Constitutive Photomorphogenic 1 Enhances ER Stress Tolerance in Arabidopsis
Abstract
:1. Introduction
2. Results
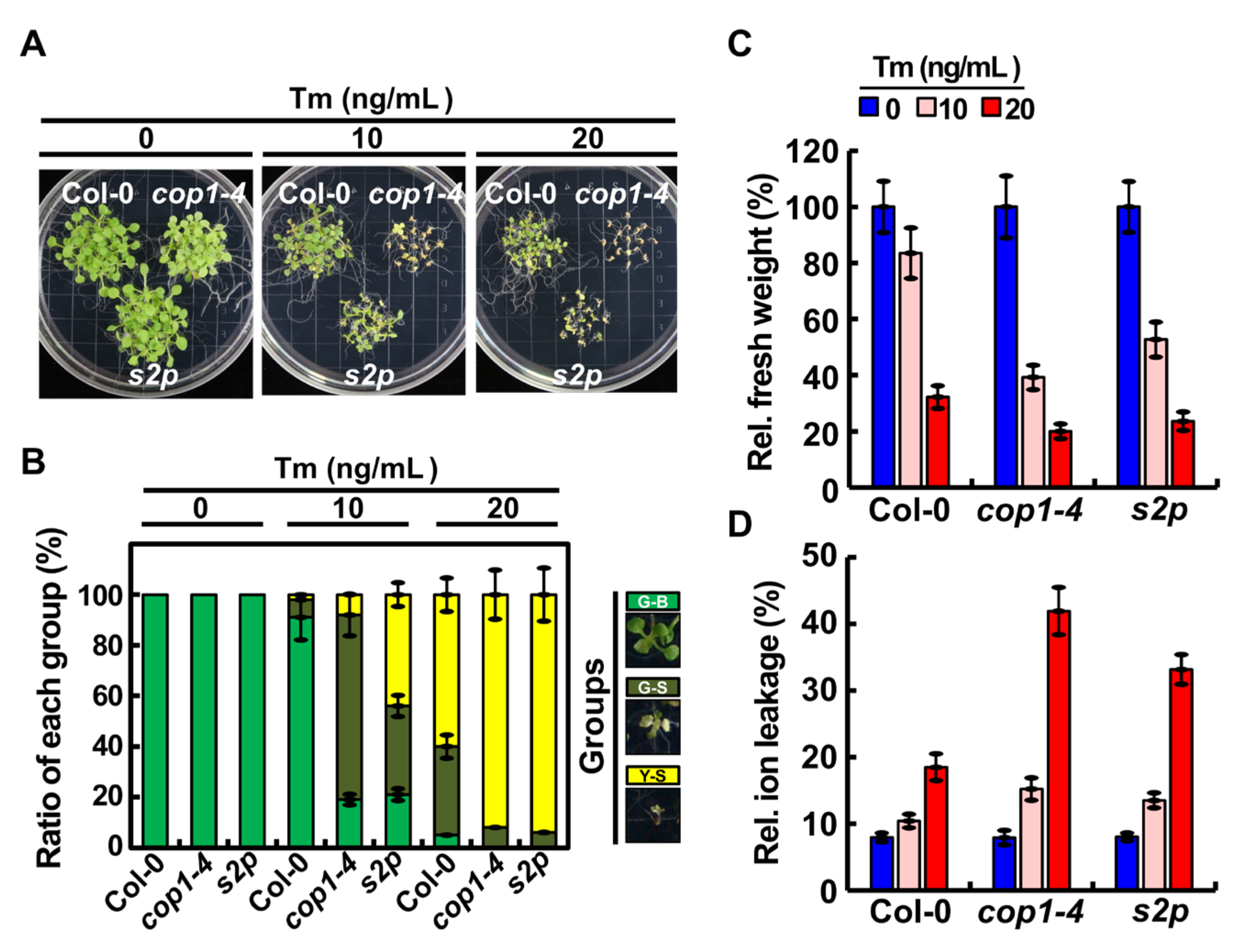
2.1. COP1 Mediated ER Stress Tolerance in Arabidopsis
2.2. COP1 Was Enriched in the Nucleus under ER Stress Conditions
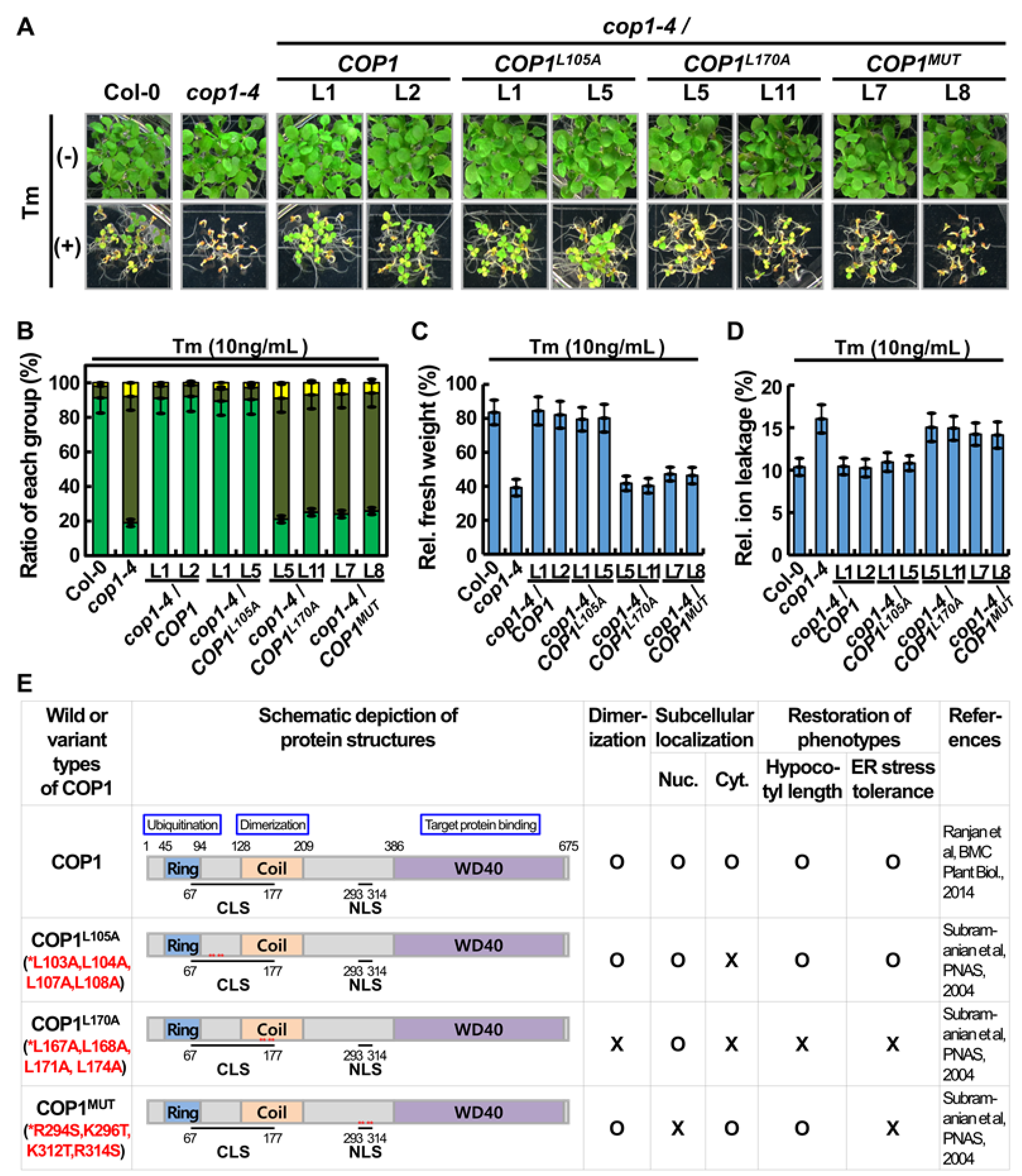
2.3. COP1 Nuclear Localization and Dimerization Were Essential for Its Role in ER Stress Tolerance
2.4. COP1 Mediated Partial Degradation of HY5 under ER Stress Conditions
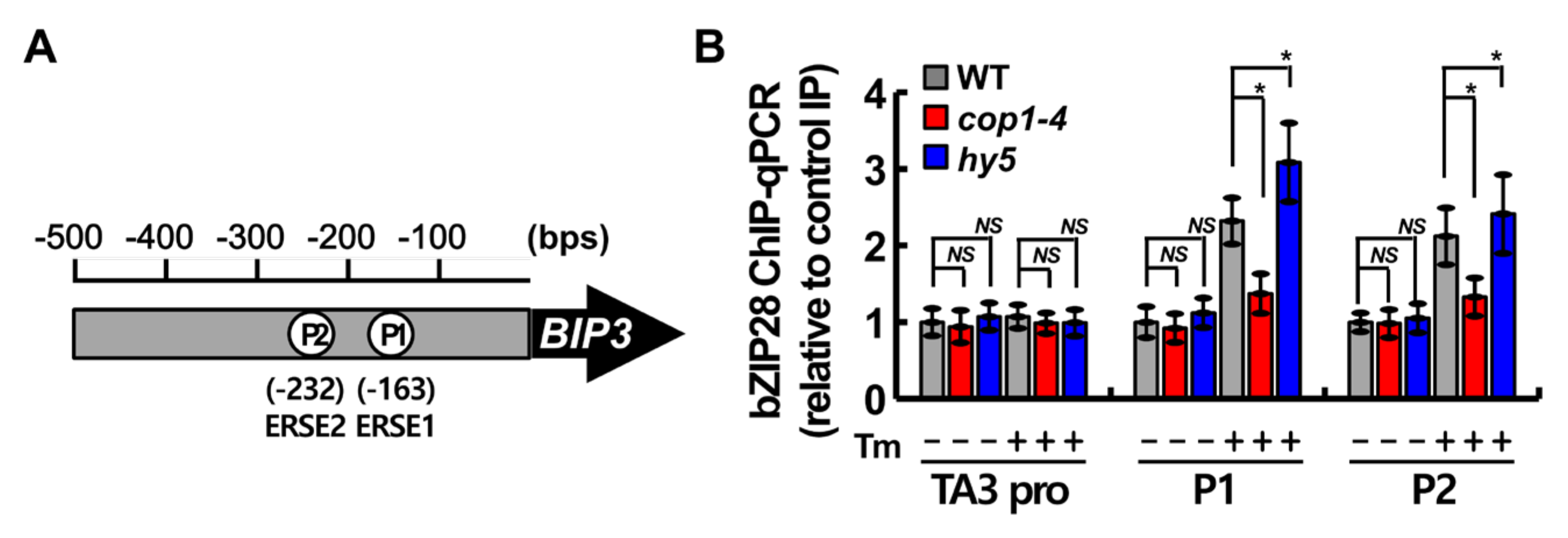
2.5. COP1 Facilitated the Binding of bZIP28 to ER Stress Response Element (ERSE) under ER Stress
2.6. hy5 Rescued the ER Stress-Sensitive Phenotype of cop1-4
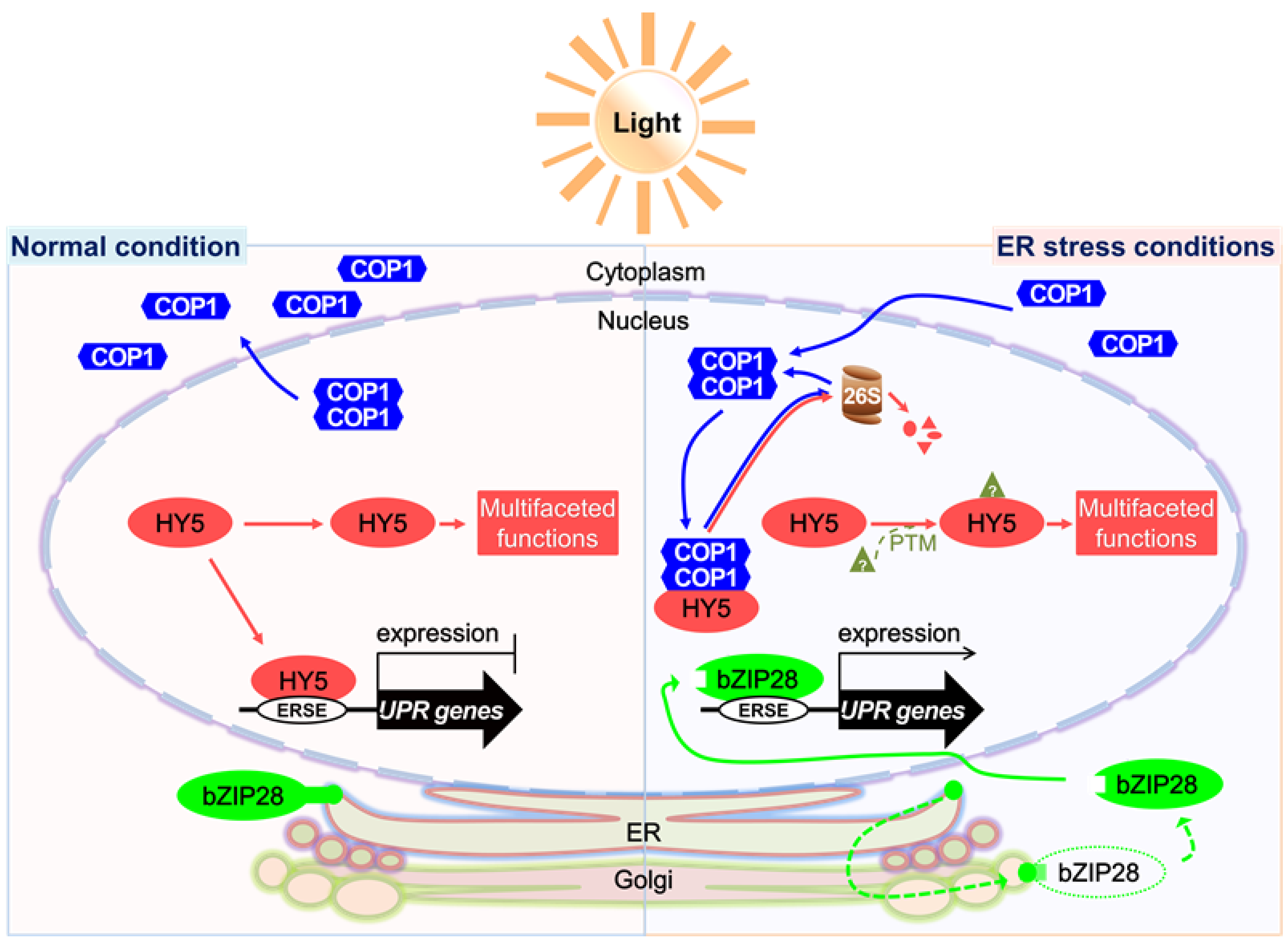
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. ER Stress Treatment and Phenotypic Analysis
4.3. Plasmid Construction and Plant Transformation
4.4. Transient Tobacco Expression Assay
4.5. Subcellular Localization
4.6. Nuclear Fractionation Experiment
4.7. HY5 Degradation Assay and Immunoblot Analysis
4.8. Total RNA Exraction and Semi-Quantitative Reverse Transcription PCR (sqRT-PCR) Analysis
4.9. Gene Expression Analysis by qRT-PCR
4.10. ChIP Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smith, H. Phytochromes and light signal perception. Nature 2000, 407, 585. [Google Scholar] [CrossRef] [PubMed]
- Lin, C. Blue light receptors and signal transduction. Plant Cell 2002, 14, 207–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizzini, L.; Favory, J.J.; Cloix, C.; Faggionato, D.; O’Hara, A.; Kaiserli, E.; Baumeister, R.; Schäfer, E.; Nagy, F.; Jenkins, G.I.; et al. Perception of UV-B by the arabidopsis UVR8 protein. Science 2011, 332, 103–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holm, M.; Deng, X.W. Structural organization and interactions of COP1, a light-regulated developmental switch. Plant Mol. Biol. 1999, 41, 151–158. [Google Scholar] [CrossRef]
