Abstract
Plants are in continuous conflict with the environmental constraints and their sessile nature demands a fine-tuned, well-designed defense mechanism that can cope with a multitude of biotic and abiotic assaults. Therefore, plants have developed innate immunity, R-gene-mediated resistance, and systemic acquired resistance to ensure their survival. Transcription factors (TFs) are among the most important genetic components for the regulation of gene expression and several other biological processes. They bind to specific sequences in the DNA called transcription factor binding sites (TFBSs) that are present in the regulatory regions of genes. Depending on the environmental conditions, TFs can either enhance or suppress transcriptional processes. In the last couple of decades, nitric oxide (NO) emerged as a crucial molecule for signaling and regulating biological processes. Here, we have overviewed the plant defense system, the role of TFs in mediating the defense response, and that how NO can manipulate transcriptional changes including direct post-translational modifications of TFs. We also propose that NO might regulate gene expression by regulating the recruitment of RNA polymerase during transcription.
1. Challenges to Plants from Pathogens
Plants are the primary producers of the ecosystem and, due to their sedentary nature, are exposed to various environmental adversities such as cold, heat, flood, salinity, and drought. However, the greatest threat that plants face is the attack from phytopathogens, herbivory, and human activity. Diseases caused by phytopathogens can drastically reduce crop productivity and yield, which affect not only the production of food but also human development [1]. Although technological advancements and scientific contributions have dramatically reduced the losses in yield and productivity, plant diseases still contribute about 20–30% loss in actual production every year [2,3]. This reflects a lack of knowledge of disease management, the mechanisms behind epidemic development, and of the causal agents.
Plant disease results from complex interactions between various biotic and abiotic stressors, including pathogens, hosts, and the environment [4]. Plant pathogens employ multiple approaches to ensure their success. Pathogenic bacteria access the host plant via stomata, hydathodes, or wounds and proliferate in the intercellular space (called the apoplast). Similarly, nematodes access the host plant by inserting a stylet directly into the host plant cell, while pathogenic and symbiotic fungi and oomycetes penetrate plant cells by inserting haustoria [5]. All these diverse pathogen types release effector molecules for their survival. Therefore, a proper understanding of the causal agents and their mechanisms of action is of paramount importance.
2. Plant’s Strategy “Guard” and “Decoy” Models
Plants have a well-defined, fine-tuned defense mechanism that responds to environmental threats and pathogen attack as demanded by their sessile nature. Recently, defense strategies in plants that are induced by pathogens were reviewed in detail [6]. Here we will briefly mention how a plant responds when a pathogen tries to invade it. Unlike mammals, plants lack portable defenders or adaptive immune system and rely solely on the innate immunity of each cell and systemic signals emerging from the site of infection [7,8]. There are two main branches of plant defense: the first uses transmembrane pattern recognition receptors (PRRs) that perceive signals from different pathogens and respond to microbial- or pathogen-associated molecular patterns (MAMPS/PAMPs) like flagellin [9]; the second branch functions inside the cell, using plant resistance (R) genes [7]. The effector molecules of pathogens are recognized by NB-LRR proteins encoded by plant R genes, leading to R-avr interactions that induce similar defense responses. This can be better explained by the zigzag model of plant defense presented by Jones and Dangl [5]. According to their model, in the first phase of plant defense response, PAMPs are recognized by plant PRRs, leading to the activation of a defense process called PAMP-triggered immunity (PTI) that can restrict further pathogen growth. In the second phase, the pathogens that succeeded in releasing their effector molecules compromise the PTI, leading to a condition called effector-triggered susceptibility. Recognition of effector molecules by the host cells causes effector-triggered immunity (ETI), resulting in disease resistance and restricted pathogen growth. This puts pressure on the pathogens to acquire additional effector molecules and diversify these effectors to suppress ETI. The last phase of the defense response is critical: if the pathogen is successful in adapting to the host R genes the plants will be unable to defend themselves against infection.
The plants seem to be smarter here by not involving the R genes directly—a strategy that is termed the “guard hypothesis” [7]. This hypothesis suggests that R proteins recognize pathogen effectors indirectly. For example, RPM1-interacting protein 4 (RIN4) is a plasma membrane–associated protein that is guarded by NUCLEOTIDE BINDING SITE LUCIN RICH REPEAT (NBS-LRR) proteins [5]. It is manipulated by three different types of bacterial effectors. Two effectors (type III), AvrRpm1 and AvrB, interact with RIN4 and induce its phosphorylation [10]. This modification induce transcription of the RPM1 NBS-LRR protein. A third effector, AvrRPt2, is a cysteine protease that cleaves RIN4 at two different sites [10,11]. This cleavage induces the RPS2 NBS-LRR protein [12]. Both RPS2 and RPM1 require NON-RACE-SPECIFIC DISEASE RESISTANCE1 (NDR1) protein, which interacts with RIN4 for resistance against Pseudomonas syringae. Interestingly, functional genomics studies have suggested that RIN4 is not the only target of these three effectors [13]. Reports suggest that an effector contributes to virulence by possible manipulation of several other host targets. However, the contact of any of these targets with the effectors is sufficient to activate plant R genes. It seems that RIN4 negatively regulates the two NBS-LRR proteins, RPS2 and RPM1. But in rpm1rps2 (knockout for RPM1 and RPS2) plants, the pathogen effectors AvrRPt2 and AvrRpm1 manipulate RIN4 to suppress PTI [14]. Therefore, the plants use R proteins to guard against pathogens by interacting with the released effectors.
Plant scientists, specifically those who have an interest in evolution, have proposed another model to explain “guard” and “effector” interactions. They called it the “decoy model” [15]. van der Hoorn and Kamoun [15] gave a realistic explanation of two opposing yet unstable naturally selective forces on the guarded effector target. They explained that plant R genes are polymorphic, suggesting the presence/absence of functional R genes in different individuals of a plant population. In the absence of a functional R gene, the binding affinity of the guardee to the effector is compelled to decrease by natural selection, thereby avoiding detection and alteration by the effector. However, natural selection is expected to favor guardees-effector interactions for better pathogen perception in the presence of a functional R gene. They suggested that these two opposing pressures on the same effector are an evolutionarily unstable situation that could be eased upon by the evolution of a host protein, which they termed the “decoy,” that could potentially perceive the effector by the R protein without functioning in development or resistance to disease [15].
3. Transcription Factors (TFs): Modulators of Gene Expression
TFs are regulatory proteins that are responsible for the mechanistic control of gene transcription. Technically, they act as the on/off switch of gene expression and are responsible for the activation and suppression of genes, thereby regulating their function. They are transcribed in the nucleus, translated in the cytoplasm, and returned to the nucleus to search for their targets in the genomic DNA; therefore, they are also called diffusible regulatory molecules [16]. Their re-entry into the nucleus is mediated by nuclear localization sites found in the protein sequences of all TFs [16]. The TFs bind to specific DNA sequences, called cis-regulatory elements or TF binding sites (TFBSs), in the promoter region of a gene and have defined DNA-binding domains. TBFs may also be located in the intron region and play regulatory roles. For example, in Arabidopsis thaliana, sequences for cis-regulatory elements of the floral homeotic gene AGAMOUS (AG) are located in the second intron [17]. The second intron contains TFBs for two direct transcriptional activators of AG, i.e., LEAFY (LFY) and WUSCHEL (WUS), and other putative regulatory elements. TFBs are usually highly conserved and are crucial for DNA binding and used to classify TFs into various groups or families [18], such as MADS, WRKY, or APETALA2/ethylene-responsive factors (AP2/ERF). TFs can also be categorized based on their three-dimensional protein structure and composition, such as basic helix-loop-helix (bHLH), helix-turn-helix, and zinc finger proteins. Sequence-specific TFs are considered vital for the regulation of genes involved in prokaryotic and eukaryotic cellular mechanisms [19]. In prokaryotes and eukaryotes, gene regulation by TFs occurs through different mechanisms: in the former, TFs role is driven by a single protein, while, in the latter, it is a combined process that requires multiple proteins to coordinate and drive gene regulation. The binding of a TF to the promoter of a gene is spatiotemporally dependent. Phillips [18] quoted an interesting example of β-globin (a protein responsible for oxygen exchange in red blood cells) to explain this: the β-globin gene is present in every human cell, but no cell type other than red blood cells expresses this gene. Reddy, et al. [20] studied the beta-globin promoters of different cell types using DNA footprinting. They found that TFs that could bind to beta-globin promoters were only expressed in erythroblasts (immature red blood cells).
TFs have two domains: a DNA-binding domain and an effector domain that regulates interactions with other TFs or proteins necessary for transcription. Most DNA-binding domains are highly conserved within the members of the same family of TFs, while the effector domains evolve more rapidly. TFs mediate many functions, including gene induction, gene repression, and response to signal transduction under various environmental conditions. In this study, we will focus on the regulatory role of TFs in plant defense and how NO plays a role in translating its bioactivity to recruit these TFs.
4. Regulatory Role of TFs in Plant Defense
The two interconnected branches of plant defense, PTI and ETI, are the major defense strategies that plants use immediately after pathogen perception [6]. These strategies require well-communicated signal transduction and fine-tuned regulation of gene expression [21,22,23]. TFs play a key role in innate plant immunity, primarily by regulating genes involved in PTI, ETI, and hormone and phytoalexin synthesis and pathways. One of the immediate responses to pathogen infection is transcriptional reprogramming. A study using high resolution temporal transcriptomic analyses in Arabidopsis demonstrated that approximately one-third of the genome showed differential expression in response to the necrotrophic pathogen Botrytis cinerea immediately after infection [24]. Thus, transcriptional reprogramming of the plant cell demands significant changes in gene expression to favor defense over other metabolic processes such as growth and development [23]. Recent studies also suggested that a metabolic shift is required to mediate the trade-off between growth and immunity to ensure proper resource allocation for plant survival [25,26,27]. Many TF families have been reported to play key roles in transcriptional reprogramming. WRKY, bHLH, AP2/ERF, NAM/ATAF/CUC (NAC), and MYB are the major plant TF families [28] regulating various biological processes including plant defense.
4.1. WRKY TFs
The WRKY TFs often called “jack-of-various-trades” [29], are one of the largest TF families in plants [30]. The detailed composition and mode of action of WRKYs are well explored [30,31,32,33]. Here, we will focus on their functional roles, particularly in plant defense.
The regulatory role of WRKYs in plant defense has been extensively studied, particularly in the model plant Arabidopsis thaliana, and are reported to have both negative and positive roles in the regulation of plant defense [34]. Reports suggested that WRKYs regulate PAMP-signaling downstream of the mitogen-activated protein kinase (MAPK) signaling cascade [35]. The MAPK cascade plays a vital role in various defense responses—particularly, in sensing PAMPs or ETI [36]. For example, WRKY33 in Arabidopsis is reported to have a role in resistance to necrotrophic fungal pathogens B. cinerea and Alternaria brassicicola [37]. Recent reports using functional genomics revealed that WRKY33 is required for MPK3/MPK6-induced camalexin biosynthesis [38]. They also showed that WRKY33- and pathogen-induced camalexin production was compromised in wrky33 mutants. They further suggested that WRKY33 is a pathogen-inducible TF that acts as a substrate for MPK3/MPK6 to undergo phosphorylation and mutation. WRKY33 also binds to the promoter of phytoalexin deficient 3 (PAD3), which catalyzes the final step in camalexin biosynthesis [28], and to the promoters of 1-aminocyclopropane-1-carboxylic acid synthases 2 and 6 (ACS2 and ACS6) in response to B. cinerea [39]. Global expression profiling of wild type and susceptible wrky33 mutants in response to B. cinerea indicated differential transcriptional reprogramming, suggesting that unidentified targets for WRKY33 might be critical for establishing immunity to this necrotrophic pathogen [40]. Similarly, the closest homolog of WRKY33 in Nicotiana benthamiana WRKY8 (NbWRKY8) is also phosphorylated by MAPKs, resulting in the induction of defense-related genes. Furthermore, silencing causes increased susceptibility to the oomycete Phytophthora infestans and the ascomycete fungus Colletotrichum orbiculare [41].
