of mta1 in cancer progression and metastasis. Cancer Metastasis Rev. 2014, 33, 879–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murai, S.; Ando, A.; Ebara, S.; Hirayama, M.; Satomi, Y.; Hara, T. Inhibition of malic enzyme 1 disrupts cellular metabolism and leads to vulnerability in cancer cells in glucose-restricted conditions. Oncogenesis 2017, 6, e329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, C.; Lu, W.; Du, Y.; Yan, W.; Bao, Q.; Tian, Y.; Wang, G.; Ye, H.; Hao, H. Cytosolic me1 integrated with mitochondrial idh2 supports tumor growth and metastasis. Redox Biol. 2020, 36, 101685. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Xu, P.; Guo, S.; Yi, X.; Wang, H.; Yang, Y.; Liu, L.; Shi, Q.; Gao, T.; Li, C. Metastatic melanoma cells rely on sestrin2 to acquire anoikis resistance via detoxifying intracellular ros. J. Investig. Dermatol. 2020, 140, 666–675.e2. [Google Scholar] [CrossRef]
- Franzetti, G.A.; Laud-Duval, K.; van der Ent, W.; Brisac, A.; Irondelle, M.; Aubert, S.; Dirksen, U.; Bouvier, C.; de Pinieux, G.; Snaar-Jagalska, E.; et al. Cell-to-cell heterogeneity of ewsr1-fli1 activity determines proliferation/migration choices in ewing sarcoma cells. Oncogene 2017, 36, 3505–3514. [Google Scholar] [CrossRef] [Green Version]
- Chaturvedi, A.; Hoffman, L.M.; Jensen, C.C.; Lin, Y.C.; Grossmann, A.H.; Randall, R.L.; Lessnick, S.L.; Welm, A.L.; Beckerle, M.C. Molecular dissection of the mechanism by which ews/fli expression compromises actin cytoskeletal integrity and cell adhesion in ewing sarcoma. Mol. Biol. Cell 2014, 25, 2695–2709. [Google Scholar] [CrossRef]
- Muzumdar, M.D.; Chen, P.Y.; Dorans, K.J.; Chung, K.M.; Bhutkar, A.; Hong, E.; Noll, E.M.; Sprick, M.R.; Trumpp, A.; Jacks, T. Survival of pancreatic cancer cells lacking kras function. Nat. Commun. 2017, 8, 1090. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.; Li, Y.; Lian, G.; Lin, H.; Shang, C.; Zeng, L.; Chen, S.; Li, J.; Huang, C.; Huang, K.; et al. Kras promotes tumor metastasis and chemoresistance by repressing rkip via the mapk-erk pathway in pancreatic cancer. Int. J. Cancer 2018, 142, 2323–2334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahmood, N.; Mihalcioiu, C.; Rabbani, S.A. Multifaceted role of the urokinase-type plasminogen activator (upa) and its receptor (upar): Diagnostic, prognostic, and therapeutic applications. Front. Oncol. 2018, 8, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mauro, C.D.; Pesapane, A.; Formisano, L.; Rosa, R.; D’Amato, V.; Ciciola, P.; Servetto, A.; Marciano, R.; Orsini, R.C.; Monteleone, F.; et al. Urokinase-type plasminogen activator receptor (upar) expression enhances invasion and metastasis in ras mutated tumors. Sci. Rep. 2017, 7, 9388. [Google Scholar] [CrossRef]
- Madsen, C.D.; Ferraris, G.M.; Andolfo, A.; Cunningham, O.; Sidenius, N. Upar-induced cell adhesion and migration: Vitronectin provides the key. J. Cell Biol. 2007, 177, 927–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, M.; Eastman, B.M.; Webb, D.L.; Stoletov, K.; Klemke, R.; Gonias, S.L. Cell signaling by urokinase-type plasminogen activator receptor induces stem cell-like properties in breast cancer cells. Cancer Res. 2010, 70, 8948–8958. [Google Scholar] [CrossRef] [Green Version]
- Alfano, D.; Iaccarino, I.; Stoppelli, M.P. Urokinase signaling through its receptor protects against anoikis by increasing bcl-xl expression levels. J. Biol. Chem. 2006, 281, 17758–17767. [Google Scholar] [CrossRef] [Green Version]
- Guo, B.; Wang, W.; Zhao, Z.; Li, Q.; Zhou, K.; Zhao, L.; Wang, L.; Yang, J.; Huang, C. Rab14 act as oncogene and induce proliferation of gastric cancer cells via akt signaling pathway. PLoS ONE 2017, 12, e0170620. [Google Scholar] [CrossRef]
- Chao, H.; Deng, L.; Xu, F.; Fu, B.; Zhu, Z.; Dong, Z.; Liu, Y.N.; Zeng, T. Rab14 activates mapk signaling to promote bladder tumorigenesis. Carcinogenesis 2019, 40, 1341–1351. [Google Scholar] [CrossRef]
- Hata, T.; Mokutani, Y.; Takahashi, H.; Inoue, A.; Munakata, K.; Nagata, K.; Haraguchi, N.; Nishimura, J.; Hata, T.; Matsuda, C.; et al. Identification of microRNA-487b as a negative regulator of liver metastasis by regulation of kras in colorectal cancer. Int. J. Oncol. 2017, 50, 487–496. [Google Scholar] [CrossRef]
- Xin, H.; Li, C.; Wang, M. Dixdc1 promotes the growth of acute myeloid leukemia cells by upregulating the wnt/beta-catenin signaling pathway. Biomed. Pharmacother. 2018, 107, 1548–1555. [Google Scholar] [CrossRef]
