PARN-like Proteins Regulate Gene Expression in Land Plant Mitochondria by Modulating mRNA Polyadenylation
Abstract
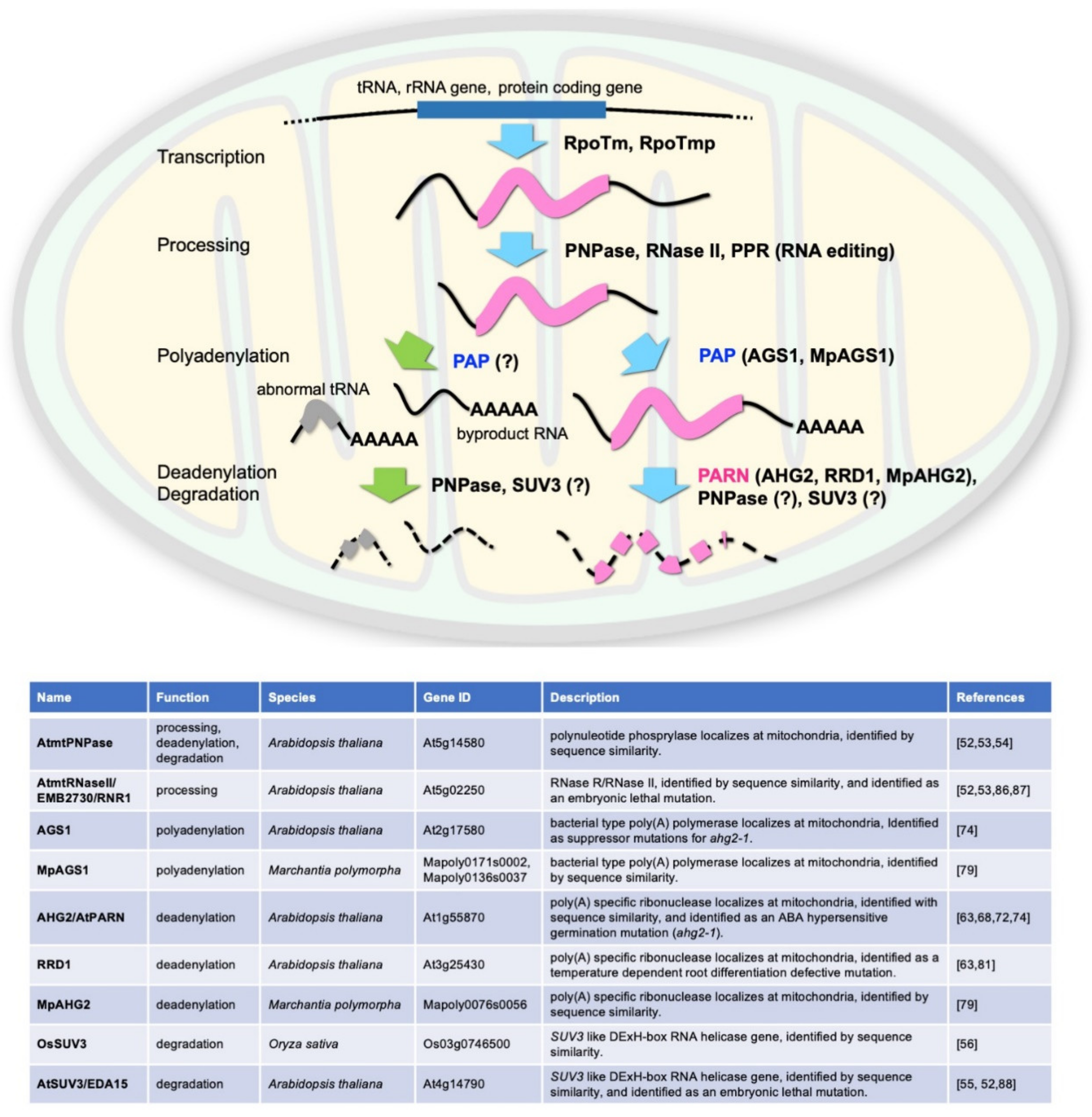
1. Introduction: Overall View of Mitochondrial Transcription
2. Polyadenylation of Mitochondrial mRNAs: Regulation and Functions Differ across Species
3. Factors Possibly Regulating the Poly(A) Status of Mitochondrial mRNAs in Plants
4. Poly(A) Removal Machinery of Eukaryotes
5. Arabidopsis PARN Regulates the Poly(A) Status of Mitochondrial mRNAs
6. The AHG2/AtPARN–AGS1 Regulatory System Is Conserved in Liverwort
7. Another PARN-Like Arabidopsis Protein Also Functions in Mitochondria
8. AHG2/AtPARN and RRD1 Appear to Have Distinct Physiological Functions
9. Physiological Role of the Polyadenylation of Plant Mitochondrial mRNA
10. Concluding Remarks and Future Prospects
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Galluzzi, L.; Kepp, O.; Kroemer, G. Mitochondria: Master Regulators of Danger Signalling. Nat. Rev. Mol. Cell Biol. 2012, 13, 780–788. [Google Scholar] [CrossRef]
- Liberatore, K.L.; Dukowic-Schulze, S.; Miller, M.E.; Chen, C.; Kianian, S.F. The Role of Mitochondria in Plant Development and Stress Tolerance. Free Radic. Biol. Med. 2016, 100, 238–256. [Google Scholar] [CrossRef]
- Mattson, M.P.; Gleichmann, M.; Cheng, A. Mitochondria in Neuroplasticity and Neurological Disorders. Neuron 2008, 60, 748–766. [Google Scholar] [CrossRef]
- Su, B.; Wang, X.; Zheng, L.; Perry, G.; Smith, M.A.; Zhu, X. Abnormal Mitochondrial Dynamics and Neurodegenerative Diseases. Biochim. Biophys. Acta BBA—Mol. Basis Dis. 2010, 1802, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Russell, O.M.; Gorman, G.S.; Lightowlers, R.N.; Turnbull, D.M. Mitochondrial Diseases: Hope for the Future. Cell 2020, 181, 168–188. [Google Scholar] [CrossRef]
- Morley, S.A.; Nielsen, B.L. Plant Mitochondrial DNA. Front. Biosci. Landmark Ed. 2017, 22, 1023–1032. [Google Scholar] [CrossRef] [PubMed]
- Burger, G.; Gray, M.W.; Franz Lang, B. Mitochondrial Genomes: Anything Goes. Trends Genet. 2003, 19, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.S.P.; Salvato, F.; Thal, B.; Eubel, H.; Thelen, J.J.; Møller, I.M. The Proteome of Higher Plant Mitochondria. Mitochondrion 2017, 33, 22–37. [Google Scholar] [CrossRef] [PubMed]
- Morgenstern, M.; Stiller, S.B.; Lübbert, P.; Peikert, C.D.; Dannenmaier, S.; Drepper, F.; Weill, U.; Höß, P.; Feuerstein, R.; Gebert, M.; et al. Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep. 2017, 19, 2836–2852. [Google Scholar] [CrossRef] [PubMed]