WRKY TFs are also reportedly involved in ETI and interact with plant R proteins. For example, in barley, mildew resistance locus A10 (MLA10) NB-LRR protein, which confers resistance to powdery mildew, interacts with Hordeum vulgare WRKY1 (HvWRKY1) and HvWRKY2 in the presence of the AVRA10 effector [42]. Both HvWRKY1 and HvWRKY2 repress basal defenses against the virulent fungus Blumeria graminis that causes powdery mildew. Following infection by B. graminis (expressing AVRA10), MLA10 interacts with HvWRKY1 and HvWRKY2 to activate the defense. Another study reported that rice panicle blast 1 (Pb1), another NB-LRR protein, interacts with Oryza sativa WRKY45 (OsWRKY45), mediating the resistance to rice blast caused by the fungus Magnaporthe oryzae [43]. Similarly, in Arabidopsis, WRKY52, also called resistance to Ralstonia solanacearum 1 (RRS1), is a TIR-NB-LRR protein with a WRKY domain that shows resistance to the bacterial pathogen Ralsotonia solanacearum [44]. Using map-based cloning and natural variation analysis, Narusaka, et al. [45] reported that RRS1 interacts with RPS4 for dual resistance toward fungal and bacterial phytopathogens. Similarly, Arabidopsis WRKY8 (AtWRKY8) negatively regulates basal defenses to Pseudomonas syringae pathovar tomato (Pst) while positively regulating defense responses to B. cinerea [46].
4.2. bHLH TFs
The bHLH TF family reported in animals and plants in 1989 [47,48] and yeast in 1990 [49] comprised of a group of TFs characterized by the so-called “basic helix-loop-helix (bHLH)” domain. The proteins with this domain are known for a broad spectrum of functions that are reviewed in detail by Heim et al. [50]. Here we will briefly discuss their role in plant defense. The bHLH domain comprises an N-terminal stretch of hydrophilic basic amino acids followed by an HLH domain predicted to have amphipathic α-helices with an intervening loop in between, to form dimers [51]. In essence, bHLH TFs bind with E-box sequences (CANNTG) in the promoters of their target genes with variation in binding specificity [52,53]. Studies in mammals have shown that the conserved HLH structure is critical for the formation of bHLH protein dimers [54]. The specificity for a particular protein partner is determined by the α-helices. In Arabidopsis, the bHLH TF family includes about 160 members (https://www.arabidopsis.org/browse/genefamily/bHLH.jsp). However, only a few of them have been characterized in detail, which has shown that the bHLH might not be directly involved in plant defense, but they have an indirect connection by producing certain metabolites that are required during stress conditions. For example, in Arabidopsis, IAA-LEUCINE RESISTANT3 (ILR3 or BHLH105) represses the production of aliphatic glucosinolates and secondary metabolites produced in response to wounding, insects, or other microbial pathogens [55]. Furthermore, they interact with JA signaling pathway, thus regulating phytohormonal balance which is also critical for plant defense [56]. Song et al. [57] identified members of the bHLH TF family (bHLH3, bHLH13, bHLH14, and bHLH17) to be targeted by JASMONATE-ZEM-Domain (JAZs). Using the loss of function mutants for these bHLH TFs, they showed that bHLH mutants showed sensitivity to JA-inhibited root growth and an increase in JA-induced defense against pathogen infection and insect attack. The transgenic plants overexpressing bHLH13 or bHLH17 showed reduced JA-mediated responses [57]. Another bHLH TF, HBI1 negatively regulates genes that are involved in plant immunity and inhibits PAMP-induced growth arrest thus mediating the trade-off between growth and PAMP-triggered immunity [26]. Similarly, another bHLH TF, ILR3 was reported to regulate iron deficiency, glucosinolate biosynthesis, and pathogen response [55,58]. MYC2 another bHLH TF, regulates a subset of plant defense responses in Nicotiana attenuate [59]
4.3. AP2/ERF TF
The AP2/ERF is another important plant-specific TF family that regulates stress responses in plants, mostly studied for responses to abiotic stresses [60]. Members of this family are characterized by the presence of an AP2 DNA binding domain which comprises 40–70 conserved amino acids [60,61,62]. The AP2/ERF TFs regulate genes involved in various biological processes including growth and development, hormone signaling, stress responses both at transcriptional and post-translational levels [63,64,65,66]. Studies involving gene expression profiling have shown that most AP2/ERF TFs have a low basal expression and can be induced or reduced by external stress stimuli or hormonal imbalance [67,68]. Some of the important AP2/ERFs include DEHYDRATION-RESPONSIVE ELEMENT BINDING proteins (DREBs), members of the RAP2 family, and ABA INSENSITIVE 4 (ABI4), etc. Reports suggested that AP2/ERFs are induced by the cis-regulatory elements present in their promotors. These elements include HEAT SHOCK ELEMENT (HSE), ETHYLEN INSENSITVE 3 (EIN3) BINDING SITE (EBS), LOW-TEMPERATURE RESPONSIVE ELEMENT (LRT), and ABA Response Element (ABRE) [69].
Post-translational changes such as phosphorylation also affect the activity and abundance of AP2/ERFs. Other studies have shown that phosphorylation affects AP2/ERF protein stability and transactivity [69]. For example, in Arabidopsis, the positive regulator of ABA signaling pathway SNF1-related protein kinases (SnRKs) interacts and phosphorylates RAV1 to constrain its transcription repression role [70]. Similarly, ERF6 and EFR104 are phosphorylated by mitogen-activated protein kinases (MAPKs) for positive regulation of pathogen responses [71,72]. AP2/ERFs are also characterized in plant defense. Mase, et al. [73] showed in Arabidopsis thaliana, by using a structural analog of AAL, a phytotoxin produced by Alternaria alternata [74], that the MODULATOR of ALL CELL DEATH 1 (MACD1), and AP2/ERF TF, was involved in ALL-induced cell death and acted downstream of ethylene.
ERF is one of the large subfamilies of AP2/ERFs. In Arabidopsis thaliana, there are about 145 members of the AP2/ERF family [67]. Among them, about 65 members are identified as ERFs. Members of the ERF sub-family are characterized for their role in plant defense. In tomato, the Pti4 and Pti5 (ERFs) are phosphorylated by Pto protein when challenged by the virulent P. syringea. The Pst-induced phosphorylation increases Pti4 and Pti5 binding to their target sequences in defense-related genes [75]. Similarly, tomato ERFs Pti4, Pti5, and Pti6 when overexpressed in Arabidopsis, induced defense response, and contributed to resistance against P. syringae [76]. In Arabidopsis constitutive expression of ERF1 has been shown to increase resistance against several necrotrophic fungal pathogens. [77]. Besides, the ERF1 is considered a point of integration between JA and ethylene signaling pathways. A detailed review on the role of AP2/ERF TFs has reported that members of ERFs are enriched in genes regulating disease resistance pathways [78] suggesting the significant role of this subfamily in the regulation of plant defense responses.
4.4. MYB TF Family
MYB TF family is one of the largest and most functionally diverse families and is conserved among all eukaryotes. They are also diverse in their structure and are classified based on the presence of a conserved MYB domain that contains two or three imperfect repeats (R1, R2, and R3). The structure, classification, and functional diversity of MYB TFs have been well studied [79,80,81,82]. The first plant MYB TF was identified in Zea mays [83]. Since then, MYB TFs in several other plant species, including Arabidopsis [84], have been reported. Although MYB TFs are often implied to be a major player in flavonoid biosynthesis or abiotic stress [85,86,87,88,89], the first MYB gene identified was the oncogene v-myb (initially called mab or amv after the name of avian myeloblastosis virus but later renamed v-myb) from the avian myeloblastosis virus [90,91,92]. Hence, their role in disease resistance cannot be ignored.
Hypersensitive response (HR), a form of programmed cell death (PCD), is one of the most effective defense strategies of the host plant in response to pathogen infection. MYB TFs are reported to positively regulate the HR response. Daniel, et al. [93] showed that, in response to avirulent pathogens such as Xanthomonas campestris pv campestris, AtMYB30 showed a rapid and transient expression. Functional genomics study using Arabidopsis Isd mutants and their corresponding suppressor phx mutants, Daniel, et al. [94] reported that MYB30 expression is likely more responsible for the initiation of the HR than for its propagation. Furthermore, overexpression of MYB30 in transgenic plants accelerated the HR following avirulent bacterial pathogen infection and caused HR-like responses to virulent bacterial pathogens [95]. Raffaele, et al. [96] reported that AtMYB30 regulated HR using long-chain fatty acids and their derivatives. Using microarray analyses of Arabidopsis plants overexpressing MYB30 (MYB30 ox) or antisense (MYB30 as), they reported that MYB30 putatively targeted genes encoding the four enzymes forming the acyl-coA elongase complex that synthesizes very-long-chain fatty acids [96]. Reports have suggested that AtMYB60 and AtMYB96 act through an ABA-signaling cascade, while AtMYB96-mediated ABA signals induce pathogen resistance responses by inducing salicylic acid (SA) biosynthesis in Arabidopsis [97]. Similarly, AtMYB102/AtM4 and AtMYB41 regulate plant resistance toward the herbivorous insect, Pieris rape [98]. Some MYB TFs regulate both biotic and abiotic stress; for example, AtMYB108, also called the Botrytis Susceptible 1 (BOS1), which is an R2R3 type MYB [99]. MYB TFs are also reported to contribute to systemic acquired resistance (SAR), a type of plant defense in which the signals broadcast from the site of infection to systemic tissues to warn them of the pathogen attack. Segarra, et al. [100] reported that defense pathways triggered by beneficial Pseudomonas and Trichoderma spp. strains are very similar and that MYB72 functions as an early point of convergence in the signaling pathways induced by these two different species of microorganisms.
However, it seems that MYB TFs are less studied for their role in plant defense compared to other TF families. Microarray- and RNA-seq-mediated studies can be used to identify the candidate MYB TFs that induce defense responses.
4.5. NAC TF Family
The NAC TF family is a key plant-specific TF family. NAC TFs are characterized by the NAC domain, which has a 150 amino acid conserved domain at the N-terminus, and a diversified C-terminal transcription regulatory region (TR) [101]. Some NAC TFs also have a transmembrane domain within the TR domain. The NAC domain has been sub-divided into five sub-groups from A to E [102]. Genome-wide identification of TFs suggested the presence of NAC TFs in many plant species [103,104,105,106,107].
Like other TFs, NACs also have the DNA-binding ability and can regulate abiotic and biotic stresses, growth, and development. For example, cold-induced NTL6, a plasma membrane-bound NAC TF that is involved in the proteolytic activation of the plasma membrane in Arabidopsis [108], is reported to bind directly to the promoter of PR genes to induce resistance against pathogens. Similarly, another NAC TF, ATAF1, that is induced by drought, high salinity, ABA, methyl jasmonate, and wounding has multiple functions in Arabidopsis [109]. Reports suggested that overexpression of ATAF1 not only enhances drought tolerance but also increases susceptibility to B. cinerea, suggesting possible crosstalk between the stressors. Similarly, in rice, JASMONIC ACID 2 (JA2) and JA2-like (JA2L), the two homologous NAC TFs are reported to mediate pathogen-induced stomatal regulation [110], which are considered to be SA- and ABA-dependent processes. These studies suggest that NAC TFs act as interlinking entities in signaling cascades in response to multiple stressors.