- Tan, C.; Qiao, F.; Wei, P.; Chi, Y.; Wang, W.; Ni, S.; Wang, Q.; Chen, T.; Sheng, W.; Du, X.; et al. Dixdc1 activates the wnt signaling pathway and promotes gastric cancer cell invasion and metastasis. Mol. Carcinog. 2016, 55, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Liu, D.; Fan, C.; Luan, L.; Zhang, X.; Wang, E. Dixdc1 increases the invasion and migration ability of non-small-cell lung cancer cells via the pi3k-akt/ap-1 pathway. Mol. Carcinog. 2014, 53, 917–925. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, C.; Rani, S.; O’Brien, K.; O’Neill, A.; Prencipe, M.; Sheikh, R.; Webb, G.; McDermott, R.; Watson, W.; Crown, J.; et al. Docetaxel-resistance in prostate cancer: Evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS ONE 2012, 7, e50999. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Kim, J.H.; Kim, O.; Ahn, E.J.; Oh, S.J.; Akanda, M.R.; Oh, I.J.; Jung, S.; Kim, K.K.; Lee, J.H.; et al. Caveolin-1 enhances brain metastasis of non-small cell lung cancer, potentially in association with the epithelial-mesenchymal transition marker snail. Cancer Cell Int. 2019, 19, 171. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Zhang, L.; Zhou, Y.; Yi, X. Caveolin-1 regulates cell apoptosis and invasion ability in paclitaxel-induced multidrug-resistant a549 lung cancer cells. Int. J. Clin. Exp. Pathol. 2015, 8, 8937–8947. [Google Scholar] [PubMed]
- Chunhacha, P.; Pongrakhananon, V.; Rojanasakul, Y.; Chanvorachote, P. Caveolin-1 regulates mcl-1 stability and anoikis in lung carcinoma cells. Am. J. Physiol. Cell Physiol. 2012, 302, 1284–1292. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.F.; Li, K.S.; Shen, Y.H.; Gao, P.T.; Dong, Z.R.; Cai, J.B.; Zhang, C.; Huang, X.Y.; Tian, M.X.; Hu, Z.Q.; et al. Galectin-1 induces hepatocellular carcinoma emt and sorafenib resistance by activating fak/pi3k/akt signaling. Cell Death Dis. 2016, 7, e2201. [Google Scholar] [CrossRef]
- Spano, D.; Russo, R.; Di Maso, V.; Rosso, N.; Terracciano, L.M.; Roncalli, M.; Tornillo, L.; Capasso, M.; Tiribelli, C.; Iolascon, A. Galectin-1 and its involvement in hepatocellular carcinoma aggressiveness. Mol. Med. 2010, 16, 102–115. [Google Scholar] [CrossRef]
- Lo, H.W.; Hsu, S.C.; Xia, W.; Cao, X.; Shih, J.Y.; Wei, Y.; Abbruzzese, J.L.; Hortobagyi, G.N.; Hung, M.C. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of twist gene expression. Cancer Res. 2007, 67, 9066–9076. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.J.; Park, J.H.; Oh, S.M. Topk promotes epithelial-mesenchymal transition and invasion of breast cancer cells through upregulation of tbx3 in tgf-beta1/smad signaling. Biochem. Biophys. Res. Commun. 2020, 522, 270–277. [Google Scholar] [CrossRef]
- Krstic, M.; Kolendowski, B.; Cecchini, M.J.; Postenka, C.O.; Hassan, H.M.; Andrews, J.; MacMillan, C.D.; Williams, K.C.; Leong, H.S.; Brackstone, M.; et al. Tbx3 promotes progression of pre-invasive breast cancer cells by inducing emt and directly up-regulating slug. J. Pathol. 2019, 248, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Dong, Q.; Chen, Y.; Li, Y.; Zhang, B.; Zhou, F.; Lyu, X.; Chen, G.G.; Lai, P.; Kung, H.F.; et al. Novel hdac5-interacting motifs of tbx3 are essential for the suppression of e-cadherin expression and for the promotion of metastasis in hepatocellular carcinoma. Signal Transduct. Target. Ther. 2018, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Kim, J.Y.; Cho, Y.; An, H.; Lee, N.; Jo, H.; Ban, C.; Seo, J.H. Overexpression of angiotensin ii type 1 receptor in breast cancer cells induces epithelial-mesenchymal transition and promotes tumor growth and angiogenesis. Biochim. Biophys. Acta 2016, 1863, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yu, S.; Lam, M.M.T.; Poon, T.C.W.; Sun, L.; Jiao, Y.; Wong, A.S.T.; Lee, L.T.O. Angiotensin ii promotes ovarian cancer spheroid formation and metastasis by upregulation of lipid desaturation and suppression of endoplasmic reticulum stress. J. Exp. Clin. Cancer Res. 2019, 38, 116. [Google Scholar] [CrossRef] [Green Version]
- Tawinwung, S.; Ninsontia, C.; Chanvorachote, P. Angiotensin ii increases cancer stem cell-like phenotype in lung cancer cells. Anticancer Res. 2015, 35, 4789–4797. [Google Scholar]
- Yuan, X.; Zhang, T.; Zheng, X.; Zhang, Y.; Feng, T.; Liu, P.; Sun, Z.; Qin, S.; Liu, X.; Zhang, L.; et al. Overexpression of set oncoprotein is associated with tumor progression and poor prognosis in human gastric cancer. Oncol. Rep. 2017, 38, 1733–1741. [Google Scholar] [CrossRef]
- Ying, X.; Wei, K.; Lin, Z.; Cui, Y.; Ding, J.; Chen, Y.; Xu, B. MicroRNA-125b suppresses ovarian cancer progression via suppression of the epithelial-mesenchymal transition pathway by targeting the set protein. Cell. Physiol. Biochem. 2016, 39, 501–510. [Google Scholar] [CrossRef]
