Transcriptome Analyses Identify Potential Key microRNAs and Their Target Genes Contributing to Ovarian Reserve
Abstract
1. Introduction
| Gene | Biological Category | Functions | Reference |
|---|---|---|---|
| AMH(19q13.3) | Anti-Müllerian hormone | Control of the formation of primary follicles by inhibiting excessive follicular recruitment by FSH | [21] |
| AMHR2(12q13) | AMH receptor | AMH signal transduction | [22] |
| BMPR2 | BMP receptor | Signal transduction between oocytes and somatic cells | [23] |
| BMP15(Xq11.2) | Growth factor | Growth and differentiation of granulosa cells (GCs) | [24] |
| ESR1 | Estrogen receptor | Regulation of follicle growth and maturation and oocyte release | [25] |
| FIGLA(2q13.3) | bHLH transcription factor | Regulation of multiple oocyte-specific genes, including genes involved in folliculogenesis and those that encode the zona pellucida | [26] |
| FMR1(Xq27) | Highly polymorphic CGG repeat in the 5′ untranslated region (UTR) of the exon 1 | Transcriptional regulation | [27] |
| FSHR(2q21-p16) | Receptor | Follicular development and ovarian steroidogenesis | [28] |
| FOXL2(3q23) | Transcription factor | Differentiation and growth of granulosa cells | [29] |
| FOXO3A | Transcription factor | Regulating primordial follicle growth activation | [30] |
| GDF9(5q31.1) | Growth factor | Growth and differentiation of granulosa cells proliferation | [31] |
| INHA variants | Growth factor | Maturation of ovarian follicles by FSH inhibition | [32] |
| KHDRBS1 | Signal transduction activator | Alter mRNA expression level and alternative splicing | [33] |
| LHR | Lutropin-choriogonadotropic hormone receptor | Regulation of ovarian follicle maturation, steroidogenesis, and ovulation | [34] |
| LHX8 | Transcription factor | Germ-cell-specific critical regulator of early oogenesis | [35] |
| NOBOX(7q35) | Transcription factor | Follicle development | [36] |
| PGRMC1(Xq22-q24) | Heme-binding protein | Regulation of apoptosis | [37] |
| POLR3H | RNA polymerase III subunit H | Regulation of cell cycle, cell growth, and differentiation | [38] |
| SOHLH1* | Transcription factor | Early folliculogenesis | [39] |
| WT1(11q13) | Transcription factor | Granulosa cell differentiation and oocyte-granulosa cell interaction | [40] |
2. Results
2.1. Identification of Transcripts in Different Reproductive Stages of Mouse Ovaries
2.2. Gene Ontology (GO) Enrichment of DEGs
2.3. KEGG Analysis of DEGs
2.4. Protein-Protein Interaction (PPI) Network Construction and Clusters Analyses
2.5. Identification of Key Target Gene and Their Validation
2.6. Correlation with miRNAs
2.7. Interaction with Piwil Gene Family
3. Discussion
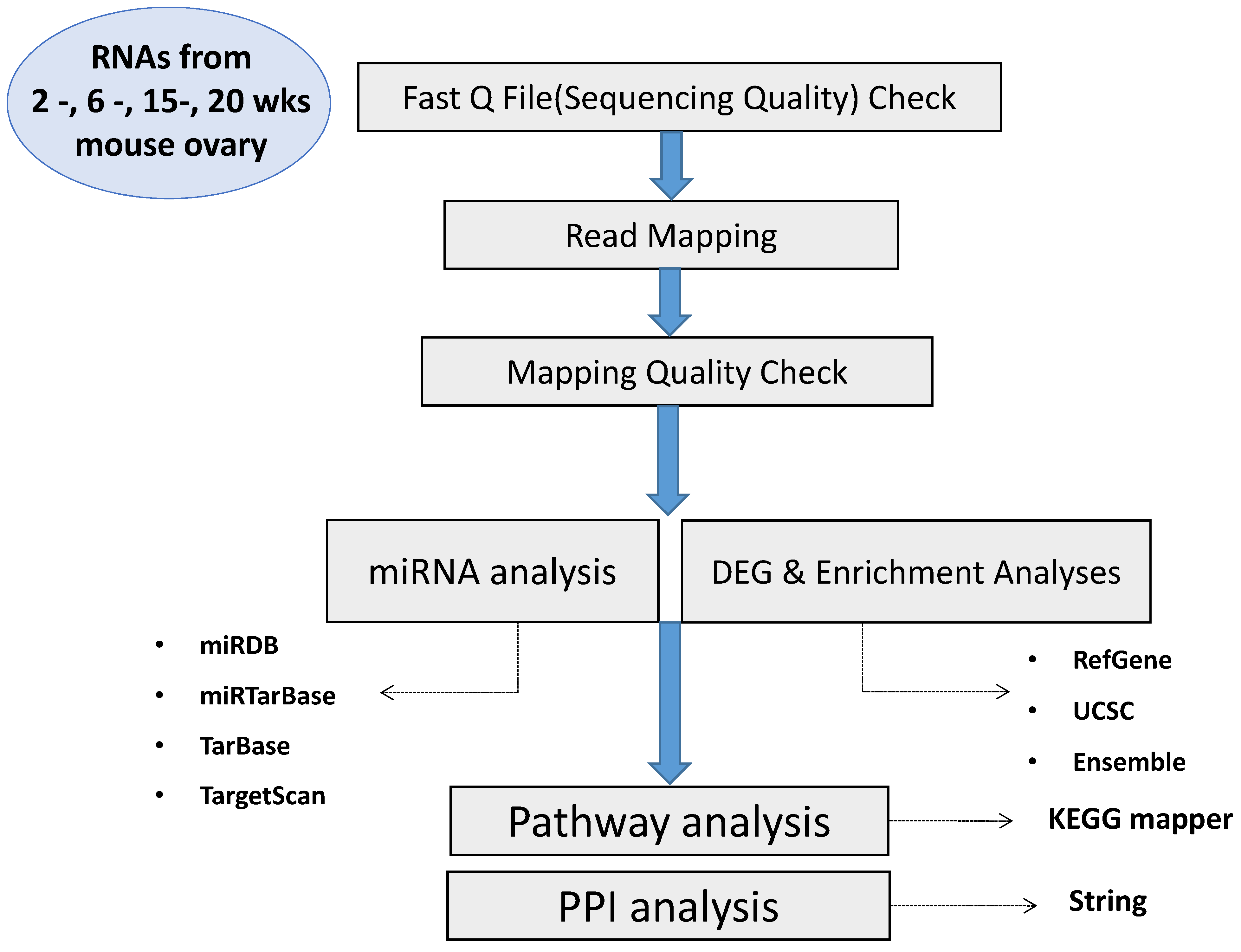
4. Materials and Methods
4.1. Ethics
4.2. Preparation of Ovaries and Extraction of Total RNA
4.3. Total Transcriptome (RNA-Seq) Analyses
4.3.1. mRNA Sequencing
4.3.2. microRNA Sequencing
4.4. Read Quality of the Experiment
4.5. Data Processing
4.6. Differentially Expressed Genes (DEGs)
4.7. KEGG and GO Enrichment Analyses of DEGs
4.8. Gene Set Enrichment Analysis (GSEA)
4.9. miRNA Profiling
4.10. Protein Interaction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Kim, Y.Y.; Min, H.; Kim, H.; Choi, Y.M.; Liu, H.C.; Ku, S.Y. Differential MicroRNA expression profile of human embryonic stem cell-derived cardiac lineage cells. Tissue Eng. Regen. Med. 2017, 14, 163–169. [Google Scholar] [CrossRef]
- Kim, Y.J.; Ku, S.Y.; Rosenwaks, Z.; Liu, H.C.; Chi, S.W.; Kang, J.S.; Lee, W.J.; Jung, K.C.; Kim, S.H.; Choi, Y.M.; et al. MicroRNA expression profiles are altered by gonadotropins and vitamin C status during in vitro follicular growth. Reprod. Sci. 2010, 17, 1081–1089. [Google Scholar] [CrossRef]
- Kim, Y.J.; Ku, S.Y.; Kim, Y.Y.; Liu, H.C.; Chi, S.W.; Kim, S.H.; Choi, Y.M.; Kim, J.G.; Moon, S.Y. MicroRNAs transfected into granulosa cells may regulate oocyte meiotic competence during in vitro maturation of mouse follicles. Hum. Reprod. 2013, 28, 3050–3061. [Google Scholar] [CrossRef]
- Kim, Y.Y.; Tamadon, A.; Ku, S.Y. Potential Use of Antiapoptotic Proteins and Noncoding RNAs for Efficient In Vitro Follicular Maturation and Ovarian Bioengineering. Tissue Eng. Part B Rev. 2017, 23, 142–158. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Sun, J.; Lai, D. Role of microRNAs in premature ovarian insufficiency. Reprod. Biol. Endocrinol. 2017, 15, 38. [Google Scholar] [CrossRef] [PubMed]