Some NAC TFs also act as negative regulators in plant defense responses and are targeted by pathogens to increase susceptibility. As an example, HopD1, a type III effector from P. syringae, interacts with NTL9, a membrane-tethered protein at the endoplasmic reticulum, to suppress ETI responses [111]. Similarly, in a study involving potato (Solanum tuberosum), Block, Toruno, Elowsky, Zhang, Steinbrenner, Beynon and Alfano [111] showed that two ER-associated Solanum tuberosum NTPs, StNTP1, and StNTP2, interact with an RxLR effector from P. infestans to prevent the movement of TFs from the ER to the nucleus and, in doing so, suppress defense responses. Similarly, a type III effector from P. syringae, HopD1, interacts with membrane-tethered NTL9 to suppress ETI responses [111]. A similar situation was also found in viral pathogenicity, where the TMV replicase protein interreacted with ATAF2, which is an NAC TF, to suppress the basal host defense [112].
Other reports suggested positive regulation of plant defense by NAC TFs. Studies involving RNAi, knockout (KO), and overexpression of genes suggested the role of NAC TFs in various plant-pathogen interactions. NAC TFs are reported to positively regulate plant defense responses by activating PR-related genes and inducing HR at the infection site [108,113,114,115]. The ATFAF1 and its ortholog in barley, HvNAC6, is reported to positively regulate penetration resistance toward the biotrophic fungus Blumeria graminis [114,115]. Thus, NAC TFs appear to be key elements in connecting signal transduction cues from different stressors and can be used to relay between various stresses in plants.
5. Evolution of Signaling Molecules
The evolution of plants from unicellular organisms to complex multicellular structures demanded the evolution of aerobic metabolisms such as respiration and photosynthesis. These metabolic processes resulted in the generation of reactive oxygen species (ROS) commonly known for causing oxidative damage to proteins, DNA, and other macromolecules such as lipids [116]. However, recent studies have indicated that ROS can act as signaling molecules for regulating various physiological responses such as growth and development [117], abiotic stress responses [118], plant responses to pathogens [119], and stomatal regulation [120]. ROS are produced by the activation or reduction of oxygen and includes the singlet oxygen (1O2), hydrogen peroxide (H2O2), superoxide radical (O2−), and hydroxyl radical (HO·) [121]. Other plant-like organisms constantly produce ROS in organelles like chloroplast, mitochondria, and peroxisomes, as they are the sites for aerobic metabolism. The generation of different ROS in plants is triggered by various environmental and biotic stressors such as drought, salinity, extreme temperature, nutrient deficiency, and pathogen attack [121].
Production of reactive oxygen intermediates (ROIs), primarily O2− and H2O2, collectively termed as the oxidative burst at the site of attempted invasion, is one of the most rapid responses of the plant following pathogen perception [122]. In plants, the oxidative burst was first reported by Doke [123], who noticed the generation of O2− following inoculation with an avirulent strain of the fungal pathogen, Phytophthora infestans. However, a virulent strain of the same pathogen was unable to induce O2− generation. Since then, O2− production has been identified in various plant-pathogen interactions involving avirulent bacteria, fungi, and viruses [124]. The most important aspect of these redox molecules is the high reactivity caused by their short half-life. For example, the half-life of O2− is less than a second and is quickly dismutated either enzymatically by superoxide dismutase [125] to H2O2 (a relatively stable molecule) or non-enzymatically [124]. Similarly, protonation of O2− can result in the production of hydroperoxyl radicals (HO2−) that can convert fatty acids to toxic lipid peroxides, resulting in membrane injury. Furthermore, H2O2 can undergo Fenton reactions in the presence of divalent metal ions such as Fe2+, thereby producing the hydroxyl radical (OH•), which is the most reactive ROI that can induce lipid peroxidation and damage to nucleic acids and proteins [124].
Plants have a sophisticated antioxidant system that involves antioxidants and antioxidant enzymes along with other small molecules to detoxify these ROS or expel them from the cell. Thus, continuous ROS generation and scavenging events are in operation in plants. ROS scavenging is carried out by induction of non-enzymatic antioxidants such as glutathione (GSH), ascorbate, flavonoids, and alkaloids—primarily by ascorbate and GSH [116]. Reverse genetics studies have shown that mutants with perturbed levels of ascorbic acid or GSH are hypersensitive to stress conditions [124]. Thus, a homeostatic status is important for normal metabolism in the plant. An imbalance will lead to oxidative damage.
6. The Era of Nitric Oxide (NO)
Initially reported in the 1980s as an endothelium-relaxing factor (EDRF) in the animal system, NO, quickly gained the attention of scientists due to its tremendous signaling and regulatory roles. The identification of NO as a potent endogenous vasodilator by Schmidt and Walter [126] was a point of excitement and interest for biologists. Subsequent investigations revealed that NO is a multifunctional effector that regulates various physiological processes in mammals, including the relaxation of smooth muscles, neural communication, immune regulation, and inhibition of platelet aggregation [126]. Further insights into the functions of NO came after NO synthase (NOS), the enzyme responsible for NO production was identified [127]. Moreover, studies on its chemical properties and chemistry have contributed to understanding the mechanism of NO signaling.
The use of NO is not restricted to animals: in the last couple of decades, extensive research has established the regulatory and signaling role of NO in plants as well. Initially identified in potato tuber tissue to induce phytoalexin accumulation, NO has been known as the main player orchestrating various cellular processes, including regulation of stomatal closure [128]; inhibition of the activity of certain enzymes [129]; reduction of seed dormancy [130]; repression of floral transition [131]; activation of MAPK signaling cascades [132]; stimulation of seed germination [133]; plant growth and pollen tube re-orientation [134]; modulation of the cell cycle [135]); photorespiration and photosynthesis [136]; regulation of plant responses to drought, salinity, and heavy metal stress (reviewed by [137]); and regulation of phytohormonal signaling in plants. For example, NO regulates gene expression involved in the JA signaling pathway [138]. Similarly, ethylene and auxin interact with NO to regulate root growth and development [139,140]. In addition, NO’s role in the SA pathway has been reviewed in detail [141]. The most important regulatory role of NO is during plant defense [142], which we will discuss in detail later. However, the list of NO functions is ever-growing with the understanding of its chemistry and signaling behavior.
7. NO Biochemical Properties, Synthesis, and Signaling
NO, one of the smallest diatomic molecules is a gaseous free radical with a comparatively short half-life. Combined with its neutral charge, NO promotes rapid membrane diffusion and has several features that make it perfectly suited for cellular signaling [143,144]. NO has an unpaired electron that supports its high reactivity with oxygen (O2), transition metals, thiols, and superoxide (O2−). NO reacts with oxygen to produce various nitrogen oxide molecules with different profiles [145]). The removal of the unpaired electron in NO produces the nitrosonium cation (NO+), while the addition of an electron forms the nitroxyl anion (NO–). These different forms of NO have different chemical reactivities [146]). NO in the form of peroxynitrite (ONOO−) a particularly destructive molecule within biological systems, reacts with ROIs in the presence of O2 [147].
The production of NO in animal systems is well understood. The major route for NO production in animal systems is the conversion of L-arginine to citrulline in the presence of NADPH and O2 by three isoforms of the nitric oxide synthase (NOS) enzyme (reviewed by Alderton et al. [148]. It has been known for a long time that plants release NO [149,150]. Reports suggested that NO release correlates with the tissue nitrite level; therefore, it was thought that NO is generated from the reaction between nitrites and plant metabolites [150]. Subsequently, researchers have shown that the release of NO is attributed to in vivo nitrate reductase (NR) activity [151]. Several experiments exploring the idea of NO generation concluded that NR reduces nitrite to NO [152,153,154]. Further research on the chemistry of NR has shown that, in maize, the Km for nitrite is 100 µM and nitrate is a competitive inhibitor with a Ki of 50 µM [154], suggesting that, under normal conditions where nitrate levels are high and nitrite levels are low, NO production from NR would be low. However, under anaerobic conditions when the nitrite levels are increased, NO production can be increased 100-fold [154]. As most of the focus related to NO production at that time was on NR, it was considered the only enzyme involved in NO production and signaling [152,155,156]. However, there were also arguments against it [157,158].
L-Arginine analogs like N-nitro-L-arginine methyl ester (L-NAME) are inhibitors used to block animal NOS. Similar approaches have shown that deploying L-NAME in plants significantly reduces the production of NO, suggesting the presence of a similar enzyme in plant systems [159]. This hints towards the presence of an arginine-dependent NO production mechanism in plants analogous to the one present in animals [160,161,162,163,164]. This school of thought was supported by immunological experiments that suggested that anti-mammalian NOS antibodies cross-reacted with plant proteins; however, proteomic analyses revealed that these proteins are not related to NOS but are heat shock proteins and glycolytic enzymes [163,165]. Although standard animal-like NOS enzymes have been found in lower pants such as the alga Ostreocuccus tauri [166]), despite the completion of several plant genomes and decades of research, a canonical plant NOS could not be identified in higher plants. A gene in Arabidopsis (At3g47450) was reported to encode AtNOS1 and had 16% similarity with a snail NOS [167]. A functional genomics study of this gene using a T-DNA insertion mutant showed that this protein has a key role in NO synthesis in Arabidopsis [167]. However, subsequent investigations showed that AtNOS1 was not directly involved in NO synthesis; rather, it was shown to be a GTPase and was renamed as AtNOA1 for “NO-associated 1” [168]. The mystery remains and is a point of interest for many plant biologists.
8. The Role of NO in Plant Defense
It is now a widely accepted fact that the most effective weapon of the plant against pathogen attack is the intentional execution of infected cells, termed the HR [5,169,170]. This phenomenon is thought to restrict biotrophic pathogens’ invasion into other parts of the host. However, despite the potential importance of this defense tactic, the underlying mechanism is largely unknown. Emerging evidence suggests that one of the immediate responses of plants after pathogen perception is the generation of NO bursts [142,171]. In plants, this phenomenon was first recorded in soybean during resistance (R) gene-mediated defense against Pseudomonas syringe pv. glycinea expressing the avrA avirulence gene in a soybean suspension culture [142]. Kinetic studies suggested that, during plant-pathogen interaction, maximal NO accumulation occurred 4 to 6 h after R gene recognition [142,171]. Furthermore, the use of animal NOS inhibitors finally revoked pathogen-triggered NO production [142,171]. Several reports suggested that, in plants, NO plays a major role in the development of hypersensitive cell death and plant disease resistance. It is suggested that HR-mediated cell death is dependent upon the balanced production of NO and ROS [172].
To elaborate on the HR, plant pathologists have compared it to the mechanistic commonalities with the well-explored process of PCD, termed apoptosis. It would dilute the subject matter to extensively discuss apoptosis here; however, several good reviews have discussed it in detail [173,174,175]. Briefly, the key to apoptosis is the activation of cysteine-dependent aspartate-specific proteases (or caspases) that have a wide range of cellular targets [176]. Mur, et al. [177] have explained the relationship between NO and HR. NO signaling is sometimes mediated by ROS; for example, NO in the presence of oxidative damage may associate with the formation of potent peroxynitrite (ONOO−). Thus, NO can influence apoptosis in several ways, including through ONOO−. Reports suggested that high ONOO− levels can cause severe damage to nucleic acids [178] and that NO has the potential to bind reversibly with the heme group in cytochrome oxidase to restrict electron transport [179], resulting in increased O2− and ONOO− production, which culminates in cellular damage [180].
In plants, R/avr interactions leading to the HR share several commonalities with animal apoptosis [181]. HR-mediated cell death and the associated calcium influxes result in permeability transition pores and the release of cytochrome c in the mitochondria [182]). Like in animals, balanced production of NO and ROIs is important for the induction of cell death in plants [172]. However, unlike in animals where ONOO− has a key role in apoptosis, reports suggested that plants are relatively resistant to this molecule [172] and that H2O2 plays a key role in developing cell death during HR. In plants, NO interacts with H2O2 rather than O2− due to the acceleration of O2− dismutation to H2O2 by superoxide dismutase (SOD) [172]. This was confirmed by Zago et al. [183] who performed experiments using transgenic tobacco lines with reduced catalase activity. They showed that, after infiltration of NO under moderate light intensities, these transgenics accumulated H2O2, and showed significantly increased cell death compared to wild type lines. In another study, the transgenic lines containing a bacterial NO dioxygenase transgene that converts NO to NO3 accumulated significantly less H2O2, suggesting that NO is required for H2O2 accumulation during HR [184]. However, the molecular mechanisms underlying the interaction between NO and H2O2 remain unknown.