- Liu, H.; Gu, Y.; Yin, J.; Zheng, G.; Wang, C.; Zhang, Z.; Deng, M.; Liu, J.; Jia, X.; He, Z. Set-mediated ndrg1 inhibition is involved in acquisition of epithelial-to-mesenchymal transition phenotype and cisplatin resistance in human lung cancer cell. Cell Signal. 2014, 26, 2710–2720. [Google Scholar] [CrossRef]
- D’Amato, N.C.; Rogers, T.J.; Gordon, M.A.; Greene, L.I.; Cochrane, D.R.; Spoelstra, N.S.; Nemkov, T.G.; D’Alessandro, A.; Hansen, K.C.; Richer, J.K. A tdo2-ahr signaling axis facilitates anoikis resistance and metastasis in triple-negative breast cancer. Cancer Res. 2015, 75, 4651–4664. [Google Scholar] [CrossRef] [Green Version]
- Duan, Z.; Li, Y.; Li, L. Promoting epithelial-to-mesenchymal transition by d-kynurenine via activating aryl hydrocarbon receptor. Mol. Cell. Biochem. 2018, 448, 165–173. [Google Scholar] [CrossRef]
- Berezovskaya, O.; Schimmer, A.D.; Glinskii, A.B.; Pinilla, C.; Hoffman, R.M.; Reed, J.C.; Glinsky, G.V. Increased expression of apoptosis inhibitor protein xiap contributes to anoikis resistance of circulating human prostate cancer metastasis precursor cells. Cancer Res. 2005, 65, 2378–2386. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Lu, X.; Wang, M.; Zhao, X.; Xue, L. X-linked inhibitor of apoptosis protein accelerates migration by inducing epithelial-mesenchymal transition through tgf-beta signaling pathway in esophageal cancer cells. Cell Biosci. 2019, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Wu, Y.; He, X.; Zhang, C.; Zhu, M.; Chen, B.; Liu, Q.; Qu, X.; Li, W.; Wen, S.; et al. MicroRNA-204-5p inhibits invasion and metastasis of laryngeal squamous cell carcinoma by suppressing forkhead box c1. J. Cancer 2017, 8, 2356–2368. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Yu, P.; Xie, N.; Wu, Y.; Liu, H.; Zhang, M.; Tao, Y.; Wang, W.; Yin, H.; Zou, B.; et al. MicroRNA-204-5p is a tumor suppressor and potential therapeutic target in head and neck squamous cell carcinoma. Theranostics 2020, 10, 1433–1453. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Zhang, B.; Wang, W.; Fei, B.; Quan, C.; Zhang, J.; Song, M.; Bian, Z.; Wang, Q.; Ni, S.; et al. MiR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating rab22a. Clin. Cancer Res. 2014, 20, 6187–6199. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Xing, L.; Xu, H.; Wang, K.; She, J.; Shi, F.; Wu, H.; Sun, Y.; Gao, J.; He, S. MiR-204-5p suppress lymph node metastasis via regulating cxcl12 and cxcr4 in gastric cancer. J. Cancer 2020, 11, 3199–3206. [Google Scholar] [CrossRef] [Green Version]
- Kochetkova, M.; Kumar, S.; McColl, S.R. Chemokine receptors cxcr4 and ccr7 promote metastasis by preventing anoikis in cancer cells. Cell Death Differ. 2009, 16, 664–673. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Zhang, C.D.; Zhang, C.; Dai, D.Q. Dlx6-as1/miR-204-5p/oct1 positive feedback loop promotes tumor progression and epithelial-mesenchymal transition in gastric cancer. Gastric Cancer 2020, 23, 212–227. [Google Scholar] [CrossRef]
- Kang, J.; Shen, Z.; Lim, J.M.; Handa, H.; Wells, L.; Tantin, D. Regulation of oct1/pou2f1 transcription activity by O-glcnacylation. FASEB J. 2013, 27, 2807–2817. [Google Scholar] [CrossRef] [Green Version]
- An, Y.; Wang, B.; Wang, X.; Dong, G.; Jia, J.; Yang, Q. Sirt1 inhibits chemoresistance and cancer stemness of gastric cancer by initiating an ampk/foxo3 positive feedback loop. Cell Death Dis. 2020, 11, 115. [Google Scholar] [CrossRef]
- Dong, G.; Wang, B.; An, Y.; Li, J.; Wang, X.; Jia, J.; Yang, Q. Sirt1 suppresses the migration and invasion of gastric cancer by regulating arhgap5 expression. Cell Death Dis. 2018, 9, 977. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Lee, G.; Han, J.; Song, K.; Choi, J.S.; Choi, Y.L.; Shin, Y.K. Ube2c overexpression aggravates patient outcome by promoting estrogen-dependent/independent cell proliferation in early hormone receptor-positive and her2-negative breast cancer. Front. Oncol. 2019, 9, 1574. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Jin, D.; Wang, X.; Du, J.; Di, W.; An, J.; Shao, C.; Guo, J. Ube2c induces cisplatin resistance via zeb1/2-dependent upregulation of abcg2 and ercc1 in nsclc cells. J. Oncol. 2019, 2019, 8607859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, F.; Zhi, C.; Wang, M.; Li, T.; Khan, S.A.; Ma, Z.; Jing, Z.; Bo, C.; Zhou, Q.; Xia, S.; et al. Dysregulated nf-kappab signal promotes the hub gene pclaf expression to facilitate nasopharyngeal carcinoma proliferation and metastasis. Biomed. Pharmacother. 2020, 125, 109905. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jung, Y.S.; Jun, S.; Lee, S.; Wang, W.; Schneider, A.; Sun Oh, Y.; Lin, S.H.; Park, B.J.; Chen, J.; et al. Paf-wnt signaling-induced cell plasticity is required for maintenance of breast cancer cell stemness. Nat. Commun. 2016, 7, 10633. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Minchin, R.F.; Butcher, N.J. Arylamine n-acetyltransferase 1 protects against reactive oxygen species during glucose starvation: Role in the regulation of p53 stability. PLoS ONE 2018, 13, e0193560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiang, J.M.; Butcher, N.J.; Minchin, R.F. Effects of human arylamine N-acetyltransferase i knockdown in triple-negative breast cancer cell lines. Cancer Med. 2015, 4, 565–574. [Google Scholar] [CrossRef]