- Nie, M.; Yu, S.; Peng, S.; Fang, Y.; Wang, H.; Yang, X. miR-23a and miR-27a promote human granulosa cell apoptosis by targeting SMAD5. Biol. Reprod. 2015, 93, 98. [Google Scholar] [CrossRef]
- Kim, Y.Y.; Kim, W.O.; Liu, H.C.; Rosenwaks, Z.; Kim, J.W.; Ku, S.Y. Effects of paclitaxel and cisplatin on in vitro ovarian follicle development. Arch. Med. Sci. 2019, 15, 1510–1519. [Google Scholar] [CrossRef]
- Armstrong, D.G.; McEvoy, T.G.; Baxter, G.; Robinson, J.J.; Hogg, C.O.; Woad, K.J.; Webb, R.; Sinclair, K.D. Effect of dietary energy and protein on bovine follicular dynamics and embryo production in vitro: Associations with the ovarian insulin-like growth factor system. Biol. Reprod. 2001, 64, 1624–1632. [Google Scholar] [CrossRef] [PubMed]
- McCallie, B.; Schoolcraft, W.B.; Katz-Jaffe, M.G. Aberration of blastocyst microRNA expression is associated with human infertility. Fertil. Steril. 2010, 93, 2374–2382. [Google Scholar] [CrossRef] [PubMed]
- Miles, J.R.; McDaneld, T.G.; Wiedmann, R.T.; Cushman, R.A.; Echternkamp, S.E.; Vallet, J.L.; Smith, T.P. MicroRNA expression profile in bovine cumulus-oocyte complexes: Possible role of let-7 and miR-106a in the development of bovine oocytes. Anim. Reprod. Sci. 2012, 130, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Coller, J. Molecular Biology of Long Non-Coding RNAs; Springer: New York, NY, USA, 2013; p. 227. [Google Scholar]
- Morris, K.V. Long Non-coding RNAs in Human Disease. In Current Topics in Microbiology and Immunology, 1st ed.; Springer International Publishing: Cham, Switzerland, 2016; Volume 8, 262p. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Hawkins, S.M.; Matzuk, M.M. Oocyte-somatic cell communication and microRNA function in the ovary. Ann. Endocrinol. 2010, 71, 144–148. [Google Scholar] [CrossRef]
- McGinnis, L.K.; Luense, L.J.; Christenson, L.K. MicroRNA in ovarian biology and disease. Cold Spring. Harb. Perspect. Med. 2015, 5, a022962. [Google Scholar] [CrossRef]
- Rah, H.; Jeon, Y.J.; Lee, B.E.; Kim, J.O.; Shim, S.H.; Lee, W.S.; Choi, D.H.; Kim, J.H.; Kim, N.K. Association of polymorphisms in microRNA machinery genes (DROSHA, DICER1, RAN, and XPO5) with risk of idiopathic primary ovarian insufficiency in Korean women. Menopause 2013, 20, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhou, Y.; Peng, S.; Wu, L.; Lin, H.Y.; Wang, S.; Wang, H. Differentially expressed plasma microRNAs in premature ovarian failure patients and the potential regulatory function of mir-23a in granulosa cell apoptosis. Reproduction 2012, 144, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.J.; Zhao, S.D.; Qin, Y.Y.; Han, T.; Li, W.P.; Chen, Z.J. MicroRNA-22-3p is down-regulated in the plasma of Han Chinese patients with premature ovarian failure. Fertil. Steril. 2015, 103, U802–U807. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, E.; Forabosco, A.; Schlessinger, D. Genetics of the ovarian reserve. Front. Genet. 2015, 6, 308. [Google Scholar] [CrossRef]
- Grive, K.J.; Freiman, R.N. The developmental origins of the mammalian ovarian reserve. Development 2015, 142, 2554–2563. [Google Scholar] [CrossRef] [PubMed]
- Alvaro Mercadal, B.; Imbert, R.; Demeestere, I.; Gervy, C.; De Leener, A.; Englert, Y.; Costagliola, S.; Delbaere, A. AMH mutations with reduced in vitro bioactivity are related to premature ovarian insufficiency. Hum. Reprod. 2015, 30, 1196–1202. [Google Scholar] [CrossRef]
- Yoon, S.H.; Choi, Y.M.; Hong, M.A.; Kim, J.J.; Lee, G.H.; Hwang, K.R.; Moon, S.Y. Association study of anti-Mullerian hormone and anti-Mullerian hormone type II receptor polymorphisms with idiopathic primary ovarian insufficiency. Hum. Reprod. 2013, 28, 3301–3305. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Patino, L.C.; Silgado, D.; Laissue, P. A potential functional association between mutant BMPR2 and primary ovarian insufficiency. Syst. Biol. Reprod. Med. 2017, 63, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Di Pasquale, E.; Beck-Peccoz, P.; Persani, L. Hypergonadotropic ovarian failure associated with an inherited mutation of human bone morphogenetic protein-15 (BMP15) gene. Am. J. Hum. Genet. 2004, 75, 106–111. [Google Scholar] [CrossRef]