- Wasilewska, A.; Vlad, F.; Sirichandra, C.; Redko, Y.; Jammes, F.; Valon, C.; dit Frey, N.F.; Leung, J. An Update on Abscisic Acid Signaling in Plants and More…. Mol. Plant 2008, 1, 198–217. [Google Scholar] [CrossRef]
- Butow, R.A.; Avadhani, N.G. Mitochondrial Signaling: The Retrograde Response. Mol. Cell 2004, 14, 1–15. [Google Scholar] [CrossRef]
- Liu, Z.; Butow, R.A. Mitochondrial Retrograde Signaling. Annu. Rev. Genet. 2006, 40, 159–185. [Google Scholar] [CrossRef]
- Woodson, J.D.; Chory, J. Coordination of Gene Expression between Organellar and Nuclear Genomes. Nat. Rev. Genet. 2008, 9, 383–395. [Google Scholar] [CrossRef]
- Da Cunha, F.M.; Torelli, N.Q.; Kowaltowski, A.J. Mitochondrial Retrograde Signaling: Triggers, Pathways, and Outcomes. Oxid. Med. Cell. Longev. 2015, 2015, e482582. [Google Scholar] [CrossRef]
- Møller, I.M. What Is Hot in Plant Mitochondria? Physiol. Plant. 2016, 157, 256–263. [Google Scholar] [CrossRef]
- Kotiadis, V.N.; Duchen, M.R.; Osellame, L.D. Mitochondrial Quality Control and Communications with the Nucleus Are Important in Maintaining Mitochondrial Function and Cell Health. Biochim. Biophys. Acta BBA—Gen. Subj. 2014, 1840, 1254–1265. [Google Scholar] [CrossRef] [PubMed]
- Posse, V.; Gustafsson, C.M. Human Mitochondrial Transcription Factor B2 Is Required for Promoter Melting during Initiation of Transcription*. J. Biol. Chem. 2017, 292, 2637–2645. [Google Scholar] [CrossRef]
- Hillen, H.S.; Morozov, Y.I.; Sarfallah, A.; Temiakov, D.; Cramer, P. Structural Basis of Mitochondrial Transcription Initiation. Cell 2017, 171, 1072–1081.e10. [Google Scholar] [CrossRef]
- Gustafsson, C.M.; Falkenberg, M.; Larsson, N.-G. Maintenance and Expression of Mammalian Mitochondrial DNA. Annu. Rev. Biochem. 2016, 85, 133–160. [Google Scholar] [CrossRef] [PubMed]
- Kühn, K.; Weihe, A.; Börner, T. Multiple Promoters Are a Common Feature of Mitochondrial Genes in Arabidopsis. Nucleic Acids Res. 2005, 33, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Emanuel, C.; von Groll, U.; Müller, M.; Börner, T.; Weihe, A. Development- and Tissue-Specific Expression of the RpoT Gene Family of Arabidopsis Encoding Mitochondrial and Plastid RNA Polymerases. Planta 2006, 223, 998–1009. [Google Scholar] [CrossRef]
- Kuhn, K.; Richter, U.; Meyer, E.H.; Delannoy, E.; Falcon de Longevialle, A.; O’Toole, N.; Borner, T.; Millar, A.H.; Small, I.D.; Whelan, J. Phage-Type RNA Polymerase RPOTmp Performs Gene-Specific Transcription in Mitochondria of Arabidopsis thaliana. Plant. Cell 2009, 21, 2762–2779. [Google Scholar] [CrossRef] [PubMed]
- Planchard, N.; Bertin, P.; Quadrado, M.; Dargel-Graffin, C.; Hatin, I.; Namy, O.; Mireau, H. The Translational Landscape of Arabidopsis Mitochondria. Nucleic Acids Res. 2018, 46, 6218–6228. [Google Scholar] [CrossRef] [PubMed]