- Stepp, M.W.; Doll, M.A.; Carlisle, S.M.; States, J.C.; Hein, D.W. Genetic and small molecule inhibition of arylamine N-acetyltransferase 1 reduces anchorage-independent growth in human breast cancer cell line mda-mb-231. Mol. Carcinog. 2018, 57, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, Q.; Zhang, Q.; Liu, L. MicroRNA-30a-5p suppresses proliferation, invasion and tumor growth of hepatocellular cancer cells via targeting foxa1. Oncol. Lett. 2017, 14, 5018–5026. [Google Scholar] [CrossRef] [Green Version]
- He, R.; Yang, L.; Lin, X.; Chen, X.; Lin, X.; Wei, F.; Liang, X.; Luo, Y.; Wu, Y.; Gan, T.; et al. MiR-30a-5p suppresses cell growth and enhances apoptosis of hepatocellular carcinoma cells via targeting aeg-1. Int. J. Clin. Exp. Pathol. 2015, 8, 15632–15641. [Google Scholar]
- Zhu, W.J.; Yan, Y.; Zhang, J.W.; Tang, Y.D.; Han, B. Effect and mechanism of miR-26a-5p on proliferation and apoptosis of hepatocellular carcinoma cells. Cancer Manag. Res. 2020, 12, 3013–3022. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Li, K.; Guo, T. MiR-26a-5p suppresses tumor metastasis by regulating emt and is associated with prognosis in hcc. Clin. Transl. Oncol. 2017, 19, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, B.; Zhao, X.; Zhao, N.; Sun, R.; Zhu, D.; Zhang, Y.; Li, Y.; Gu, Q.; Dong, X.; et al. Twist1-related miR-26b-5p suppresses epithelial-mesenchymal transition, migration and invasion by targeting smad1 in hepatocellular carcinoma. Oncotarget 2016, 7, 24383–24401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zouq, N.K.; Keeble, J.A.; Lindsay, J.; Valentijn, A.J.; Zhang, L.; Mills, D.; Turner, C.E.; Streuli, C.H.; Gilmore, A.P. Fak engages multiple pathways to maintain survival of fibroblasts and epithelia: Differential roles for paxillin and p130cas. J. Cell Sci. 2009, 122, 357–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subauste, M.C.; Pertz, O.; Adamson, E.D.; Turner, C.E.; Junger, S.; Hahn, K.M. Vinculin modulation of paxillin-fak interactions regulates erk to control survival and motility. J. Cell Biol. 2004, 165, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Mei, Y.; Mi, Y.; Li, C.; Sun, X.; Zhao, X.; Liu, J.; Cao, W.; Li, Y.; Wang, Y. Downregulation of lncRNA anril suppresses growth and metastasis in human osteosarcoma cells. OncoTargets Ther. 2018, 11, 4893–4899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, Z.W.; Ma, C.; Medoro, L.; Chen, L.; Wang, B.; Gupta, R.; Liu, T.; Yang, X.Z.; Chen, T.T.; Wang, R.Z.; et al. LncRNA ANRIL is up-regulated in nasopharyngeal carcinoma and promotes the cancer progression via increasing proliferation, reprograming cell glucose metabolism and inducing side-population stem-like cancer cells. Oncotarget 2016, 7, 61741–61754. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.H.; Tang, J.M.; Li, J.; Li, X.W. Upregulation of sox2-activated lncRNA anril promotes nasopharyngeal carcinoma cell growth. Sci. Rep. 2018, 8, 3333. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Zhang, Y.; Cai, J.; Zhang, J.; Liu, X.; Yin, J.; Bai, Z.; Yao, H.; Zhang, Z. LncRNA-ANRIL promotes gastric cancer progression by enhancing NF-kB signaling. Exp. Biol. Med. 2019, 244, 953–959. [Google Scholar] [CrossRef]
- Lu, Y.; Zhou, X.; Xu, L.; Rong, C.; Shen, C.; Bian, W. Long noncoding RNA ANRIL could be transactivated by c-Myc and promote tumor progression of non-small-cell lung cancer. OncoTargets Ther. 2016, 9, 3077–3084. [Google Scholar]
- Wang, X.; Zhang, X.; Han, Y.; Wang, Q.; Ren, Y.; Wang, B.; Hu, J. Silence of lncRNA ANRIL represses cell growth and promotes apoptosis in retinoblastoma cells through regulating miR-99a and c-Myc. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2265–2273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, W.; Tian, C.; Jin, S. Effect of lncRNA ANRIL silencing on anoikis and cell cycle in human glioma via microRNA-203a. OncoTargets Ther. 2018, 11, 5103–5109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Fu, Q.; Li, S.; Liang, N.; Li, F.; Li, C.; Sui, C.; Dionigi, G.; Sun, H. LncRNA FOXD2-AS1 functions as a competing endogenous RNA to regulate TERT expression by sponging miR-7-5p in thyroid cancer. Front. Endocrinol. 2019, 10, 207. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Yamashita, K.; Harada, H.; Ushiku, H.; Tanaka, T.; Nishizawa, N.; Yokoi, K.; Washio, M.; Ema, A.; Mieno, H.; et al. The h19-peg10/igf2bp3 axis promotes gastric cancer progression in patients with high lymph node ratios. Oncotarget 2017, 8, 74567–74581. [Google Scholar] [CrossRef]