- Ang, L.H.; Chattopadhyay, S.; Wei, N.; Oyama, T.; Okada, K.; Batschauer, A.; Deng, X.W. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1998, 1, 213–222. [Google Scholar] [CrossRef]
- Oyama, T.; Shimura, Y.; Okada, K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997, 11, 2983–2995. [Google Scholar] [CrossRef] [Green Version]
- Osterlund, M.T.; Hardtke, C.S.; Wei, N.; Deng, X.W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Science 2000, 405, 462–466. [Google Scholar] [CrossRef]
- Wang, Q.; Lin, C. Photoreceptor signaling: When COP1 meets VPs. EMBO J. 2019, 38, 1–2. [Google Scholar] [CrossRef]
- Podolec, R.; Ulm, R. Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol. 2018, 45, 18–25. [Google Scholar] [CrossRef]
- Vitale, A.; Boston, R.S. Endoplasmic reticulum quality control and the unfolded protein response: Insights from plants. Traffic 2008, 9, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Howell, S.H. Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 2010, 22, 2930–2942. [Google Scholar] [CrossRef] [Green Version]
- Beaugelin, I.; Chevalier, A.; D’Alessandro, S.; Ksas, B.; Havaux, M. Endoplasmic reticulum-mediated unfolded protein response is an integral part of singlet oxygen signalling in plants. Plant J. 2020, 102, 1266–1280. [Google Scholar] [CrossRef]
- Angelos, E.; Ruberti, C.; Kim, S.J.; Brandizzi, F. Maintaining the factory: The roles of the unfolded protein response in cellular homeostasis in plants. Plant J. 2017, 90, 671–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, R.; Deng, Y.; Shah, S.; Rao, A.G.; Howell, S.H. Binding protein is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis. Plant Cell 2013, 25, 1416–1429. [Google Scholar] [CrossRef] [Green Version]
- Iwata, Y.; Ashida, M.; Hasegawa, C.; Tabara, K.; Mishiba, K.I.; Koizumi, N. Activation of the Arabidopsis membrane-bound transcription factor bZIP28 is mediated by site-2 protease, but not site-1 protease. Plant J. 2017, 91, 408–415. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.X.; Howell, S.H. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 2010, 22, 782–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.H.; Kang, C.H.; Nawkar, G.M.; Lee, E.S.; Paeng, S.K.; Chae, H.B.; Chi, Y.H.; Kim, W.Y.; Yun, D.J.; Lee, S.Y. EMR, a cytosolic-abundant ring finger E3 ligase, mediates ER-associated protein degradation in Arabidopsis. New Phytol. 2018, 220, 163–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, B.; Verchot, J.; Dickman, M.B. When supply does not meet demand-ER stress and plant programmed cell death. Front. Plant Sci. 2014, 5, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, G.; Wang, S.; Han, S.; Xie, K.; Wang, Y.; Li, J.; Liu, Y. Plant Bax Inhibitor-1 interacts with ATG6 to regulate autophagy and programmed cell death. Autophagy 2017, 13, 1161–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawkar, G.M.; Kang, C.H.; Maibam, P.; Park, J.H.; Jung, Y.J.; Chae, H.B.; Chi, Y.H.; Jung, I.J.; Kim, W.Y.; Yun, D.J.; et al. HY5, a positive regulator of light signaling, negatively controls the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 2084–2089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNellis, T.W.; von Arnim, A.G.; Wang, D.X. Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: Evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell 1994, 6, 1391–1400. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.W.; Quail, P.H. Genetic and phenotypic characterization of cop1 mutants of Arabidopsis thaliana. Plant J. 1992, 2, 83–95. [Google Scholar] [CrossRef]
- McNellis, T.W.; von Arnim, A.G.; Araki, T.; Komeda, Y.; Misera, S.; Wang, D.X. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 1994, 6, 487–500. [Google Scholar] [CrossRef] [Green Version]
- Chi, Y.H.; Melencion, S.M.B.; Alinapon, C.V.; Kim, M.J.; Lee, E.S.; Paeng, S.K.; Park, J.H.; Nawkar, G.M.; Jung, Y.J.; Chae, H.B.; et al. The membrane-tethered NAC transcription factor, AtNTL7, contributes to ER-stress resistance in Arabidopsis. Biochem. Biophys. Res. Commun. 2017, 488, 641–647. [Google Scholar] [CrossRef]