- Giegé, P.; Brennicke, A. RNA Editing in Arabidopsis Mitochondria Effects 441 C to U Changes in ORFs. Proc. Natl. Acad. Sci. USA 1999, 96, 15324–15329. [Google Scholar] [CrossRef]
- Small, I.D.; Schallenberg-Rüdinger, M.; Takenaka, M.; Mireau, H.; Ostersetzer-Biran, O. Plant Organellar RNA Editing: What 30 Years of Research Has Revealed. Plant. J. 2020, 101, 1040–1056. [Google Scholar] [CrossRef] [PubMed]
- Perrin, R.; Meyer, E.H.; Zaepfel, M.; Kim, Y.-J.; Mache, R.; Grienenberger, J.-M.; Gualberto, J.M.; Gagliardi, D. Two Exoribonucleases Act Sequentially to Process Mature 3’-Ends of Atp9 MRNAs in Arabidopsis Mitochondria. J. Biol. Chem. 2004, 279, 25440–25446. [Google Scholar] [CrossRef]
- Perrin, R.; Lange, H.; Grienenberger, J.M.; Gagliardi, D. AtmtPNPase Is Required for Multiple Aspects of the 18S RRNA Metabolism in Arabidopsis thaliana Mitochondria. Nucleic Acids Res. 2004, 32, 5174–5182. [Google Scholar] [CrossRef]
- Holec, S.; Lange, H.; Kuhn, K.; Alioua, M.; Borner, T.; Gagliardi, D. Relaxed Transcription in Arabidopsis Mitochondria Is Counterbalanced by RNA Stability Control Mediated by Polyadenylation and Polynucleotide Phosphorylase. Mol. Cell Biol. 2006, 26, 2869–2876. [Google Scholar] [CrossRef]
- Bollenbach, T.J.; Lange, H.; Gutierrez, R.; Erhardt, M.; Stern, D.B.; Gagliardi, D. RNR1, a 3′–5′ Exoribonuclease Belonging to the RNR Superfamily, Catalyzes 3′ Maturation of Chloroplast Ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res. 2005, 33, 2751–2763. [Google Scholar] [CrossRef]
- Tzafrir, I.; Pena-Muralla, R.; Dickerman, A.; Berg, M.; Rogers, R.; Hutchens, S.; Sweeney, T.C.; McElver, J.; Aux, G.; Patton, D.; et al. Identification of Genes Required for Embryo Development in Arabidopsis. Plant. Physiol. 2004, 135, 1206–1220. [Google Scholar] [CrossRef]
- Hirayama, T.; Shinozaki, K. Perception and Transduction of Abscisic Acid Signals: Keys to the Function of the Versatile Plant Hormone ABA. Trends Plant. Sci. 2007, 12, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Reverdatto, S.V.; Dutko, J.A.; Chekanova, J.A.; Hamilton, D.A.; Belostotsky, D.A. MRNA Deadenylation by PARN Is Essential for Embryogenesis in Higher Plants. RNA 2004, 10, 1200–1214. [Google Scholar] [CrossRef]