- Endo, H.; Shiroki, T.; Nakagawa, T.; Yokoyama, M.; Tamai, K.; Yamanami, H.; Fujiya, T.; Sato, I.; Yamaguchi, K.; Tanaka, N.; et al. Enhanced expression of long non-coding RNA hotair is associated with the development of gastric cancer. PLoS ONE 2013, 8, e77070. [Google Scholar] [CrossRef] [Green Version]
- Okugawa, Y.; Toiyama, Y.; Hur, K.; Toden, S.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Kusunoki, M.; Boland, C.R.; et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis 2014, 35, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Sun, T.; Liu, Z.; Zhang, R.; Ma, S.; Lin, T.; Li, Y.; Yang, S.; Zhang, W.; Wang, Y. Long non-coding RNA LEF1-AS1 promotes migration, invasion and metastasis of colon cancer cells through miR-30-5p/sox9 axis. OncoTargets Ther. 2020, 13, 2957–2972. [Google Scholar] [CrossRef] [Green Version]
- Braconi, C.; Kogure, T.; Valeri, N.; Huang, N.; Nuovo, G.; Costinean, S.; Negrini, M.; Miotto, E.; Croce, C.M.; Patel, T. MicroRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011, 30, 4750–4756. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Zhao, W.; Zhang, Y.; Liang, B. Long non-coding RNA-NEAT1 promotes cell migration and invasion via regulating miR-124/NF-κB pathway in cervical cancer. OncoTargets Ther. 2020, 13, 3265–3276. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Zhou, W.; Du, S.Q.; Gong, D.X.; Li, J.; Bi, J.B.; Li, Z.H.; Zhang, Z.; Li, Z.L.; Liu, X.K.; et al. Overexpression of SNHG12 regulates the viability and invasion of renal cell carcinoma cells through modulation of HIF1α. Cancer Cell Int. 2019, 19, 128. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Du, Y.; Hu, X.; Zhao, L.; Xia, W. Up-regulation of ceRNA TINCR by SP1 contributes to tumorigenesis in breast cancer. BMC Cancer 2018, 18, 367. [Google Scholar] [CrossRef] [PubMed]
- Hannen, R.; Bartsch, J.W. Essential roles of telomerase reverse transcriptase hTERT in cancer stemness and metastasis. FEBS Lett. 2018, 592, 2023–2031. [Google Scholar] [CrossRef]
- Fleisig, H.B.; Wong, J.M. Telomerase promotes efficient cell cycle kinetics and confers growth advantage to telomerase-negative transformed human cells. Oncogene 2012, 31, 954–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshimura, H.; Matsuda, Y.; Yamamoto, M.; Michishita, M.; Takahashi, K.; Sasaki, N.; Ishikawa, N.; Aida, J.; Takubo, K.; Arai, T.; et al. Reduced expression of the H19 long non-coding RNA inhibits pancreatic cancer metastasis. Lab. Investig. 2018, 98, 814–824. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, X.; Li, X.; Li, X.; Chen, Z. LncRNA H19 promotes epithelial-mesenchymal transition (EMT) by targeting miR-484 in human lung cancer cells. J. Cell. Biochem. 2018, 119, 4447–4457. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, L.; Lu, S. LncRNA H19 promotes viability and epithelial-mesenchymal transition of lung adenocarcinoma cells by targeting miR-29b-3p and modifying STAT3. Int. J. Oncol. 2019, 54, 929–941. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Liu, Z.; Zeng, W.; Huang, T. Down-regulation of long non-coding RNA HOTAIR sensitizes breast cancer to trastuzumab. Sci. Rep. 2019, 9, 19881. [Google Scholar] [CrossRef]
- Wang, S.L.; Huang, Y.; Su, R.; Yu, Y.Y. Silencing long non-coding RNA HOTAIR exerts anti-oncogenic effect on human acute myeloid leukemia via demethylation of HOXA5 by inhibiting Dnmt3b. Cancer Cell Int. 2019, 19, 114. [Google Scholar] [CrossRef]
- Cantile, M.; Di Bonito, M.; Cerrone, M.; Collina, F.; De Laurentiis, M.; Botti, G. Long non-coding RNA HOTAIR in breast cancer therapy. Cancers 2020, 12, 1197. [Google Scholar] [CrossRef]
- Botti, G.; Scognamiglio, G.; Aquino, G.; Liguori, G.; Cantile, M. LncRNA HOTAIR in tumor microenvironment: What role? Int. J. Mol. Sci. 2019, 20, 2279. [Google Scholar] [CrossRef] [Green Version]