- Subramanian, C.; Kim, B.H.; Lyssenko, N.N.; Xu, X.; Johnson, C.H.; Von Arnim, A.G. The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: Mutational analysis by bioluminescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 2004, 101, 6798–6802. [Google Scholar] [CrossRef] [Green Version]
- Osterlund, M.T.; Deng, X.W. Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 1998, 16, 201–208. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Wang, J.; Shi, H.; Gu, J.; Dong, J.; Deng, X.W.; Huang, R. Salt stress and ethylene antagonistically regulate nucleocytoplasmic partitioning of COP1 to control seed germination. Plant Physiol. 2016, 170, 2340–2350. [Google Scholar] [CrossRef] [Green Version]
- Catalá, R.; Medina, J.; Salinas, J. Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 16475–16480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karayekov, E.; Sellaro, R.; Legris, M.; Yanovsky, M.J.; Casal, J.J. Heat shock-induced fluctuations in clock and light signaling enhance phytochrome B-mediated arabidopsis deetiolation. Plant Cell 2013, 25, 2892–2906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, B.D.; Kim, M.R.; Kang, M.Y.; Cha, J.Y.; Han, S.H.; Nawkar, G.M.; Sakuraba, Y.; Lee, S.Y.; Imaizumi, T.; McClung, C.R.; et al. The F-box protein FKF1 inhibits dimerization of COP1 in the control of photoperiodic flowering. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stacey, M.G.; Kopp, O.R.; Kim, T.H.; Von Arnim, A.G. Modular domain structure of Arabidopsis COP1. Reconstitution of activity by fragment complementation and mutational analysis of a nuclear localization signal in planta. Plant Physiol. 2000, 124, 979–989. [Google Scholar] [CrossRef] [Green Version]
- Stacey, M.G.; Hicks, S.N.; Von Arnim, A.G. Discrete domains mediate the light-responsive nuclear and cytoplasmic localization of Arabidopsis COP1. Plant Cell 1999, 11, 349–363. [Google Scholar] [CrossRef] [Green Version]