- Chiba, Y.; Johnson, M.A.; Lidder, P.; Vogel, J.T.; van Erp, H.; Green, P.J. AtPARN Is an Essential Poly(A) Ribonuclease in Arabidopsis. Gene 2004, 328, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, N.; Kitahata, N.; Seki, M.; Narusaka, Y.; Narusaka, M.; Kuromori, T.; Asami, T.; Shinozaki, K.; Hirayama, T. Analysis of ABA Hypersensitive Germination2 Revealed the Pivotal Functions of PARN in Stress Response in Arabidopsis. Plant J. 2005, 44, 972–984. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, K.; Mamiya, A.; Konishi, M.; Nozaki, M.; Kinoshita, A.; Tamaki, H.; Arita, M.; Saito, M.; Yamamoto, K.; Hachiya, T.; et al. Temperature-Dependent Fasciation Mutants Provide a Link between Mitochondrial RNA Processing and Lateral Root Morphogenesis. eLife 2021, 10, e61611. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, M.; Ikeda, Y.; Nishihama, R.; Yamaoka, S.; Lee, N.-H.; Yamato, K.T.; Kohchi, T.; Hirayama, T. Regulation of the Poly(A) Status of Mitochondrial MRNA by Poly(A)-Specific Ribonuclease Is Conserved among Land Plants. Plant. Cell Physiol. 2020, 61, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, D.; Kuhn, J.; Spadinger, U.; Brennicke, A.; Leaver, C.J.; Binder, S. An RNA Helicase (AtSUV3) Is Present in Arabidopsis thaliana Mitochondria. FEBS Lett. 1999, 458, 337–342. [Google Scholar] [CrossRef]
- Tuteja, N.; Sahoo, R.K.; Garg, B.; Tuteja, R. OsSUV3 Dual Helicase Functions in Salinity Stress Tolerance by Maintaining Photosynthesis and Antioxidant Machinery in Rice (Oryza sativa L. Cv. IR64). Plant. J. 2013, 76, 115–127. [Google Scholar] [CrossRef]
- Pagnussat, G.C.; Yu, H.-J.; Ngo, Q.A.; Rajani, S.; Mayalagu, S.; Johnson, C.S.; Capron, A.; Xie, L.-F.; Ye, D.; Sundaresan, V. Genetic and Molecular Identification of Genes Required for Female Gametophyte Development and Function in Arabidopsis. Development 2005, 132, 603–614. [Google Scholar] [CrossRef]
- Parker, R.; Song, H. The Enzymes and Control of Eukaryotic MRNA Turnover. Nat. Struct. Mol. Biol. 2004, 11, 121–127. [Google Scholar] [CrossRef]
- Houseley, J.; LaCava, J.; Tollervey, D. RNA-Quality Control by the Exosome. Nat. Rev. Mol. Cell Biol. 2006, 7, 529–539. [Google Scholar] [CrossRef]
- Garneau, N.L.; Wilusz, J.; Wilusz, C.J. The Highways and Byways of MRNA Decay. Nat. Rev. Mol. Cell Biol. 2007, 8, 113–126. [Google Scholar] [CrossRef]
- Houseley, J.; Tollervey, D. The Many Pathways of RNA Degradation. Cell 2009, 136, 763–776. [Google Scholar] [CrossRef] [PubMed]
- Condon, C. Maturation and Degradation of RNA in Bacteria. Curr. Opin. Microbiol. 2007, 10, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Belasco, J.G. All Things Must Pass: Contrasts and Commonalities in Eukaryotic and Bacterial MRNA Decay. Nat. Rev. Mol. Cell Biol. 2010, 11, 467–478. [Google Scholar] [CrossRef]
- Gagliardi, D.; Stepien, P.P.; Temperley, R.J.; Lightowlers, R.N.; Chrzanowska-Lightowlers, Z.M.A. Messenger RNA Stability in Mitochondria: Different Means to an End. Trends Genet. 2004, 20, 260–267. [Google Scholar] [CrossRef]