- de Mattos, C.S.; Trevisan, C.M.; Peluso, C.; Adami, F.; Cordts, E.B.; Christofolini, D.M.; Barbosa, C.P.; Bianco, B. ESR1 and ESR2 gene polymorphisms are associated with human reproduction outcomes in Brazilian women. J. Ovarian Res. 2014, 7, 114. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chen, Z.J.; Qin, Y.Y.; Shi, Y.H.; Wang, S.; Choi, Y.; Simpson, J.L.; Rajkovic, A. Transcription factor FIGLA is mutated in patients with Premature Ovarian Failure. Am. J. Hum. Genet. 2008, 82, 1342–1348. [Google Scholar] [CrossRef]
- Oostra, B.A.; Willemsen, R. FMR1: A gene with three faces. Biochim. Biophys. Acta 2009, 1790, 467–477. [Google Scholar] [CrossRef]
- Welt, C.K. Primary ovarian insufficiency: A more accurate term for premature ovarian failure. Clin. Endocrinol. 2008, 68, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Bouilly, J.; Beau, I.; Barraud, S.; Bernard, V.; Azibi, K.; Fagart, J.; Fevre, A.; Todeschini, A.L.; Veitia, R.A.; Beldjord, C.; et al. Identification of multiple gene mutations accounts for a new genetic architecture of primary ovarian insufficiency. J. Clin. Endocrinol. Metab. 2016, 101, 4541–4550. [Google Scholar] [CrossRef] [PubMed]
- Watkins, W.J.; Umbers, A.J.; Woad, K.J.; Harris, S.E.; Winship, I.M.; Gersak, K.; Shelling, A.N. Mutational screening of FOXO3A and FOXO1A in women with premature ovarian failure. Fertil. Steril. 2006, 86, 1518–1521. [Google Scholar] [CrossRef] [PubMed]
- Patino, L.C.; Walton, K.L.; Mueller, T.D.; Johnson, K.E.; Stocker, W.; Richani, D.; Agapiou, D.; Gilchrist, R.B.; Laissue, P.; Harrison, C.A. BMP15 Mutations Associated With Primary Ovarian Insufficiency Reduce Expression, Activity, or Synergy With GDF9. J. Clin. Endocr. Metab. 2017, 102, 1009–1019. [Google Scholar] [CrossRef]
- Dixit, H.; Deendayal, M.; Singh, L. Mutational analysis of the mature peptide region of inhibin genes in Indian women with ovarian failure. Hum. Reprod. 2004, 19, 1760–1764. [Google Scholar] [CrossRef][Green Version]
- Wang, B.B.; Li, L.; Zhu, Y.; Zhang, W.; Wang, X.; Chen, B.L.; Li, T.Y.; Pan, H.; Wang, J.; Kee, K.; et al. Sequence variants of KHDRBS1 as high penetrance susceptibility risks for primary ovarian insufficiency by mis-regulating mRNA alternative splicing. Hum. Reprod. 2017, 32, 2138–2146. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.L. Genetic and phenotypic heterogeneity in ovarian failure: Overview of selected candidate genes. Ann. N. Y. Acad. Sci. 2008, 1135, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Rossetti, R.; Ferrari, I.; Bonomi, M.; Persani, L. Genetics of primary ovarian insufficiency. Clin. Genet. 2017, 91, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Rajkovic, A.; Pangas, S.A.; Ballow, D.; Suzumori, N.; Matzuk, M.M. NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science 2004, 305, 1157–1159. [Google Scholar] [CrossRef]
- Venturella, R.; De Vivo, V.; Carlea, A.; D’Alessandro, P.; Saccone, G.; Arduino, B.; Improda, F.P.; Lico, D.; Rania, E.; De Marco, C.; et al. The Genetics of Non-Syndromic Primary Ovarian Insufficiency: A Systematic Review. Int. J. Fertil. Steril. 2019, 13, 161–168. [Google Scholar] [CrossRef]
- Franca, M.M.; Han, X.F.; Funari, M.F.A.; Lerario, A.M.; Nishi, M.Y.; Fontenele, E.G.P.; Domenice, S.; Jorge, A.A.L.; Garcia-Galiano, D.; Elias, C.F.; et al. Exome sequencing reveals the POLR3H gene as a novel cause of primary ovarian insufficiency. J. Clin. Endocr. Metab. 2019, 104, 2827–2841. [Google Scholar] [CrossRef]
- Zhao, S.; Li, G.; Dalgleish, R.; Vujovic, S.; Jiao, X.; Li, J.; Simpson, J.L.; Qin, Y.; Ivanisevic, M.; Ivovic, M.; et al. Transcription factor SOHLH1 potentially associated with primary ovarian insufficiency. Fertil. Steril. 2015, 103, 548–553. [Google Scholar] [CrossRef]
- Gao, F.; Zhang, J.; Wang, X.; Yang, J.; Chen, D.; Huff, V.; Liu, Y.X. Wt1 functions in ovarian follicle development by regulating granulosa cell differentiation. Hum. Mol. Genet. 2014, 23, 333–341. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Pashaiasl, M.; Ebrahimi, M.; Ebrahimie, E. Identification of the key regulating genes of diminished ovarian reserve (DOR) by network and gene ontology analysis. Mol. Biol. Rep. 2016, 43, 923–937. [Google Scholar] [CrossRef]
- Samartzis, E.P.; Labidi-Galy, S.I.; Moschetta, M.; Uccello, M.; Kalaitzopoulos, D.R.; Perez-Fidalgo, J.A.; Boussios, S. Endometriosis-associated ovarian carcinomas: Insights into pathogenesis, diagnostics, and therapeutic targets-a narrative review. Ann. Transl. Med. 2020, 8, 1712. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Ahn, E.H.; An, H.J.; Kim, J.H.; Ko, J.J.; Kim, Y.R.; Lee, W.S.; Kim, N.K. Association of miR-938G > A Polymorphisms with Primary Ovarian Insufficiency (POI)-Related Gene Expression. Int. J. Mol. Sci. 2017, 18, 1255. [Google Scholar] [CrossRef] [PubMed]