- Torii, K.U.; McNellis, T.W.; Deng, X.W. Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J. 1998, 17, 5577–5587. [Google Scholar] [CrossRef] [Green Version]
- Ranjan, A.; Dickopf, S.; Ullrich, K.K.; Rensing, S.A.; Hoecker, U. Functional analysis of COP1 and SPA orthologs from Physcomitrella and rice during photomorphogenesis of transgenic Arabidopsis reveals distinct evolutionary conservation. BMC Plant Biol. 2014, 14, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; He, K.; Stolc, V.; Lee, H.; Figueroa, P.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Xing, W.D. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 2007, 19, 731–749. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; He, H.; Wang, X.; Wang, X.; Yang, X.; Li, L.; Deng, X.W. Genome-wide mapping of the HY5-mediated genenetworks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 2011, 65, 346–358. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Winter, C.M.; Wu, M.-F.; Kwon, C.S.; William, D.A.; Wagner, D. PROTOCOL: Chromatin Immunoprecipitation from Arabidopsis Tissues. Arab. B. 2014, 12, e0170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawkar, G.M.; Lee, E.S.; Shelake, R.M.; Park, J.H.; Ryu, S.W.; Kang, C.H.; Lee, S.Y. Activation of the transducers of unfolded protein response in plants. Front. Plant Sci. 2018, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nieto, C.; Luengo, L.M.; Prat, S. Regulation of COP1 Function by Brassinosteroid Signaling. Front. Plant Sci. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huai, J.; Shang, F.; Xu, G.; Tang, W.; Jing, Y.; Lin, R. A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol. 2017, 174, 2487–2500. [Google Scholar] [CrossRef] [Green Version]
- Ponnu, J.; Hoecker, U. Illuminating the COP1/SPA Ubiquitin Ligase: Fresh Insights Into Its Structure and Functions During Plant Photomorphogenesis. Front. Plant Sci. 2021, 12, 1–19. [Google Scholar] [CrossRef]