Emerging evidence suggests that NO not only helps in developing HR but also in the establishment of disease resistance. The first direct link in this context was provided by Delledonne et al. [142], who reported that infiltration of the NOS inhibitors L-NNA and PBITU increased growth of the avirulent bacterial pathogens P. syringae pv. tomato (Pst) DC3000 expressing the avrRPm1 avirulent effector, suggesting the role of NO in R-gene-mediated disease resistance against pathogenic bacteria.
The controlled use of NO donors, in cell suspension cultures of tobacco plants, induces the expression of defense-related genes, encoding pathogenesis-related protein 1 (PR1), phenylalanine ammonia-lyase marker for phenylpropanoid biosynthesis, and SA mediated signaling. Both genes play a valuable role in the growth and development of plants’ disease resistance [142,171]. R proteins in plants, produced on pathogen recognition, trigger the inducible defense response [7]. In the absence of R gene recognition, plants depend on their basal resistance responses.
It has been proposed that NO functions in basal disease resistance that is triggered by the recognition of lipopolysaccharides (LPS) [185], which exhibit a pathogen-associated molecular pattern (PAMP) [186]. Loss of AtNOA function diminished NO accumulation in response to LPS, reduced defense-related transcript accumulation, and, most significantly, compromised basal disease resistance against Pst DC3000 [187]. Collectively, these data argue that NO has an important signaling function in basal disease resistance—at least against bacterial pathogens.
9. NO and TFs
Due to its high reactivity and unique chemistry, NO and its derived redox-active species are excellent biological messengers in plants and animals. Despite the importance of NO in various cellular processes, its mode of action remains poorly understood. Scientists have tried to explain it using NO-mediated redox modifications that have the potential to regulate protein function. One such mechanism is the post-translational modification, in which an NO moiety is covalently attached to exposed cysteine thiols, making S-nitrosothiols (SNOs) [188]. Reports suggested that, contrary to other signaling cascades, NO functions by transferring its bioactivity through S-nitrosation (previously called S-nitrosylation). After cGMP signaling, S-nitrosation is the most important feature of NO [189] and plays a key role in cellular processes that modulate enzyme activity, protein localization, and protein-protein interactions [190]. Several proteins regulating key physiological processes have been reported to be S-nitrosated by NO, including NPR1 [190], AtSABP3 [191], NADPH oxidase [192], and the auxin receptor TIR1 [193]. Besides, Lindermayr, et al. [194] identified more than 100 other proteins as potential candidates for S-nitrosation in Arabidopsis. Similarly, a site-specific proteomic study of atgsnor1–3 having perturbations in Arabidopsis S-nitrosoglutathione reductase (AtGSNOR), thus having higher SNO levels [170], showed 926 proteins and 1195 peptides that were S-nitrosated [195].
However, the question remains: how does NO regulate expression? The eukaryotic gene expression is modulated by Pol II that require GTFs to bind to the promoter of a gene to enhance or repress its expression. Thus, Pol II recruitment is one of the key events of transcription processes. The TFs may also interact with other proteins and bind to the promoter as a protein complex [189]. However, the DNA-binding affinity of TFs can be altered by redox-mediated post-translational modifications (such as S-nitrosation or phosphorylation) that have the potential to bring conformational changes into the protein structure and alter its function. For instance, S-nitrosation affects the structure and DNA-binding activity of AtMYB30 in Arabidopsis [196]. Similarly, OxyR, a thiol-containing transcriptional activator that, upon oxidation, regulates the expression of genes involved in H2O2 detoxification, is modulated by S-nitrosation [197]. Studies by others have supported our argument by showing that S-nitrosation directly modifies several transcription factors, including NF-κB, HIF-1 [198] zinc finger transcription factor SRG1 [199] bZIP TF TGA1 [200]. Studies involving NO-mediated transcriptional changes have shown that a substantial number of genes and TFs are regulated by NO. Changes in cellular redox tone mediated by NO can regulate the expression of important genes and TFs such as HY5, MYB, and Trx [189], suggesting that NO plays a role in the regulation of various cellular processes via mechanistic control of transcriptional machinery. In the model plant A. thaliana, transcriptional changes in response to NO have been studied using cDNA-amplified fragment length polymorphisms [201], microarrays, real-time PCR [202,203], and RNA-seq [204,205]. Transcriptome analyses in response to different NO donors have shown differential expression of numerous genes. In an RNA-seq-based transcriptomic approach using Arabidopsis roots and leaves, Begara-Morales et al. [204] showed a differential expression of 3263 genes and 35 TFs after 3 h of 1 mM GSNO application. It was interesting that, among the 35 differentially expressed TFs, 25 were from roots and only 10 from leaves. Similarly, in response to 0.1 mM and 1 mM sodium nitroprusside (SNP), Parani, et al. [203] showed differential expression of 422 genes in A. thaliana using whole-genome microarray analysis. Recently, using high throughput RNA sequencing, changes in the expression of about 6,436 Arabidopsis genes 6 h after infiltration of 1 mM S-nitrocysteine (CySNO) were reported [205]. These included about 673 TFs representing a broad range of TF families. Gene ontology and MapMan analyses showed that these genes were enriched in pathways like hormone signaling, protein degradation, and biotic and abiotic stresses [206]. A list of top 20 differentially expressed TFs in response to 1 mM CySNO is given in Table 1 which shows various biologically important TFs such as ABR1 that is expressed in response to ABA or osmotic stress, DREB2C involved in drought stress, AtMYB3, that represses phenylpropanoid biosynthesis gene expression, and AtMYB48 involved in cold stress acclimation (Table 1). The differential expression of this huge number of genes by a single molecule could only be explained by the co-operation of a set of TFs that could bind to a common region in the promoter of the regulated genes [189]. To find this, Palmieri et al. [189] searched for a common TFBS in the promoter region of NO-regulated genes based on microarray analyses using Genomatix, Gene2Promoter, and MatInspector. They found that eight families of TFBSs occur at least 15% more often in the promoter region of NO-responsive genes compared to more than 28,000 Arabidopsis genes. Among these, the majority were ocs element-like-sequences and WRKYs. The above-mentioned evidence establishes the mechanistic control of gene transcription by direct pos-translational modification of TFs, thereby affecting their DNA-binding affinity. Table 1. List of top-20 up- and down-regulated transcription factors (TFs) that were differentially regulated in response to 1 mM CySNO. Red and green color represents up- and down-regulated TFs respectively [206].

Table 1.
List of top-20 up- and down-regulated transcription factors that showed differential expression in response to CySNO in RNA-seq based transcriptome.
10. Conclusions and Future Recommendations
Plants, the primary producers of the ecosystem, are under continuous threat from several environmental adversities such as cold, heat, flood, salinity, and drought. Besides, attack from phytopathogens is a serious problem that demands the immediate attention of plant scientists. As part of their adaptation to their sedentary nature, plants have a fine-tuned defense mechanism that responds to environmental constraints and pathogen attack. The NO, initially reported in animal systems as an endothelium-relaxing factor, gained the attention of scientists due to its tremendous signaling and regulatory roles. Research reports in the last couple of decades have unraveled the role of NO in plant defense. After pathogen perception, redox bursts result in the production of NO and ROS inducing a downstream signaling cascade including the induction of HR response and activation of pathogen-induced SA pathway (Figure 1). But how does NO regulate gene expression? One of the mechanisms includes direct S-nitrosation of transcription factor proteins by nitric oxide that results in significant changes in the structure of these proteins, thereby affecting their ability to bind at their specific sites in the promoters of target genes.
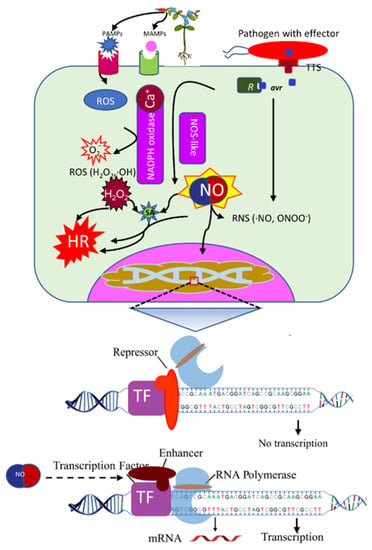
Figure 1.
Production of nitric oxide (NO) in response to virulent and avirulent pathogens. Putative model showing the production of NO in response to pathogens, and its role in the recruitment of RNA polymerase under stress conditions. TFs can enhance or repress the expression of genes. PAMP: pathogen-associated molecular patterns, MAMP: Microbe-associated molecular patterns, TTS: type three secretion system, ROS: reactive oxygen species, RNS: reactive nitrogen species, NOS, nitric oxide synthase, NADPH: Nicotinamide adenine dinucleotide phosphate, HR: hypersensitive cell-death response.
Another possible mechanism may be via modification of the RNA polymerase II by NO. All the eukaryotic genes are transcribed by RNA polymerase II (Pol-II) which is a complex of 12 subunits (Rpb1-Rpb12). However, Pol-II cannot recognize the promoter sequence on its own, rather it requires general TFs (GTFs)—TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH [207,208] and are conserved across the eukaryotic species including plants [209]. These GTFs along with Pol II assemble in a defined order on the promoter of a target gene to make a pre-initiation complex [210]. Histone integrity is crucial for maintaining these complexes (to maintain its binding to the promoter) for proper functioning. Post-translational modification (PTMs) including acetylation, phosphorylation, and methylation can cause histone modification [211] and hence can hinder transcription. NO can also modulate PTMs, the chief among them is S-nitrosation, therefore NO has the potential to mediate RNA polymerase binding and regulate transcription possibly through S-nitrosation of one or more of the Pol II subunits or GTFs. To support our argument, we analyzed the protein sequence of Arabidopsis Rpb9, a core subunit of Pol II through GPS-SNO 1.0 [212] and found that even using a high threshold, there was a strong prediction for possible S-nitrosation of the cysteine residue (Cys 07) (Figure 2A). Further studying the 3D structure of Rpb9, we found that the target Cys07 was also solvent-exposed (Figure 2B) making it a potential target for S-nitrosation. However, detailed in vitro and in vivo investigations are required to confirm this hypothesis. A combined approach, using genomics, transcriptomics, proteomics, and metabolomics may be required to unravel the unexplored roles of NO in gene transcription.

Figure 2.
Prediction of possible S-nitrosation site in Rpb9 subunit of Pol II. (A) The peptide sequence of Rpb9 was analyzed using GPS SNO 1.0 with maximum threshold. Cys07 was predicted as site for S-nitrosation (B) The 3D structure of Rpb9 was retrieved from the uniport database and analyzed using PyMOL 2.4. The Cys07 was highlighted as red.
Author Contributions
Conceptualization, B.-W.Y. and Q.M.I.; investigation, Q.M.I. and N.F.; resources, B.-W.Y.; writing—original draft preparation, N.F. and Q.M.I.; writing—review and editing, A.H.; supervision, B.-W.Y.; funding acquisition, B.-W.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Kyungpook National University Development Project Research Fund, 2018 and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (Grant number 2020R1I1A3073247).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article
Conflicts of Interest
The authors declare no conflict of interest.