- Hammani, K.; Giegé, P. RNA Metabolism in Plant Mitochondria. Trends Plant. Sci. 2014, 19, 380–389. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, A.R.; Minczuk, M. Mitochondrial Transcription and Translation: Overview. Essays Biochem. 2018, 62, 309–320. [Google Scholar] [CrossRef]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.L.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and Organization of the Human Mitochondrial Genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef]
- Nagaike, T.; Suzuki, T.; Ueda, T. Polyadenylation in Mammalian Mitochondria: Insights from Recent Studies. Biochim. Biophys. Acta BBA—Gene Regul. Mech. 2008, 1779, 266–269. [Google Scholar] [CrossRef]
- Levy, S.; Schuster, G. Polyadenylation and Degradation of RNA in the Mitochondria. Biochem. Soc. Trans. 2016, 44, 1475–1482. [Google Scholar] [CrossRef]
- Tomecki, R.; Dmochowska, A.; Gewartowski, K.; Dziembowski, A.; Stepien, P.P. Identification of a Novel Human Nuclear-Encoded Mitochondrial Poly(A) Polymerase. Nucl. Acids Res. 2004, 32, 6001–6014. [Google Scholar] [CrossRef]
- Piechota, J.; Tomecki, R.; Gewartowski, K.; Szczesny, R.; Dmochowska, A.; Kudła, M.; Dybczyńska, L.; Stepien, P.P.; Bartnik, E. Differential Stability of Mitochondrial MRNA in HeLa Cells. Acta Biochim. Pol. 2006, 53, 157–167. [Google Scholar] [CrossRef]
- Slomovic, S.; Laufer, D.; Geiger, D.; Schuster, G. Polyadenylation and Degradation of Human Mitochondrial RNA: The Prokaryotic Past Leaves Its Mark. Mol. Cell Biol. 2005, 25, 6427–6435. [Google Scholar] [CrossRef]
- Toompuu, M.; Tuomela, T.; Laine, P.; Paulin, L.; Dufour, E.; Jacobs, H.T. Polyadenylation and Degradation of Structurally Abnormal Mitochondrial TRNAs in Human Cells. Nucleic Acids Res. 2018, 46, 5209–5226. [Google Scholar] [CrossRef] [PubMed]
- Osinga, K.A.; De Vrries, E.; Van der Horst, G.; Tabak, H.F. Processing of Yeast Mitochondrial Messenger RNAs at a Conserved Dodecamer Sequence. EMBO J. 1984, 3, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Butow, R.A.; Zhu, H.; Perlman, P.; Conrad-Webb, H. The Role of a Conserved Dodecamer Sequence in Yeast Mitochondrial Gene Expression. Genome 1989, 31, 757–760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zassenhaus, H.P. Purification and Characterization of an RNA Dodecamer Sequence Binding Protein from Mitochondria of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1999, 261, 740–745. [Google Scholar] [CrossRef] [PubMed]