- Larsimont, J.C.; Youssef, K.K.; Sanchez-Danes, A.; Sukumaran, V.; Defrance, M.; Delatte, B.; Liagre, M.; Baatsen, P.; Marine, J.C.; Lippens, S.; et al. Sox9 controls self-renewal of oncogene targeted cells and links tumor initiation and invasion. Cell Stem Cell 2015, 17, 60–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrasco-Garcia, E.; Lopez, L.; Aldaz, P.; Arevalo, S.; Aldaregia, J.; Egana, L.; Bujanda, L.; Cheung, M.; Sampron, N.; Garcia, I.; et al. Sox9-regulated cell plasticity in colorectal metastasis is attenuated by rapamycin. Sci. Rep. 2016, 6, 32350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khurana, N.; Sikka, S.C. Interplay between SOX9, wnt/β-catenin and androgen receptor signaling in castration-resistant prostate cancer. Int. J. Mol. Sci. 2019, 20, 2066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, W.; Vanderbilt, D.B.; Lin, C.C.; Martin, K.H.; Brundage, K.M.; Ruppert, J.M. SOX9 inhibits β-TRcP-mediated protein degradation to promote nuclear GLI1 expression and cancer stem cell properties. J. Cell Sci. 2015, 128, 1123–1138. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Gao, Y. Long non-coding RNA MEG3 inhibits cervical cancer cell growth by promoting degradation of P-STAT3 protein via ubiquitination. Cancer Cell Int. 2019, 19, 175. [Google Scholar] [CrossRef] [Green Version]
- Dan, J.; Wang, J.; Wang, Y.; Zhu, M.; Yang, X.; Peng, Z.; Jiang, H.; Chen, L. LncRNA-MEG3 inhibits proliferation and metastasis by regulating miRNA-21 in gastric cancer. Biomed. Pharmacother. 2018, 99, 931–938. [Google Scholar] [CrossRef]
- Zheng, Q.; Lin, Z.; Xu, J.; Lu, Y.; Meng, Q.; Wang, C.; Yang, Y.; Xin, X.; Li, X.; Pu, H.; et al. Long noncoding RNA MEG3 suppresses liver cancer cells growth through inhibiting β-catenin by activating PKM2 and inactivating PTEN. Cell Death Dis. 2018, 9, 253. [Google Scholar] [CrossRef]
- Terashima, M.; Tange, S.; Ishimura, A.; Suzuki, T. MEG3 long noncoding RNA contributes to the epigenetic regulation of epithelial-mesenchymal transition in lung cancer cell lines. J. Biol. Chem. 2017, 292, 82–99. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Li, J.; Chen, C.; Zhang, R.; Wang, K. Pan-cancer analysis of long non-coding RNA NEAT1 in various cancers. Genes Dis. 2018, 5, 27–35. [Google Scholar] [CrossRef]
- Liu, D.; Zou, Z.; Li, G.; Pan, P.; Liang, G. Long noncoding RNA NEAT1 suppresses proliferation and promotes apoptosis of glioma cells via downregulating miR-92b. Cancer Control 2020, 27, 1073274819897977. [Google Scholar] [CrossRef]
- Dong, P.; Xiong, Y.; Yue, J.; Xu, D.; Ihira, K.; Konno, Y.; Kobayashi, N.; Todo, Y.; Watari, H. Long noncoding RNA NEAT1 drives aggressive endometrial cancer progression via miR-361-regulated networks involving STAT3 and tumor microenvironment-related genes. J. Exp. Clin. Cancer Res. 2019, 38, 295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Liang, S.; Yu, Y.; Shi, Y.; Zheng, H. Knockdown of SNHG12 suppresses tumor metastasis and epithelial-mesenchymal transition via the Slug/ZEB2 signaling pathway by targeting miR-218 in NSCLC. Oncol. Lett. 2019, 17, 2356–2364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, S.; Qu, W.; Jiao, Y.; Zhang, J.; Zhang, C.; Dang, S. LncRNA SNHG12 promotes the proliferation and metastasis of papillary thyroid carcinoma cells through regulating wnt/β-catenin signaling pathway. Cancer Biomark. 2018, 22, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.X.; Bradbury, R.; Flamini, V.; Wu, B.; Jordan, N.; Jiang, W.G. MicroRNA-7 suppresses the homing and migration potential of human endothelial cells to highly metastatic human breast cancer cells. Br. J. Cancer 2017, 117, 89–101. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Pan, M.; Wang, J.; You, C.; Zhao, F.; Zheng, D.; Guo, M.; Xu, H.; Wu, D.; Wang, L.; et al. MiR-7 reduces breast cancer stem cell metastasis via inhibiting RELA to decrease ESAM expression. Mol. Ther. Oncolytics 2020, 18, 70–82. [Google Scholar] [CrossRef]
- Okuda, H.; Xing, F.; Pandey, P.R.; Sharma, S.; Watabe, M.; Pai, S.K.; Mo, Y.Y.; Iiizumi-Gairani, M.; Hirota, S.; Liu, Y.; et al. MiR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 2013, 73, 1434–1444. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.N.; Wei, C.C.; Wang, Z.X.; Sun, M. Long non-coding RNAs in anti-cancer drug resistance. Oncotarget 2017, 8, 1925–1936. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Li, T.; Yan, F.; Cai, W.; Zheng, J.; Jiang, X.; Sun, J. Effect of simvastatin and microRNA-21 inhibitor on metastasis and progression of human salivary adenoid cystic carcinoma. Biomed. Pharmacother. 2018, 105, 1054–1061. [Google Scholar] [CrossRef]
- Pedini, F.; De Luca, G.; Felicetti, F.; Puglisi, R.; Boe, A.; Arasi, M.B.; Fratini, F.; Mattia, G.; Spada, M.; Caporali, S.; et al. Joint action of miR-126 and MAPK/PI3K inhibitors against metastatic melanoma. Mol. Oncol. 2019, 13, 1836–1854. [Google Scholar] [CrossRef]