References
- Palmgren, M.G.; Edenbrandt, A.K.; Vedel, S.E.; Andersen, M.M.; Landes, X.; Østerberg, J.T.; Falhof, J.; Olsen, L.I.; Christensen, S.B.; Sandøe, P. Are we ready for back-to-nature crop breeding? Trends Plant Sci. 2015, 20, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Oerke, E.C. Crop losses to pests. J. Agric. Sci 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Oerke, E.C.; Dehne, H.W. Safeguarding production—losses in major crops and the role of crop protection. Crop. Prot. 2004, 23, 275–285. [Google Scholar] [CrossRef]
- He, D.C.; Zhan, J.S.; Xie, L.H. Problems, challenges and future of plant disease management: From an ecological point of view. J. Integr. Agric. 2016, 15, 705–715. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Imran, Q.M.; Yun, B.-W. Pathogen-induced Defense Strategies in Plants. J. Crop Sci. Biotechnol. 2020, 23, 97–105. [Google Scholar] [CrossRef]
- Dangl, J.L.; Jones, J.D.G. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef]
- Ausubel, F.M. Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 2005, 6, 973–979. [Google Scholar] [CrossRef]
- Zipfel, C.; Felix, G. Plants and animals: A different taste for microbes? Curr. Opin. Plant Biol. 2005, 8, 353–360. [Google Scholar] [CrossRef]
- Mackey, D.; Holt, B.F., III; Wiig, A.; Dangl, J.L. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 2002, 108, 743–754. [Google Scholar] [CrossRef]
- Coaker, G.; Falick, A.; Staskawicz, B. Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 2005, 308, 548–550. [Google Scholar] [CrossRef]
- Mackey, D.; Belkhadir, Y.; Alonso, J.M.; Ecker, J.R.; Dangl, J.L. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 2003, 112, 379–389. [Google Scholar] [CrossRef]
- Day, B.; Dahlbeck, D.; Staskawicz, B.J. NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis. Plant Cell 2006, 18, 2782–2791. [Google Scholar] [CrossRef] [PubMed]
- Day, B.; Dahlbeck, D.; Huang, J.; Chisholm, S.T.; Li, D.H.; Staskawicz, B.J. Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell 2005, 17, 1292–1305. [Google Scholar] [CrossRef] [PubMed]
- van der Hoorn, R.A.L.; Kamoun, S. From Guard to Decoy: A new model for perception of plant pathogen effectors. Plant Cell 2008, 20, 2009–2017. [Google Scholar] [CrossRef] [PubMed]
- Peter, I.S.; Davidson, E.H. Genomic Control Process: Development and Evolution; Academic Press: London, UK; San Diego, CA, USA, 2015; p. xii. 448p. [Google Scholar]
- Hong, R.L.; Hamaguchi, L.; Busch, M.A.; Weigel, D. Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 2003, 15, 1296–1309. [Google Scholar] [CrossRef]
- Phillips, T. Regulation of transcription and gene expression in eukaryotes. Nat. Educ. 2008, 1, 199. [Google Scholar]
- Pulverer, B. Sequence-specific DNA-binding transcription factors. Nat. Cell Biol. 2005, 13, 1–2. [Google Scholar]
- Reddy, P.M.S.; Stamatoyannopoulos, G.; Papayannopoulou, T.; Shen, C.K.J. Genomic Footprinting and Sequencing of Human Beta-Globin Locus—Tissue-Specificity and Cell-Line Artifact. J. Biol. Chem. 1994, 269, 8287–8295. [Google Scholar] [CrossRef]
- Jalali, B.L.; Bhargava, S.; Kamble, A. Signal transduction and transcriptional regulation of plant defence responses. J. Phytopathol. 2006, 154, 65–74. [Google Scholar] [CrossRef]
- Van Verk, M.C.; Gatz, C.; Linthorst, H.J.M. Transcriptional Regulation of Plant Defense Responses. Adv. Bot. Res. 2009, 51, 397–438. [Google Scholar] [CrossRef]
- Buscaill, P.; Rivas, S. Transcriptional control of plant defence responses. Curr. Opin. Plant Biol. 2014, 20, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Windram, O.; Madhou, P.; McHattie, S.; Hill, C.; Hickman, R.; Cooke, E.; Jenkins, D.J.; Penfold, C.A.; Baxter, L.; Breeze, E.; et al. Arabidopsis Defense against Botrytis cinerea: Chronology and Regulation Deciphered by High-Resolution Temporal Transcriptomic Analysis. Plant Cell 2012, 24, 3530–3557. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Duran, R.; Macho, A.P.; Boutrot, F.; Segonzac, C.; Somssich, I.E.; Zipfel, C. The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. Elife 2013, 2, e00983. [Google Scholar] [CrossRef]
- Fan, M.; Bai, M.Y.; Kim, J.G.; Wang, T.N.; Oh, E.; Chen, L.; Park, C.H.; Son, S.H.; Kim, S.K.; Mudgett, M.B.; et al. The bHLH Transcription Factor HBI1 Mediates the Trade-Off between Growth and Pathogen-Associated Molecular Pattern-Triggered Immunity in Arabidopsis. Plant Cell 2014, 26, 828–841. [Google Scholar] [CrossRef]
- Malinovsky, F.G.; Batoux, M.; Schwessinger, B.; Youn, J.H.; Stransfeld, L.; Win, J.; Kim, S.K.; Zipfel, C. Antagonistic Regulation of Growth and Immunity by the Arabidopsis Basic Helix-Loop-Helix Transcription Factor HOMOLOG OF BRASSINOSTEROID ENHANCED EXPRESSION2 INTERACTING WITH INCREASED LEAF INCLINATION1 BINDING bHLH1. Plant Physiol. 2014, 164, 1443–1455. [Google Scholar] [CrossRef]
- Seo, E.; Choi, D. Functional studies of transcription factors involved in plant defenses in the genomics era. Brief. Funct. Genom. 2015, 14, 260–267. [Google Scholar] [CrossRef]
- Imran, Q.M.; Lee, S.-U.; Mun, B.-G.; Hussain, A.; Asaf, S.; Lee, I.-J.; Yun, B.-W. WRKYs, the Jack-of-various-Trades, Modulate Dehydration Stress in Populus davidiana—A Transcriptomic Approach. Int. J. Mol. Sci. 2019, 20, 414. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.X.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Cai, M.; Qiu, D.Y.; Yuan, T.; Ding, X.H.; Li, H.J.; Duan, L.; Xu, C.G.; Li, X.H.; Wang, S.P. Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ. 2008, 31, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.X.; Palmqvist, S.; Olsson, H.; Boren, M.; Ahlandsberg, S.; Jansson, C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 2003, 15, 2076–2092. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.P.; Somssich, I.E. The Role of WRKY Transcription Factors in Plant Immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.Z.; Zhang, S.Q. MAPK Cascades in Plant Disease Resistance Signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef]
- Zheng, Z.Y.; Qamar, S.A.; Chen, Z.X.; Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef]
- Mao, G.H.; Meng, X.Z.; Liu, Y.D.; Zheng, Z.Y.; Chen, Z.X.; Zhang, S.Q. Phosphorylation of a WRKY Transcription Factor by Two Pathogen-Responsive MAPKs Drives Phytoalexin Biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1639–1653. [Google Scholar] [CrossRef]
- Li, G.J.; Meng, X.Z.; Wang, R.G.; Mao, G.H.; Han, L.; Liu, Y.D.; Zhang, S.Q. Dual-Level Regulation of ACC Synthase Activity by MPK3/MPK6 Cascade and Its Downstream WRKY Transcription Factor during Ethylene Induction in Arabidopsis. PLoS Genet. 2012, 8, e1002767. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Diezel, C.; Somssich, I.E. Arabidopsis WRKY33 Is a Key Transcriptional Regulator of Hormonal and Metabolic Responses toward Botrytis cinerea Infection. Plant Physiol. 2012, 159, 266–285. [Google Scholar] [CrossRef]
- Ishihama, N.; Yamada, R.; Yoshioka, M.; Katou, S.; Yoshioka, H. Phosphorylation of the Nicotiana benthamiana WRKY8 Transcription Factor by MAPK Functions in the Defense Response. Plant Cell 2011, 23, 1153–1170. [Google Scholar] [CrossRef]
- Shen, Q.H.; Saijo, Y.; Mauch, S.; Biskup, C.; Bieri, S.; Keller, B.; Seki, H.; Ulker, B.; Somssich, I.E.; Schulze-Lefert, P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 2007, 315, 1098–1103. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Hayashi, N.; Matsushita, A.; Liu, X.Q.; Nakayama, A.; Sugano, S.; Jiang, C.J.; Takatsuji, H. Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc. Natl. Acad. Sci. USA 2013, 110, 9577–9582. [Google Scholar] [CrossRef]
- Deslandes, L.; Olivier, J.; Theulieres, F.; Hirsch, J.; Feng, D.X.; Bittner-Eddy, P.; Beynon, J.; Marco, Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA 2002, 99, 2404–2409. [Google Scholar] [CrossRef] [PubMed]
- Narusaka, M.; Shirasu, K.; Noutoshi, Y.; Kubo, Y.; Shiraishi, T.; Iwabuchi, M.; Narusaka, Y. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 2009, 60, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.G.; Zhang, L.P.; Yu, D.Q. Wounding-Induced WRKY8 Is Involved in Basal Defense in Arabidopsis. Mol. Plant Microbe Interact. 2010, 23, 558–565. [Google Scholar] [CrossRef]
- Murre, C.; Mccaw, P.S.; Baltimore, D. A New DNA-Binding and Dimerization Motif in Immunoglobulin Enhancer Binding, Daughterless, Myod, and Myc Proteins. Cell 1989, 56, 777–783. [Google Scholar] [CrossRef]
- Ludwig, S.R.; Habera, L.F.; Dellaporta, S.L.; Wessler, S.R. Lc, a Member of the Maize R-Gene Family Responsible for Tissue-Specific Anthocyanin Production, Encodes a Protein Similar to Transcriptional Activators and Contains the Myc-Homology Region. Proc. Natl. Acad. Sci. USA 1989, 86, 7092–7096. [Google Scholar] [CrossRef]
- Berben, G.; Legrain, M.; Gilliquet, V.; Hilger, F. The Yeast Regulatory Gene Pho4 Encodes a Helix-Loop-Helix Motif. Yeast 1990, 6, 451–454. [Google Scholar] [CrossRef]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef]
- Murre, C.; Bain, G.; Vandijk, M.A.; Engel, I.; Furnari, B.A.; Massari, M.E.; Matthews, J.R.; Quong, M.W.; Rivera, R.R.; Stuiver, M.H. Structure and Function of Helix-Loop-Helix Proteins. BBA Gene Struct. Exp. 1994, 1218, 129–135. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martinez-Garcia, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-Wide Classification and Evolutionary Analysis of the bHLH Family of Transcription Factors in Arabidopsis, Poplar, Rice, Moss, and Algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Calvo, P.; Chini, A.; Fernandez-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH Transcription Factors MYC3 and MYC4 Are Targets of JAZ Repressors and Act Additively with MYC2 in the Activation of Jasmonate Responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef] [PubMed]
- Ferredamare, A.R.; Pognonec, P.; Roeder, R.G.; Burley, S.K. Structure and Function of the B/Hlh/Z Domain of Usf. EMBO J. 1994, 13, 180–189. [Google Scholar] [CrossRef]
- Samira, R.; Li, B.H.; Kliebenstein, D.; Li, C.Y.; Davis, E.; Gillikin, J.W.; Long, T.A. The bHLH transcription factor ILR3 modulates multiple stress responses in Arabidopsis. Plant Mol. Biol. 2018, 97, 297–309. [Google Scholar] [CrossRef]
- Goossens, J.; Mertens, J.; Goossens, A. Role and functioning of bHLH transcription factors in jasmonate signalling. J. Exp. Bot. 2017, 68, 1333–1347. [Google Scholar] [CrossRef]