- Norling, A.; Hirschberg, A.L.; Rodriguez-Wallberg, K.A.; Iwarsson, E.; Wedell, A.; Barbaro, M. Identification of a duplication within the GDF9 gene and novel candidate genes for primary ovarian insufficiency (POI) by a customized high-resolution array comparative genomic hybridization platform. Hum. Reprod. 2014, 29, 1818–1827. [Google Scholar] [CrossRef]
- Tripurani, S.K.; Lee, K.B.; Wee, G.; Smith, G.W.; Yao, J. MicroRNA-196a regulates bovine newborn ovary homeobox gene (NOBOX) expression during early embryogenesis. BMC Dev. Biol. 2011, 11, 25. [Google Scholar] [CrossRef]
- Cenik, E.S.; Zamore, P.D. Argonaute proteins. Curr. Biol. 2011, 21, R446–R449. [Google Scholar] [CrossRef]
- Kawamura, Y.; Saito, K.; Kin, T.; Ono, Y.; Asai, K.; Sunohara, T.; Okada, T.N.; Siomi, M.C.; Siomi, H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 2008, 453, 793–797. [Google Scholar] [CrossRef]
- Kuramochi-Miyagawa, S.; Kimura, T.; Yomogida, K.; Kuroiwa, A.; Tadokoro, Y.; Fujita, Y.; Sato, M.; Matsuda, Y.; Nakano, T. Two mouse piwi-related genes: Miwi and mili. Mech. Dev. 2001, 108, 121–133. [Google Scholar] [CrossRef]
- Yang, Q.; Hua, J.; Wang, L.; Xu, B.; Zhang, H.; Ye, N.; Zhang, Z.; Yu, D.; Cooke, H.J.; Zhang, Y.; et al. MicroRNA and piRNA profiles in normal human testis detected by next generation sequencing. PLoS ONE 2013, 8, e66809. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Wang, P.J. Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet. 2012, 8, e1003038. [Google Scholar] [CrossRef]
- Malki, S.; van der Heijden, G.W.; O’Donnell, K.A.; Martin, S.L.; Bortvin, A. A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev. Cell 2014, 29, 521–533. [Google Scholar] [CrossRef]
- Yu, G.C.; Wang, L.G.; Han, Y.Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Gauthier, L.; Baibakov, B.; Jimenez-Movilla, M.; Dean, J. FIGLA, a basic helix-loop-helix transcription factor, balances sexually dimorphic gene expression in postnatal oocytes. Mol. Cell. Biol. 2010, 30, 3661–3671. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huntriss, J.; Gosden, R.; Hinkins, M.; Oliver, B.; Miller, D.; Rutherford, A.J.; Picton, H.M. Isolation, characterization and expression of the human Factor In the Germline alpha (FIGLA) gene in ovarian follicles and oocytes. Mol. Hum. Reprod. 2002, 8, 1087–1095. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Sun, S.; Charkraborty, T.; Wu, L.; Sun, L.; Wei, J.; Nagahama, Y.; Wang, D.; Zhou, L. Figla Favors Ovarian Differentiation by Antagonizing Spermatogenesis in a Teleosts, Nile Tilapia (Oreochromis niloticus). PLoS ONE 2015, 10, e0123900. [Google Scholar] [CrossRef]
- Reilly, J.N.; McLaughlin, E.A.; Stanger, S.J.; Anderson, A.L.; Hutcheon, K.; Church, K.; Mihalas, B.P.; Tyagi, S.; Holt, J.E.; Eamens, A.L.; et al. Characterisation of mouse epididymosomes reveals a complex profile of microRNAs and a potential mechanism for modification of the sperm epigenome. Sci. Rep. 2016, 6, 31794. [Google Scholar] [CrossRef] [PubMed]
- Mikaeili, S.; Rashidi, B.H.; Safa, M.; Najafi, A.; Sobhani, A.; Asadi, E.; Abbasi, M. Altered FoxO3 expression and apoptosis in granulosa cells of women with polycystic ovary syndrome. Arch. Gynecol. Obstet. 2016, 294, 185–192. [Google Scholar] [CrossRef]
- Ezzati, M.M.; Baker, M.D.; Saatcioglu, H.D.; Aloisio, G.M.; Pena, C.G.; Nakada, Y.; Cuevas, I.; Carr, B.R.; Castrillon, D.H. Regulation of FOXO3 subcellular localization by Kit ligand in the neonatal mouse ovary. J. Assist. Reprod. Genet. 2015, 32, 1741–1747. [Google Scholar] [CrossRef][Green Version]
- Albamonte, M.I.; Calabro, L.Y.; Albamonte, M.S.; Zuccardi, L.; Stella, I.; Halperin, J.; Vitullo, A.D. PTEN and FOXO3 expression in the prenatal and postnatal human ovary. J. Assist. Reprod. Genet. 2020, 37, 1613–1622. [Google Scholar] [CrossRef]
- Li, C.; Zhang, Y.; Zhao, W.; Cui, S.; Song, Y. miR-153-3p regulates progression of ovarian carcinoma in vitro and in vivo by targeting MCL1 gene. J. Cell. Biochem. 2019, 120, 19147–19158. [Google Scholar] [CrossRef]
- Nandedkar, T.D.; Moodbidri, S.B.; Raghavan, V.P. FSH binding to granulosa cells of normal and atretic follicles of mouse ovary in vivo and in vitro. Indian J. Exp. Biol. 1985, 23, 685–688. [Google Scholar]