- Chen, Z.; Zhuang, Q.; Cheng, K.; Ming, Y.; Zhao, Y.; Ye, Q.; Zhang, S. Long non-coding RNA TCL6 enhances preferential toxicity of paclitaxel to renal cell carcinoma cells. J. Cancer 2020, 11, 1383–1392. [Google Scholar] [CrossRef] [Green Version]
- Moeng, S.; Son, S.W.; Lee, J.S.; Lee, H.Y.; Kim, T.H.; Choi, S.Y.; Kuh, H.J.; Park, J.K. Extracellular vesicles (EVs) and pancreatic cancer: From the role of EVs to the interference with EV-mediated reciprocal communication. Biomedicines 2020, 8, 267. [Google Scholar] [CrossRef] [PubMed]
- Catalano, M.; O’Driscoll, L. Inhibiting extracellular vesicles formation and release: A review of EV inhibitors. J. Extracell. Vesicles 2020, 9, 1703244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawaz, M. Extracellular vesicle-mediated transport of non-coding RNAs between stem cells and cancer cells: Implications in tumor progression and therapeutic resistance. Stem Cell Investig. 2017, 4, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, F.; Li, L.; Wang, G.; Deng, X.; Li, Z.; Kong, X. VDR signaling inhibits cancer-associated-fibroblasts’ release of exosomal miR-10a-5p and limits their supportive effects on pancreatic cancer cells. Gut 2019, 68, 950–951. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jiang, P.; Li, J.; Peng, M.; Zhao, X.; Zhang, X.; Chen, K.; Zhang, Y.; Liu, H.; Gan, L.; et al. Tumor-derived exosomal lnc-sox2ot promotes emt and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene 2018, 37, 3822–3838. [Google Scholar] [CrossRef]
- Wei, F.; Ma, C.; Zhou, T.; Dong, X.; Luo, Q.; Geng, L.; Ding, L.; Zhang, Y.; Zhang, L.; Li, N.; et al. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol. Cancer 2017, 16, 132. [Google Scholar] [CrossRef] [Green Version]
- Donnarumma, E.; Fiore, D.; Nappa, M.; Roscigno, G.; Adamo, A.; Iaboni, M.; Russo, V.; Affinito, A.; Puoti, I.; Quintavalle, C.; et al. Cancer-associated fibroblasts release exosomal microRNAs that dictate an aggressive phenotype in breast cancer. Oncotarget 2017, 8, 19592–19608. [Google Scholar] [CrossRef] [Green Version]
- Schaller, J.; Agudo, J. Metastatic colonization: Escaping immune surveillance. Cancers 2020, 12, 3385. [Google Scholar] [CrossRef]
- Wang, X.; Luo, G.; Zhang, K.; Cao, J.; Huang, C.; Jiang, T.; Liu, B.; Su, L.; Qiu, Z. Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kγ to promote pancreatic cancer metastasis. Cancer Res. 2018, 78, 4586–4598. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Xu, J.; Lan, H. Tumor-associated macrophages in tumor metastasis: Biological roles and clinical therapeutic applications. J. Hematol. Oncol. 2019, 12, 76. [Google Scholar] [CrossRef]
- Zhou, M.; Chen, J.; Zhou, L.; Chen, W.; Ding, G.; Cao, L. Pancreatic cancer derived exosomes regulate the expression of TLR4 in dendritic cells via miR-203. Cell. Immunol. 2014, 292, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Q.; Huang, Z.P.; Li, H.F.; Ou, Y.L.; Huo, F.; Hu, L.K. MicroRNA-520f-3p inhibits proliferation of gastric cancer cells via targeting SOX9 and thereby inactivating wnt signaling. Sci. Rep. 2020, 10, 6197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
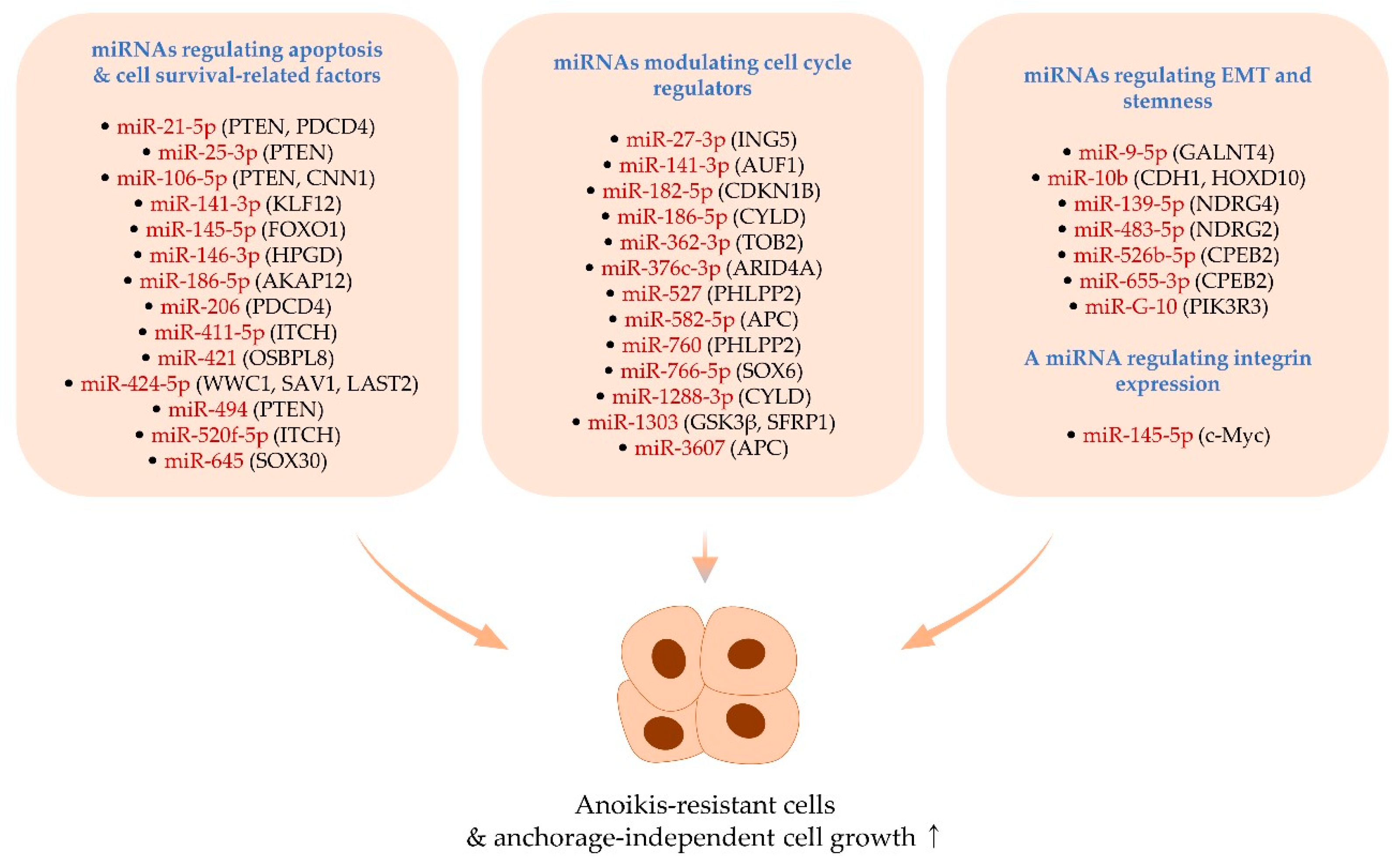
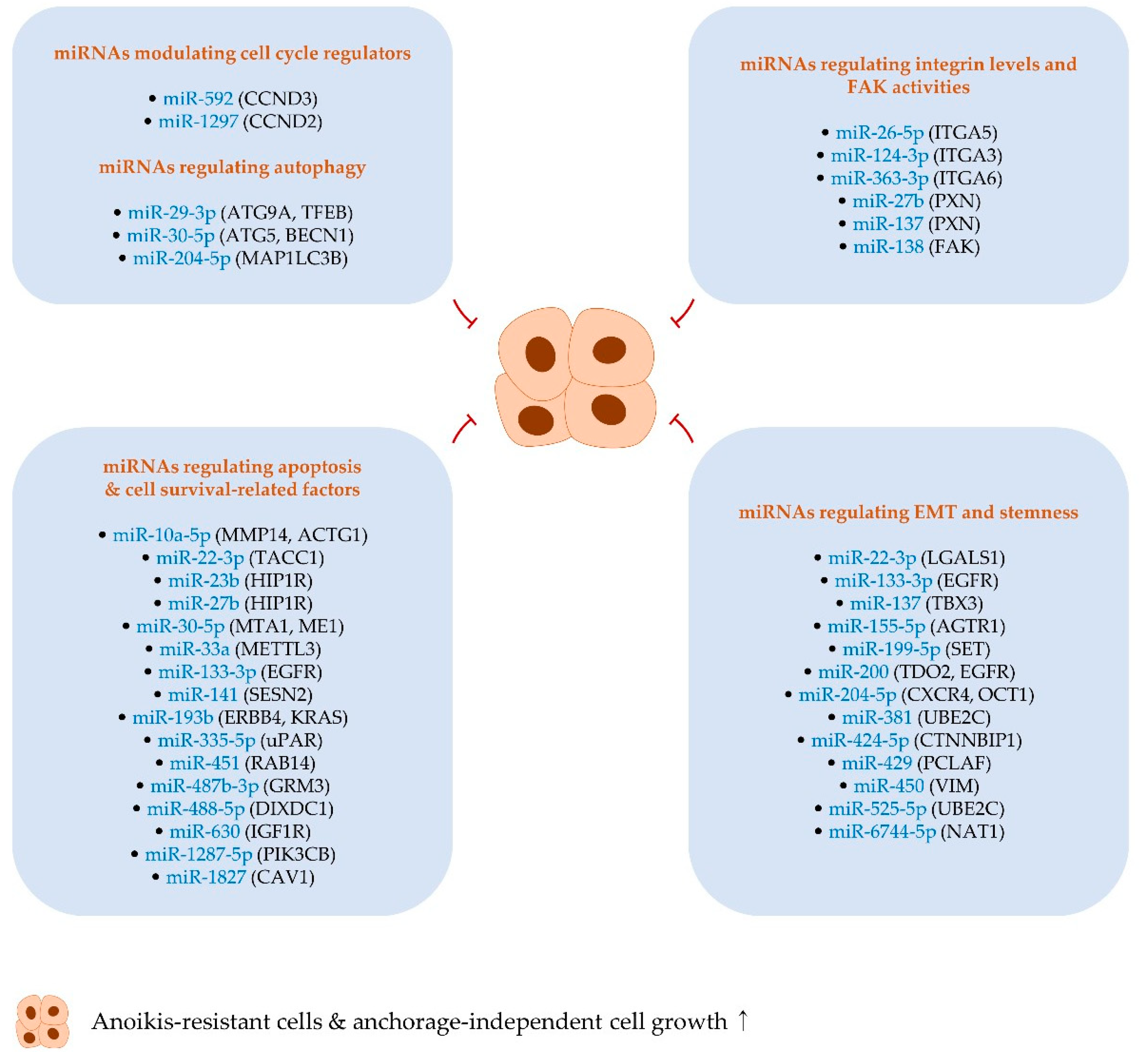
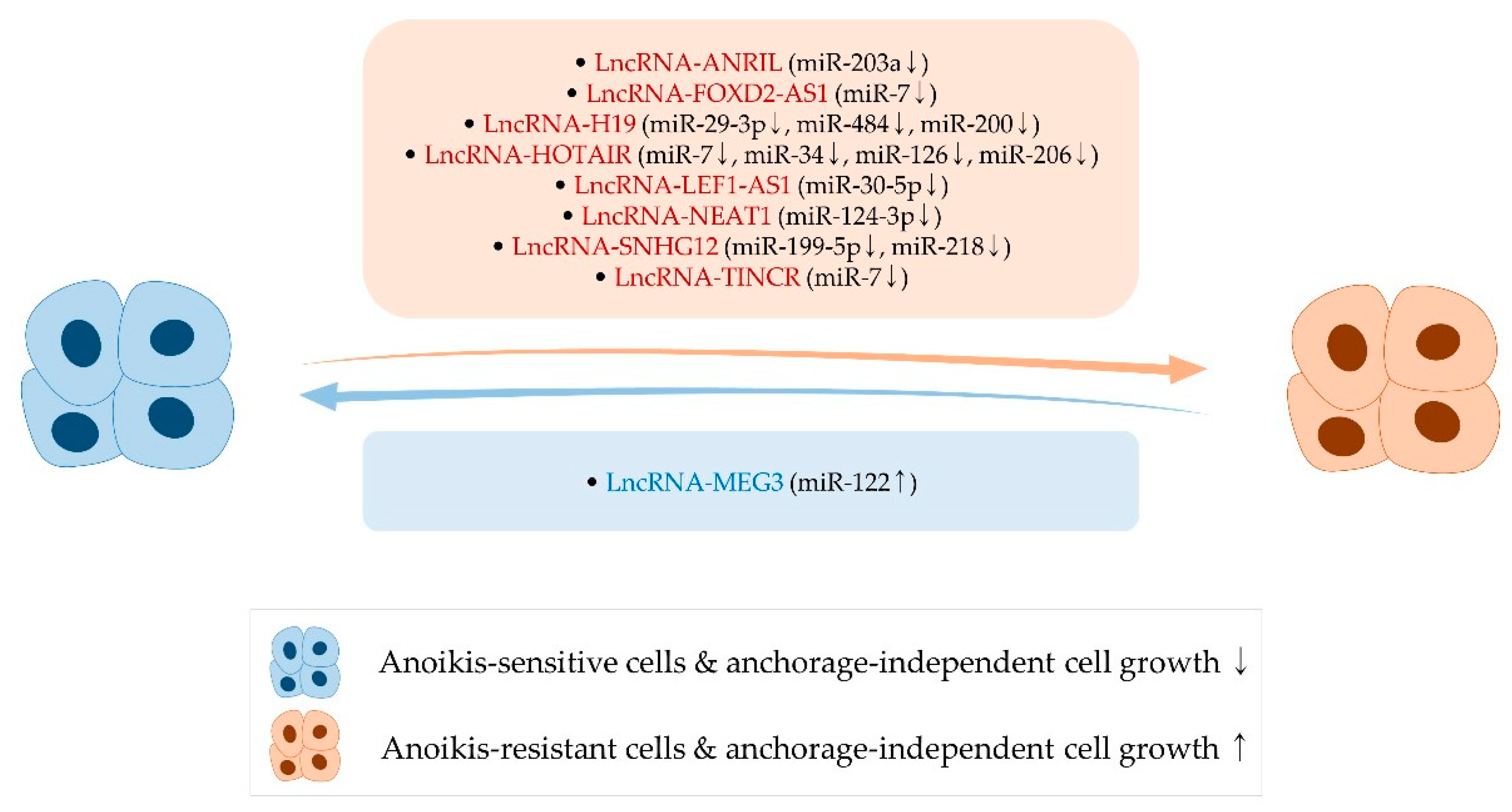
| miRNA | Type of Cancer | In Vitro Experiment | In Vivo Experiment | Ref. |
|---|---|---|---|---|
| miR-9-5p | Hepatocellular carcinoma | Plating cells on polyHEMA-coated plates + annexin-V/7-AAD analysis * | Subcutaneous injection of SK-Hep1 cells and Huh7 stably overexpressing and knocking down GALNT4, respectively | [29] |
| miR-10b | Melanoma | Soft agar assay ** | - | [30] |
| miR-21-5p | Esophageal cancer | Plating cells on polyHEMA-coated plates + annexin-V/propidium iodide analysis * | Tail vein injection of OE33 cells stably overexpressing or knocking down miR-21 | [27] |
| miR-25-3p | Retinoblastoma | Soft agar assay ** | Subcutaneous injection of WERI-RB-1 cells transduced with miR-25-3p lentiviral plasmids | [28] |
| miR-27-3p | Osteosarcoma | Soft agar assay ** | - | [31] |
| miR-106-5p | Pituitary adenoma | Agarose clonal formation assay ** | - | [32] |
| miR-139-5p | Adrenocortical cancer | Soft agar colony formation assay ** | - | [33] |
| miR-141-3p | Ovarian cancer | Plating cells on 1% agarose gel coated plates + TUNEL assay * | Intraperitoneal injection of SKOV3 cells stably expressing miR-141-3p | [34] |
| miR-141-3p | Bladder cancer | Soft agar assay ** | Subcutaneous injection of TAp63α-expressing UMUC3 cells | [35] |
| miR-145-5p | Esophageal cancer | Plating cells on low adhesion plates + Western blotting for detecting PARP cleavage * | Flank injection of SK-GT-4 cells overexpressing miR-145 | [36] |