- Song, S.S.; Qi, T.C.; Fan, M.; Zhang, X.; Gao, H.; Huang, H.; Wu, D.W.; Guo, H.W.; Xie, D.X. The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and Development. PLoS Genet. 2013, 9, e1003653. [Google Scholar] [CrossRef]
- Aparicio, F.; Pallas, V. The coat protein of Alfalfa mosaic virus interacts and interferes with the transcriptional activity of the bHLH transcription factor ILR3 promoting salicylic acid-dependent defence signalling response. Mol. Plant Pathol. 2017, 18, 173–186. [Google Scholar] [CrossRef]
- Woldemariam, M.G.; Dinh, S.T.; Oh, Y.; Gaquerel, E.; Baldwin, I.T.; Galis, I. NaMYC2 transcription factor regulates a subset of plant defense responses in Nicotiana attenuata. BMC Plant Biol. 2013, 13, 73. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Meyerowitz, E.M. The AP2/EREBP family of plant transcription factors. Biol. Chem. 1998, 379, 633–646. [Google Scholar]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.J.; Vogel, M.O.; Viehhauser, A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 2010, 245, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. BBA Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Conde, J.V.; Berckhan, S.; Prasad, G.; Mendiondo, G.M.; Holdsworth, M.J. Group VII Ethylene Response Factors Coordinate Oxygen and Nitric Oxide Signal Transduction and Stress Responses in Plants. Plant Physiol. 2015, 169, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.X.; Liu, D.; Pan, Y.; Gong, W.; Ma, L.G.; Luo, J.C.; Deng, X.W.; Zhu, Y.X. An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol. Biol. 2005, 59, 853–868. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Wu, M.; Li, L.H.; Li, C.; Han, Z.P.; Yuan, J.Y.; Chen, C.B.; Song, W.Q.; Wang, C.G. Genome-Wide Identification of AP2/ERF Transcription Factors in Cauliflower and Expression Profiling of the ERF Family under Salt and Drought Stresses. Front. Plant Sci. 2017, 8, 946. [Google Scholar] [CrossRef]
- Xie, Z.L.; Nolan, T.M.; Jiang, H.; Yin, Y.H. AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef]
- Feng, C.Z.; Chen, Y.; Wang, C.; Kong, Y.H.; Wu, W.H.; Chen, Y.F. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 2014, 80, 654–668. [Google Scholar] [CrossRef] [PubMed]
- Bethke, G.; Unthan, T.; Uhrig, J.F.; Poschl, Y.; Gust, A.A.; Scheel, D.; Lee, J. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 2009, 106. [Google Scholar] [CrossRef]
- Meng, X.Z.; Xu, J.; He, Y.X.; Yang, K.Y.; Mordorski, B.; Liu, Y.D.; Zhang, S.Q. Phosphorylation of an ERF Transcription Factor by Arabidopsis MPK3/MPK6 Regulates Plant Defense Gene Induction and Fungal Resistance. Plant Cell 2013, 25, 1126–1142. [Google Scholar] [CrossRef] [PubMed]
- Mase, K.; Ishihama, N.; Mori, H.; Takahashi, H.; Kaminaka, H.; Kodama, M.; Yoshioka, H. Ethylene-Responsive AP2/ERF Transcription Factor MACD1 Participates in Phytotoxin-Triggered Programmed Cell Death. Mol. Plant Microbe Interact. 2013, 26, 868–879. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, J.; Bostock, R.M.; Gilchrist, D.G. Apoptosis: A functional paradigm for programmed plant cell death induced by a host-selective phytotoxin and invoked during development. Plant Cell 1996, 8, 375–391. [Google Scholar] [CrossRef]
- Zhou, J.; Tang, X.; Martin, G.B. The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 1997, 16, 3207–3218. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.Q.; Wildermuth, M.C.; Chakravarthy, S.; Loh, Y.T.; Yang, C.M.; He, X.H.; Han, Y.; Martin, G.B. Tomato transcription factors Pti4, Pti5, and Pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 2002, 14, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Berrocal-Lobo, M.; Molina, A.; Solano, R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 2002, 29, 23–32. [Google Scholar] [CrossRef]
- Gutterson, N.; Reuber, T.L. Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 2004, 7, 465–471. [Google Scholar] [CrossRef]
- Kanei-Ishii, C.; Sarai, A.; Sawazaki, T.; Nakagoshi, H.; He, D.N.; Ogata, K.; Nishimura, Y.; Ishii, S. The tryptophan cluster: A hypothetical structure of the DNA-binding domain of the myb protooncogene product. J. Biol. Chem. 1990, 265, 19990–19995. [Google Scholar] [CrossRef]
- Jia, L.; Clegg, M.T.; Jiang, T. Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol. 2004, 134, 575–585. [Google Scholar] [CrossRef]
- Rosinski, J.A.; Atchley, W.R. Molecular evolution of the Myb family of transcription factors: Evidence for polyphyletic origin. J. Mol. Evol. 1998, 46, 74–83. [Google Scholar] [CrossRef]
- Du, H.; Zhang, L.; Liu, L.; Tang, X.F.; Yang, W.J.; Wu, Y.M.; Huang, Y.B.; Tang, Y.X. Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry 2009, 74, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Paz-Ares, J.; Ghosal, D.; Wienand, U.; Peterson, P.A.; Saedler, H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987, 6, 3553–3558. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Ravaglia, D.; Espley, R.V.; Henry-Kirk, R.A.; Andreotti, C.; Ziosi, V.; Hellens, R.P.; Costa, G.; Allan, A.C. Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 2013, 13, 68. [Google Scholar] [CrossRef]
- Huang, W.J.; Lv, H.Y.; Wang, Y. Functional Characterization of a Novel R2R3-MYB Transcription Factor Modulating the Flavonoid Biosynthetic Pathway from Epimedium sagittatum. Front. Plant Sci. 2017, 8, 1274. [Google Scholar] [CrossRef]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB Transcription Factors as Regulators of Secondary Metabolism in Plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.Q.; Zhao, Y.L.; Wang, Y.C.; Liu, Z.Y.; Gao, C.Q. Comprehensive Analysis of MYB Gene Family and Their Expressions Under Abiotic Stresses and Hormone Treatments in Tamarix hispida. Front. Plant Sci. 2018, 9, 1303. [Google Scholar] [CrossRef]
- Roy, S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 2016, 11. [Google Scholar] [CrossRef]
- Duesberg, P.H.; Bister, K.; Moscovici, C. Genetic structure of avian myeloblastosis virus, released from transformed myeloblasts as a defective virus particle. Proc. Natl. Acad. Sci. USA 1980, 77, 5120–5124. [Google Scholar] [CrossRef]
- Gonda, T.J.; Sheiness, D.K.; Bishop, J.M. Transcripts from the cellular homologs of retroviral oncogenes: Distribution among chicken tissues. Mol. Cell. Biol. 1982, 2, 617–624. [Google Scholar] [CrossRef]
- Souza, L.M.; Strommer, J.N.; Hillyard, R.L.; Komaromy, M.C.; Baluda, M.A. Cellular sequences are present in the presumptive avian myeloblastosis virus genome. Proc. Natl. Acad. Sci. USA 1980, 77, 5177. [Google Scholar] [CrossRef] [PubMed]
- Daniel, P.T.; Scholz, C.; Essmann, F.; Westermann, J.; Pezzutto, A.; Dorken, B. Dendritic cells inhibit CD95/Fas-triggered apoptosis of activated T lymphocytes by a mechanism upstream of caspase-activation. Blood 1999, 94, 688A. [Google Scholar]
- Daniel, X.; Lacomme, C.; Morel, J.B.; Roby, D. A novel myb oncogene homologue in Arabidopsis thaliana related to hypersensitive cell death. Plant J. 1999, 20, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Vailleau, F.; Daniel, X.; Tronchet, M.; Montillet, J.L.; Triantaphylides, C.; Roby, D. A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc. Natl. Acad. Sci. USA 2002, 99, 10179–10184. [Google Scholar] [CrossRef]
- Raffaele, S.; Vailleau, F.; Leger, A.; Joubes, J.; Miersch, O.; Huard, C.; Blee, E.; Mongrand, B.; Domergue, F.; Roby, D. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 2008, 20, 752–767. [Google Scholar] [CrossRef]
- Seo, P.J.; Park, C.M. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 2010, 186, 471–483. [Google Scholar] [CrossRef]
- De Vos, M.; Denekamp, M.; Dicke, M.; Vuylsteke, M.; Van Loon, L.; Smeekens, S.C.; Pieterse, C.M. The Arabidopsis thaliana Transcription Factor AtMYB102 Functions in Defense Against the Insect Herbivore Pieris rapae. Plant Signal. Behav. 2006, 1, 305–311. [Google Scholar] [CrossRef]
- Mengiste, T.; Chen, X.; Salmeron, J.; Dietrich, R. The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 2003, 15, 2551–2565. [Google Scholar] [CrossRef]
- Segarra, G.; Van der Ent, S.; Trillas, I.; Pieterse, C.M.J. MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol. 2009, 11, 90–96. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.H.; Wang, S.; Wu, M.S.; Guo, R.Z.; Xue, Z.H.; Meng, N.; Tao, X.M.; Chen, M.M.; Zhang, Y.F. Molecular Characterization and Expression Analysis of NAC Family Transcription Factors in Tomato. Plant Mol. Biol. Rep. 2014, 32, 501–516. [Google Scholar] [CrossRef]
- Hu, R.B.; Qi, G.A.; Kong, Y.Z.; Kong, D.J.; Gao, Q.A.; Zhou, G.K. Comprehensive Analysis of NAC Domain Transcription Factor Gene Family in Populus trichocarpa. BMC Plant Biol. 2010, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Pinheiro, G.L.; Marques, C.S.; Costa, M.D.B.L.; Reis, P.A.B.; Alves, M.S.; Carvalho, C.M.; Fietto, L.G.; Fontes, E.P.B. Complete inventory of soybean NAC transcription factors: Sequence conservation and expression analysis uncover their distinct roles in stress response. Gene 2009, 444, 10–23. [Google Scholar] [CrossRef]
- Seo, P.J.; Kim, M.J.; Park, J.Y.; Kim, S.Y.; Jeon, J.; Lee, Y.H.; Kim, J.; Park, C.M. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 2010, 61, 661–671. [Google Scholar] [CrossRef]
- Wu, Y.R.; Deng, Z.Y.; Lai, J.B.; Zhang, Y.Y.; Yang, C.P.; Yin, B.J.; Zhao, Q.Z.; Zhang, L.; Li, Y.; Yang, C.W.; et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009, 19, 1279–1290. [Google Scholar] [CrossRef]
- Du, M.M.; Zhai, Q.Z.; Deng, L.; Li, S.Y.; Li, H.S.; Yan, L.H.; Huang, Z.; Wang, B.; Jiang, H.L.; Huang, T.T.; et al. Closely Related NAC Transcription Factors of Tomato Differentially Regulate Stomatal Closure and Reopening during Pathogen Attack. Plant Cell 2014, 26, 3167–3184. [Google Scholar] [CrossRef]
- Block, A.; Toruno, T.Y.; Elowsky, C.G.; Zhang, C.; Steinbrenner, J.; Beynon, J.; Alfano, J.R. The Pseudomonas syringae type III effector HopD1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9. New Phytol. 2014, 201, 1358–1370. [Google Scholar] [CrossRef]