- Ahn, H.W.; Morin, R.D.; Zhao, H.; Harris, R.A.; Coarfa, C.; Chen, Z.J.; Milosavljevic, A.; Marra, M.A.; Rajkovic, A. MicroRNA transcriptome in the newborn mouse ovaries determined by massive parallel sequencing. Mol. Hum. Reprod. 2010, 16, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Mineno, J.; Okamoto, S.; Ando, T.; Sato, M.; Chono, H.; Izu, H.; Takayama, M.; Asada, K.; Mirochnitchenko, O.; Inouye, M.; et al. The expression profile of microRNAs in mouse embryos. Nucl. Acids Res. 2006, 34, 1765–1771. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.Y.; Kim, Y.J.; Cho, K.M.; Kim, S.H.; Park, K.E.; Kang, B.C.; Jung, K.C.; Kim, M.S.; Ku, S.Y. The expression profile of angiotensin system on thawed murine ovaries. Tissue Eng. Regen. Med. 2016, 13, 724–731. [Google Scholar] [CrossRef]
- Kim, Y.J.; Kim, Y.Y.; Kang, B.C.; Kim, M.S.; Ko, I.K.; Liu, H.C.; Rosenwaks, Z.; Ku, S.Y. Induction of multiple ovulation via modulation of angiotensin II receptors in in vitro ovarian follicle culture models. J. Tissue Eng. Regen. Med. 2017, 11, 3100–3110. [Google Scholar] [CrossRef] [PubMed]
- Rajapakse, S.; Yamano, N.; Ogiwara, K.; Hirata, K.; Takahashi, S.; Takahashi, T. Estrogen-dependent expression of the tissue kallikrein gene (Klk1) in the mouse uterus and its implications for endometrial tissue growth. Mol. Reprod. Dev. 2007, 74, 1053–1063. [Google Scholar] [CrossRef]
- Dedio, J.; Wiemer, G.; Rutten, H.; Dendorfer, A.; Scholkens, B.A.; Muller-Esterl, W.; Wohlfart, P. Tissue kallikrein KLK1 is expressed de novo in endothelial cells and mediates relaxation of human umbilical veins. Biol. Chem. 2001, 382, 1483–1490. [Google Scholar] [CrossRef]
- Gomes Fernandes, M.; He, N.; Wang, F.; Van Iperen, L.; Eguizabal, C.; Matorras, R.; Roelen, B.A.J.; Chuva De Sousa Lopes, S.M. Human-specific subcellular compartmentalization of P-element induced wimpy testis-like (PIWIL) granules during germ cell development and spermatogenesis. Hum. Reprod. 2018, 33, 258–269. [Google Scholar] [CrossRef]
- Jin, Y.H.; Davie, A.; Migaud, H. Expression pattern of nanos, piwil, dnd, vasa and pum genes during ontogenic development in Nile tilapia Oreochromis niloticus. Gene 2019, 688, 62–70. [Google Scholar] [CrossRef]
- Yang, X.G.; Yue, H.M.; Ye, H.; Shan, X.S.; Xie, X.; Li, C.J.; Wei, Q.W. Identification and characterization of two piwi genes and their expression in response to E2 (17 beta-estradiol) in Dabry’s sturgeon Acipenser dabryanus. Fish. Sci. 2020, 86, 307–317. [Google Scholar] [CrossRef]
- Yiew, N.K.H.; Chatterjee, T.K.; Tang, Y.L.; Pellenberg, R.; Stansfield, B.K.; Bagi, Z.; Fulton, D.J.; Stepp, D.W.; Chen, W.; Patel, V.; et al. A novel role for the Wnt inhibitor APCDD1 in adipocyte differentiation: Implications for diet-induced obesity. J. Biol. Chem. 2017, 292, 6312–6324. [Google Scholar] [CrossRef]
- Fang, X.; Ni, N.; Lydon, J.P.; Ivanov, I.; Bayless, K.J.; Rijnkels, M.; Li, Q. Enhancer of zeste 2 polycomb repressive complex 2 subunit is required for uterine epithelial integrity. Am. J. Pathol. 2019, 189, 1212–1225. [Google Scholar] [CrossRef]
- Cousminer, D.L.; Stergiakouli, E.; Berry, D.J.; Ang, W.; Groen-Blokhuis, M.M.; Korner, A.; Siitonen, N.; Ntalla, I.; Marinelli, M.; Perry, J.R.; et al. Genome-wide association study of sexual maturation in males and females highlights a role for body mass and menarche loci in male puberty. Hum. Mol. Genet. 2014, 23, 4452–4464. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.R.; Wang, A.; Lv, Z.; Yuan, Y.; Xu, Q. Mitochondria-related core genes and TF-miRNA-hub mrDEGs network in breast cancer. Biosci. Rep. 2021, 41, 1–17. [Google Scholar] [CrossRef]
- Yun, J.W.; Kim, Y.Y.; Ahn, J.H.; Kang, B.C.; Ku, S.Y. Use of nonhuman primates for the development of bioengineered female reproductive organs. Tissue Eng. Regen. Med. 2016, 13, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.Y.; Yun, J.W.; Kim, J.M.; Park, C.G.; Rosenwaks, Z.; Liu, H.C.; Kang, B.C.; Ku, S.Y. Gonadotropin ratio affects the in vitro growth of rhesus ovarian preantral follicles. J. Investig. Med. 2016, 64, 888–893. [Google Scholar] [CrossRef]
- Kim, Y.Y.; Kang, B.C.; Yun, J.W.; Ahn, J.H.; Kim, Y.J.; Kim, H.; Rosenwaks, Z.; Ku, S.Y. Expression of transcripts in marmoset oocytes retrieved during follicle isolation without gonadotropin induction. Int. J. Mol. Sci. 2019, 20, 1133. [Google Scholar] [CrossRef] [PubMed]