| miR-145-5p | Bladder cancer | Soft agar assay ** | Subcutaneous injection of T24T cells stably overexpressing miR-145-5p | [37] |
| miR-146-3p | Cervical cancer | Soft agar assay ** | - | [38] |
| miR-182-5p | Bladder cancer | Soft agar colony formation assay ** | - | [39] |
| miR-186-5p | Prostate cancer | Soft agar assay ** | - | [40] |
| miR-186-5p | Melanoma | Soft agar assay ** | - | [41] |
| miR-206 | Breast cancer | Cell culture using 1% methylcellulose-based media + propidium iodide staining/trypan blue exclusion * | Orthotopic injection of MDA-MB-231 cells treated with miR-206 inhibitors | [42] |
| miR-362-3p | Hepatocellular carcinoma | Soft agar colony formation assay ** | - | [43] |
| miR-376c-3p | Gastric cancer | Soft agar assay ** | - | [44] |
| miR-411-5p | Hepatocellular carcinoma | Soft agar assay ** | - | [45] |
| miR-421 | Lung cancer | Soft agar assay ** | - | [46] |
| miR-424-5p | Thyroid cancer | Plating cells on polyHEMA-coated plates + annexin-V/propidium iodide analysis * | Tail vein injection of K1 cells stably overexpressing or knocking down miR-424-5p | [47] |
| miR-483-5p | Adrenocortical cancer | Soft agar colony formation assay ** | - | [33] |
| miR-494 | Colorectal cancer | Soft agar assay ** | - | [48] |
| miR-520f-5p | Melanoma | Soft agar assay ** | - | [49] |
| miR-526b-5p | Breast cancer | Spheroid formation assay ** | Intravenous or subcutaneous injection of CPEB2A-silencing MCF10A cells | [50] |
| miR-527 | Esophageal cancer | Soft agar assay ** | - | [51] |
| miR-582-5p | Colorectal cancer | Soft agar assay ** | - | [52] |
| miR-645 | Colorectal cancer | Soft agar assay ** | Subcutaneous injection of WiDr and EB cells stably knocking down miR-645 | [53] |
| miR-655-3p | Breast cancer | Spheroid formation assay ** | Intravenous or subcutaneous injection of CPEB2A-silencing MCF10A cells | [50] |
| miR-760 | Ovarian cancer | Soft agar assay ** | - | [54] |
| miR-766-5p | Colorectal cancer | Soft agar assay ** | - | [55] |
| miR-1288-3p | Glioblastoma | Soft agar colony formation assay ** | - | [56] |
| miR-1303 | Neuroblastoma | Soft agar assay ** | - | [57] |
| miR-3607 | Lung cancer | Soft agar assay ** | - | [58] |
| miR-G-10 | Cervical cancer | Cell–matrix adhesion assay ** | Intravenous injection of HeLa cells expressing miR-G-10 | [59] |
| LncRNA | Type of Cancer (Expression of lncRNA) | In Vitro Experiment | In Vivo Experiment | Ref. |
|---|---|---|---|---|
| LncRNA-ANRIL | Glioblastoma (↑) | Plating cells on polyHEMA-coated plates + annexin-V assay * | - | [242] |
| LncRNA-FOXD2-AS1 | Thyroid cancer (↑) | Soft agar assay ** | Subcutaneous injection of K1 cells stably silencing lncRNA-FOXD2-AS1 | [243] |
| LncRNA-H19 | Gastric cancer (↑) | Soft agar assay ** | - | [244] |
| LncRNA-HOTAIR | Gastric cancer (↑) | Soft agar assay ** | Tail vein injection of MKN74 cells expressing LncRNA-HOTAIR. Intraperitoneal injection of KATO III cells following lncRNA-HOTAIR knockdown | [245] |
| LncRNA-HOTAIR | Gastric cancer (↑) | Plating cells on ultra-low attachment plates + annexin-V and MTT assay * | Intraperitoneal injection of MKN45 cells following the transfection of lncRNA-HOTAIR-targeting siRNA | [246] |
| LncRNA-LEF1-AS1 | Colorectal cancer (↑) | Soft agar assay ** | Tail vein injection of HT29 cells following the knockdown of lncRNA-LEF1-AS1 | [247] |
| LncRNA-MEG3 | Hepatocellular carcinoma (↓) | Soft agar assay ** | - | [248] |
| LncRNA-NEAT1 | Cervical cancer (↑) | Soft agar assay ** | - | [249] |
| LncRNA-SNHG12 | Renal cell carcinoma (↑) | Soft agar assay ** | Subcutaneous injection of Caki-1 cells stably silencing lncRNA-SNHG12 | [250] |
| LncRNA-TINCR | Breast cancer (↑) | Soft agar assay ** | Subcutaneous injection of MDA-MB-468 cells following the transfection of lncRNA-TINCR-targeting siRNA | [251] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.Y.; Son, S.W.; Moeng, S.; Choi, S.Y.; Park, J.K. The Role of Noncoding RNAs in the Regulation of Anoikis and Anchorage-Independent Growth in Cancer. Int. J. Mol. Sci. 2021, 22, 627. https://doi.org/10.3390/ijms22020627
Lee HY, Son SW, Moeng S, Choi SY, Park JK. The Role of Noncoding RNAs in the Regulation of Anoikis and Anchorage-Independent Growth in Cancer. International Journal of Molecular Sciences. 2021; 22(2):627. https://doi.org/10.3390/ijms22020627
Chicago/Turabian StyleLee, Han Yeoung, Seung Wan Son, Sokviseth Moeng, Soo Young Choi, and Jong Kook Park. 2021. "The Role of Noncoding RNAs in the Regulation of Anoikis and Anchorage-Independent Growth in Cancer" International Journal of Molecular Sciences 22, no. 2: 627. https://doi.org/10.3390/ijms22020627