- Alabadí, D.; Gallego-Bartolomé, J.; Orlando, L.; García-Cárcel, L.; Rubio, V.; Martínez, C.; Frigerio, M.; Iglesias-Pedraz, J.M.; Espinosa, A.; Deng, X.W.; et al. Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 2008, 53, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Vandenbussche, F.; Habricot, Y.; Condiff, A.S.; Maldiney, R.; Van Der Straeten, D.; Ahmad, M. HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J. 2007, 49, 428–441. [Google Scholar] [CrossRef]
- Wang, W.; Paik, I.; Kim, J.; Hou, X.; Sung, S.; Huq, E. Direct phosphorylation of HY5 by SPA kinases to regulate photomorphogenesis in Arabidopsis. New Phytol. 2021, 105. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Holm, M.; Botto, J.F. Molecular interactions of BBX24 and BBX25 with HYH, HY5 HOMOLOG, to modulate Arabidopsis seedling development. Plant Signal. Behav. 2013, 8, 24–28. [Google Scholar] [CrossRef] [Green Version]
- Datta, S.; Hettiarachchi, C.; Johansson, H.; Holm, M. Salt Tolerance Homolog2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 2007, 19, 3242–3255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holtan, H.E.; Bandong, S.; Marion, C.M.; Adam, L.; Tiwari, S.; Shen, Y.; Maloof, J.N.; Maszle, D.R.; Ohto, M.; Preuss, S.; et al. Bbx32, an arabidopsis b-box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 2011, 156, 2109–2123. [Google Scholar] [CrossRef] [Green Version]
- Jeong, R.D.; Chandra-Shekara, A.C.; Barman, S.R.; Navarre, D.; Klessig, D.F.; Kachroo, A.; Kachroo, P. Cryptochrome 2 and phototropin 2 regulate resistance protein-mediated viral defense by negatively regulating an E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 2010, 107, 13538–13543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.Y.; Jang, I.C.; Seo, H.S. COP1 controls abiotic stress responses by modulating AtSIZ1 function through its E3 ubiquitin ligase activity. Front. Plant Sci. 2016, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.L.; Niu, D.; Hu, Z.L.; Kim, D.H.; Jin, Y.H.; Cai, B.; Liu, P.; Miura, K.; Yun, D.J.; Kim, W.Y.; et al. An Arabidopsis SUMO E3 Ligase, SIZ1, Negatively Regulates Photomorphogenesis by Promoting COP1 Activity. PLoS Genet. 