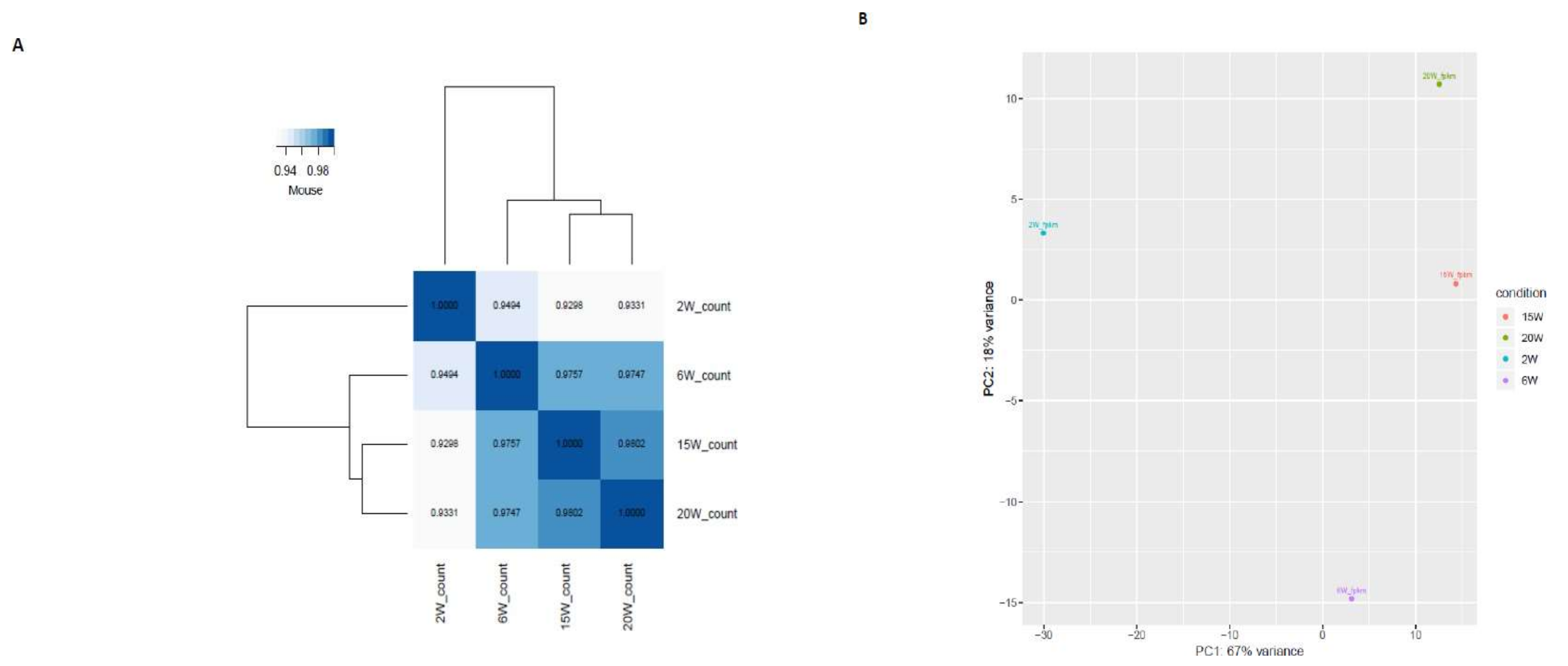

| A | 6 W vs. 2 W | 15 W vs. 2 W | 20 W vs. 2 W | 15 W vs. 6 W | 20 W vs. 6 W | 20 W vs. 15 W |
|---|---|---|---|---|---|---|
| Up-regulated | 545 | 925 | 696 | 208 | 170 | 114 |
| Down-regulated | 447 | 745 | 727 | 199 | 217 | 65 |
| Total expression | 992 | 1670 | 1423 | 407 | 387 | 179 |
 | ||||||
| (A) 6 W vs. 2 W | ||
| Up-Regulation | ||
| Gene | Category | Biological Functions |
| Bhmt | Biological process | Homocysteine catabolic process |
| Molecular Function | Oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor | |
| KEGG pathway | Cystein and methionine metabolism | |
| Spp1 | Biological process | Calcium-independent cell-matrix adhesion |
| Molecular Function | Extracellular matrix binding | |
| KEGG pathway | ECM-receptor interaction | |
| Klk1 | Biological process | Positive regulation of steroid hormone biosynthetic process |
| Molecular Function | Bradykinin receptor activity | |
| KEGG pathway | Renin-angiotensin system | |
| Serpina5 | Biological process | Blood coagulation, fibrin clot formation |
| Molecular Function | Serine-type endopeptidase activity | |
| KEGG pathway | Complement and coagulation cascades | |
| Sfrp4 | Biological process | Positive regulation of dermatome development |
| Molecular Function | Chemoattractant activity involved in axon guidance | |
| KEGG pathway | Basal cell carcinoma | |
| Plin4 | Biological process | Negative regulation of sequestering of triglyceride |
| Molecular Function | Tryglyceride lipase activity | |
| KEGG pathway | Regulation of lipolysis in adipocytes | |
| Wisp2 | Biological process | Regulation of dermatome development |
| Molecular Function | Fibronectin binding | |
| KEGG pathway | Bladder cancer | |
| GM1987 (Ccl21a) | Biological process | Lymphocyte chemotaxis across high endothelial venule |
| Molecular Function | CXCR chemokine receptor binding | |
| KEGG pathway | Chemokine signaling pathway | |
| Down-Regulation | ||
| Gene | Category | Biological Functions |
| BC100530, Stfa1 | Biological process | Negative regulation of endopeptidase activity |
| Molecular Function | Cysteine-type endopeptidase inhibitor activity | |
| Figla | Biological process | Negative regulation of fertilization |
| Molecular Function | Acrosin binding | |
| KEGG pathway | Ovarian steroidogenesis | |
| S100a9, S100a8 | Biological process | Neutrophil aggregation |
| Molecular Function | Toll-like receptor 4 binding | |
| KEGG pathway | IL-17 signaling pathway | |
| (B) 15 W vs. 2 W | ||
| Up-Regulation | ||
| Gene | Category | Biological Functions |
| Wnt10b | Biological process | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation |
| Molecular Function | Frizzled binding | |
| KEGG pathway | Basal cell carcinoma | |
| H2-T9 (Gm7030) | Biological process | Antigen processing and presentation of exogenous protein antigen via MHC class |
| Molecular Function | MHC class I protein binding | |
| KEGG pathway | Antigen processing and presentation | |
| Atp1a3 | Biological process | Positive regulation of sodium ion export across plasma membrane |
| Molecular Function | Sodium:potassium-exchanging atpase activity | |
| KEGG pathway | Proximal tubule bicarbonate reclamation | |
| Ptgfr | Biological process | Phospholipase c-activating angiotensin-activated signaling pathway |
| Molecular Function | Angiotensin type I receptor activity | |
| KEGG pathway | Renin-angiotensin system | |
| Lipg | Biological process | Triglyceride catabolic process |
| Molecular Function | Lipoprotein lipase activity | |
| KEGG pathway | Glycerolipid metabolism | |
| Abcb1b | Biological process | Drug export |
| Molecular Function | Xenobiotic transmembrane transporting atpase activity | |
| KEGG pathway | Bile secretion | |
| Hsd17b7 | Biological process | Estrogen biosynthetic process |
| Molecular Function | Estradiol 17-beta-dehydrogenase activity | |
| KEGG pathway | Steroid biosynthesis | |
| Star | Biological process | Vesicle tethering to endoplasmic reticulum |
| Molecular Function | Porin activity | |
| KEGG pathway | Cholesterol metabolism | |
| Plin4, Bhmt, Sfrp4 | Functions are indicated in previous page | |
| Down-Regulation | ||
| Gene | Category | Biological Functions |
| Atf3 | Biological process | Positive regulation of transcription from RNA polymerase |