- Holec, S.; Lange, H.; Canaday, J.; Gagliardi, D. Coping with Cryptic and Defective Transcripts in Plant Mitochondria. Biochim. Biophys. Acta BBA—Gene Regul. Mech. 2008, 1779, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Nagaike, T.; Suzuki, T.; Katoh, T.; Ueda, T. Human Mitochondrial MRNAs Are Stabilized with Polyadenylation Regulated by Mitochondria-Specific Poly(A) Polymerase and Polynucleotide Phosphorylase. J. Biol. Chem. 2005, 280, 19721–19727. [Google Scholar] [CrossRef]
- Khemici, V.; Linder, P. RNA Helicases in RNA Decay. Biochem. Soc. Trans. 2018, 46, 163–172. [Google Scholar] [CrossRef]
- Borowski, L.S.; Dziembowski, A.; Hejnowicz, M.S.; Stepien, P.P.; Szczesny, R.J. Human Mitochondrial RNA Decay Mediated by PNPase–HSuv3 Complex Takes Place in Distinct Foci. Nucleic Acids Res. 2013, 41, 1223–1240. [Google Scholar] [CrossRef]
- Wang, D.D.-H.; Guo, X.E.; Modrek, A.S.; Chen, C.-F.; Chen, P.-L.; Lee, W.-H. Helicase SUV3, Polynucleotide Phosphorylase, and Mitochondrial Polyadenylation Polymerase Form a Transient Complex to Modulate Mitochondrial MRNA Polyadenylated Tail Lengths in Response to Energetic Changes*. J. Biol. Chem. 2014, 289, 16727–16735. [Google Scholar] [CrossRef]
- Chujo, T.; Ohira, T.; Sakaguchi, Y.; Goshima, N.; Nomura, N.; Nagao, A.; Suzuki, T. LRPPRC/SLIRP Suppresses PNPase-Mediated MRNA Decay and Promotes Polyadenylation in Human Mitochondria. Nucleic Acids Res. 2012, 40, 8033–8047. [Google Scholar] [CrossRef] [PubMed]
- Schuster, G.; Stern, D. Chapter 10 RNA Polyadenylation and Decay in Mitochondria and Chloroplasts. In Molecular Biology of RNA Processing and Decay in Prokaryotes; Academic Press: Cambridge, MA, USA, 2009; Volume 85, pp. 393–422. ISBN 1877-1173. [Google Scholar]
- Wahle, E.; Winkler, G.S. RNA Decay Machines: Deadenylation by the Ccr4–Not and Pan2–Pan3 Complexes. Biochim. Biophys. Acta BBA—Gene Regul. Mech. 2013, 1829, 561–570. [Google Scholar] [CrossRef]
- Schäfer, I.B.; Yamashita, M.; Schuller, J.M.; Schüssler, S.; Reichelt, P.; Strauss, M.; Conti, E. Molecular Basis for Poly(A) RNP Architecture and Recognition by the Pan2-Pan3 Deadenylase. Cell 2019, 177, 1619–1631.e21. [Google Scholar] [CrossRef]
- Aström, J.; Aström, A.; Virtanen, A. In Vitro Deadenylation of Mammalian MRNA by a HeLa Cell 3′ Exonuclease. EMBO J. 1991, 10, 3067–3071. [Google Scholar] [CrossRef] [PubMed]
- Korner, C.G.; Wormington, M.; Muckenthaler, M.; Schneider, S.; Dehlin, E.; Wahle, E. The Deadenylating Nuclease (DAN) Is Involved in Poly(A) Tail Removal during the Meiotic Maturation of Xenopus Oocytes. EMBO J. 1998, 17, 5427–5437. [Google Scholar] [CrossRef]
- Tourrière, H.; Chebli, K.; Tazi, J. MRNA Degradation Machines in Eukaryotic Cells. Biochimie 2002, 84, 821–837. [Google Scholar] [CrossRef]