- Wang, X.; Goregaoker, S.P.; Culver, J.N. Interaction of the Tobacco Mosaic Virus Replicase Protein with a NAC Domain Transcription Factor Is Associated with the Suppression of Systemic Host Defenses. J. Virol. 2009, 83, 9720–9730. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, T.; Taga, Y.; Takai, R.; Iwano, M.; Matsui, H.; Takayama, S.; Isogai, A.; Che, F.S. The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO J. 2009, 28, 926–936. [Google Scholar] [CrossRef]
- Jensen, M.K.; Rung, J.H.; Gregersen, P.L.; Gjetting, T.; Fuglsang, A.T.; Hansen, M.; Joehnk, N.; Lyngkjaer, M.F.; Collinge, D.B. The HvNAC6 transcription factor: A positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol. Biol. 2007, 65, 137–150. [Google Scholar] [CrossRef]
- Jensen, M.K.; Hagedorn, P.H.; de Torres-Zabala, M.; Grant, M.R.; Rung, J.H.; Collinge, D.B.; Lyngkjaer, M.F. Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp hordei in Arabidopsis. Plant J. 2008, 56, 867–880. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A.; Forzani, C.; Hirt, H. Reactive oxygen species signaling in plants. Antioxid. Redox Signal. 2006, 8, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.L.; Ullah, F.; Zhou, D.X.; Yi, M.; Zhao, Y. Mechanisms of ROS Regulation of Plant Development and Stress Responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef] [PubMed]
- Jaspers, P.; Kangasjarvi, J. Reactive oxygen species in abiotic stress signaling. Physiol. Plantarum 2010, 138, 405–413. [Google Scholar] [CrossRef]
- Torres, M.A.; Jones, J.D.G.; Dangl, J.L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006, 141, 373–378. [Google Scholar] [CrossRef]
- Qi, J.S.; Song, C.P.; Wang, B.S.; Zhou, J.M.; Kangasjarvi, J.; Zhu, J.K.; Gong, Z.Z. Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant. Biol. 2018, 60, 805–826. [Google Scholar] [CrossRef]
- Karuppanapandian, T.; Moon, J.C.; Kim, C.; Manoharan, K.; Kim, W. Reactive oxygen species in plants: Their generation, signal transduction, and scavenging mechanisms. Aust. J. Crop Sci. 2011, 5, 709–725. [Google Scholar]
- Apostol, I.; Heinstein, P.F.; Low, P.S. Rapid Stimulation of an Oxidative Burst during Elicitation of Cultured Plant Cells: Role in Defense and Signal Transduction. Plant Physiol. 1989, 90, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Doke, N. Involvement of superoxide anion generation in the hypersensitive response of potato tuber tissues to infection with an incompatible race of Phytophthora infestans and to the hyphal wall components. Physiol. Plant. Pathol. 1983, 23, 345–357. [Google Scholar] [CrossRef]
- Grant, J.J.; Loake, G.J. Role of Reactive Oxygen Intermediates and Cognate Redox Signaling in Disease Resistance. Plant Physiol. 2000, 124, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Tunc-Ozdemir, M.; Miller, G.; Song, L.H.; Kim, J.; Sodek, A.; Koussevitzky, S.; Misra, A.N.; Mittler, R.; Shintani, D. Thiamin Confers Enhanced Tolerance to Oxidative Stress in Arabidopsis. Plant Physiol. 2009, 151, 421–432. [Google Scholar] [CrossRef]
- Schmidt, H.H.; Walter, U. NO at work. Cell 1994, 78, 919–925. [Google Scholar] [CrossRef]
- Nathan, C.; Xie, Q.-W. Nitric oxide synthases: Roles, tolls, and controls. Cell 1994, 78, 915–918. [Google Scholar] [CrossRef]
- Neill, S.J.; Desikan, R.; Clarke, A.; Hancock, J.T. Nitric Oxide Is a Novel Component of Abscisic Acid Signaling in Stomatal Guard Cells. Plant Physiol. 2002, 128, 13–16. [Google Scholar] [CrossRef]
- Clarke, A.; Desikan, R.; Hurst, R.D.; Hancock, J.T.; Neill, S.J. NO way back: Nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J. 2000, 24, 667–677. [Google Scholar] [CrossRef]
- Bethke, P.C.; Libourel, I.G.L.; Jones, R.L. Nitric oxide reduces seed dormancy in Arabidopsis. J. Exp. Bot. 2006, 57, 517–526. [Google Scholar] [CrossRef]
- He, Y.; Tang, R.-H.; Hao, Y.; Stevens, R.D.; Cook, C.W.; Ahn, S.M.; Jing, L.; Yang, Z.; Chen, L.; Guo, F.; et al. Nitric Oxide Represses the Arabidopsis Floral Transition. Science 2004, 305, 1968–1971. [Google Scholar] [CrossRef]
- Kumar, D.; Klessig, D.F. Differential induction of tobacco MAP kinases by the defense signals nitric oxide, salicylic acid, ethylene, and jasmonic acid. Mol. Plant. Microbe Interact. 2000, 13, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Beligni, M.V.; Lamattina, L. Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 2000, 210, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Prado, A.M.; Porterfield, D.M.; Feijo, J.A. Nitric oxide is involved in growth regulation and re-orientation of pollen tubes. Development 2004, 131, 2707–2714. [Google Scholar] [CrossRef] [PubMed]
- Correa-Aragunde, N.; Graziano, M.; Chevalier, C.; Lamattina, L. Nitric oxide modulates the expression of cell cycle regulatory genes during lateral root formation in tomato. J. Exp. Bot. 2006, 57, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Farnese, F.S.; Oliveira, J.A.; Paiva, E.A.S.; Menezes-Silva, P.E.; da Silva, A.A.; Campos, F.V.; Ribeiro, C. The Involvement of Nitric Oxide in Integration of Plant Physiological and Ultrastructural Adjustments in Response to Arsenic. Front. Plant Sci. 2017, 8, 516, Erratum in 2017, 8, 979. [Google Scholar] [CrossRef]
- Nabi, R.B.S.; Tayade, R.; Hussain, A.; Kulkarni, K.P.; Imran, Q.M.; Mun, B.-G.; Yun, B.-W. Nitric oxide regulates plant responses to drought, salinity, and heavy metal stress. Environ. Exp. Bot. 2019, 161, 120–133. [Google Scholar] [CrossRef]
- Orozco-Cárdenas, M.L.; Ryan, C.A. Nitric Oxide Negatively Modulates Wound Signaling in Tomato Plants. Plant Physiol. 2002, 130, 487–493. [Google Scholar] [CrossRef]
- Liu, M.; Liu, X.X.; He, X.L.; Liu, L.J.; Wu, H.; Tang, C.X.; Zhang, Y.S.; Jin, C.W. Ethylene and nitric oxide interact to regulate the magnesium deficiency-induced root hair development in Arabidopsis. New Phytol. 2017, 213, 1242–1256. [Google Scholar] [CrossRef]
- Sun, H.W.; Feng, F.; Liu, J.; Zhao, Q.Z. The Interaction between Auxin and Nitric Oxide Regulates Root Growth in Response to Iron Deficiency in Rice. Front. Plant Sci. 2017, 8, 2169. [Google Scholar] [CrossRef]
- Klessig, D.F.; Durner, J.; Noad, R.; Navarre, D.A.; Wendehenne, D.; Kumar, D.; Zhou, J.M.; Shah, J.; Zhang, S.Q.; Kachroo, P.; et al. Nitric oxide and salicylic acid signaling in plant defense. Proc. Natl. Acad. Sci. USA 2000, 97, 8849–8855. [Google Scholar] [CrossRef]
- Delledonne, M.; Xia, Y.; Dixon, R.A.; Lamb, C. Nitric oxide functions as a signal in plant disease resistance. Nature 1998, 394, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N.; Chandra, M.; Tuteja, R.; Misra, M.K. Nitric Oxide as a Unique Bioactive Signaling Messenger in Physiology and Pathophysiology. J. Biomed. Biotechnol. 2004, 2004, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Goretski, J.; Hollocher, T.C. Trapping of Nitric-Oxide Produced during Denitrification by Extracellular Hemoglobin. J. Biol. Chem. 1988, 263, 2316–2323. [Google Scholar] [CrossRef]
- Henry, Y.A.; Guissani, A.; Ducastel, B. Nitric Oxide Research from Chemistry to Biology: EPR Spectroscopy of Nitrosylated Compounds; Springer Science & Business Media: Georgetown, TX, USA, 2012. [Google Scholar]
- Stamler, J.; Singel, D.; Loscalzo, J. Biochemistry of nitric oxide and its redox-activated forms. Science 1992, 258, 1898–1902. [Google Scholar] [CrossRef] [PubMed]
- Bonfoco, E.; Krainc, D.; Ankarcrona, M.; Nicotera, P.; Lipton, S.A. Apoptosis and Necrosis—2 Distinct Events Induced, Respectively, by Mild and Intense Insults with N-Methyl-D-Aspartate or Nitric-Oxide Superoxide in Cortical Cell-Cultures. Proc. Natl. Acad. Sci. USA 1995, 92, 7162–7166. [Google Scholar] [CrossRef]
- Alderton, W.K.; Cooper, C.E.; Knowles, R.G. Nitric oxide synthases: Structure, function and inhibition. Biochem. J. 2001, 357, 593–615. [Google Scholar] [CrossRef]
- Nishimura, H.; Hayamizu, T.; Yanagisawa, Y. Reduction of nitrogen oxide (NO2) to nitrogen oxide (NO) by rush and other plants. Environ. Sci. Technol. 1986, 20, 413–416. [Google Scholar] [CrossRef]
- Klepper, L. Nitric oxide (NO) and nitrogen dioxide (NO2) emissions from herbicide-treated soybean plants. Atmos. Environ. (1967) 1979, 13, 537–542. [Google Scholar] [CrossRef]
- Harper, J.E. Evolution of nitrogen oxide (s) during in vivo nitrate reductase assay of soybean leaves. Plant Physiol. 1981, 68, 1488–1493. [Google Scholar] [CrossRef]
- Yamasaki, H.; Sakihama, Y.; Takahashi, S. An alternative pathway for nitric oxide production in plants: New features of an old enzyme. Trends Plant Sci. 1999, 4, 128–129. [Google Scholar] [CrossRef]
- Yamasaki, H.; Sakihama, Y. Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: In vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett. 2000, 468, 89–92. [Google Scholar] [CrossRef]
- Rockel, P.; Strube, F.; Rockel, A.; Wildt, J.; Kaiser, W.M. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 2002, 53, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Desikan, R.; Cheung, M.K.; Bright, J.; Henson, D.; Hancock, J.T.; Neill, S.J. ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J. Exp. Bot. 2004, 55, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mata, C.; Lamattina, L. Abscisic acid, nitric oxide and stomatal closure—is nitrate reductase one of the missing links? Trends Plant Sci. 2003, 8, 20–26. [Google Scholar] [CrossRef]
- Garces, H.; Durzan, D.; Pedroso, M.C. Mechanical stress elicits nitric oxide formation and DNA fragmentation in Arabidopsis thaliana. Ann. Bot. 2001, 87, 567–574. [Google Scholar] [CrossRef]
- Zhang, C.; Czymmek, K.J.; Shapiro, A.D. Nitric oxide does not trigger early programmed cell death events but may contribute to cell-to-cell signaling governing progression of the Arabidopsis hypersensitive response. Mol. Plant Microbe Interact. 2003, 16, 962–972. [Google Scholar] [CrossRef][Green Version]
- Crawford, N.M. Mechanisms for nitric oxide synthesis in plants. J. Exp. Bot. 2006, 57, 471–478. [Google Scholar] [CrossRef]
- Cueto, M.; Hernandez-Perera, O.; Martin, R.; Bentura, M.L.; Rodrigo, J.; Lamas, S.; Golvano, M.P. Presence of nitric oxide synthase activity in roots and nodules of Lupinus albus. FEBS Lett. 1996, 398, 159–164. [Google Scholar] [CrossRef]
- Ninnemann, H.; Maier, J. Indications for the occurrence of nitric oxide synthases in fungi and plants and the involvement in photoconidiation of Neurospora crassa. Photochem. Photobiol. 1996, 64, 393–398. [Google Scholar] [CrossRef]
- Barroso, J.B.; Corpas, F.J.; Carreras, A.; Sandalio, L.M.; Valderrama, R.; Palma, J.M.; Lupianez, J.A.; del Rio, L.A. Localization of nitric-oxide synthase in plant peroxisomes. J. Biol. Chem. 1999, 274, 36729–36733. [Google Scholar] [CrossRef]