| Molecular Function | Camp response element binding protein binding | |
| KEGG pathway | Cocaine addiction | |
| Fos, Fosb | Biological process | Response to gravity |
| Molecular Function | Nad-dependent histone deacetylase activity | |
| KEGG pathway | Amphetamine addiction | |
| S100a9, S100a8 | Functions are indicated in previous page | |
| (C) 20 W vs. 2 W | ||
| Up-Regulation | ||
| Gene | Category | Biological Functions |
| Sprr1a | Biological process | Peptide cross-linking |
| Wisp2 | Biological process | Regulation of dermatome development |
| Molecular Function | Fibronectin binding | |
| KEGG pathway | Bladder cancer | |
| Inhba | Biological process | Regulation of follicle-stimulating hormone secretion |
| Molecular Function | Inhibin binding | |
| KEGG pathway | TGF-beta signaling pathway | |
| Mgst2 | Biological process | Xenobiotic catabolic process |
| Molecular Function | Glutathione disulfide oxidoreductase activity | |
| KEGG pathway | Metabolism of xenobiotics by cytochrome P450 | |
| Akr1c18 | Biological process | Polyprenol catabolic process |
| Molecular Function | Enone reductase activity | |
| KEGG pathway | Steroid hormone biosynthesis | |
| Sfrp4 | Biological process | Positive regulation of dermatome development |
| Molecular Function | Chemoattractant activity involved in axon guidance | |
| KEGG pathway | Basal cell carcinoma | |
| Wnt10b, Plin4, Lipg, Bhmt, Abcb1b, Spp1, Ptgfr, Hsd17b7 | Functions are indicated in previous page | |
| Down-Regulation | ||
| Gene | Category | Biological Functions |
| BC100530 | Biological process | Negative regulation of endopeptidase activity |
| Molecular Function | Cysteine-type endopeptidase inhibitor activity | |
| S100a8 | Functions are indicated in previous page | |
| (D) 15 W vs. 6 W | ||
| Up-Regulation | ||
| Gene | Category | Biological Functions |
| Tgoln1 (Tgoln2) | Biological process | Golgi ribbon formation |
| Molecular Function | Mannose binding | |
| KEGG pathway | SNARE interactions in vesicular transport | |
| Eif3j1 | Biological process | Viral translational termination-reinitiation |
| Molecular Function | Translation initiation factor activity | |
| KEGG pathway | RNA transport | |
| Nme2 | Biological process | ITP metabolic process |
| Molecular Function | CTP synthase activity | |
| KEGG pathway | Pyrimidine metabolism | |
| Tmem254b | Unknown | |
| H2-T9 (Gm7030) | Functions are indicated in previous page | |
| Down-Regulation | ||
| Gene | Category | Biological Functions |
| Plac9a | Unknown | |
| Atf3, Fosb, GM1987 (Ccl21a), Fos | Functions are indicated in previous page | |
| (E) 20 W vs. 6 W | ||
| Up-Regulation | ||
| Gene | Category | Biological Functions |
| Tmem254b | Unknown | |
| Eif3j1, Sprr1a | Functions are indicated in previous page | |
| Down-Regulation | ||
| Gene | Category | Biological Functions |
| Ooog1 | Biological process | Female gamete generation |
| GM1987 (Ccl21a) | Functions are indicated in previous page | |
| (F) 20 W vs. 15 W | ||
| Up-Regulation | ||
| Gene | Category | Biological Functions |
| Cxcl1 | Biological process | Interleukin-8-mediated signaling pathway |
| Molecular Function | Interleukin-8 receptor activity | |
| KEGG pathway | IL-17 signaling pathway | |
| Atf3 | Biological process | Positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress |
| Molecular Function | Camp response element binding protein binding | |
| KEGG pathway | Cocaine addiction | |
| Hspa1a, Hspa1b | Biological process | Telomerase holoenzyme complex assembly |
| Molecular Function | CTP binding | |
| KEGG pathway | Prion diseases | |
| Zfp36 | Biological process | Epithelial cell proliferation involved in salivary gland morphogenesis |
| Molecular Function | Mrna 3’-UTR au-rich region binding | |
| KEGG pathway | Antifolate resistance | |
| Sprr1a, Fosb, GM1987 (Ccl21a), Nme2 | Functions are indicated in previous page | |
| Down-Regulation | ||
| Gene | Category | Biological Functions |
| GM1987 (Ccl21a), Nme2 | Functions are indicated in previous page | |
| Genes | Bmp15 | Ccnd2 | Figla | Foxo1 | Foxo3 | Fshr | Gdf9 | Igf1 | Inha | Nobox | Smad3 | Taf4b | Pou5f1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 W | FPKM | 14.64 | 44.14 | 51.84 | 19.5 | 10.55 | 3.07 | 130.57 | 15.29 | 533.01 | 33.33 | 28.04 | 3.82 | 24.29 |
| 6 W | 20.02 | 44.18 | 1.68 | 66.92 | 7.96 | 8.71 | 52.19 | 16.04 | 1396.52 | 7.42 | 16.71 | 4.5 | 5.17 | |