- Nousch, M.; Techritz, N.; Hampel, D.; Millonigg, S.; Eckmann, C.R. The Ccr4–Not Deadenylase Complex Constitutes the Main Poly(A) Removal Activity in C. Elegans. J. Cell Sci. 2013, 126, 4274–4285. [Google Scholar] [CrossRef]
- Stuart, B.D.; Choi, J.; Zaidi, S.; Xing, C.; Holohan, B.; Chen, R.; Choi, M.; Dharwadkar, P.; Torres, F.; Girod, C.E.; et al. Exome Sequencing Links Mutations in PARN and RTEL1 with Familial Pulmonary Fibrosis and Telomere Shortening. Nat. Genet. 2015, 47, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Moon, D.H.; Segal, M.; Boyraz, B.; Guinan, E.; Hofmann, I.; Cahan, P.; Tai, A.K.; Agarwal, S. Poly(A)-Specific Ribonuclease (PARN) Mediates 3′-End Maturation of the Telomerase RNA Component. Nat. Genet. 2015, 47, 1482–1488. [Google Scholar] [CrossRef] [PubMed]
- Montellese, C.; Montel-Lehry, N.; Henras, A.K.; Kutay, U.; Gleizes, P.-E.; O’Donohue, M.-F. Poly(A)-Specific Ribonuclease Is a Nuclear Ribosome Biogenesis Factor Involved in Human 18S RRNA Maturation. Nucleic Acids Res. 2017, 45, 6822–6836. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Bjerke, G.A.; Muhlrad, D.; Yi, R.; Parker, R. The RNase PARN Controls the Levels of Specific MiRNAs That Contribute to P53 Regulation. Mol. Cell 2019, 73, 1204–1216.e4. [Google Scholar] [CrossRef]
- Johnson, J.M.; Thürich, J.; Petutschnig, E.K.; Altschmied, L.; Meichsner, D.; Sherameti, I.; Dindas, J.; Mrozinska, A.; Paetz, C.; Scholz, S.S.; et al. A Poly(A) Ribonuclease Controls the Cellotriose-Based Interaction between Piriformospora Indica and Its Host Arabidopsis. Plant. Physiol. 2018, 176, 2496–2514. [Google Scholar] [CrossRef]
- Liang, W.; Li, C.; Liu, F.; Jiang, H.; Li, S.; Sun, J.; Wu, X.; Li, C. The Arabidopsis Homologs of CCR4-Associated Factor 1 Show MRNA Deadenylation Activity and Play a Role in Plant Defence Responses. Cell Res. 2009, 19, 307–316. [Google Scholar] [CrossRef]
- Suzuki, Y.; Arae, T.; Green, P.J.; Yamaguchi, J.; Chiba, Y. AtCCR4a and AtCCR4b Are Involved in Determining the Poly(A) Length of Granule-Bound Starch Synthase 1 Transcript and Modulating Sucrose and Starch Metabolism in Arabidopsis thaliana. Plant. Cell Physiol. 2015, 56, 863–874. [Google Scholar] [CrossRef]
- Nishimura, N.; Okamoto, M.; Narusaka, M.; Yasuda, M.; Nakashita, H.; Shinozaki, K.; Narusaka, Y.; Hirayama, T. ABA Hypersensitive Germination2-1 Causes the Activation of Both Abscisic Acid and Salicylic Acid Responses in Arabidopsis. Plant. Cell Physiol. 2009, 50, 2112–2122. [Google Scholar] [CrossRef]
- Hirayama, T.; Matsuura, T.; Ushiyama, S.; Narusaka, M.; Kurihara, Y.; Yasuda, M.; Ohtani, M.; Seki, M.; Demura, T.; Nakashita, H.; et al. A Poly(A)-Specific Ribonuclease Directly Regulates the Poly(A) Status of Mitochondrial MRNA in Arabidopsis. Nat. Commun. 2013, 4, 2247. [Google Scholar] [CrossRef]