- Ribeiro, E.A.; Cunha, F.Q.; Tamashiro, W.M.S.C.; Martins, I.S. Growth phase-dependent subcellular localization of nitric oxide synthase in maize cells. FEBS Lett. 1999, 445, 283–286. [Google Scholar] [CrossRef]
- Wendehenne, D.; Pugin, A.; Klessig, D.F.; Durner, J. Nitric oxide: Comparative synthesis and signaling in animal and plant cells. Trends Plant Sci. 2001, 6, 177–183. [Google Scholar] [CrossRef]
- Butt, Y.K.C.; Lum, J.H.K.; Lo, S.C.L. Proteomic identification of plant proteins probed by mammalian nitric oxide synthase antibodies. Planta 2003, 216, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Foresi, N.; Correa-Aragunde, N.; Parisi, G.; Caló, G.; Salerno, G.; Lamattina, L. Characterization of a Nitric Oxide Synthase from the Plant Kingdom: NO Generation from the Green Alga Ostreococcus tauri Is Light Irradiance and Growth Phase Dependent. Plant Cell 2010, 22, 3816–3830. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.Q.; Okamoto, M.; Crawford, N.M. Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 2003, 302, 100–103. [Google Scholar] [CrossRef]
- Moreau, M.; Lindermayr, C.; Durner, J.; Klessig, D.F. NO synthesis and signaling in plants–where do we stand? Physiol. Plant. 2010, 138, 372–383. [Google Scholar] [CrossRef]
- Greenberg, J.T.; Yao, N. The role and regulation of programmed cell death in plant–pathogen interactions. Cell. Microbiol. 2004, 6, 201–211. [Google Scholar] [CrossRef]
- Feechan, A.; Kwon, E.; Yun, B.-W.; Wang, Y.; Pallas, J.A.; Loake, G.J. A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. USA 2005, 102, 8054–8059. [Google Scholar] [CrossRef]
- Durner, J.; Wendehenne, D.; Klessig, D.F. Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 1998, 95, 10328–10333. [Google Scholar] [CrossRef]
- Delledonne, M.; Zeier, J.; Marocco, A.; Lamb, C. Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. USA 2001, 98, 13454–13459. [Google Scholar] [CrossRef]
- Strasser, A.; O’Connor, L.; Dixit, V.M. Apoptosis signaling. Annu. Rev. Biochem. 2000, 69, 217–245. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.J.; Wang, X.D. Cytochrome C-mediated apoptosis. Annu. Rev. Biochem. 2004, 73, 87–106. [Google Scholar] [CrossRef] [PubMed]
- Boatright, K.M.; Salvesen, G.S. Mechanisms of caspase activation. Curr. Opin. Cell Biol. 2003, 15, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Creagh, E.M.; Conroy, H.; Martin, S.J. Caspase-activation pathways in apoptosis and immunity. Immunol. Rev. 2003, 193, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.A.J.; Carver, T.L.W.; Prats, E. NO way to live; the various roles of nitric oxide in plant–pathogen interactions. J. Exp. Bot. 2006, 57, 489–505. [Google Scholar] [CrossRef] [PubMed]
- Messmer, U.K.; Ankarcrona, M.; Nicotera, P.; Brune, B. P53 Expression in Nitric Oxide-Induced Apoptosis. FEBS Lett. 1994, 355, 23–26. [Google Scholar] [CrossRef]
- Brown, G.C.; Cooper, C.E. Nanomolar Concentrations of Nitric-Oxide Reversibly Inhibit Synaptosomal Respiration by Competing with Oxygen at Cytochrome-Oxidase. FEBS Lett. 1994, 356, 295–298. [Google Scholar] [CrossRef]
- Packer, M.A.; Porteous, C.M.; Murphy, M.P. Superoxide production by mitochondria in the presence of nitric oxide forms peroxynitrite. Biochem. Mol. Biol. Int. 1996, 40, 527–534. [Google Scholar] [CrossRef]
- Lam, E.; Kato, N.; Lawton, M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature 2001, 411, 848–853. [Google Scholar] [CrossRef]
- Tiwari, B.S.; Belenghi, B.; Levine, A. Oxidative stress increased respiration and generation of reactive oxygen species, resulting in ATP depletion, opening of mitochondrial permeability transition, and programmed cell death. Plant Physiol. 2002, 128, 1271–1281. [Google Scholar] [CrossRef]
- Zago, E.; Morsa, S.; Dat, J.F.; Alard, P.; Ferrarini, A.; Inze, D.; Delledonne, M.; Van Breusegem, F. Nitric oxide- and hydrogen peroxide-responsive gene regulation during cell death induction in tobacco. Plant Physiol. 2006, 141, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Zeier, J.; Pink, B.; Mueller, M.J.; Berger, S.J.P. Light conditions influence specific defence responses in incompatible plant–pathogen interactions: Uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 2004, 219, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Helps, S.C.; Sims, N.R. Inhibition of nitric oxide synthase with 7-nitroindazole does not modify early metabolic recovery following focal cerebral ischemia in rats. Neurochem. Res. 2007, 32, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef]
- Zeidler, D.; Zahringer, U.; Gerber, I.; Dubery, I.; Hartung, T.; Bors, W.; Hutzler, P.; Durner, J. Innate immunity in Arabidopsis thaliana: Lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc. Natl. Acad. Sci. USA 2004, 101, 15811–15816. [Google Scholar] [CrossRef]
- Stamler, J.S.; Lamas, S.; Fang, F.C. Nitrosylation: The prototypic redox-based signaling mechanism. Cell 2001, 106, 675–683. [Google Scholar] [CrossRef]
- Palmieri, M.C.; Sell, S.; Huang, X.; Scherf, M.; Werner, T.; Durner, J.; Lindermayr, C. Nitric oxide-responsive genes and promoters in Arabidopsis thaliana: A bioinformatics approach. J. Exp. Bot. 2008, 59, 177–186. [Google Scholar] [CrossRef]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant Immunity Requires Conformational Charges of NPR1 via S-Nitrosylation and Thioredoxins. Science 2008, 321. [Google Scholar] [CrossRef]
- Wang, Y.-Q.; Feechan, A.; Yun, B.-W.; Shafiei, R.; Hofmann, A.; Taylor, P.; Xue, P.; Yang, F.-Q.; Xie, Z.-S.; Pallas, J.A.; et al. S-Nitrosylation of AtSABP3 Antagonizes the Expression of Plant Immunity. J. Biol. Chem. 2009, 284, 2131–2137. [Google Scholar] [CrossRef]
- Yun, B.-W.; Feechan, A.; Yin, M.; Saidi, N.B.B.; Le Bihan, T.; Yu, M.; Moore, J.W.; Kang, J.-G.; Kwon, E.; Spoel, S.H.; et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 2011, 478, 264–268. [Google Scholar] [CrossRef]
- Terrile, M.C.; Paris, R.; Calderon-Villalobos, L.I.A.; Iglesias, M.J.; Lamattina, L.; Estelle, M.; Casalongue, C.A. Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 2012, 70, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Lindermayr, C.; Saalbach, G.; Durner, J. Proteomic Identification of S-Nitrosylated Proteins in Arabidopsis. Plant Physiol. 2005, 137, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.L.; Huang, X.H.; Chen, L.C.; Sun, X.W.; Lu, C.M.; Zhang, L.X.; Wang, Y.C.; Zuo, J.R. Site-Specific Nitrosoproteomic Identification of Endogenously S-Nitrosylated Proteins in Arabidopsis. Plant Physiol. 2015, 167, 1731–1746. [Google Scholar] [CrossRef] [PubMed]
- Tavares, C.P.; Vernal, J.; Delena, R.A.; Lamattina, L.; Cassia, R.; Terenzi, H. S-nitrosylation influences the structure and DNA binding activity of AtMYB30 transcription factor from Arabidopsis thaliana. Biochim. Biophys. Acta Proteins Proteom. 2014, 1844, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Hausladen, A.; Privalle, C.T.; Keng, T.; DeAngelo, J.; Stamler, J.S. Nitrosative stress: Activation of the transcription factor OxyR. Cell 1996, 86, 719–729. [Google Scholar] [CrossRef]
- Sha, Y.G.; Marshall, H.E. S-nitrosylation in the regulation of gene transcription. Bba-Gen. Subj. 2012, 1820, 701–711. [Google Scholar] [CrossRef]
- Cui, B.M.; Pan, Q.N.; Clarke, D.; Villarreal, M.O.; Umbreen, S.; Yuan, B.; Shan, W.X.; Jiang, J.H.; Loake, G.J. S-nitrosylation of the zinc finger protein SRG1 regulates plant immunity. Nat. Commun. 2018, 9, 4226. [Google Scholar] [CrossRef]
- Lindermayr, C.; Sell, S.; Muller, B.; Leister, D.; Durnera, J. Redox Regulation of the NPR1-TGA1 System of Arabidopsis thaliana by Nitric Oxide. Plant Cell 2010, 22, 2894–2907. [Google Scholar] [CrossRef]
- Polverari, A.; Molesini, B.; Pezzotti, M.; Buonaurio, R.; Marte, M.; Delledonne, M. Nitric oxide-mediated transcriptional changes in Arabidopsis thaliana. Mol. Plant. Microbe Interact. 2003, 16, 1094–1105. [Google Scholar] [CrossRef]
- Huang, X.; von Rad, U.; Durner, J. Nitric oxide induces transcriptional activation of the nitric oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta 2002, 215, 914–923. [Google Scholar] [CrossRef]
- Parani, M.; Rudrabhatla, S.; Myers, R.; Weirich, H.; Smith, B.; Leaman, D.W.; Goldman, S.L. Microarray analysis of nitric oxide responsive transcripts in Arabidopsis. Plant. Biotechnol. J. 2004, 2, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Begara-Morales, J.C.; Sánchez-Calvo, B.; Luque, F.; Leyva-Pérez, M.O.; Leterrier, M.; Corpas, F.J.; Barroso, J.B. Differential transcriptomic analysis by RNA-Seq of GSNO-responsive genes between Arabidopsis roots and leaves. Plant Cell Physiol. 2014, 55, 1080–1095. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Mun, B.G.; Imran, Q.M.; Lee, S.U.; Adamu, T.A.; Shahid, M.; Kim, K.M.; Yun, B.W. Nitric oxide mediated transcriptome profiling reveals activation of multiple regulatory pathways in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 975. [Google Scholar] [CrossRef] [PubMed]
- Imran, Q.M.; Hussain, A.; Lee, S.-U.; Mun, B.-G.; Falak, N.; Loake, G.J.; Yun, B.-W. Transcriptome profile of NO-induced Arabidopsis transcription factor genes suggests their putative regulatory role in multiple biological processes. Sci. Rep. 2018, 8, 771. [Google Scholar] [CrossRef]
- Roeder, R.G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 1996, 21, 327–335. [Google Scholar] [CrossRef]
- Orphanides, G.; Lagrange, T.; Reinberg, D. The general transcription factors of RNA polymerase II. Gene Dev. 1996, 10, 2657–2683. [Google Scholar] [CrossRef]
- Hong, J.C. General aspects of plant transcription factor families. In Plant Transcription Factors; Elsevier: Amsterdam, The Netherlands, 2016; pp. 35–56. [Google Scholar]
- Karijolich, J.J.; Hampsey, M. The Mediator complex. Curr. Biol. 2012, 22, R1030–R1031. [Google Scholar] [CrossRef]
- Wu, J.; Grunstein, M. 25 years after the nucleosome model: Chromatin modifications. Trends Biochem. Sci. 2000, 25, 619–623. [Google Scholar] [CrossRef]
- Xue, Y.; Liu, Z.X.; Gao, X.J.; Jin, C.J.; Wen, L.P.; Yao, X.B.; Ren, J.A. GPS-SNO: Computational Prediction of Protein S-Nitrosylation Sites with a Modified GPS Algorithm. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).