2016, 12, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Mishiba, K.I.; Nagashima, Y.; Suzukia, E.; Hayashi, N.; Ogata, Y.; Shimada, Y.; Koizumi, N. Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc. Natl. Acad. Sci. USA 2013, 110, 5713–5718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, C.H.; Jung, W.Y.; Kang, Y.H.; Kim, J.Y.; Kim, D.G.; Jeong, J.C.; Baek, D.W.; Jin, J.B.; Lee, J.Y.; Kim, M.O.; et al. AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differ. 2006, 13, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.D.; Grossniklaus, U. A Gateway Cloning Vector Set for High-Throughput Functional Analysis of Genes in Planta. Plant Physiol. 2003, 133, 462–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, C.H.; Lee, E.S.; Nawkar, G.M.; Park, J.H.; Wi, S.D.; Bae, S.B.; Chae, H.B.; Paeng, S.K.; Hong, J.C.; Lee, S.Y. Constitutive Photomorphogenic 1 Enhances ER Stress Tolerance in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 10772. https://doi.org/10.3390/ijms221910772
Kang CH, Lee ES, Nawkar GM, Park JH, Wi SD, Bae SB, Chae HB, Paeng SK, Hong JC, Lee SY. Constitutive Photomorphogenic 1 Enhances ER Stress Tolerance in Arabidopsis. International Journal of Molecular Sciences. 2021; 22(19):10772. https://doi.org/10.3390/ijms221910772
Chicago/Turabian StyleKang, Chang Ho, Eun Seon Lee, Ganesh M. Nawkar, Joung Hun Park, Seong Dong Wi, Su Bin Bae, Ho Byoung Chae, Seol Ki Paeng, Jong Chan Hong, and Sang Yeol Lee. 2021. "Constitutive Photomorphogenic 1 Enhances ER Stress Tolerance in Arabidopsis" International Journal of Molecular Sciences 22, no. 19: 10772. https://doi.org/10.3390/ijms221910772
APA StyleKang, C. H., Lee, E. S., Nawkar, G. M., Park, J. H., Wi, S. D., Bae, S. B., Chae, H. B., Paeng, S. K., Hong, J. C., & Lee, S. Y. (2021). Constitutive Photomorphogenic 1 Enhances ER Stress Tolerance in Arabidopsis. International Journal of Molecular Sciences, 22(19), 10772. https://doi.org/10.3390/ijms221910772