- Waltz, F.; Nguyen, T.-T.; Arrivé, M.; Bochler, A.; Chicher, J.; Hammann, P.; Kuhn, L.; Quadrado, M.; Mireau, H.; Hashem, Y.; et al. Small Is Big in Arabidopsis Mitochondrial Ribosome. Nat. Plants 2019, 5, 106. [Google Scholar] [CrossRef]
- Hirayama, T. A Unique System for Regulating Mitochondrial MRNA Poly(A) Status and Stability in Plants. Plant. Signal. Behav. 2014, 9, e973809. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Floyd, S.K.; Sakakibara, K. Green Genes—Comparative Genomics of the Green Branch of Life. Cell 2007, 129, 229–234. [Google Scholar] [CrossRef]
- Sugiyama, M. Isolation and Initial Characterization of Temperature-Sensitive Mutants of Arabidopsis thaliana That Are Impaired in Root Redifferentiation. Plant. Cell Physiol. 2003, 44, 588–596. [Google Scholar] [CrossRef] [PubMed][Green Version]
- MacIntosh, G.C.; Castandet, B. Organellar and Secretory Ribonucleases: Major Players in Plant RNA Homeostasis. Plant. Physiol. 2020, 183, 1438–1452. [Google Scholar] [CrossRef] [PubMed]
- Zimorski, V.; Ku, C.; Martin, W.F.; Gould, S.B. Endosymbiotic Theory for Organelle Origins. Curr. Opin. Microbiol. 2014, 22, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Roger, A.J.; Muñoz-Gómez, S.A.; Kamikawa, R. The Origin and Diversification of Mitochondria. Curr. Biol. 2017, 27, R1177–R1192. [Google Scholar] [CrossRef]
- Salinas-Giegé, T.; Cavaiuolo, M.; Cognat, V.; Ubrig, E.; Remacle, C.; Duchêne, A.-M.; Vallon, O.; Maréchal-Drouard, L. Polycytidylation of Mitochondrial MRNAs in Chlamydomonas Reinhardtii. Nucleic Acids Res. 2017, 45, 12963–12973. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hirayama, T. PARN-like Proteins Regulate Gene Expression in Land Plant Mitochondria by Modulating mRNA Polyadenylation. Int. J. Mol. Sci. 2021, 22, 10776. https://doi.org/10.3390/ijms221910776
Hirayama T. PARN-like Proteins Regulate Gene Expression in Land Plant Mitochondria by Modulating mRNA Polyadenylation. International Journal of Molecular Sciences. 2021; 22(19):10776. https://doi.org/10.3390/ijms221910776
Chicago/Turabian StyleHirayama, Takashi. 2021. "PARN-like Proteins Regulate Gene Expression in Land Plant Mitochondria by Modulating mRNA Polyadenylation" International Journal of Molecular Sciences 22, no. 19: 10776. https://doi.org/10.3390/ijms221910776
APA StyleHirayama, T. (2021). PARN-like Proteins Regulate Gene Expression in Land Plant Mitochondria by Modulating mRNA Polyadenylation. International Journal of Molecular Sciences, 22(19), 10776. https://doi.org/10.3390/ijms221910776