| 20 W | 9.55 | 47.87 | 1.8 | 82.89 | 10.89 | 21.87 | 33.55 | 16 | 1915.61 | 6.47 | 26.57 | 9.52 | 3.62 | |
| 2 W | TPM | 20.33 | 61.29 | 71.97 | 27.08 | 14.65 | 4.27 | 181.28 | 21.23 | 740.04 | 46.27 | 38.93 | 5.31 | 33.73 |
| 6 W | 31.03 | 68.48 | 2.61 | 103.73 | 12.34 | 13.49 | 80.89 | 24.87 | 2164.77 | 11.51 | 25.91 | 6.97 | 8.02 | |
| 20 W | 16.46 | 82.45 | 3.09 | 142.78 | 18.75 | 37.68 | 57.79 | 27.55 | 3299.65 | 11.14 | 45.76 | 16.4 | 6.23 | |
| Gene | Mature miRNAs | |
|---|---|---|
| FPKM | Ccnd2 | mmu-miR-1251-5p, mmu-miR-503-5p, mmu-miR-497a-5p, mmu-miR-195b, mmu-miR-322-5p, mmu-miR-15a-5p, mmu-miR-16-5p, mmu-miR-195a-5p, mmu-miR-195a-5p, mmu-miR-6955-3p, mmu-miR-29a-3p, mmu-miR-29b-3p, mmu-miR-29c-3p, mmu-miR-302c-3p, mmu-miR-302b-3p, mmu-miR-302d-3p, mmu-miR-302a-3p, mmu-miR-294-3p, mmu-miR-295-3p, mmu-miR-291a-3p, mmu-miR-106a-5p, mmu-miR-106b-5p, mmu-miR-93-5p, mmu-miR-20a-5p, mmu-miR-20b-5p, mmu-miR-17-5p, mmu-miR-19b-3p, mmu-miR-19a-3p, mmu-miR-503-5p, mmu-miR-195a-5p, mmu-miR-16-5p, mmu-miR-195b, mmu-miR-322-5p, mmu-miR-15a-5p, mmu-miR-497a-5p, mmu-miR-322-5p, mmu-miR-195a-5p, mmu-miR-195b, mmu-miR-15a-5p, mmu-miR-16-5p, mmu-miR-15b-5p, mmu-miR-503-5p, mmu-miR-182-5p, mmu-miR-9-3p, mmu-miR-124-3p.1, mmu-miR-1193-3p |
| Figla | mmu-miR-669d-2-3p | |
| Foxo1 | mmu-miR-370-3p, mmu-miR-183-5p, mmu-miR-183-5p.2, mmu-miR-96-5p, mmu-miR-144-3p, mmu-miR-27a-3p, mmu-miR-27b-3p, mmu-miR-411-5p, mmu-miR-145b, mmu-miR-145a-5p, mmu-miR-6715-5p, mmu-miR-135a-5p, mmu-miR-135b-5p | |
| Foxo3 | mmu-miR-153-3p | |
| Fshr | mmu-miR-670-3p | |
| Igf1 | mmu-miR-365-3p, mmu-miR-1a-3p, mmu-miR-206-3p, mmu-miR-483-3p.2, mmu-miR-18a-5p, mmu-miR-18b-5p, mmu-miR-452-5p, mmu-miR-29c-3p, mmu-miR-29b-3p, mmu-miR-29a-3p, mmu-miR-495-3p, mmu-miR-378c, mmu-miR-378a-3p, mmu-miR-1839-5p, mmu-let-7a-5p, mmu-let-7c-5p, mmu-miR-98-5p, mmu-let-7i-5p, mmu-let-7g-5p, mmu-let-7d-5p, mmu-let-7b-5p, mmu-let-7k, mmu-let-7f-5p, mmu-let-7e-5p, mmu-miR-192-5p, mmu-miR-215-5p, mmu-miR-29b-3p, mmu-miR-29a-3p, mmu-miR-29c-3p, mmu-miR-142a-5p, mmu-miR-340-5p, mmu-miR-489-3p, mmu-miR-425-5p, mmu-miR-186-5p | |
| Smad3 | mmu-miR-129-2-3p, mmu-miR-129-1-3p, mmu-miR-145a-5p, mmu-miR-145b, mmu-miR-3154 | |
| Taf4b | mmu-miR-148a-3p, mmu-miR-152-3p, mmu-miR-148b-3p | |
| Pou5f1 | mmu-miR-218-5p, mmu-miR-1955-5p, mmu-miR-881-3p, mmu-miR-186-5p |
| Gene | Target Genes and Mature miRNAs | |
| FPKM | Target genes | |
| FOXL2, FOXO3, NANOG, POU5F1, LIN28A, SOX2, SOX3, KLF4, PRDM1, NANOS3, UTF1, CD38, DAZL, DDX4, SYCP3, RNF17, TDRD5, TDRD9, SOX17, GATA4, TEAD4, TFAP2C, LHX9, WT1 | ||
| Interactive miRNAs | ||
| Piwil1 | mmu-miR-1249-5p, mmu-miR-147-5p, mmu-miR-1892, mmu-miR-3084-3p, mmu-miR-330-5p, mmu-miR-6540-3p, mmu-miR-7116-3p, mmu-miR-7669-5p | |
| Piwil2 | mmu-miR-103-2-5p, mmu-miR-1198-5p, mmu-miR-139-5p, mmu-miR-145a-5p, mmu-miR-149-5p, mmu-miR-1b-5p, mmu-miR-216b-5p, mmu-miR-223-5p, mmu-miR-23a-3p, mmu-miR-26a-5p, mmu-miR-26b-3p, mmu-miR-346-5p, mmu-miR-425-3p, mmu-miR-466d-3p, mmu-miR-466f-3p, mmu-miR-466g, mmu-miR-7065-3p, mmu-miR-760-3p, mmu-miR-7647-3p, mmu-miR-7661-5p, mmu-miR-935 | |
| Piwil4 | mmu-miR-210-5p, mmu-miR-3470b | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.-Y.; Kim, K.-S.; Kim, Y.-J.; Kim, S.-W.; Kim, H.; Ku, S.-Y. Transcriptome Analyses Identify Potential Key microRNAs and Their Target Genes Contributing to Ovarian Reserve. Int. J. Mol. Sci. 2021, 22, 10819. https://doi.org/10.3390/ijms221910819
Kim Y-Y, Kim K-S, Kim Y-J, Kim S-W, Kim H, Ku S-Y. Transcriptome Analyses Identify Potential Key microRNAs and Their Target Genes Contributing to Ovarian Reserve. International Journal of Molecular Sciences. 2021; 22(19):10819. https://doi.org/10.3390/ijms221910819
Chicago/Turabian StyleKim, Yoon-Young, Kwang-Soo Kim, Yong-Jin Kim, Sung-Woo Kim, Hoon Kim, and Seung-Yup Ku. 2021. "Transcriptome Analyses Identify Potential Key microRNAs and Their Target Genes Contributing to Ovarian Reserve" International Journal of Molecular Sciences 22, no. 19: 10819. https://doi.org/10.3390/ijms221910819
APA StyleKim, Y.-Y., Kim, K.-S., Kim, Y.-J., Kim, S.-W., Kim, H., & Ku, S.-Y. (2021). Transcriptome Analyses Identify Potential Key microRNAs and Their Target Genes Contributing to Ovarian Reserve. International Journal of Molecular Sciences, 22(19), 10819. https://doi.org/10.3390/ijms221910819








